Abstract
Galectin-3 has been reported to regulate the functions of a number of immune cell types. We previously reported that galectin-3 is translocated to immunological synapses in T cells upon T-cell receptor engagement, where it associates with ALG-2-interacting protein X (Alix). Alix is known to coordinate with the endosomal sorting complex required for transport (ESCRT) to promote human immunodeficiency virus (HIV)-1 virion release. We hypothesized that galectin-3 plays a role in HIV-1 viral budding. Cotransfection of cells of the Jurkat T line with galectin-3 and HIV-1 plasmids resulted in increased HIV-1 budding, and suppression of galectin-3 expression by RNAi in Hut78 and primary CD4+ T cells led to reduced HIV-1 budding. We used immunofluorescence microscopy to observe the partial colocalization of galectin-3, Alix and Gag in HIV-1-infected cells. Results from co-immunoprecipitation experiments indicate that galectin-3 expression promotes Alix-Gag p6 association, whereas the results of Alix knockdown suggest that galectin-3 promotes HIV-1 budding through Alix. HIV-1 particles released from galectin-3-expressing cells acquire the galectin-3 protein in an Alix-dependent manner, with proteins primarily residing inside the virions. We also found that the galectin-3 N-terminal domain interacts with the proline-rich region of Alix. Collectively, these results suggest that endogenous galectin-3 facilitates HIV-1 budding by promoting the Alix-Gag p6 association.
Keywords: Alix, galectin-3, HIV-1, viral budding
Introduction
Human immunodeficiency virus (HIV) triggers decreases in both the number and function of CD4+ T lymphocytes. Its replication cycle consists of three stages: attachment and entry, gene and protein expression and assembly and budding. Currently available anti-HIV-1 drugs target viral or host proteins involved in these stages (Takeuchi and Matano 2008; Martin-Serrano and Neil 2011). The plasma membrane is the final barrier that HIV-1 must cross during its egress from infected cells (Martin-Serrano and Neil 2011). Viral Gag proteins (which drive HIV-1 assembly and release and which are associated with the inner leaflet of the plasma membrane) oligomerize into spherical protein shells by deforming the attached membrane. The buds grow and pinch off from the cell surface, leading to the release of immature virions (Martin-Serrano and Neil 2011; Weiss and Gottlinger 2011). HIV-1 is believed to seize components of endosomal sorting complexes required for transport (ESCRT), thereby promoting membrane scission from the cytosolic side of bud necks via late-domain motifs (Weiss and Gottlinger 2011). The ESCRT machinery is primarily associated with endosomes, where it recognizes membrane proteins modified by ubiquitination and sorts cargoes into membrane domains for the purpose of forming the intralumenal vesicles (ILVs) of multivesicular bodies (Hurley and Emr 2006). Topologically, ILV formation is identical to enveloped virus budding (Martin-Serrano and Marsh 2007). The ESCRT pathway is also localized at the plasma membrane and facilitates plasma membrane vesicle formation and cytokinesis (Caballe and Martin-Serrano 2011; Elia et al. 2011; McCullough et al. 2013).
HIV-1 Gag p6 contains two late domains, Pro-Thr/Ser-Ala-Pro (PTAP) and Tyr-Pro-Xn-Leu (YPXL), that interact with Tsg101 and ALG-2-interacting protein X (Alix), respectively; both are essential for efficient viral budding (Garrus et al. 2001; Fisher et al. 2007; Dussupt et al. 2009; Fujii et al. 2009). Alix consists of three domains: (i) an N-terminal Bro1 domain that is homologous to yeast Bro1 and contains a binding site for the ESCRT-III component-charged multivesicular body protein 4; (ii) a central V domain that binds to YPXL motifs of p6; and (iii) a proline-rich region (PRR) that binds to a number of cellular factors, including endophilins, ALG2 and the ESCRT-I component Tsg101. Deletion of Alix or transfection of dominant-negative Alix has an inhibitory effect, although a modest one, on HIV-1 budding in model cell lines. Alix overexpression rescues the release and infectivity of PTAP-truncated HIV-1 (Fisher et al. 2007; Usami et al. 2007).
Galectin-3 (Gal3), a member of the β-galactoside-binding lectin family (Liu et al. 2002; Liu and Rabinovich 2005; Rabinovich et al. 2007; Vasta 2009), has a carbohydrate-recognition-binding domain (CRD) that contains ∼130 amino acids connected to a nonlectin N-terminal region (∼120 amino acids), which is responsible for lectin oligomerization and ligand cross-linking. Galectin-3 is known to function extracellularly by interacting with cell-surface glycoconjugates. However, it is also capable of acting intracellularly in a manner that may be independent of its lectin properties (Liu et al. 2002; Yang et al. 2008). Galectin-3 has also been described as a component of intracellular vesicles, such as phagosomes and exosomes (Thery et al. 2001; Liu et al. 2002; Yang et al. 2008). We previously used a yeast two-hybrid system to identify Alix as a galectin-3-binding partner (Chen et al. 2009). Moreover, we observed galectin-3 translocation to the cytosolic face of the immunological synapse in T cells following T-cell receptor engagement, as well as galectin-3 association with Alix (Chen et al. 2009).
Exogenous addition of recombinant galectin-1 has been shown to promote HIV-1 infectivity via extracellular stabilization of viral adhesion to host cells, including CD4+ T lymphocytes and macrophages (Ouellet et al. 2005; Mercier et al. 2008; St-Pierre et al. 2011). However, no data have been published regarding the involvement of endogenous galectin in HIV-1 expansion or propagation. HIV-1 infection is known to increase galectin-3 (CBP35) mRNA levels in MOLT-3T cells (Schroder et al. 1995), and the HIV-1 Tat protein has been shown to be responsible for galectin-3 upregulation in several human cell types (Fogel et al. 1999). Thus, we hypothesized that an intracellular association between endogenous galectin-3 and Alix may be involved in the regulation of HIV-1 functions in infected cells. We found that endogenous galectin-3 is capable of promoting HIV-1 budding via stabilization of the Alix-Gag p6 complex. Furthermore, we observed an association between galectin-3 and the Alix PRR domain via its N-terminal region. Our results suggest that inhibitors of the galectin-3-Alix interaction may have anti-HIV-1 therapeutic effects.
Results
Endogenous galectin-3 promotes HIV-1 budding
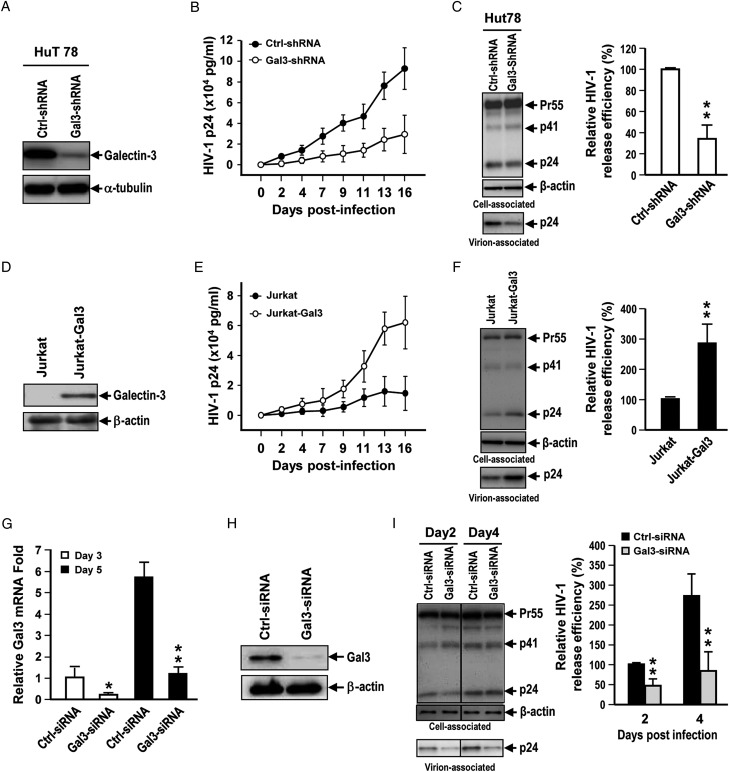
To understand the effects of galectin-3 on HIV-1 release kinetics, we infected galectin-3 knockdown and control Hut78T cells with HIV-1, and then measured the quantities of HIV-1 particles released over time (Figure 1A and B). We observed significantly lower release kinetics in galectin-3-knockdown cells (Figure 1B). The long-term HIV-1 release process entails progeny virus budding, reinfection and replication. We assessed the short-term virus release kinetics to evaluate the effect of galectin-3 on HIV-1 budding. As shown in Figure 1C, galectin-3 knockdown in Hut78T cells significantly reduced HIV-1 release efficiency on day 2 postinfection (P < 0.01). To confirm the effects of galectin-3 on HIV-1 release, we infected galectin-3-overexpressing Jurkat (Jurkat-Gal3) and parental Jurkat T cells (Figure 1D) with HIV-1 and quantified the release of HIV-1 particles. The data indicated that galectin-3 expression enhanced HIV-1 release kinetics (Figure 1E). We also found that galectin-3 expression in Jurkat T cells significantly promoted HIV-1 release efficiency on day 2 postinfection (P < 0.01) (Figure 1F). Additionally, our data showed that neither galectin-3 knockdown nor overexpression affected HIV-1 viral protein expression, cell proliferation or Gag processing (the proteolytic cleavage of Gag by the viral protease) (Supplementary data, Figure S1A–D).
Fig. 1.
Endogenous Galectin-3 enhances HIV-1 virus release. (A) Lentiviral shRNA-mediated knockdown of galectin-3 was performed in Hut78 cells and galectin-3 levels were determined by immunoblotting. (B) Control and galectin-3-knockdown Hut78 cells were infected with NL4-3 virions. Supernatants were collected at different time points for HIV-1 p24 measurement by enzyme-linked immunosorbent assay (ELISA). (C) Viral supernatants and cell lysates were collected for immunoblotting analysis and ELISA for HIV-1 p24 on day 2 postinfection. Relative HIV-1 release efficiency was calculated by dividing the amount of Gag(p24) in viral lysates by the total amount of Gag(p24) in cell and viral lysates. (D) Lentivirus-mediated galectin-3 expression was performed in Jurkat cells and galectin-3 levels were determined by immunoblotting. (E) Jurkat and Jurkat-Gal-3 cells were infected with NL4-3 viruses. Supernatants were collected at different time points for HIV-1 p24 measurement by ELISA. (F) Viral supernatants and cell lysates were collected for immunoblotting analysis and ELISA for HIV-1 p24 on day 2 postinfection. Relative HIV-1 release efficiency was calculated in (C). (G) Human primary CD4+ T cells were subjected to galectin-3 knockdown by treatment with siRNAs. Relative mRNA levels of galectin-3 in control and galectin-3-siRNA-treated primary CD4+ T cells cultured for 3 or 5 days were analyzed by quantitative RT-PCR. (H) Galectin-3 protein expression in control and galectin-3-siRNA-treated primary CD4+ T cells was analyzed by immunoblotting. (I) Control and galectin-3-siRNA-treated primary CD4+ T cells were infected with HIV-1; supernatants and cell lysates were collected for immunoblotting analysis and ELISA for HIV-1 p24, and relative HIV-1 release efficiency was calculated in (C). Quantitative data represent the means ± SD of results from three independent experiments. Significance values were calculated using two-tailed Student's t-tests (*P < 0.05; **P < 0.01).
We also cotransfected HEK293T cells with vectors expressing galectin-3 and HIV-1 NL4-3 (pNL4-3), and collected virus-containing supernatants for HIV-1 p24 ELISAs. These same supernatants were used to infect JLTRG cells (Jurkat cells containing the GFP reporter gene controlled by the HIV-1 LTR promoter). A correlation was noted between the amount of viral budding and the level of galectin-3 expression (Supplementary data, Figure S2A–C). Similar results were observed with Magi-5 cells (Supplementary data, Figure S2D–G).
Last, we confirmed the role of galectin-3 in HIV budding in human CD4+ T lymphocytes isolated from healthy donors and activated with PHA and IL-2, which contain galectin-3 (Supplementary data, Figure S3A). When galectin-3 expression was suppressed by siRNA prior to HIV infection (Figure 1G and H), we observed significantly reduced HIV-1 release from cells (Figure 1). In these experiments, we confirmed that galectin-3 levels did not significantly affect cell viability within 7 days postinfection (data not shown).
Galectin-3 is associated with Alix in HIV-1-infected cells
Alix and Tsg101 have been described as facilitating HIV-1 budding via interaction with HIV-1 Gag p6 (Strack et al. 2003; Martin-Serrano and Marsh 2007). We previously reported an association between galectin-3 and Alix in the immunological synapses of activated T cells following TCR engagement (Chen et al. 2009). Co-immunoprecipitation assays were performed to confirm the association between Alix and galectin-3; the results indicated that Alix was pulled down when galectin-3 was immunoprecipitated, and galectin-3 was pulled down when Alix was immunoprecipitated (Figure 2A). We also found that galectin-3 was not associated with Tsg101 (Figure 2A). The results of immunofluorescent staining from the present study indicated partial colocalization of HIV-1 Gag, Alix and galectin-3 in both HIV-1-infected Magi-5 cells (Figure 2B) and human primary CD4+ T cells (Figure 2C). Total internal reflection fluorescence (TIRF) data combined with super resolution (SR) analyses also indicated partial Alix colocalization with HIV-1 Gag and galectin-3 on the membranes of HIV-1-infected T cells (Figure 2D).
Fig. 2.
Galectin-3 association with Alix in HIV-1-infected cells. (A) pFlag-Gal3 and pNL4-3 vectors were cotransfected into HEK293T cells, followed by incubation for 48 h at 37°C. After three washes with PBS containing 30 mM lactose, cells were treated with the chemical cross-linker DSP, lysed and immunoprecipitated with rabbit anti-Flag (left and right panel) or rabbit anti-Alix (middle panel) antibodies. Precipitated proteins were analyzed by immunoblotting for Flag, galectin-3, Alix and Tsg101 proteins. H-chain indicates IgG heavy chain; C and T refer to control (nontransfected) and transfected groups, respectively. Magi-5 (B) and primary human CD4+ cells (C) were infected with HIV-1 NL4-3 viruses. After 24 h, cells were fixed and stained with antibodies against the indicated proteins and observed using confocal microscopy. Representative cells are shown. Colocalization values for Gag (green) with Alix (red) and for Alix (red) with galectin-3 (blue) in 50 stained cells of each type were measured using Global Pearson's Correlation Coefficient statistics, in Volocity version 6.1.1 (PerkinElmer). (D) HIV-1-infected Jurkat-Gal3T cells were fixed and stained with antibodies against the indicated proteins. Stained cells were analyzed by TIRF and SR fluorescence microscopy. The images were recorded from the lower membrane of the cells and each dot in the SR images represents a protein molecule. The colocalization coefficients of two proteins on the lower membranes of single cells were determined by Manders' analysis (Manders et al. 1993). Protein colocalization was quantified using two-dimensional spatial histogram maps of two fluorescence channels, with fluorescence backgrounds removed during the intensity-based colocalization analyses. Results from one representative experiment of three independently performed experiments are shown.
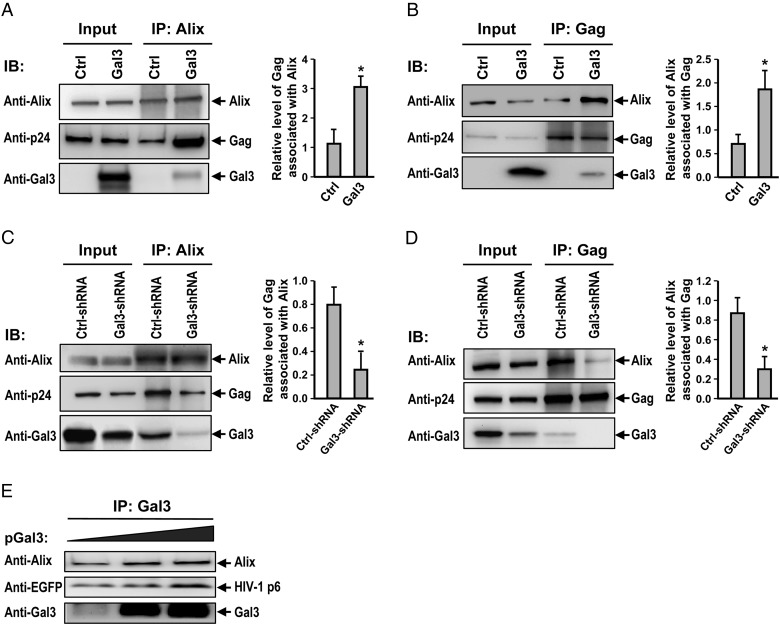
Galectin-3 promotes Alix-Gag association
We performed co-immunoprecipitation assays to test our hypothesis that galectin-3 promotes HIV-1 budding by enhancing Alix-Gag association. All transfected cells were pretreated with a membrane-permeable chemical cross-linker to stabilize the existing complexes. Although the isotype control antibody did not pull-down Alix or Gag proteins, in galectin-3-overexpression and knockdown cells (data not shown), specific antibodies resulted in co-immunoprecipitation of the relevant components. We found that galectin-3 expression led to a 3-fold increase in the levels of Gag co-precipitated with Alix (P < 0.05) (Figure 3A), and a 2.6-fold increase in the levels of Alix co-precipitated with Gag (P < 0.05) (Figure 3B). Galectin-3 knockdown was used to confirm the impact of galectin-3 on Alix-Gag interaction. We noted that the quantity of Gag associated with Alix was reduced by 60% (P < 0.05) (Figure 3C) and that the amount of Alix associated with Gag was reduced by 70% (P < 0.05) (Figure 3D). We also observed that an increase in galectin-3 expression triggered (o) a stronger association between Alix and Gag and (ii) a positive correlation between levels of transfected galectin-3 and levels of Alix and Gag co-precipitated with galectin-3 (Figure 3E). Regarding the effect of galectin-3 on HIV-1 Gag processing, we found that galectin-3 expression and knockdown did not significantly affect Gag processing in HEK293T and Magi-5 cells (Supplementary data, Figure S3B).
Fig. 3.
Galectin-3 promotes Alix-Gag association. HEK293T cells were cotransfected with pNL4-3, pEF1-Gal3 or a control pEF-1 vector, lysed, immunoprecipitated with anti-Alix (A) or anti-Gag (B) antibodies, and immunoblotted with the indicated antibodies. Immunoblot band intensities were quantified by densitometry. Amounts of co-immunoprecipitated Gag (A) or Alix (B) were normalized to pulled down Alix (A) or Gag (B) levels, subsequently. (C) Control and galectin-3-knockdown Magi-5 cells were transfected with pNL4-3, lysed, immunoprecipitated with anti-Alix (C) or anti-Gag (D) antibodies, and subjected to immunoblotting with the indicated antibodies. Immunoblot band intensities were quantified by densitometry. Amounts of co-immunoprecipitated Gag (C) or Alix (D) were subsequently normalized to pulled down Alix levels (C) or Gag (D) levels. (E) pEGFP-p6 (2 μg) was cotransfected with different concentrations of pEF1-Gal3 (0–2 μg) into HEK293T cells. Cell lysates were subjected to co-immunoprecipitation assays with anti-galectin-3, followed by immunoblotting with the indicated antibodies. Representative immunoblotting results are shown. Quantitative data represent the means ± SD of results from three independent experiments. Significance values were calculated using two-tailed Student's t-tests (*P < 0.05; **P < 0.01).
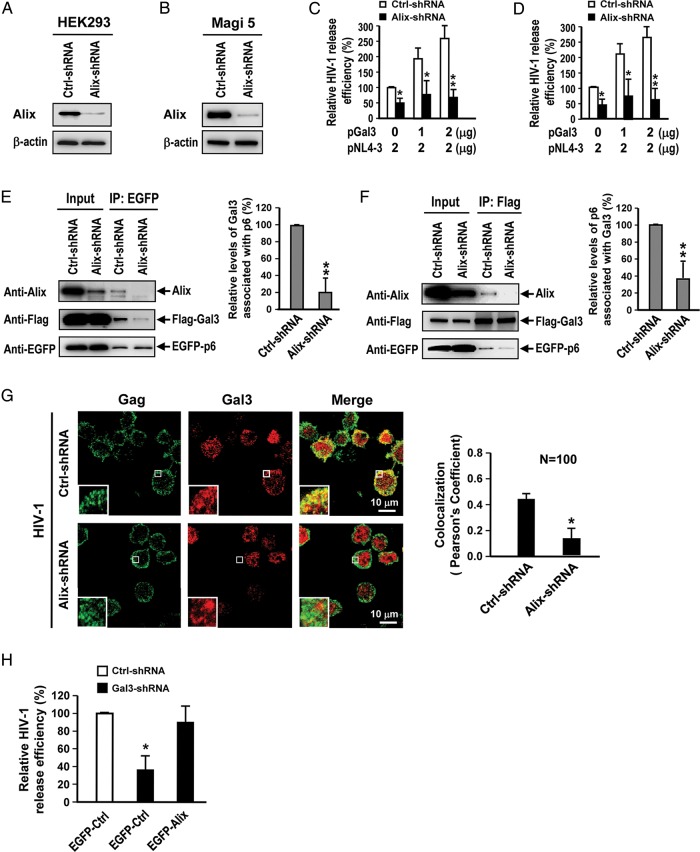
Galectin-3 interaction with Alix is required to enhance HIV-1 budding
Our next task was to determine whether the promotion of HIV-1 budding by galectin-3 is Alix dependent. After suppressing Alix expression in HEK293T and Magi-5 cells with lentiviral shRNA (Figure 4A and B), we cotransfected cells with galectin-3 and HIV-1 plasmids. The data indicated that the effect of galectin-3 on promotion of HIV-1 budding was significantly attenuated by Alix knockdown in both cell lines (Figure 4C and D). To further examine the role of Alix in the galectin-3-mediated promotion of HIV-1 budding, we cotransfected Alix knockdown HEK293T cells with pFlag-Gal3 and pEGFP-p6, and then performed co-immunoprecipitation assays to detect galectin-3, Alix and Gag p6 complexes after cross-linker pretreatment. Results indicated that Alix knockdown substantially reduced the amounts of galectin-3 associated with p6 and that the levels of galectin-3 co-immunoprecipitated with p6 were reduced by 80% (P < 0.01) (Figure 4E). Further, Alix knockdown reduced the amount of p6 associated with galectin-3, and levels of p6 co-immunoprecipitated with galectin-3 were reduced by 70% (P < 0.01) (Figure 4F). In addition, the results of immunofluorescent staining showed that Alix knockdown significantly reduced the colocalization of galectin-3 and Gag in HIV-1-infected CD4+ T cells (Figure 4G). Finally, we also demonstrated that Alix overexpression rescued the effect of galectin-3 knockdown in T cells (Figure 4H).
Fig. 4.
Galectin-3 depends on Alix to facilitate HIV-1 budding. (A and B) Lentivirus shRNA-mediated Alix knockdown in HEK293T and Magi-5 cells was analyzed by immunoblotting. (C and D) Control and Alix-knockdown HEK293T and Magi-5 cells were cotransfected with different ratios of pNL4-3 and pEF1-Gal3. Supernatants and cell lysates were collected for HIV-1 p24 ELISA. Relative HIV-1 release efficiency was calculated by dividing the amount of Gag(p24) in viral lysates by the total amount of Gag(p24) in cell and viral lysates. (E) Control and Alix-knockdown HEK293T cells were cotransfected with pEGFP-p6 and pFlag-Gal3. The lysates were subjected to a co-immunoprecipitation assay followed by immunoblot analysis; the band intensities were quantified by densitometry. Amounts of co-immunoprecipitated galectin-3 were normalized to the levels of pulled down p6, and the normalized quantities of galectin-3 for Alix-knockdown cells relative to the control group are shown. (F) Control and Alix-knockdown HEK293T cells were cotransfected with pEGFP-p6 and pFlag-Gal3. The lysates were subjected to a co-immunoprecipitation assay followed by immunoblot analysis; the band intensities were quantified by densitometry. Amounts of co-immunoprecipitated p6 were normalized to the pulled down galectin-3 levels and the normalized quantities of p6 for Alix-knockdown cells relative to the control group are shown. (G) HIV-1 infected control and Alix-knockdown Hut78 cells were stained with antibodies against HIV-1 Gag and galectin-3 prior to observation using confocal microscopy. Colocalization values for Gag (green) with galectin-3 (red) in 100 control or Alix-knockdown cells were measured using Global Pearson's Correlation Coefficient statistics. (H) pEGFP (EGFP-Ctrl) or pEGFP-Alix (EGFP-Alix) was transfected into control and galectin-3 knockdown Hut78T cells. Following HIV-1 infection, supernatants and cell lysates were collected for p24 ELISAs, and relative HIV-1 release efficiency was calculated by dividing the amount of Gag(p24) in viral lysates by the total amount of Gag(p24) in cell and viral lysates. In Quantitative data represent the means ± SD of results from three independent experiments. Significance values were calculated using two-tailed Student's t-tests (*P < 0.05; **P < 0.01).
Cells transfected with the L41A mutant, which contains a mutation at the amino acid L41 within the Alix-binding motif of Gag p6, were used to further establish the association among Alix, Gag and galectin-3. A co-immunoprecipitation assay confirmed a much weaker association between Alix and this mutant, independently of the presence of galectin-3 (Supplementary data, Figure S3C). In an immunofluorescence staining assay, the colocalization of galectin-3 and Gag was significantly lower in HIV-1 L41A mutant-infected CD4+ T cells than in wild-type NL4-3-infected cells (P < 0.01) (Supplementary data, Figure S3D). Collectively, these results suggest that galectin-3 depends on Alix to promote HIV-1 budding.
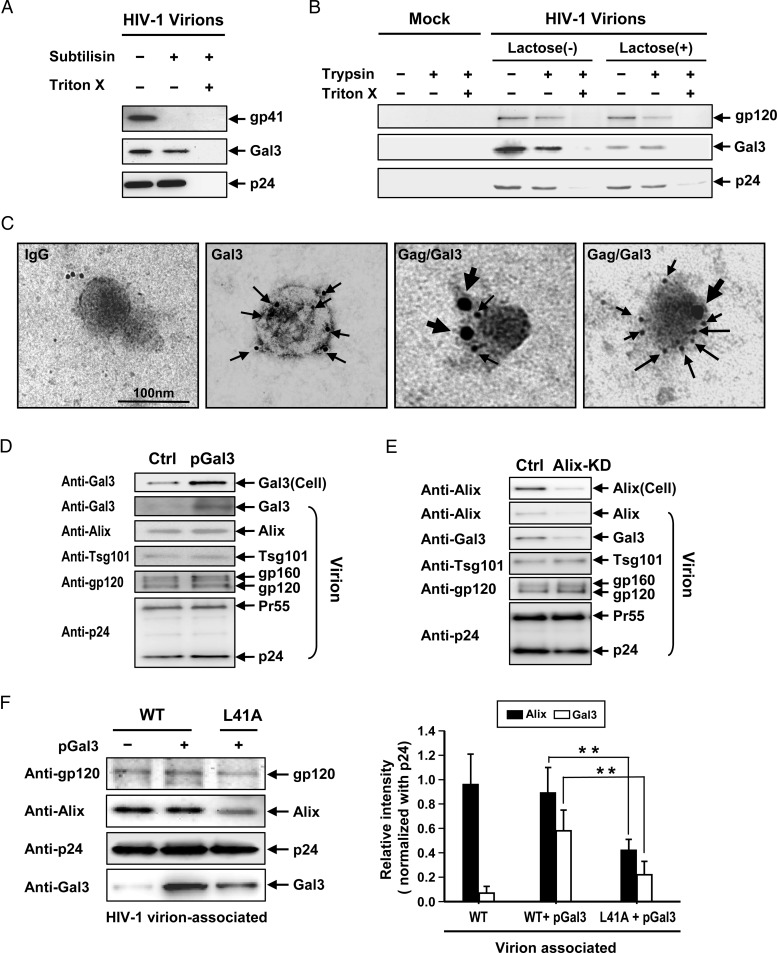
Galectin-3 is acquired by HIV-1 and primarily resides inside virions
Because some host proteins involved in intracellular transport (e.g. those associated with the ESCRT machinery) are capable of incorporation into HIV-1 virions (Booth et al. 2006; Chertova et al. 2006), we searched for galectin-3 in HIV-1 virions released from galectin-3-expressing cells. Virions collected from HEK293T cells cotransfected with galectin-3 and pNL4-3 were purified and treated with subtilisin to digest extracellular proteins displayed on virions and to digest other microvesicles possibly co-purified with the virions. Immunoblotting results indicated that galectin-3 was contained in HIV-1 virions and was largely resistant to digestion by subtilisin (Figure 5A). Complete galectin-3 protein digestion occurred when virions were treated with a mixture of Triton X-100 and subtilisin (Figure 5A). This finding was confirmed by washing the virions with or without lactose (a galectin-3 inhibitor known to elute galectin-3 present on the exterior surfaces of cells; Frigeri and Liu 1992), prior to treating them with trypsin in the presence or absence of Triton X-100 (Figure 5B). Our data indicated that complete digestion of galectin-3 by trypsin only occurred following virion permeabilization with Triton X-100 (Figure 5B). Together, these results suggest that galectin-3 is enclosed within the viral envelope. However, the combination of lactose treatment and trypsin digestion in the absence of Triton X-100 did result in an apparent decrease in galectin-3, suggesting that a fraction of the galectin-3 proteins associated with the virions was present on the exterior of the virions (Figure 5B). Immunoelectron microscopy data supported these conclusions (Figure 5C). Immunoblot results indicated that other host proteins, Alix and Tsg101, are also acquired by HIV-1 virions (Figure 5D). A semi-quantitative flow cytometry assay (using anti-gp120 antibody-coated beads to capture virions for further detection of galectin-3 using anti-galectin-3 antibodies) (Supplementary data, Figure S4A) also confirmed the presence of galectin-3 in HIV-1 virions at high levels (Supplementary data, Figure S4B), as well as the presence of a small quantity of galectin-3 on the exterior surfaces of virions, which could be eliminated by lactose treatment (Supplementary data, Figure S4C).
Fig. 5.
Presence of Galectin-3 in HIV-1 virions. pEF1-Gal3 and pNL4-3 plasmids were cotransfected into HEK293T cells. Supernatants were collected for HIV-1 virion purification via sucrose gradients. (A) Sucrose gradient-purified HIV-1 virions (100 ng, as quantified by p24 ELISAs) were left untreated or were treated with subtilisin (1 mg/mL) at room temperature for 1 h in the absence or presence of Triton X-100 (1.5%). Treated and untreated virions were subjected to immunoblotting analyses for each indicated protein. (B) Purified HIV-1 virions (100 ng, as quantified by p24 ELISAs) were left untreated or were treated with lactose and subjected to trypsin digestion (150 μg/mL) at room temperature for 1 h in the absence or presence of Triton X-100 (1.5%). Treated and untreated virions were subjected to immunoblotting analyses for each indicated protein. (C) Purified virions were loaded on grids, fixed and stained with goat anti-galectin-3 (Gal3), mouse anti-p24 (Gag) or control IgG (IgG), followed by Donkey anti-goat-labeled 6-nm gold particles and Donkey anti-mouse-labeled 18-nm gold particles. Following 4% uranyl acetate staining, grids were examined by TEM (scale bar =100 nm; thin and thick arrows indicate galectin-3 and Gag protein localization, respectively). (D) Purified HIV-1 virions were analyzed by immunoblotting for each indicated protein. (E) Alix-knockdown Magi-5 cells were transfected with pNL4-3 and incubated at 37°C for 48 h. The supernatants were collected for HIV-1 virion purification. Purified virions and cell lysates were subjected to immunoblotting assays for the indicated proteins. (F) HEK293T cells were transfected with pNL4-3 or pL41A together with pGal3. The supernatants were collected for HIV-1 virion purification. Purified HIV-1 wild-type or L41A mutant virions were analyzed by immunoblotting for each indicated protein.
In addition, immunoblotting results indicated that HIV-1 virions released from Alix-knockdown HEK293T cells contained a significantly lower amount of galectin-3 (Figure 5E). A semi-quantitative assay also indicated a significant decrease in galectin-3 in HIV-1 virions released from Alix-knockdown CD4+ T cells (Supplementary data, Figure S4D). The L41A mutant was used to further confirm this observation. Immunoblotting results indicated that L41A virions acquired smaller quantities of Alix and galectin-3 than wild-type virions (Figure 5F). Semi-quantitative flow cytometry analysis produced similar results (Supplementary data, Figure S5A and B). These results indicate that galectin-3 is acquired by virions through interaction with Alix.
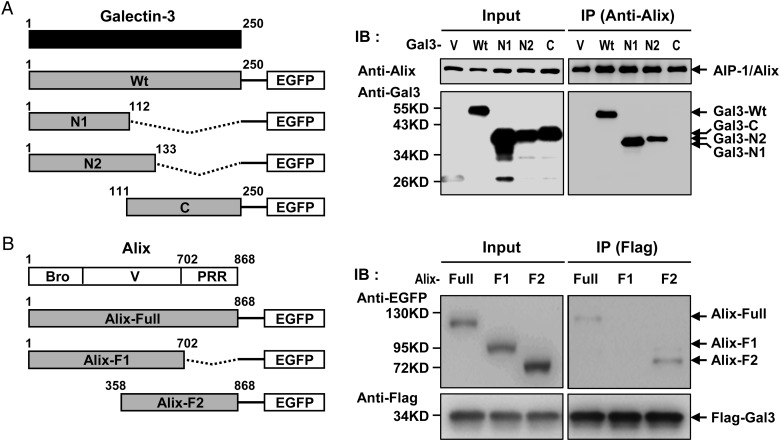
The galectin-3 N-terminal domain interacts with the Alix PRR
To identify the molecular domains involved in Alix binding by galectin-3, we cotransfected various plasmids containing truncated forms of galectin-3 plus HIV-1 plasmids into HEK293T cells. The results of a co-immunoprecipitation assay suggested that galectin-3 interacted with Alix via the N-terminal region (Figure 6A). We also tested Alix mutants to map its binding domain, and found that galectin-3 binds to the Alix PRR (Figure 6B).
Fig. 6.
(A) Galectin-3 N-terminal region interacts with the PRR of Alix. Various truncated forms of galectin-3 (wild type, N1, N2 and C domains) were cloned into pEFGP-N1 and used to cotransfect HEK293T cells with pNL4-3. Transfected cell lysates were immunoprecipitated with anti-Alix and precipitated proteins were subjected to immunoblotting with anti-galectin-3 antibodies. V, vector control; Wt, wild type; N1, N-terminal region (residues 1–112) of galectin-3; N2, N-terminal region (residues 1–133) of galectin-3; C, C-terminal region containing the entire CBD domain (residues 111–250) of galectin-3. (B) Various truncated forms of Alix (including Full, F1 and F2) were cloned into pEFGP-N1 and used to cotransfect HEK293T cells with pFlag-Gal3 and pNL4-3. Cell lysates were immunoprecipitated with antibodies against Flag; precipitated proteins were immunoblotted with anti-Alix antibodies.
Discussion
Exogenous addition of galectins can stabilize the attachment of viruses (HTLV-1, HCV, EBV and HIV-1) to cells via interaction with glycans on viruses or cell surfaces (Gauthier et al. 2008; Mercier et al. 2008; King et al. 2009; Shim et al. 2011; Yang et al. 2011). However, to the best of our knowledge, no reports have been published on the function of endogenous galectins in viral expansion and propagation. In the present study, we found that endogenous galectin-3 plays an important role in HIV-1 budding in T cell lines and primary CD4+ T cells via intracellular functions. Using cell lines that express limited quantities of galectin-3 (e.g. HEK293T and Jurkat), we examined the effects of increased galectin-3 expression, induced by gene transfection, on HIV-1 budding. In cell lines expressing higher levels of galectin-3 (e.g. Magi-5 and Hut78), we analyzed the effects of knockdown of galectin-3 expression. In both cases, we noted a strong positive correlation between the galectin-3 expression level and HIV-1 budding, suggesting that galectin-3 promotes HIV-1 propagation. In several independent experiments, galectin-3 was observed to promote HIV-1 budding, inducing an increase of 2- to 4-fold in various cell lines. It should also be noted that galectin-3 substantially promoted (>5-fold) HIV-1 propagation in longer-term cultures (Figure 1B and E).
HIV-1 typically assembles at and buds from the plasma membranes of T and macrophage-lineage cells. Previous studies have suggested that it engages the Gag p6 structural protein to subvert host Tsg101 and Alix in the vacuolar protein sorting pathways to facilitate viral budding (Garrus et al. 2001; Strack et al. 2003; Stuchell et al. 2004). The release of HIV-1 from T-cells involves two distinct and independent pathways. The first is controlled by TSG101 (Garrus et al. 2001), which plays an important role in both viral budding and infectivity; the second is controlled by ALIX, which plays a role in budding and also in other discrete functions in infectivity (Fisher et al. 2007). In the present study, our data showed that galectin-3 is associated with Alix, but not Tsg101. We suggest that the ability of galectin-3 to promote HIV-1 budding is primarily dependent on its interaction with Alix.
Proteomic data reported by Chertova et al. (2006) indicated that Alix and Tsg101 are incorporated into HIV-1 virions. Here, we demonstrated that galectin-3 is incorporated into virions released from galectin-3-expressing cells, and confirmed the presence of both Alix and Tsg101 in these virions (Figure 5). According to past reports, galectin-3 is present in dendritic cell-derived exosomes (Thery et al. 2001), and exosomes and HIV Gag both bud from the endosome-like domains of T-cell plasma membranes (Booth et al. 2006). Additionally, Jouvenet et al. (2006) reported that HIV-1 assembly is initiated and completed at the plasma membrane and not at the endosomal membrane. Our data suggest that galectin-3 is involved in the ESCRT-mediated release pathway and is destined for transport to the cell membrane during the HIV-1 budding process, and that this mechanism occurs via associations with Alix and Gag.
Our galectin-3 immunoprecipitation and immunofluorescence staining results indicated an association between galectin-3 and Alix in HIV-1-infected cells (Figure 2). As an accessory protein in the ESCRT pathway, Alix plays a role in endocytic membrane trafficking, cell adhesion and budding of viruses such as HIV-1 (Strack et al. 2003; Fujii et al. 2007). Alix interacts with the Gag p6 domain (Chen et al. 2005; Odorizzi 2006; Fujii et al. 2007) but is also capable of interacting with Tsg101, leading to association with ESCRT-I and CbI-interacting protein 85 (CIN85) (Odorizzi 2006). Other researchers have reported that Gag p6–Alix interaction is essential during HIV-1 replication and that mutations at the Alix-binding site of p6 resulted in impaired HIV-1 replication and decreased efficiency of viral release in a number of cell types (Fisher et al. 2007; Fujii et al. 2009). In the present study, we observed that the association between galectin-3 and Alix resulted in enhanced HIV-1 budding, perhaps via stabilization of the Alix–Gag p6 interaction. We also found that when Alix expression was suppressed by shRNA, both the promotion of HIV-1 budding by galectin-3 and the association between galectin-3 and Gag were reduced (Figure 4). These results suggest that galectin-3 depends on Alix to interact with Gag p6 and on the ESCRT complex to regulate viral budding.
Here, we established that it is the N-terminal region of galectin-3 (residues 1–112, a region containing proline-rich tandem repeats) and not the C-terminal carbohydrate-recognition domain that binds to Alix. Accordingly, galectin-3 interacts with Alix via protein–protein rather than lectin–glycoconjugate interactions, analogous to its interactions with a number of other cellular proteins such as Gemin4, CBP70, Chrp and Bcl-2 (Liu et al. 2002; Yang et al. 2008; Chen et al. 2009). Alix consists of three major regions (the Bro1 domain, the V domain and a PRR) and engages in multiple cellular processes via its domain architecture (Chen et al. 2005; Odorizzi 2006; Fisher et al. 2007). The C-terminal PRR contains binding sites for several proteins that support the participation of Alix in diverse cellular processes that facilitate viral budding (Fisher et al. 2007; Fujii et al. 2007; Gottlinger 2007; Martin-Serrano and Marsh 2007). It is noteworthy that the Tsg101-binding site in the Alix PRR provides a potential mechanism for linking with HIV-1 late-domain-binding partners (Fisher et al. 2007). Previously, researchers have reported that some factors (e.g. Tsg101, CEP55, endophilins, ALG-2 and Src kinase) that bind to the PRR contribute to Alix activation by releasing the PRR from the Bro1-V domain and exposing the YPXL late-domain-binding site (Carlton et al. 2008; Zhai et al. 2011). According to Usami et al. (2007), the extreme C-terminus of the PRR is essential for the ability of Alix to promote HIV-1 budding and may connect Alix to a yet-to-be-identified cofactor that is required to support its viral budding function. Based on our finding that galectin-3 binds to the PRR domain of Alix, we suggest that this link contributes to Alix activation and to the stable binding of Alix to Gag p6.
We previously reported that Jurkat cells transfected with galectin-3 cDNA constitutively overexpressed galectin-3 protein and exhibited higher growth rates than control transfectants (Yang et al. 1996). However, in the present study we found that, following short-term HIV infection, galactin-3 knockdown or overexpression did not affect cell proliferation in Hut78 and Jurkat cells (Figure 1A–F). We believe that this may be due, at least in part, to differences in the serum concentration of the culture medium (10% FBS in the present study vs. a low serum concentration in the previous study). In addition, some viral factors (such as HIV-1 Vpr and Tat) that display anti-apoptosis functions may affect cell proliferation or viability regulated by galectin-3 in T cells. However, it should be noted that although we did not observe any significant differences between control and galectin-3-overexpression or knockdown CD4+ T cells in T cell viability during short-term HIV-1 infection, the long-term HIV-1 infection data showed that galectin-3 expression significantly reduced virus-induced cell death (data not shown).
Currently, two known mechanisms underlying infection of target cells by HIV are (i) cell-free viral infection and (ii) viral transmission through cell-to-cell transfer involving the virological synapse (Piguet and Sattentau 2004). Previously, our group found that galectin-3 colocalizes with Alix at the immunological synapse of activated T cells (Chen et al. 2009) and that galectin-3 localizes to membrane lipid rafts in dendritic cells (Hsu et al. 2009). Because the virological synapse was reported to be functionally similar to the immunological synapse (Piguet and Sattentau 2004; Haller and Fackler 2008) and to require lipid raft integrity for cell-to-cell transfer of HIV (Jolly and Sattentau 2005), it is possible that galectin-3 may also participate in the formation of the virological synapse and influence this mode of HIV transfer.
The majority of functional studies of galectins have relied on the use of recombinant proteins to analyze extracellular activities. In this study, we focused on identifying the role of endogenous galectin-3 in HIV-1 infection, and we found that endogenous galectin-3 promotes HIV-1 budding by intracellular action through its association with Alix. We conclude that intracellular galectin-3 promotes HIV-1 infection and therefore may serve as a target molecule for the development of therapeutic drugs.
Materials and methods
Ethics statement
Written informed consent was obtained from all study participants, and approval was received from the Institutional Ethics Committee of National Yang-Ming University (approval number 1000065). All procedures were conducted according to committee rules.
Cell lines and viruses
Six cell lines were used in this research: HEK293T (a human kidney cell line), Jurkat (a human T-cell line, ATCC No. TIB-152), Hut78 (a human T-cell line, ATCC No. TIB-161), U87.CD4.CXCR4 (a human glioblastoma cell line expressing human CD4 and CXCR4), Magi-5 (HeLa cells expressing CD4, CXCR4 and CCR5, and containing the β-galactosidase gene controlled by HIV-1 LTR), and JLTRG (Jurkat cells containing the GFP reporter gene controlled by HIV-1 LTR). U87.CD4.CXCR4, Magi-5 and JLTRG cells were obtained from the US NIH AIDS Research and Reference Reagent program. X4 tropism HIV-1 NL4-3 viruses were generated by transfecting pNL4-3 (catalog no. 114) from the same reagent program into HEK293T cells and propagating HIV-1 viruses into U87.CD4.CXCR4 and Hut78 cells. HIV-1 virions were purified by sucrose gradient and subtilisin digestion according to previously described procedures (Ott 2009; Waheed et al. 2009).
Plasmids
For a detailed description of pEF1-Gal3 (pGal3) preparation see Yang et al. (1996). Briefly, galectin-3 cDNA was excised using EcoRI from clone 2.2 (Robertson et al. 1990) and cloned into pEF1-neo, which was prepared by replacing the CMV promoter in the pIRES1-neo bicistronic vector (Clontech, Palo Alto, CA). Truncated galectin-3 clones were prepared using PCR to amplify full-length galectin-3 (nt 1–750) and its N1 (nt 1–336), N2 (nt 1–399) and C fragments (nt 333–750), and then cloning these into pEGFP-N1 (Clontech, GenBank Accession No. U55762). Truncated Alix plasmids were prepared by cloning full-length Alix (nt 1–2604) and its F1 (nt 1–2160) or F2 fragments (nt 1074–2604) (Sakaguchi et al. 2005; Irie et al. 2007) into pEGFP-N1. The p6 L41A mutant NL4-3 clone (Munshi et al. 2007; Fujii et al. 2009) was kindly provided by Dr. Eric O. Freed.
Infectivity assays
Jurkat, JLTRG and Magi-5 cells (1 × 105 cells/well) were used in direct infection assays. Briefly, HIV-1 viruses (MOI = 0.1–0.01) were incubated with the cells for 2 h at 37°C in serum-free medium and subsequently washed out by PBS. Following incubation, 2 mL of RPMI or DMEM medium containing 10% serum, antibiotics and polybrene (8 µg/mL) were added, followed by incubation in 5% CO2 at 37°C for 48 h. Viral supernatants were collected and subjected to HIV-1 p24 measurements using an Alliance HIV-1 P24 Antigen ELISA Kit (PerkinElmer Life Sciences, Waltham, MA, USA; Cat. No. NEK050B). For JLTRG, HIV-1-infected cells were observed using confocal microscopy or flow cytometry. Mean fluorescent densities were measured using the FlowJo or Image-Pro Plus software packages.
HIV-1 replication kinetics assay
Briefly, Jurkat or Hut78T cells (5 × 105 cells/well) were infected with HIV-1 (MOI = 0.1) at 37°C for 2 h. After washing with PBS three times, HIV-1 infected T cells were incubated at 37°C for 16 days. Viral supernatants (200 μL) were collected and the RPMI 1640 culture medium (200 μL) was refreshed at different time points (from day 2 to day 16). The collected supernatants were subjected to p24 ELISA.
shRNA lentiviral system knockdown of galectin-3 and Alix
Galectin-3 shRNA (Gal3-shRNA) and Alix-shRNA were expressed in cells using lentiviral vectors. Gal3-shRNA cloned into the pLKO.1 plasmid was prepared by the National RNAi Core Facility, Academia Sinica, Taiwan. Gal3-shRNA-5′-CCGGGCAG TACAATCATCGGGTTAACTCGAGTTAACCCGATGATTGTACTGCTTTTT-3′ (TRCN 0000029307) and 5′-CCGGCCTGAATTACTGCAACGAAATCTCGAGATTTCGTTG CAGTAATTCAGGTTTTT-3′ (TRCN 0000029304) were used in Magi-5 and Hut78 cells, respectively. Alix-shRNA-5′-CCGGGCTGCTAAACATTACCAGTTTCTCGAGAAACTGG TAATGTTTAGCAGCTTTTT-3′ (TRCN 0000029394) and 5′-CCGGCCTGAATTACTGC AACGAAATCTCGAGATTTCGTTGCAGTAATTCAGGTTTTT-3′ (TRCN 0000029395) were used in HEK293 and Magi-5 cells, respectively. Luciferase shRNA-5′-CCGGA TCACAGAATCGTCGTATGCACTCGAGTGCATACGACGATTCTGTGATTTTTTG-3′ (TRCN 0000072244) was used as a negative control. For the lentiviral shRNA knockdown experiments, lentiviral Gal3-shRNAs or Alix-shRNAs were used to infect HEK293T, Magi-5 or Hut78 cells (MOI = 5) in the presence of 8 μg/mL polybrene. Cells were harvested 5 days posttransduction, and levels of galectin-3, Alix mRNA and proteins were measured.
Primary CD4+ T-cell purification and culture
Peripheral blood mononuclear cells were culled from venous blood collected from healthy adult HIV-1(−) donors by Ficoll-Paque™ PLUS density gradient centrifugation (GE Healthcare). CD4+ T cells were purified by negative selection using a Miltenyi Biotech magnetic-activated cell sorting system. Purified CD4+ T cells (2 × 106) were cultured in RPMI 1640 medium with l-glutamine (Invitrogen) containing 10% FBS (Sigma, Munich, Germany) and penicillin-G/streptomycin (Invitrogen). Cells were stimulated by adding 5 μg/mL phytohemagglutinin (PHA) for 3 days and culturing with IL-2 for an additional 2 days. Activated CD4+ T cells were used for further infectivity assays.
siRNA knockdown of galectin-3
Accell SMARTpool siRNA targeting the human galectin-3 gene and Accell nontargeting pool siRNA were purchased from Dharmacon Accell siRNA Library, Thermo Fisher Scientific, Lafayette, CO. The target sequences for Accell SMARTpool (LGALS3) were A-010606-14, 5′-CGGUGAAGCCCAAUGCAAA-3′; A-010606-15, 5′-UUUCGCUCCAUGAUGCGUU-3′; A-010606-16, 5′-CUCGCAUGCUG AUAACAAU-3′; and A-010606-17, 5′-GUACAAUCAUCGGGUUAAA-3′. To produce galectin-3 knockdown in human primary CD4+ T cells for infectivity assays, purified cells were cultured in Accell siRNA delivery medium (3% FBS and PHA) plus galectin-3 Accell SMARTpool siRNA and held for 3 days at 37°C, followed by culturing for another 2 days with IL-2. The truncated galectin-3 plasmids were transfected into primary CD4+ by electroporation using a BTX ECM830 under optimized conditions (200 V/10 ms).
Immunotransmission electron microscopy
Standard Immuno-TEM is described in detail in Waheed et al. (2009). Briefly, HEK293T cells were transfected with pNL4-3 and pEF1-Gal3 vectors (2 : 1 ratio) and incubated at 37°C for 48 h prior to collecting viral supernatants and cell pellets for immuno-TEM staining. Supernatants were filtered (0.22-mm mesh), placed on 20% sucrose and ultracentrifuged at 100,000 × g for 2 h; cells were resuspended in 50 μL PBS. Viral droplets were placed on grids and fixed with 0.5% glutaraldehyde and 4% paraformaldehyde for 20 min. Grids were washed three times with PBS, treated with 0.05% Triton X-100 for 15 min, blocked with 1% fish gelatin and held at room temperature for 30 min. Immunolabeling was performed overnight at 4°C using primary goat anti-Gal3 polyclonal Ab antibodies. After three PBS washes, grids were incubated with secondary antibodies labeled with 10-nm gold particles (rabbit anti-goat polyclonal Ab) and held for 1 h at 37°C. After three additional PBS washes, grids were stained with 4% uranyl acetate and lead citrate. Electron micrographs were obtained using a Hitachi H-7000 Transmission Electron Microscope.
Immunoblotting
Lysates were collected from cells transfected with pEF1-Gal3 or truncated forms of galectin-3 and pEGFP-p6 with pNL4-3 plasmids, galectin-3 knockdown or Alix knockdown. Pull-down proteins were collected by immunoprecipitation with anti-galectin-3 or anti-Alix. For viral protein analysis, a portion of the viral pool was treated with subtilisin (2 mg/mL) in 2× digestion buffer (1 : 1 dilution) at 37°C for 18 h. Reactions were stopped by adding 1 mM phenylmethylsulfonyl fluoride (Sigma) for 20 min at room temperature and subjected to sucrose gradient purification (Ott 2009). Analyzed proteins were quantified and used for immunoblotting analyses. These proteins or equal volumes of immunoprecipitated proteins were analyzed by SDS–PAGE. Separated proteins were transferred to nitrocellulose membranes (PolyScreen, PerkinElmer). Transferred antigens were incubated with the following antibodies: rabbit anti-galectin-3, mouse anti-galectin-3, rabbit anti-Alix, goat anti-HIV-1 gp120 (AbD Serotec), Rabbit anti-Flag (Sigma), mouse anti-HIV-1 p24 (Millipore), mouse anti-GFP (eBioscience), mouse anti-Tsg101 (Abgene), mouse anti-β-actin (Sigma) or rabbit anti-α-tubulin (Epitomics) and held for 1 h at 37°C. After three washes with PBST, membranes were incubated with HRP-conjugated antibodies (goat anti-mouse IgG, goat anti-human IgG or goat anti-rabbit IgG) (all purchased from Amersham Biosciences) for 1 h at 37°C. Hybridized protein bands were created with an Immobilon™ Western ECL protein detection system (Millipore).
Immunofluorescence assays
Detailed procedures are described in Chen et al. (2009). Briefly, Magi-5, Hut78, Jurkat or primary human CD4+ cells (5 × 104) were infected with HIV-1 NL4-3 viruses (MOI = 0.05). After 48 h, cells were added to slides treated with poly-l-lysine and fixed with 4% paraformaldehyde for 15 min. Next, cells were stained with mouse anti-HIV-1 p24, rabbit anti-AIP-1/Alix and mouse anti-galectin-3 antibodies (1:200 dilution in PBS, 37°C incubation for 1 h), followed by the addition of FITC-conjugated goat anti-mouse IgG (Chemicon), Alexa 555-conjugated goat anti-rabbit IgG (Invitrogen) or Cy5-conjugated goat anti-rabbit IgG (Abcam) (1:200 in PBS; incubated at 37°C for 1 h). After mounting, stained cells were observed using a Carl Zeiss LSM 700 Laser Scanning Confocal Microscope.
TIRF and SR
TIRF and SR fluorescence localization imaging with 20-nm spatial uncertainty were used to record the fluorescence images of samples with a commercial SR microscope (Leica SR GSD). Imaging fields were magnified using a 100× oil objective (Leica) with a 1.47 numerical aperture and 1.6× optical magnification. The penetration depth of the excitation laser source for TIRF and SR fluorescence imaging was 200 nm. TIRF fluorescence image stacks consisting of over 30,000 frames were used to calculate SR fluorescence images. Using immunostaining, galectin-3 and Gag were labeled with Alexa 488 and Alix was labeled with Alexa 647. Phosphate-buffered saline containing 100 mM β-mercaptoethylamine was used for SR fluorescence localization imaging. A two-dimensional spatial histogram map in each fluorescence channel was calculated using the SR images with an effective pixel size of 20 nm. The colocalization coefficients of the two proteins were quantified by a combination of Manders analysis (Manders et al. 1993; Spira et al. 2012) and two-dimensional spatial histogram maps of two fluorescence channels, with the fluorescence background removed during the intensity-based colocalization analysis.
Co-immunoprecipitation
HEK293T or Magi-5 cells were transfected with plasmids (pEF1, pEF1-Gal3, pNL4-3 and pEGFP-p6) or truncated galectin-3 (pEGFP-Gal3-WT, -WL mutant,- N1, -N2 and C) and truncated Alix (pEGFP-Alix-Full, -F1 and -F2) using Effectene (Qiagen), and then incubated for 48 h at 37°C. After three washes with lactose (30 mM) to remove extracellular and cell-surface-bound galectins, cells were treated with 8 μg/mL of the cross-linker DSP (3,3′-dithiodipropionic acid di[N-hydroxysuccinimide ester]) (Sigma) at room temperature for 30 min. This was followed by adding one-tenth volume of 1 M Tris–HCl (pH 7.5) and incubation at room temperature for 15 min. The cells were then lysed in NP-40 buffer (150 mM NaCl, 25 mM Tris–HCl, pH 7.4, 1 mM EDTA, 0.5% NP-40 and 5% glycerol containing a protease inhibitor cocktail) (Calbiochem) and the lysates were centrifuged at 16,000 × g. Five micrograms of each antibody, including anti-Flag (Sigma), anti-EGFP (GeneCopoiea), anti-Alix, anti-Tsg101 (Abcam) and anti-galectin-3, was incubated with 5 μL protein A/G-Sepharose beads at 4°C overnight, and then the beads were pelleted and washed with NP-40 buffer to remove unbound antibodies. Supernatants containing 500–700 μg total proteins were immunoprecipitated with the antibody-conjugated beads (such as anti-Flag, anti-EGFP, anti-Alix, and anti-galectin-3 antibody-conjugated beads) and incubated at 4°C overnight. After five washes with NP-40 buffer, the beads were subjected to 30 μL of 2× sample loading dye and incubated at 95°C for 10 min. After pelleting the beads, the 30-μL supernatants containing precipitated proteins were collected. Thirty micrograms of input proteins and 5 μL of precipitated proteins were analyzed by SDS–PAGE and immunoblotting, using antibodies against EGFP, Flag, Alix, TSg101, galectin-3, HIV-1 p24 or HIV-1 p6.
Immunoadsorption and detection of galectin-3 in HIV particles by flow cytometry
Immunoadsorption combined with flow cytometry analysis is widely used to detect the surfaces or internal components of microvesicles and virions (Zhang et al. 1994; Ostrowski et al. 2010). The assay scheme is shown in Supplementary data, Figure S4A. Briefly, 10 µg of anti-HIV-1 gp120 antibodies (Fitzgerald, Lot#G-825) was coupled with 20 µL aldehyde-sulfate latex beads (3.9-μm) and blocked with PBS containing 5% BSA. HIV particles were incubated with anti-gp120-coupled beads overnight at 4°C. After three washes with PBS, HIV-1 virions captured on beads were fixed with 4% paraformaldehyde for 15 min, and then permeabilized with 0.05% Triton X-100 for 10 min. The HIV-1 bead complexes were stained with Alexa 488-coated rabbit anti-galectin-3 antibodies for 30 min on ice and analyzed using flow cytometer (BD FACSCalibur™ system; BD Biosciences). Data were analyzed using the FlowJo software (Tree Star).
Quantitative PCR
Total RNA was extracted from primary CD4+ T cells using Tri-Reagent (Sigma-Aldrich), according to the manufacturer's instructions. The cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase (Amersham; Piscataway, NJ). Briefly, 2 μg of RNA was added to 200 U enzyme in the presence of 1 μL of each of 20 mM deoxynucleotide triphosphates and 1 μL random hexamers (10 μM) in a total reaction volume of 20 μL. After incubation at 37°C for 50 min, the mixture was heated to 95°C and cooled to 4°C. Quantitative PCR was performed using a TaqMan assay and analyzed using an Applied Biosystems 7500 Real-Time PCR System. Relative mRNA expression was calculated using glyceraldehyde 3-phosphate dehydrogenase gene expression as an internal reference. The primers for human galectin-3 were forward: 5′-CTTCTGGACAGCCAAGTG C-3′ and reverse: 5′-AAAGGCAGGTTATAAG GCACAA-3′.
Sulforhodamine B assay
A sulforhodamine B (SRB), which is an acidic compound that can bind to protonized basic amino acids and form electrostatic complexes, assay was used to determine cell growth. For standard curve establishment, Magi-5 cells were seeded in 2-fold serial dilutions in 96-well plates ranging from 625 to 2 × 104 cells/well. For the experimental group, control and galectin-3 knockdown Magi-5 cells were seeded in a 96-well plate (2 × 103/well). At the indicated time points, 10% trichloroacetic acid (TCA) was added to fix the cells in the standard and experimental groups for 15 min, and then any superfluous TCA was washed away. After fixation, SRB in 1% acetic acid was added for 30 min to stain cellular protein. Extra SRB dye was washed with 1% acetic acid. Finally, 10 mM Tris-based buffer (pH 7.4) was added and the absorbance at 515 nm was measured.
Statistical analysis
All experiments were performed at least three times. The GraphPad Prism software was used for all statistical analyses. Statistical significance (P < 0.05) was calculated using unpaired Student's t-tests.
Supplementary data
Supplementary data for this article are available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported by grants from the Republic of China National Science Council (NSC 101-2320-B-001-015-MY2) and the National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIH/NIAMS RO1 AR056343).
Conflict of interest statement
None declared.
Abbreviations
Alix, ALG-2-interacting protein X; CIN85, CbI-interacting protein 85; CRD, carbohydrate-recognition-binding domain; ESCRT, endosomal sorting complex required for transport; Gal3-shRNA, galectin-3 shRNA; HIV, human immunodeficiency virus; ILVs, intralumenal vesicles; pGal3, pEF1-Gal3; PHA, phytohaemagglutinin; PRR, proline-rich region; PTAP, Pro-Thr/Ser-Ala-Pro; SR, super resolution; SRB, sulforhodamine B; TCA, trichloroacetic acid; TIRF, total internal reflection fluorescence; YPXL, Tyr-Pro-Xn-Leu
Supplementary Material
Acknowledgements
The authors thank the Glycoscience Core, Institute of Biomedical Sciences at Academia Sinica, Taiwan, and Dr An-Li Huang from the same institute for her technical assistance with electron microscopy. We also thank Frank Chuang and Deanna Thompson from the NSF Center for Biophotonics Science and Technology, University of California at Davis and our colleagues at the AIDS Prevention and Research Center of National Yang-Ming University and Center of Infectious Disease and Cancer Research of Kaohsiung Medical University for their assistance and useful discussions. Finally, we thank Dr. Eric Freed of the HIV Drug Resistance Program, National Cancer Institute of the Frederick National Laboratory for Cancer Research, for providing the L41A mutant clone and valuable comments.
References
- Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JE, Gould SJ. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol. 2006;172:923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballe A, Martin-Serrano J. ESCRT machinery and cytokinesis: The road to daughter cell separation. Traffic. 2011;12:1318–1326. doi: 10.1111/j.1600-0854.2011.01244.x. [DOI] [PubMed] [Google Scholar]
- Carlton JG, Agromayor M, Martin-Serrano J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc Natl Acad Sci USA. 2008;105:10541–10546. doi: 10.1073/pnas.0802008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Fermin A, Vardhana S, Weng IC, Lo KF, Chang EY, Maverakis E, Yang RY, Hsu DK, Dustin ML, et al. Galectin-3 negatively regulates TCR-mediated CD4+ T-cell activation at the immunological synapse. Proc Natl Acad Sci USA. 2009;106:14496–14501. doi: 10.1073/pnas.0903497106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Vincent O, Jin J, Weisz OA, Montelaro RC. Functions of early (AP-2) and late (AIP1/ALIX) endocytic proteins in equine infectious anemia virus budding. J Biol Chem. 2005;280:40474–40480. doi: 10.1074/jbc.M509317200. [DOI] [PubMed] [Google Scholar]
- Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, Sowder RC, II, Barsov E, Hood BL, Fisher RJ, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussupt V, Javid MP, Abou-Jaoude G, Jadwin JA, de La Cruz J, Nagashima K, Bouamr F. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 2009;5:e1000339. doi: 10.1371/journal.ppat.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci USA. 2011;108:4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Fogel S, Guittaut M, Legrand A, Monsigny M, Hebert E. The tat protein of HIV-1 induces galectin-3 expression. Glycobiology. 1999;9:383–387. doi: 10.1093/glycob/9.4.383. [DOI] [PubMed] [Google Scholar]
- Frigeri LG, Liu FT. Surface expression of functional IgE binding protein, an endogenous lectin, on mast cells and macrophages. J Immunol. 1992;148:861–867. [PubMed] [Google Scholar]
- Fujii K, Hurley JH, Freed EO. Beyond Tsg101: The role of Alix in ‘ESCRTing’ HIV-1. Nat Rev Microbiol. 2007;5:912–916. doi: 10.1038/nrmicro1790. [DOI] [PubMed] [Google Scholar]
- Fujii K, Munshi UM, Ablan SD, Demirov DG, Soheilian F, Nagashima K, Stephen AG, Fisher RJ, Freed EO. Functional role of Alix in HIV-1 replication. Virology. 2009;391:284–292. doi: 10.1016/j.virol.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gauthier S, Pelletier I, Ouellet M, Vargas A, Tremblay MJ, Sato S, Barbeau B. Induction of galectin-1 expression by HTLV-I Tax and its impact on HTLV-I infectivity. Retrovirology. 2008;5:105. doi: 10.1186/1742-4690-5-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlinger HG. How HIV-1 hijacks ALIX. Nat Struct Mol Biol. 2007;14:254–256. doi: 10.1038/nsmb0407-254. [DOI] [PubMed] [Google Scholar]
- Haller C, Fackler OT. HIV-1 at the immunological and T-lymphocytic virological synapse. Biol Chem. 2008;389:1253–1260. doi: 10.1515/BC.2008.143. [DOI] [PubMed] [Google Scholar]
- Hsu DK, Chernyavsky AI, Chen HY, Yu L, Grando SA, Liu FT. Endogenous galectin-3 is localized in membrane lipid rafts and regulates migration of dendritic cells. J Invest Dermatol. 2009;129:573–583. doi: 10.1038/jid.2008.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Emr SD. The ESCRT complexes: Structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomol Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie T, Shimazu Y, Yoshida T, Sakaguchi T. The YLDL sequence within Sendai virus M protein is critical for budding of virus-like particles and interacts with Alix/AIP1 independently of C protein. J Virol. 2007;81:2263–2273. doi: 10.1128/JVI.02218-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 virological synapse formation in T cells requires lipid raft integrity. J Virol. 2005;79:12088–12094. doi: 10.1128/JVI.79.18.12088-12094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJ, Bess C, Johnson MC, Virgen CA, Simon SM, Bieniasz PD. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006;4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RD, Lubinski JM, Friedman HM. Herpes simplex virus type 1 infection increases the carbohydrate binding activity and the secretion of cellular galectin-3. Arch Virol. 2009;154:609–618. doi: 10.1007/s00705-009-0351-7. [DOI] [PubMed] [Google Scholar]
- Liu FT, Patterson RJ, Wang JL. Intracellular functions of galectins. Biochim Biophys Acta. 2002;1572:263–273. doi: 10.1016/s0304-4165(02)00313-6. [DOI] [PubMed] [Google Scholar]
- Liu FT, Rabinovich GA. Galectins as modulators of tumour progression. Nat Rev Cancer. 2005;5:29–41. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- Manders EM, Verbeek FJ, JA A. Measurement of co-localization of objects in dual-colour confocal images. J Microsc. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Marsh M. ALIX catches HIV. Cell Host Microbe. 2007;1:5–7. doi: 10.1016/j.chom.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Neil SJ. Host factors involved in retroviral budding and release. Nat Rev Microbiol. 2011;9:519–531. doi: 10.1038/nrmicro2596. [DOI] [PubMed] [Google Scholar]
- McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier S, St-Pierre C, Pelletier I, Ouellet M, Tremblay MJ, Sato S. Galectin-1 promotes HIV-1 infectivity in macrophages through stabilization of viral adsorption. Virology. 2008;371:121–129. doi: 10.1016/j.virol.2007.09.034. [DOI] [PubMed] [Google Scholar]
- Munshi UM, Kim J, Nagashima K, Hurley JH, Freed EO. An Alix fragment potently inhibits HIV-1 budding: Characterization of binding to retroviral YPXL late domains. J Biol Chem. 2007;282:3847–3855. doi: 10.1074/jbc.M607489200. [DOI] [PubMed] [Google Scholar]
- Odorizzi G. The multiple personalities of Alix. J Cell Sci. 2006;119:3025–3032. doi: 10.1242/jcs.03072. [DOI] [PubMed] [Google Scholar]
- Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. sup pp 11–13. [DOI] [PubMed] [Google Scholar]
- Ott DE. Purification of HIV-1 virions by subtilisin digestion or CD45 immunoaffinity depletion for biochemical studies. Methods Mol Biol. 2009;485:15–25. doi: 10.1007/978-1-59745-170-3_2. [DOI] [PubMed] [Google Scholar]
- Ouellet M, Mercier S, Pelletier I, Bounou S, Roy J, Hirabayashi J, Sato S, Tremblay MJ. Galectin-1 acts as a soluble host factor that promotes HIV-1 infectivity through stabilization of virus attachment to host cells. J Immunol. 2005;174:4120–4126. doi: 10.4049/jimmunol.174.7.4120. [DOI] [PubMed] [Google Scholar]
- Piguet V, Sattentau Q. Dangerous liaisons at the virological synapse. J Clin Invest. 2004;114:605–610. doi: 10.1172/JCI22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich GA, Toscano MA, Jackson SS, Vasta GR. Functions of cell surface galectin-glycoprotein lattices. Curr Opin Struct Biol. 2007;17:513–520. doi: 10.1016/j.sbi.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson MW, Albrandt K, Keller D, Liu FT. Human IgE-binding protein: A soluble lectin exhibiting a highly conserved interspecies sequence and differential recognition of IgE glycoforms. Biochemistry. 1990;29:8093–8100. doi: 10.1021/bi00487a015. [DOI] [PubMed] [Google Scholar]
- Sakaguchi T, Kato A, Sugahara F, Shimazu Y, Inoue M, Kiyotani K, Nagai Y, Yoshida T. AIP1/Alix is a binding partner of Sendai virus C protein and facilitates virus budding. J Virol. 2005;79:8933–8941. doi: 10.1128/JVI.79.14.8933-8941.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder HC, Ushijima H, Theis C, Seve AP, Hubert J, Muller WE. Expression of nuclear lectin carbohydrate-binding protein 35 in human immunodeficiency virus type 1-infected Molt-3 cells. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;9:340–348. [PubMed] [Google Scholar]
- Shim JA, Park S, Lee ES, Niki T, Hirashima M, Sohn S. Galectin-9 ameliorates herpes simplex virus-induced inflammation through apoptosis. Immunobiology. 2011;217:657–666. doi: 10.1016/j.imbio.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Spira F, Mueller NS, Beck G, von Olshausen P, Beig J, Wedlich-Soldner R. Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat Cell Biol. 2012;14:640–648. doi: 10.1038/ncb2487. [DOI] [PubMed] [Google Scholar]
- St-Pierre C, Manya H, Ouellet M, Clark GF, Endo T, Tremblay MJ, Sato S. Host-soluble galectin-1 promotes HIV-1 replication through a direct interaction with glycans of viral gp120 and host CD4. J Virol. 2011;85:11742–11751. doi: 10.1128/JVI.05351-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Stuchell MD, Garrus JE, Muller B, Stray KM, Ghaffarian S, McKinnon R, Krausslich HG, Morham SG, Sundquist WI. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J Biol Chem. 2004;279:36059–36071. doi: 10.1074/jbc.M405226200. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Matano T. Host factors involved in resistance to retroviral infection. Microbiol Immunol. 2008;52:318–325. doi: 10.1111/j.1348-0421.2008.00040.x. [DOI] [PubMed] [Google Scholar]
- Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: A secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81:6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasta GR. Roles of galectins in infection. Nat Rev Microbiol. 2009;7:424–438. doi: 10.1038/nrmicro2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waheed AA, Ono A, Freed EO. Methods for the study of HIV-1 assembly. Methods Mol Biol. 2009;485:163–184. doi: 10.1007/978-1-59745-170-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss ER, Gottlinger H. The role of cellular factors in promoting HIV budding. J Mol Biol. 2011;410:525–533. doi: 10.1016/j.jmb.2011.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ML, Chen YH, Wang SW, Huang YJ, Leu CH, Yeh NC, Chu CY, Lin CC, Shieh GS, Chen YL, et al. Galectin-1 binds to influenza virus and ameliorates influenza virus pathogenesis. J Virol. 2011;85:10010–10020. doi: 10.1128/JVI.00301-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang RY, Hsu DK, Liu FT. Expression of galectin-3 modulates T-cell growth and apoptosis. Proc Natl Acad Sci USA. 1996;93:6737–6742. doi: 10.1073/pnas.93.13.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang RY, Rabinovich GA, Liu FT. Galectins: Structure, function and therapeutic potential. Expert Rev Mol Med. 2008;10:e17. doi: 10.1017/S1462399408000719. [DOI] [PubMed] [Google Scholar]
- Zhai Q, Landesman MB, Chung HY, Dierkers A, Jeffries CM, Trewhella J, Hill CP, Sundquist WI. Activation of the retroviral budding factor ALIX. J Virol. 2011;85:9222–9226. doi: 10.1128/JVI.02653-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bagasra O, Niikura M, Poiesz BJ, Pomerantz RJ. Intravirion reverse transcripts in the peripheral blood plasma on human immunodeficiency virus type 1-infected individuals. J Virol. 1994;68:7591–7597. doi: 10.1128/jvi.68.11.7591-7597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.