This study reports on the use of rapamycin to enhance transduction in adult granulocyte colony-stimulating factor-mobilized hematopoietic stem and progenitor cells (HSPCs) using in vitro and in vivo hematopoietic cell assays to optimize the process. These methods will be used to manufacture cell products for our subsequent gene therapy studies but are applicable to other lentiviral vector-based HSPC gene therapy studies and thus represent an important advancement in the field of human gene therapy.
Keywords: Hematopoietic stem cells, Mice, SCID, HIV, Transduction, Genetic, Transplantation, Sirolimus
Abstract
Genetic modification of adult human hematopoietic stem and progenitor cells (HSPCs) with lentiviral vectors leads to long-term gene expression in the progeny of the HSPCs and has been used to successfully treat several monogenic diseases. In some cases, the gene-modified cells have a selective growth advantage over nonmodified cells and eventually are the dominant engrafted population. However, in disease indications for which the gene-modified cells do not have a selective advantage, optimizing transduction of HSPC is paramount to successful stem cell-based gene therapy. We demonstrate here that transduction of adult CD34+ HSPCs with lentiviral vectors in the presence of rapamycin, a widely used mTORC1 inhibitor, results in an approximately threefold increase in stable gene marking with minimal effects on HSPC growth and differentiation. Using this approach, we have demonstrated that we can enhance the frequency of gene-modified HSPCs that give rise to clonogenic progeny in vitro without excessive increases in the number of vector copies per cell or changes in integration pattern. The genetic marking of HSPCs and expression of transgenes is durable, and transplantation of gene-modified HSPCs into immunodeficient mice results in high levels of gene marking of the lymphoid and myeloid progeny in vivo. The prior safe clinical history of rapamycin in other applications supports the use of this compound to generate gene-modified autologous HSPCs for our HIV gene therapy clinical trials.
Introduction
The successful use of lentiviral vectors to genetically modify hematopoietic stem and progenitor cells (HSPCs) for the treatment of a number of monogenic disease conditions including adrenoleukodystrophy [1] and β-thalassemia [2] has been reported, and the number of trials demonstrating general proof of principle continues to expand [3, 4]. We previously reported on a clinical trial in HIV+ patients in whom autologous CD34+ HSPCs were genetically modified using a lentiviral vector encoding a short hairpin RNA (shRNA) targeting the HIV genome, a nucleolar localizing transactivation response (TAR) decoy, and a CCR5-specific hammerhead ribozyme previously shown to effectively inhibit HIV infection in vitro [5]. We were successful in preparing gene-modified HSPC (GM-HSPC) products for infusion in four of five patients, and all patients transplanted had detectable gene marking and expression at one or more time points for up to 2 years at the time of the report [6]. In one patient (UPN0306), we have now observed long-term (>3 years) gene marking and expression of anti-HIV RNA in peripheral blood cells [7], demonstrating the potential for long-term provision of an HIV-resistant immune system. Unfortunately, the gene marking of the more primitive hematopoietic stem cells was low (∼1%), resulting in a low frequency of gene-modified progeny cells (0.1%–0.3%) in the peripheral blood and bone marrow of these patients. Thus, there were insufficient HIV-resistant blood cells in these patients to see a clinical benefit.
Since this trial, one of the major goals of our laboratory has been to improve the level of genetic modification of HSPCs using lentiviral vectors. Previously reported methods for enhancing transduction of CD34+ HSPCs with viral vectors include duration of prestimulation, number of transductions, multiplicity of infection, centrifugation, use of ABC transport inhibitors, fragments of fibronectin, culture media, and time [8–13]. Most of these methods, however, result in only incremental effects on overall transduction efficiency, are not scalable (centrifugation), are highly dependent on vector quality (infectious titer), and are subject to patient-specific variability. Recently, however, Wang and Torbett [14] reported that rapamycin enhanced lentiviral transduction of cord blood and bone marrow-derived HSPCs from several species. Rapamycin (sirolimus) is a nonantibiotic macrolide with pleiotropic activity, including immunosuppression and regulation of cell cycling and autophagy [15–18]. The drug has been used clinically to prevent solid organ rejection and graft versus host disease, as an anticancer agent, and to coat coronary stents to prevent restenosis [19–23]. Moreover, engraftment of stem cells in immunodeficient mice has been shown to be enhanced by treatment with rapamycin [24] although this effect has not been tested in humans. Thus, we reasoned that treatment of HSPCs with rapamycin during transduction would potentially improve clinical outcome (in our current gene therapy application) via multiple mechanisms. The history of safe clinical use of rapamycin is supportive of this approach.
We report here on the use of rapamycin to enhance transduction in adult granulocyte colony-stimulating factor (G-CSF)-mobilized HSPCs using in vitro and in vivo hematopoietic cell assays to optimize the process. The methods developed in this study will be used to manufacture cell products for our subsequent gene therapy studies but are applicable to other lentiviral vector-based HSPC gene therapy studies and thus represent an important advancement in the field of human gene therapy.
Materials and Methods
Lentiviral Vectors
Self-inactivating HIV1-based lentiviral (LV1, rHIV7-U6-sh1-U6-TAR-VA1-CCR5RZ-CMV-GFP-P2A-MGMT; and LV2, rHIV7-CMV-GFP-P2A-MGMT) vectors were produced with modified packaging plasmids developed at City of Hope [25]. Batches of viral vector were produced by calcium phosphate precipitation and transient transfection of HEK293T cells as previously described [7]. The lentiviral vectors were pseudotyped with vesicular stomatitis virus G (VSV-G) protein to extend host range to include HSPCs [26–28]. LV3 rRSCSMPGW2 vector was a gift from Dr. Hans Peter Kiem and has been previously described [29, 30]. LV4 rHIV-shI-TAR-CCR5 is a self-inactivating VSV-G pseudotyped lentiviral vector described in [5]. Additional methodology is detailed in the supplemental online data.
CD34+ HSPC Isolation
CD34+ HSPCs were isolated from G-CSF-mobilized peripheral blood from healthy donors provided by Progenitor Cell Therapy (Allendale, NJ, http://pctcelltherapy.com). Apheresis products were washed, concentrated, labeled with CliniMACS CD34 MicroBeads (Miltenyi Biotec, Auburn, CA, http://www.miltenyibiotec.com), and enriched with the CliniMACS Cell Separation System (Miltenyi Biotec). The purity of selected cells was higher than 95%. The purified CD34+ cells were either used freshly for transduction or cryopreserved in CryoStor CS5 solution (BioLife Solutions, Bothell, WA, http://biolifesolutions.com) using a controlled rate freezer and stored in the vapor phase of a liquid nitrogen dewar for later use.
Expansion Culture
For expansion of hematopoietic cells, HSPCs at 16–24 hours after transduction were seeded at 5 × 104 cells per milliliter in expansion medium, StemSpan serum-free expansion medium (SFEM) (StemCell Technologies Inc., Vancouver, BC, Canada, http://www.stemcell.com), containing 50–100 ng/ml recombinant human stem cell factor, 50–100 ng/ml FMS-like tyrosine kinase-3 ligand (Flt-3L), 10–50 ng/ml thrombopoietin (CellGenix, Freiburg, Germany, http://www.cellgenix.com), 50 ng/ml interleukin-6 (Invitrogen, Carlsbad, CA, http://www.invitrogen.com) (SFT6) and with (0.75 μM) or without StemRegenin-1 (SR-1; Cellagen Technology, San Diego, CA, http://www.cellagentech.com), hereinafter referred to as expansion medium, unless otherwise indicated. Cultures were maintained by feeding fresh medium every 2–3 days as described by Boitano et al. [31] for up to 2 weeks or as specified. Cells were taken weekly for cell count and viability and used to set up colony-forming unit (CFU) assays (at day 7 of culture) or to be used for phenotype analysis and quantitative polymerase chain reaction (qPCR).
Lentiviral Transduction
Freshly isolated or frozen CD34+ HSPCs (1–2 × 106 cells per milliliter) were prestimulated for 16–20 hours before transduction in SFT6. After prestimulation, the cells were transduced with lentivirus at an multiplicity of infection (MOI) of 10–50 on RetroNectin-coated (Takara, Shiga, Japan, http://www.takara-bio.com) non-tissue culture-treated plates according to the manufacturer’s instructions in the presence or absence of rapamycin (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com) at the indicated concentrations. After 24 hours of incubation at 37°C with 5% CO2, the cells were washed three times in phosphate-buffered saline (PBS) before performance of cell culture assays.
Myeloid Differentiation Culture (Bulk Culture)
For myeloid hematopoietic potential, cells were collected 16–24 hours after transduction, washed, and seeded at 1 × 105 cells per milliliter in myeloid differentiation culture medium as described [32]. Cultures were maintained by demidepletion of media every 3–5 days. Cells were collected weekly for up to 4 weeks for cell count and viability using Guava PCA-96 (EMD Millipore, Danvers, MA, http://www.millipore.com) and used for performance of phenotypic analysis and qPCR.
Flow Cytometric Analysis of In Vitro Cultures
Aliquots of cells from in vitro cultures were harvested weekly for phenotypic analysis using antibodies to lineage-specific cell surface antigens. Antibodies CD33-PE, CD15-FITC, GlyA-PC5 (BD Biosciences, San Jose, CA, CA, http://www.bdbiosciences.com), and CD14-APC-Alexa750 (Invitrogen) were used to identify progenitors, granulocyte, erythrocyte, and monocyte subpopulations, respectively, in myeloid differentiation cultures as previously described [32]. Cells from expansion culture were also collected weekly to determine GFP expression and phenotype using CD90-PE or CD90-APC, CD49f-PE (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com), and CD34-PC7 (Beckman Coulter, Brea, CA, http://www.beckmancoulter.com). Phenotypic data were collected on a Gallios flow cytometer (Beckman Coulter) and analyzed with FCS Express software (DeNovo Software, Los Angeles, CA, http://www.denovosoftware.com).
Fluorescence-Activated Cell Sorting
CD34+ HSPCs from three different donors were prestimulated in SFEM SFT6 overnight followed by LV1 transduction (MOI 10) in the presence or absence of 20 μg/ml rapamycin (Sigma-Aldrich). Transduced cells were cultured in expansion culture medium for 1 week and sorted for GFP+ and GFP− using a fluorescence-activated cell sorting (FACS) Aria II (BD Biosciences). Sorted cells were monitored for growth expansion or plated for CFUs as described above.
Colony-Forming Unit Assay
Untransduced controls and transduced cells were harvested 16–24 hours after transduction or after 7 days of growth in expansion culture medium. A total of 500 cells were plated per plate in triplicate in MethoCult H4435-enriched methylcellulose medium according to the manufacturer’s instructions (StemCell Technologies). Total colonies and differential lineage colonies were enumerated under inverted microscope 12–14 days after incubation.
qPCR
Samples from myeloid differentiation culture, expansion culture or CFU colonies were analyzed for copy number of integrated viral vector using specific primers by qPCR using previously described methods [6]. The number of copies of integrated viral vector (woodchuck hepatitis virus posttranslational regulatory element [WPRE]) detected was normalized to cell number (copies of apolipoprotein B) to calculate the average copy number/cell. To determine the absolute copy number per cell of clones, individual colonies derived from methylcellulose cultures were isolated, and qPCR was performed using same methods described above.
Mice
NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ (NSG) were originally obtained from the Jackson Laboratory (Bar Harbor, ME, http://www.jax.org) and then bred for these studies at the City of Hope National Medical Center Animal Resources Center. The mice were housed in an Association for Assessment and Accreditation of Laboratory Animal Care-accredited barrier facility under specific pathogen-free environment. All experimentation with mice was performed under protocols approved by the Institutional Animal Care and Use Committee (IACUC) of City of Hope National Medical Center/Beckman Research Institute and the Guide for the Care and Use of Laboratory Animals [33].
Mouse Transplantation
All animal transplant experiments were conducted according to an IACUC-reviewed and -approved protocol held by D.L.D. Adult (8–10 weeks old) NSG mice were irradiated at 270 cGy 24 hours prior to transplantation. The mice were injected intravenously with 1 × 106 CD34+ HSPCs per animal in saline for injection (APP Pharmaceuticals, Lake Zurich, IL, http://www.fresenius-kabi.us) in cohorts of 10 or 11 mice per condition. The mice were either transplanted with nontransduced CD34+ HSPCs, HSPCs transduced without rapamycin, or HSPCs transduced with 20 μg/ml rapamycin, each following overnight stimulation and 24 hours of transduction. Animals were maintained on sulfamethoxazole and trimethoprim (Hi-Tech Pharmacal, Amityville, NY, http://www.hitechpharm.com) water and autoclaved food with subcutaneous hydration (0.9% NaCl solution) as required after transplantation. All animal husbandry was performed according to the IACUC standard procedures. At 16 weeks post-transplant, bone marrow and spleens from mice transplanted with HSPCs were harvested and analyzed by FACS.
Flow Cytometric Analysis of Engraftment
Mice were necropsied 16 weeks post-transplantation for analysis of engraftment. Single cell suspensions of bone marrow (femurs) and spleen were prepared by mechanical dissociation and red cells lysed using red blood cell lysis buffer (Sigma-Aldrich). All cell suspensions were pretreated with human immunoglobulin (GammaGard; Baxter, Westlake Village, CA, https://www.baxter.com) for 30 minutes to block nonspecific antibody staining. Spleen cell suspensions were stained with a human pan-leukocyte antibody to CD45-PC7 (BioLegend, San Diego, CA, http://www.biolegend.com) and lineage-specific anti-human CD3-ECD and CD4-APC (Invitrogen) for 20 minutes and washed twice with 1 ml of PBS containing 0.1% bovine serum albumin (Sigma-Aldrich). Bone marrow cells were stained with anti-human antibodies to CD45-PC7 (Beckman Coulter) and CD14-APC-Alexa 750 (Invitrogen). For isotype controls, we used matching Ig isotypes conjugated to APC from (BD Biosciences), ECD, APC-Alexa 750, and PC7 (Beckman Coulter). Samples were analyzed using Gallios cytometer as described before.
Statistical Analysis
Statistical analysis was carried out using GraphPad Prism 6.03 software (GraphPad Software, Inc., La Jolla, CA, http://www.graphpad.com). Pairs of data sets were analyzed for statistical significance using Student’s t test (95% confidence interval, two-tailed, paired or unpaired test); multiple data set comparisons were performed with one-way or two-way analysis of variance as specified in figure legends. The results are presented as means ± SEM. SEMs are shown as error bars in figures. Significance is shown as follows: ∗, p < .05; ∗∗, p < .01; ∗∗∗, p < .001; and ∗∗∗∗, p < .0001.
Results
Rapamycin Enhances Genetic Modification of Adult HSPCs With Lentiviral Vectors
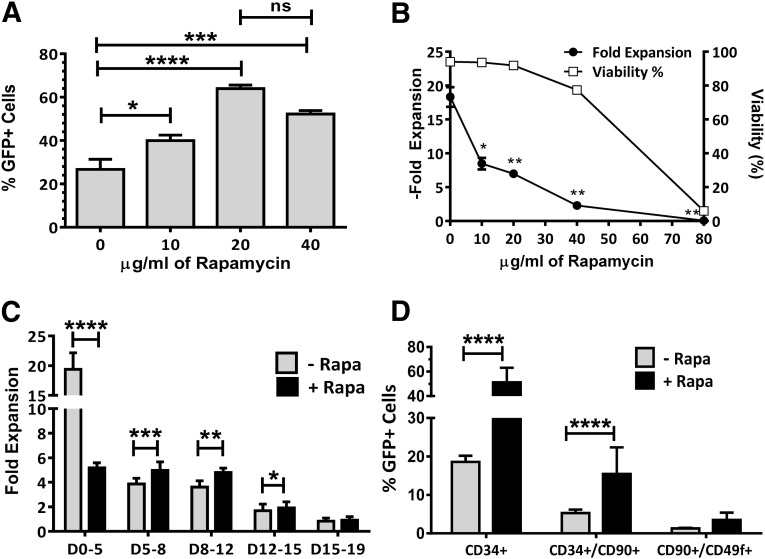
For these studies, we used G-CSF-mobilized apheresis products from healthy donors because this is the most commonly used source of HSPCs for gene therapy clinical studies. CD34+ HSPCs were isolated using a magnetic selection device (CliniMACS; Miltenyi Biotec) using methods previously developed in our laboratory [34]. CD34-enriched HSPCs were prestimulated in serum-free expansion medium (StemSpan SFEM) containing hematopoietic cytokines (SFT6) and an aryl-hydrocarbon receptor antagonist (SR-1) because these conditions were previously shown to maintain the stem cell contents of HSPCs [31]. After prestimulation, cells were transduced with a lentiviral vector (LV2) encoding enhanced green fluorescent protein (GFP) in the presence of 0–80 μg/ml rapamycin. Cells were incubated with virus for 24 hours and then washed and placed in stem cell expansion cultures (SFEM + SFT6 + SR-1) for 5 days, after which they were counted and analyzed for viability, cell growth, and GFP expression. Treatment of cells with up to 40 μg/ml rapamycin resulted in significant enhancement of gene marking with little to no toxicity, but the optimal enhancement of gene marking was observed using 20 μg/ml rapamycin (Fig. 1A, 1B). The use of 80 μg/ml rapamycin induced significant cell death, and it was not possible to analyze transduction in these samples. The growth of cells during the first 5 days of culture was significantly inhibited (p < .05) at all rapamycin concentrations, but this was expected because rapamycin is a known cell cycle inhibitor (Fig. 1B). Cell cycle analysis of HSPCs from three donors 20 hours after treatment with rapamycin demonstrated that the percentage of HSPCs going through the S phase was reduced from an average of 39% to <1% with a significant portion (60%–70%) of the cells arrested in either G0 or G1 of the cell cycle (supplemental online Fig. 1).
Figure 1.
Rapamycin dose effect on transduction efficiency, cell viability, and cell growth. (A): LV2 transduction of hematopoietic stem and progenitor cells (HSPCs) (percentage of GFP expression) over a range of rapamycin concentrations (n = 3). (B): Cell growth and viability of the same samples shown in (A). (C): Incremental in vitro HPSC proliferation following transduction with or without rapamycin (n = 7). (D): Phenotypic analysis of GFP expression in subpopulations of HSPCs 7 days after transduction in the presence or absence of rapamycin (n = 9 donors for CD34+ and CD34+/CD90+, n = 3 for CD90+/CD49f+). Height of bars represents the average value, and error bars represent SEM. ∗, p < .05; ∗∗, p < .01; ∗∗∗, p < .001; ∗∗∗∗, p < .0001. Abbreviations: D, days; GFP, green fluorescent protein; HSPC, hematopoietic stem-progenitor cells; LV, lentivirus; ns, not significant; Rapa, rapamycin.
Evaluation of cell growth over 19 days of culture following treatment with 20 μg/ml rapamycin demonstrated that the growth inhibition was reversible and that rapamycin-treated cells grew better than controls between days 5 and 15 of culture, after which growth was similar between treated and untreated cultures (Fig. 1C).
CD34+ HSPCs are known to be a mixture of multipotent hematopoietic progenitors and more primitive hematopoietic stem cells. CD34+ cells that coexpress the CD90 antigen are highly enriched in primitive hematopoietic stem cells [35], and this population can be further enriched in CD90+ cells that also coexpress CD49f [36]. In order to determine the gene marking of these more primitive cell populations, we performed 19 transductions on HSPCs from 9 healthy volunteers in 5 independent experiments and assayed after 7 days for the level of GFP expression in various (phenotypically defined) stem and progenitor cell compartments. We observed enhancement of gene transduction of CD34+ HSPCs, as well as the CD34+/CD90+ and CD34+/CD90+/CD49f+ primitive hematopoietic stems in the presence of rapamycin (supplemental online Fig. 2). This enhancement was highly reproducible and averaged 3-fold in CD34+ (n = 9 donors, 19 transductions), 3.3-fold in CD34+/CD90+ (n = 9 donors, 19 transductions), and 1.5-fold in CD34+/CD90+/CD49f+ (n = 3 donors and transductions) cell populations, although the enhancement did not reach statistical significance in the CD34+/CD90+/CD49f+ cells (Fig. 1D).
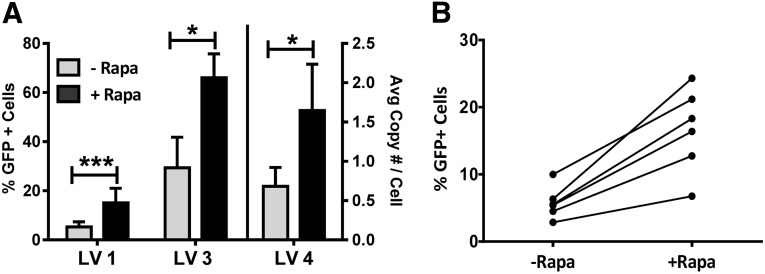
We also evaluated whether the transduction was reproducible using other vectors and different donors or in the absence of SR-1. CD34+ HSPCs from multiple donors were prestimulated and transduced in the presence or absence of rapamycin with three different lentiviral vectors (LV1, LV3, and LV4) in the absence of SR-1, and the percentage transduction was measured for each group after 7 days of expansion culture (Fig. 2A). We observed a similar enhancement of transduction by rapamycin with all three vectors in the absence of SR-1. Interestingly, LV4 did not contain a GFP gene, and the analysis of the level of transduction was performed by quantitative PCR for the WPRE element in the vector. Thus, the magnitude of observed enhancement of transduction with rapamycin treatment (threefold) was not dependent on method of analysis. We also observed enhancement of transduction in HSPCs from each of six independent donors, although the absolute level of enhancement by rapamycin did vary with donor (Fig. 2B).
Figure 2.
Rapamycin enhances genetic modification of adult hematopoietic stem and progenitor cells (HSPCs) with multiple lentiviral vectors. (A): Enhancement of genetic modification of adult HSPCs in the presence of rapamycin with multiple vectors. GFP+ percentage analysis from LV1 (n = 9) and LV3 (n = 3) transduced cells (left y-axis) and transgene copy number analysis by quantitative polymerase chain reaction from LV4 (n = 3) transduced cells (right y-axis) after 7 days of expansion culture. (B): Increase in transduction of individual donor hematopoietic stem and progenitor cells in response to rapamycin treatment. CD34+ cells from six donors were transduced with LV1 and analyzed for GFP+ expression after 7 days of expansion culture. Treatment groups were analyzed for significance using the Student’s paired, two-tailed t test. Height of bars represents the average value, and error bars represent SEM. ∗∗∗, p < .001; ∗, p < .05. Abbreviations: Avg, average; GFP, green fluorescent protein; HSPC, hematopoietic stem/progenitor cells; LV, lentivirus; Rapa, rapamycin.
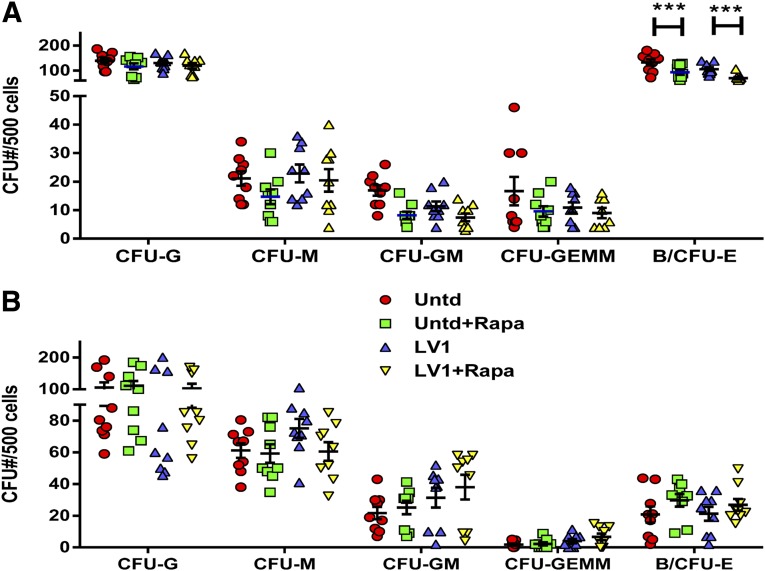
Lentiviral Transduction in the Presence of Rapamycin Has Minimal but Reversible Effects on Hematopoietic Colony Formation
We used in vitro colony formation assays to assess the effect of rapamycin treatment on hematopoietic potential of transduced HSPCs. CD34+ HSPCs from three donors were transduced with lentiviral vector (LV1) in the presence or absence of 20 μg/ml rapamycin and then plated 12–16 hours after transduction in methylcellulose cultures in triplicate. Alternatively, HSPCs were transduced as described, cultured in SFEM + SFT6 + SR-1 for 7 days, and then plated in methylcellulose cultures in triplicate. All cultures were scored for the number of CFU-G, CFU-M, CFU-GM, CFU-GEMM, and B/CFU-E 2 weeks after plating. Cells plated immediately after transduction showed no differences in the number of myeloid colonies produced but had a small but statistically significant reduction in the number of erythroid colonies formed (p < .001) (Fig. 3A). When the cells were cultured for 7 days in expansion medium prior to plating for CFU, no significant differences in the number of colonies of any type between rapamycin-treated and untreated cultures were observed (Fig. 3B). These data further support the reversibility of the growth inhibitory effects of rapamycin treatment.
Figure 3.
Rapamycin effect on hematopoietic potential. CFU assays were conducted as described in Materials and Methods. (A, B): 500 cells from each sample were plated in methylcellulose (in triplicate) 1 day after transduction with LV1 (A) or after 7 days of expansion in SFT6 + SR-1 (B). Colony number for each colony type was scored on day 14. The average number of colonies from each sample is shown with a bar for each colony type. Statistical analysis was performed using paired t tests (n = 3 donors). Height of bars represents the average value, and error bars represent SEM. ∗∗∗, p < .001. Abbreviations: B/CFU-E, late erythrocyte; CFU, colony-forming unit; G, granulocytes; GEMM, granulocyte, erythrocyte, monocyte, megakaryocyte; GM, granulocyte-macrophage progenitor; LV, lentivirus; M, monocytes; Rapa, rapamycin; SFT6, stem cell factor, flt-3 ligand, thrombopoietin, and interleukin-6; Untd, untransduced.
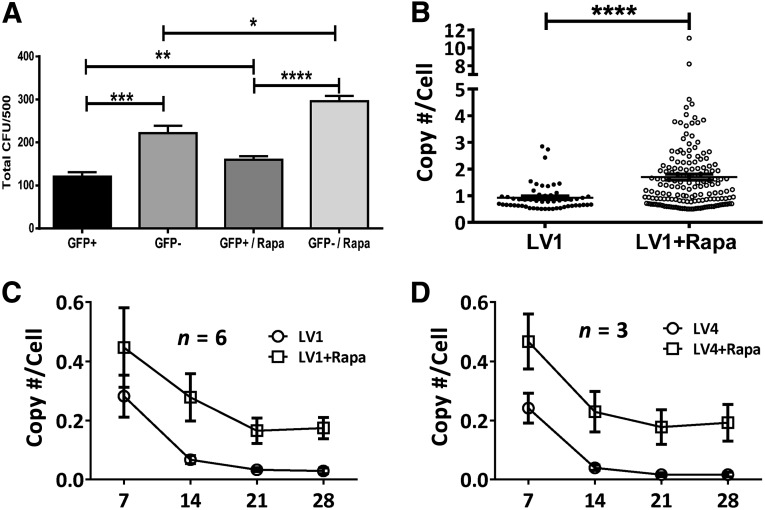
Previous studies, including those conducted in our laboratory, have shown that lentiviral vectors transiently decrease the growth of cells [37, 38] and (D.L.D., unpublished observation). To distinguish between the effects of transduction and rapamycin treatment on hematopoietic colony formation, we fractionated cells into genetic modified (GFP+) and nonmodified (GFP−) populations derived from transduction in the presence or absence of rapamycin by fluorescence-activated cell sorting. Treated cell samples were all cultured for 7 days in expansion medium prior to cell sorting to maintain the frequency of primitive hematopoietic cells. Sorted cells were plated in triplicate in methylcellulose, and total colonies were counted after 14 days. Among the groups tested, GFP+ cells gave rise to significantly fewer colonies than GFP− cells regardless of whether they were derived from transductions in the presence or absence of rapamycin (Fig. 4A). However, transduction in the presence of rapamycin resulted in an increase in the number of colonies from both the GFP+ and GFP− populations. This is possibly due to the expression of GFP protein or high levels of shRNA expressed from these vectors and is consistent with results reported by our group and others [39, 40].
Figure 4.
Analysis of colony-forming potential and transgene copy number following transduction in rapamycin. (A): Rapamycin enhanced colony-forming potential of genetically modified cells. CD34+ cells were transduced with LV1 (multiplicity of infection, 10) after overnight prestimulation in SFT6 followed by 7 days of expansion in SFEM SFT6 SR1. GFP+ and GFP− cells were sorted and plated for CFU. Statistical analysis was performed with paired t test (n = 3). (B): Methylcellulose colonies of LV1-transduced cells with or without rapamycin treatment were picked, lysed, and analyzed by quantitative polymerase chain reaction with WPRE- and apolipoprotein B (ApoB)-specific primers as described in Materials and Methods. The ratio of transgene copy number determined by WPRE and cell number determined by ApoB were calculated and presented. The data sets were analyzed for statistical significance by paired t test. (C, D): Stability of transgene average copy number per cell in 4 weeks differentiation cultures of transduced hematopoietic stem and progenitor cells using LV1 (C) and LV4 (D). Height of bars represents the average value, and error bars represent SEM. ∗, p < .05; ∗∗, p < .01; ∗∗∗, p < .001; and ∗∗∗∗, p < .0001. Abbreviations: CFU, colony-forming unit; GFP, green fluorescent protein; LV, lentivirus; Rapa, rapamycin; WPRE, woodchuck hepatitis virus post-translational regulatory element.
Transduction of HSPCs With Rapamycin Results in a Nominal Increase in the Average Number of Integrated Copies of Lentiviral Vector and Is Stable
We placed the HSPCs into methylcellulose cultures following transduction in the presence or absence of rapamycin to generate colonies for analysis of the average lentiviral vector copy number per cell using previously described methods [6]. Although there was an increase in the average copy number per cell in the presence versus the absence of rapamycin (1.7 vs. 0.92, respectively), 160 of 162 (98.7%) of colonies had <5 integrated copies per cell in the rapamycin-treated group, with only 2 colonies carrying 8 and 11 copies per cell, respectively (Fig. 4B). These data suggest that rapamycin treatment increases the frequency of transduction more than the magnitude of transduction.
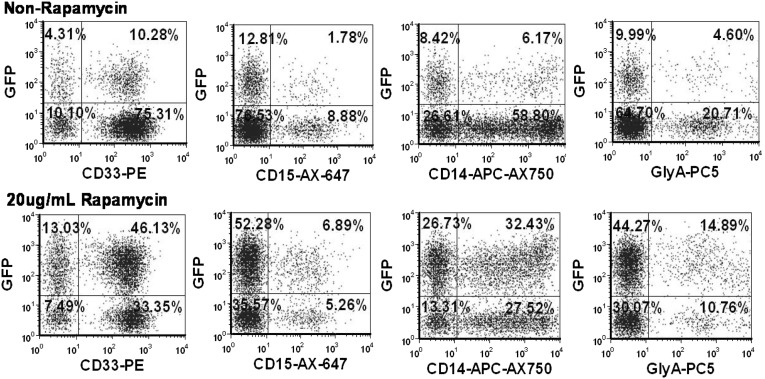
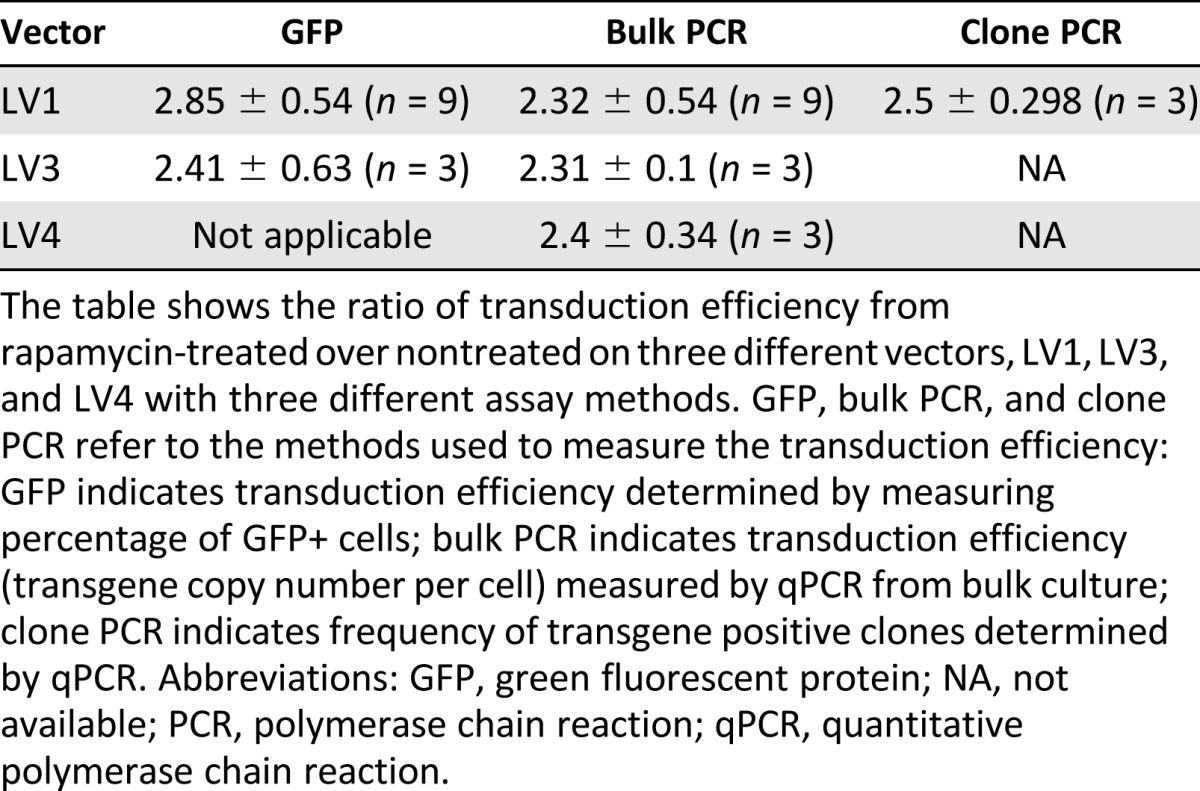
In order to understand the stability of the enhanced lentiviral vector integration, we also performed 4-week bulk cultures on cells transduced with 2 different lentiviral vectors (LV1 and LV4) and again used PCR analysis of vector sequences to determine the average number of copies per cell of integrated vector. Although we observed an overall reduction in the average copy number per cell over the first 2–3 weeks of the culture period, the rapamycin-treated cells maintained a higher overall level of integrated vector for each of two different vectors (Fig. 4C, 4D). No changes in the level of gene marking were observed in the cultures of cells treated with rapamycin between weeks 3 and 4, suggesting that transgenes were integrated and stable after 3 weeks and that gene marking was enhanced in CD33+, CD14+, and CD15+ myeloid cells as well as glycophorin A+ erythroid progenitor cells (Fig. 5). These results suggest that the subpopulations of cells that maintain long-term cultures are also more highly transduced in the presence of rapamycin and are consistent with the phenotypic analysis data of transduction described above that suggests that the more primitive stem cell compartment is transduced at a higher level in the presence of rapamycin. Overall, irrespective of method of measurement, our data indicate that CD34+ HSPCs are transduced two- to threefold higher in the presence of rapamycin with minimal impact on in vitro hematopoietic activity (Table 1).
Figure 5.
Rapamycin-enhanced gene marking in differentiated lineage progenies. Phenotypic analysis of GFP+ cells in CD33+, CD14+, and CD15+ myeloid cells, as well as glycophorin A+ erythroid population. CD34+ cells were prestimulated overnight in SFT6 and transduced with LV3 (multiplicity of infection, 10) in the absence (top) and presence (bottom) of rapamycin during transduction. Transduced cells were harvested after overnight transduction and plated in myeloid differentiation bulk culture as described in Materials and Methods. The plots shown result from day 14 of culture. Abbreviation: GFP, green fluorescent protein; LV, lentivirus; SFT6, stem cell factor, flt-3 ligand, thrombopoietin, and interleukin-6.
Table 1.
Summary of enhanced transduction efficiency by rapamycin with different methods
Transduction of HSPCs in the Presence of Rapamycin Does Not Change the Integration Pattern of Vector
We performed integration site analysis on genomic DNA samples isolated from HSPCs transduced in the presence or absence of rapamycin to determine whether there were changes in the patterns of integration. Because our vectors were designed to PCR from within the lentiviral long terminal repeat (LTR), and there are two LTRs in reverse orientation in the integrated proviral construct, we detected both vector and genomic sequences. Those results that showed proper linker sequences showing homology to human DNA and with more than 5 read coverage were reported to ensure authenticity and were further analyzed for integration site. Similar (random) patterns and frequencies of integration into exons, introns, and intergenic sequences were observed for the samples transduced in the presence or absence of rapamycin (supplemental online Table 1). A complete listing of integration sites meeting the criteria described above is provided in supplemental online Tables 2 and 3. Therefore, rapamycin treatment during transduction does not appear to alter the integration pattern of the lentiviral vector.
Rapamycin-Enhanced Transduction of HSPCs Results in Improved Gene Marking of Myeloid and Lymphoid Lineages In Vivo
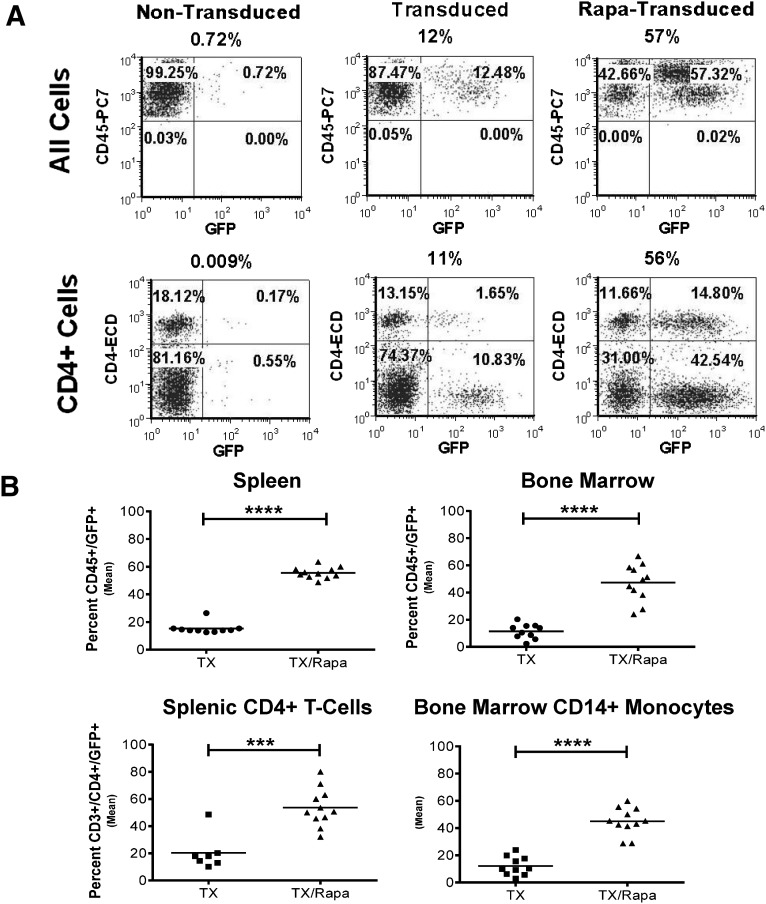
In order to better assess the effects of rapamycin on engraftment on long-term engrafting cells, we transplanted cohorts of immunodeficient NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ (NSG) mice with HSPCs transduced in the presence or absence of rapamycin and measured engraftment and gene marking in multiple blood cell lineages at 16 weeks after transplant. Although we did not observe an enhancement of overall engraftment of human cells (% CD45+) following treatment with rapamycin (data not shown), as has been previously reported [24], we did detect a significant increase in the level of GFP expression in the CD45+ and CD45+/CD4+ cell populations in the spleen of these mice (p < .0001) when the cells were transduced in the presence of rapamycin (Fig. 6A, 6B). Additionally, we observed equivalent levels of enhancement of gene marking and expression in the CD45+ cells and CD45+/CD14+ monocytes (p < .001) in the bone marrow of these mice. No tumors were evident in any of the mice tested, and no mice died as a result of the treatment, which supports the safety of the treatment. The relative level of genetic modification within control and rapamycin-treated groups was similar in all mice tested. These data demonstrate that enhancement of gene marking with rapamycin seen in vitro is also seen in the mature progeny of the HSPCs in vivo without any demonstrable loss of engrafting activity.
Figure 6.
Rapamycin treatment on CD34+ cells greatly enhanced engraftment of GFP+ cells in NSG mice spleen and bone marrow at 16 weeks. Cells were prestimulated overnight in SFT6 StemSpan SFEM medium before 24 hours of transduction, and then they were transplanted. (A): Fluorescence-activated cell sorting phenotypic analysis of spleens from NSG mice transplanted with 1 × 106 nontransduced CD34+ cells, transduced CD34+ cells without rapamycin, or transduced CD34+ cells with rapamycin treatment. Upper row: CD45+/GFP+ cells. Lower row: CD4+/GFP+ cells. (B): Statistical analysis of spleens and bone marrows from NSG mice transplanted with 1 × 106 transduced CD34+ cells without rapamycin (n = 10) or with rapamycin treatment (n = 11). The data were analyzed with unpaired two-tailed Student’s t test. ∗∗∗, p < .001; ∗∗∗∗, p < .0001. Abbreviations: GFP, green fluorescent protein; NSG, NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ; Rapa, rapamycin; SFEM, serum free expansion media; SFT6, stem cell factor, flt-3 ligand, thrombopoietin, and interleukin-6; Tx, transduction.
Mechanism of Action of Rapamycin
In order to understand the mechanism of action of rapamycin, we performed several experiments. First, we measured the effect of treatment with rapamycin on the expression of low-density lipoprotein (LDL) receptor, which has been shown to be the receptor for the VSV-G protein used to pseudotype our lentiviral vectors [41]. Our data demonstrate that the LDL receptor is down-regulated in the presence of rapamycin despite the higher level of transduction (supplemental online Fig. 3). We also evaluated another mTOR inhibitor (KU0063794), which has been shown to inhibit both mTORC1 and mTORC2, leading to arrest of cell cycling and induction of autophagy in renal cell carcinoma cell lines [42]. HSPCs were transduced under standard conditions, in the presence of 20 μg/ml rapamycin or KU0063794 ranging from 5 to 80 μM. The proliferation and GFP marking of the HSPCs was measured for each condition after 7 days of culture. Proliferation of HSPCs was inhibited in the presence of rapamycin and at all concentrations of KU0063794 tested (p < .05 for all groups compared with untreated control) (supplemental online Fig. 4). However, enhancement of transduction only occurred in the presence of rapamycin. Therefore, inhibition of cell proliferation is not sufficient to result in enhanced transduction of HSPCs.
Finally, we performed PCR analysis for WPRE DNA in transduced cells at 3 and 16 hours post-transduction to assess the average number of reverse transcribed vector copies per cell. There was no increase in the number of copies of vector per cell at 3 hours, but at 16 hours we observed a 2.5-fold increase in (presumably unintegrated) copies of vector per cell compared with transduction in the absence of rapamycin (p < .01) (supplemental online Fig. 5). These data support the enhancement of intracellular trafficking, leading to an enhanced level of reverse transcription of RNA vector as a major mechanism of action for rapamycin.
Discussion
Genetic modification of HSPCs has been successfully used to treat a significant number of monogenic diseases, leading to increased interest in this form of therapy for other indications. In two recently reported cases, high levels of genetic modification were observed in vivo following transplantation of gene-modified CD34+ HSPCs from patients with Wiscott-Aldrich syndrome or metachromatic leukodystrophy [3, 4]. These results were obtained using one to two transductions of HSPCs with very high doses of lentiviral vector each time (MOI = 100 per transduction). The amount of vector required for these studies is prohibitively expensive and not amenable to treatments of large cohorts of patients. Additionally, although not observed in these studies, the increased concentration of vector has the potential for increasing the number of viral integrants, and these methods may create significant safety issues in other indications or with other vectors. In other indications (adenosine deaminase [AD]/severe combined immunodeficiency [SCID]), high levels of genetically modified cells were observed because the target cell population (T and NK cells) had a natural growth advantage over nonmodified cells [43]. In other cases, however, in which the cells do not enjoy a growth advantage, modifying the majority of the HSPCs with a low number of integrated copies of vector is desirable and may be necessary to observe a clinical effect.
In our previous clinical trial, we observed levels of gene marking of HSPCs of between 10% and 20% immediately after transduction, but those levels dropped to approximately 1% after 4 weeks of culture and were similarly low in patients transplanted with the gene-modified HSPCs [6]. In order to improve the level of gene-modified cells in vivo for proof of concept studies, we sought a method for increasing transduction of adult HSPCs that was safe and reproducible and not dependent upon vector titer or composition. The process needed to be scalable and cGMP-compliant and ideally would not require the manufacturing or use of vast amounts of viral vector. The process would necessarily have to result in similarly high levels of genetic modification of the CD4+ T-cell and monocytic progeny of the HSPCs to be useful for our HIV gene therapy program, and we did not wish to use reagents with no prior clinical history.
The results presented here demonstrate that treatment of adult HSPCs with rapamycin results in gene-modified HSPCs that meet all of the above requirements. Moreover, rapamycin treatment at the time of transduction resulted in robust enhancement of the level of gene modification of CD34+ cells, as well as the more primitive CD34+/CD90+ stem cell populations from nine healthy donors irrespective of the viral vector used. Although rapamycin is a cell cycle inhibitor and we observed an initial delay in cell growth and erythroid colony formation in vitro, these differences were reversible and did not alter the overall hematopoietic potential of the HSPCs. The average copy number per cell was limited to approximately two in clones analyzed from CFU assays and bulk liquid culture analysis demonstrated a stable, higher level of transduction of monocytic, granulocytic, and erythroid progenitor cells in those cultures initiated with rapamycin-treated HSPCs. The prolonged marking of CD33+ myeloid progenitors that persist in these cultures for 4 weeks supports the phenotypic data that rapamycin enhances the transduction of more primitive hematopoietic stem cells that initiate long-term cultures, as does the engraftment of NSG mice with an increased frequency of gene-modified cells.
Use of an alternative mTOR inhibitor (KU0063794), which inhibits mTORC1 and mTORC2, also resulted in the inhibition of proliferation but did not result in the enhancement of transduction. Similar to studies in renal cell carcinoma lines, KU0063794 abrogates the effects of inhibition of mTORC1 by simultaneous inhibition of mTORC2. Because this inhibitor has also been shown to induce autophagy, it is reasonable to assume that induction of autophagy also occurs in HSPCs and that neither cell cycle inhibition nor autophagy induction are sufficient mechanisms for the enhancement of transduction induced by rapamycin. The increase in the average number of copies per cell of reverse transcribed vector measured in our PCR assays indicates that intracellular trafficking of internalized virus after rapamycin treatment leads to a higher rate of conversion of vector RNA to reverse transcribed DNA sequences. Because this process is observed as early as 16 hours, it is likely that little integration of the vector has taken place and that the mechanism of enhancement may entirely precede proviral integration. We are currently conducting additional testing in our laboratory to further elucidate the mechanism of action.
Conclusion
Importantly, we were also able to demonstrate the enhancement of transduction of CD4+ T cells and CD14+ monocytes in vivo using an immunodeficient mouse model. Although we did not observe an enhancement of the level of engraftment of human cells following treatment of HSPCs with rapamycin as has been previously reported for cord blood [24], we did not see any reduction in engraftment or skewing of lineages. Overall our data suggest that the enhancement of rapamycin acts on progenitor and long-term engrafting stem cells without altering the engraftment or lineage potential of HSPCs but does not lead to an unacceptable level of copies per cell of integrated vector or change the integration pattern of vector in the host cell genomic DNA. The ability to enhance transduction of our target populations (CD4+ T cell and monocytes) suggests that this is clinically relevant method for enhancing transduction of HSPCs with in vivo engrafting ability and multilineage potential. We are currently exploring the use of these methods in our clinical trials in stem cell-based gene therapy for HIV.
Supplementary Material
Acknowledgments
We thank Drs. John Rossi and John Zaia for critical review of the manuscript. We thank the staff of the City of Hope Animal Resource Center for animal breeding and husbandry and the Analytical Cytometry Core for cell sorting. We also thank Dr. Bruce Torbett for helpful discussions on the use of rapamycin to enhance lentiviral genetic modification of adult HSPCs and discerning mechanism of action. This research was supported by California Institute for Regenerative Medicine Grant TR2-01771 (to D.L.D.).
Author Contributions
L.L. and M.T.-C.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; A.G.: conception and design, collection and/or assembly of data; A.R., A.M.G., E.W.E., C.-A.T., and J.-H.W.: collection and/or assembly of data; N.G.: manuscript writing, final approval of manuscript; X.W.: collection and/or assembly of data, data analysis and interpretation; D.L.D.: conception and design, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 2.Cavazzana-Calvo M, Payen E, Negre O, et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiuti A, Biasco L, Scaramuzza S, et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biffi A, Montini E, Lorioli L, et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 5.Li MJ, Kim J, Li S, et al. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- 6.DiGiusto DL, Krishnan A, Li L, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiGiusto DL, Stan R, Krishnan A, et al. Development of hematopoietic stem cell based gene therapy for HIV-1 infection: Considerations for proof of concept studies and translation to standard medical practice. Viruses. 2013;5:2898–2919. doi: 10.3390/v5112898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Millington M, Arndt A, Boyd M, et al. Towards a clinically relevant lentiviral transduction protocol for primary human CD34 hematopoietic stem/progenitor cells. PLoS One. 2009;4:e6461. doi: 10.1371/journal.pone.0006461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee HJ, Lee YS, Kim HS, et al. Retronectin enhances lentivirus-mediated gene delivery into hematopoietic progenitor cells. Biologicals. 2009;37:203–209. doi: 10.1016/j.biologicals.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Hangoc G, Campbell TB, et al. Identification of parameters required for efficient lentiviral vector transduction and engraftment of human cord blood CD34(+) NOD/SCID-repopulating cells. Exp Hematol. 2008;36:947–956. doi: 10.1016/j.exphem.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis BM, Humeau L, Slepushkin V, et al. ABC transporter inhibitors that are substrates enhance lentiviral vector transduction into primitive hematopoietic progenitor cells. Blood. 2004;104:364–373. doi: 10.1182/blood-2003-07-2363. [DOI] [PubMed] [Google Scholar]
- 12.Donahue RE, Sorrentino BP, Hawley RG, et al. Fibronectin fragment CH-296 inhibits apoptosis and enhances ex vivo gene transfer by murine retrovirus and human lentivirus vectors independent of viral tropism in nonhuman primate CD34+ cells. Mol Ther. 2001;3:359–367. doi: 10.1006/mthe.2001.0269. [DOI] [PubMed] [Google Scholar]
- 13.Haas DL, Case SS, Crooks GM, et al. Critical factors influencing stable transduction of human CD34(+) cells with HIV-1-derived lentiviral vectors. Mol Ther. 2000;2:71–80. doi: 10.1006/mthe.2000.0094. [DOI] [PubMed] [Google Scholar]
- 14.Wang CX, Sather BD, Wang X, et al. Rapamycin relieves lentiviral vector transduction resistance in human and mouse hematopoietic stem cells. Blood. doi: 10.1182/blood-2013-12-546218. 2014 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menendez JA, Vellon L, Oliveras-Ferraros C, et al. mTOR-regulated senescence and autophagy during reprogramming of somatic cells to pluripotency: A roadmap from energy metabolism to stem cell renewal and aging. Cell Cycle. 2011;10:3658–3677. doi: 10.4161/cc.10.21.18128. [DOI] [PubMed] [Google Scholar]
- 16.Jung CH, Ro SH, Cao J, et al. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webster AC, Lee VW, Chapman JR, et al. Target of rapamycin inhibitors (sirolimus and everolimus) for primary immunosuppression of kidney transplant recipients: A systematic review and meta-analysis of randomized trials. Transplantation. 2006;81:1234–1248. doi: 10.1097/01.tp.0000219703.39149.85. [DOI] [PubMed] [Google Scholar]
- 18.Sehgal SN. Rapamune (RAPA, rapamycin, sirolimus): Mechanism of action immunosuppressive effect results from blockade of signal transduction and inhibition of cell cycle progression. Clin Biochem. 1998;31:335–340. doi: 10.1016/s0009-9120(98)00045-9. [DOI] [PubMed] [Google Scholar]
- 19.Grube E, Buellesfeld L. Rapamycin analogs for stent-based local drug delivery. Everolimus- and tacrolimus-eluting stents. Herz. 2004;29:162–166. doi: 10.1007/s00059-004-2556-6. [DOI] [PubMed] [Google Scholar]
- 20.Holdaas H, Midtvedt K, Åsberg A. A drug safety evaluation of everolimus in kidney transplantation. Expert Opin Drug Saf. 2012;11:1013–1022. doi: 10.1517/14740338.2012.722993. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen SA, Walker D, Gillespie MB, et al. mTOR inhibitors and its role in the treatment of head and neck squamous cell carcinoma. Curr Treat Options Oncol. 2012;13:71–81. doi: 10.1007/s11864-011-0180-2. [DOI] [PubMed] [Google Scholar]
- 22.Voss MH, Molina AM, Motzer RJ. mTOR inhibitors in advanced renal cell carcinoma. Hematol Oncol Clin North Am. 2011;25:835–852. doi: 10.1016/j.hoc.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abouelnasr A, Roy J, Cohen S, et al. Defining the role of sirolimus in the management of graft-versus-host disease: From prophylaxis to treatment. Biol Blood Marrow Transplant. 2013;19:12–21. doi: 10.1016/j.bbmt.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 24.Rohrabaugh SL, Campbell TB, Hangoc G, et al. Ex vivo rapamycin treatment of human cord blood CD34+ cells enhances their engraftment of NSG mice. Blood Cells Mol Dis. 2011;46:318–320. doi: 10.1016/j.bcmd.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung J, Scherer LJ, Gu A, et al. Optimized vectors for HIV gene therapy: Multiplexed expression of small RNAs and inclusion of MGMT (P140K) drug resistance gene. Mol Ther. 2014;22:952–963. doi: 10.1038/mt.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Escarpe P, Zayek N, Chin P, et al. Development of a sensitive assay for detection of replication-competent recombinant lentivirus in large-scale HIV-based vector preparations. Mol Ther. 2003;8:332–341. doi: 10.1016/s1525-0016(03)00167-9. [DOI] [PubMed] [Google Scholar]
- 27.Manilla P, Rebello T, Afable C, et al. Regulatory considerations for novel gene therapy products: A review of the process leading to the first clinical lentiviral vector. Hum Gene Ther. 2005;16:17–25. doi: 10.1089/hum.2005.16.17. [DOI] [PubMed] [Google Scholar]
- 28.Sastry L, Xu Y, Johnson T, et al. Certification assays for HIV-1-based vectors: Frequent passage of gag sequences without evidence of replication-competent viruses. Mol Ther. 2003;8:830–839. doi: 10.1016/j.ymthe.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 29.Trobridge GD, Beard BC, Gooch C, et al. Efficient transduction of pigtailed macaque hematopoietic repopulating cells with HIV-based lentiviral vectors. Blood. 2008;111:5537–5543. doi: 10.1182/blood-2007-09-115022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beard BC, Trobridge GD, Ironside C, et al. Efficient and stable MGMT-mediated selection of long-term repopulating stem cells in nonhuman primates. J Clin Invest. 2010;120:2345–2354. doi: 10.1172/JCI40767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boitano AE, Wang J, Romeo R, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Krymskaya L, Wang J, et al. Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther. 2013;21:1259–1269. doi: 10.1038/mt.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. National Research Council. Guide for the Care and Use of Laboratory Animals. 8th ed. Washington, D.C.: The National Academies Press, 2011. [Google Scholar]
- 34.Tran CA, Torres-Coronado M, Gardner A, et al. Optimized processing of growth factor mobilized peripheral blood CD34+ products by counterflow centrifugal elutriation. Stem Cells Translational Medicine. 2012;1:422–429. doi: 10.5966/sctm.2011-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baum CM, Weissman IL, Tsukamoto AS, et al. Isolation of a candidate human hematopoietic stem-cell population. Proc Natl Acad Sci USA. 1992;89:2804–2808. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Notta F, Doulatov S, Laurenti E, et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- 37.Lee CI, Kohn DB, Ekert JE, et al. Morphological analysis and lentiviral transduction of fetal monkey bone marrow-derived mesenchymal stem cells. Mol Ther. 2004;9:112–123. doi: 10.1016/j.ymthe.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 38.Nayak SK, McCallister T, Han LJ, et al. Transduction of human renal carcinoma cells with human gamma-interferon gene via retroviral vector. Cancer Gene Ther. 1996;3:143–150. [PubMed] [Google Scholar]
- 39.An DS, Qin FX, Auyeung VC, et al. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol Ther. 2006;14:494–504. doi: 10.1016/j.ymthe.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung J, Scherer LJ, Gu A, et al. Optimized lentiviral vectors for HIV gene therapy: Multiplexed expression of small RNAs and inclusion of MGMT(P140K) drug resistance gene. Mol Ther. 2014;22:952–963. doi: 10.1038/mt.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amirache F, Lévy C, Costa C, et al. Mystery solved: VSV-G-LVs do not allow efficient gene transfer into unstimulated T cells, B cells, and HSCs because they lack the LDL receptor. Blood. 2014;123:1422–1424. doi: 10.1182/blood-2013-11-540641. [DOI] [PubMed] [Google Scholar]
- 42.Zhang H, Berel D, Wang Y, et al. A comparison of Ku0063794, a dual mTORC1 and mTORC2 inhibitor, and temsirolimus in preclinical renal cell carcinoma models. PLoS One. 2013;8:e54918. doi: 10.1371/journal.pone.0054918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaspar HB, Bjorkegren E, Parsley K, et al. Successful reconstitution of immunity in ADA-SCID by stem cell gene therapy following cessation of PEG-ADA and use of mild preconditioning. Mol Ther. 2006;14:505–513. doi: 10.1016/j.ymthe.2006.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.