Abstract
A number of quantitative trait loci (QTLs) recently have been discovered that affect various activity traits in mice, but their collective impact does not appear to explain the consistently moderate to high heritabilities for these traits. We previously suggested interactions of genes, or epistasis, might account for additional genetic variability of activity, and tested this for the average distance, duration and speed run by mice during a 3 week period. We found abundant evidence for epistasis affecting these traits, although, recognized that epistatic effects may well vary within individuals over time. We therefore conducted a full genome scan for epistatic interactions affecting these traits in each of seven three-day intervals. Our intent was to assess the extent and trends in epistasis affecting these traits in each of the intervals. We discovered a number of epistatic interactions of QTLs that influenced the activity traits in the mice, the majority of which were not previously found and appeared to affect the activity traits (especially distance and speed) primarily in the early or in the late age intervals. The overall impact of epistasis was considerable, its contribution to the total phenotypic variance varying from an average of 22–35% in the three traits across all age intervals. It was concluded that epistasis is more important than single-locus effects of genes on activity traits at specific ages and it is therefore an essential component of the genetic architecture of physical activity.
Keywords: Physical activity traits, House mice, Within-individual variation, Epistasis, Pleiotropy
Introduction
Might epistasis, the interactions between genes, be a crucial component of the genetic architecture of physical activity? In mice, a number of quantitative trait loci (QTLs) recently have been discovered that affect various activity traits such as the distance or speed run on exercise wheels (Lightfoot et al. 2007, 2008; Kas et al. 2009; Yang et al. 2009; Nehrenberg et al. 2010; Leamy et al. 2010). However, the collective impact of the single-locus effects of these QTLs has tended to be low to moderate in magnitude (average contribution around 20%), considerably less than the broad-sense heritability estimates of 50% and higher for these kinds of traits (Houle-Leroy et al. 2000; Lerman et al. 2002; Lightfoot et al. 2004). Lightfoot et al. (2008) suggested that epistatic genetic variance might account for this disparity.
We tested this idea by conducting a genome scan for epistasis affecting the average distance, duration and speed run by mice during a 3 week testing period. In fact we found many significant epistatic interactions of QTLs affecting the activity traits, the total impact (average of 26%) of which actually exceeded that of single-locus effects of QTLs (Leamy et al. 2008). If this kind of result proves to be general, it would suggest that we cannot simply screen for single effects of genes to fully understand the genetic basis of physical activity. On the other hand, our study (Leamy et al. 2008) appears to be the only one to comprehensively scan the mouse genome for epistatic effects on activity traits, so more information clearly is needed before we can be more confident about the potential impact of epistasis on physical activity.
Our previous investigation (Leamy et al. 2008) made use of activity traits that were measured during a single time frame (average over a 3 week period), but epistatic effects may well vary over time. Recently we divided the daily activity data in this mouse population into seven 3 day intervals, and conducted a search for QTLs affecting the traits at each age. We uncovered a number of new QTLs not seen in the genome scan of the 3 week average data, some affecting the activity traits only in the early or in the late age intervals (Leamy et al. 2010). In this study, we made use of these same activity data to search for epistatic effects on the distance, duration, and time run by mice during each of these seven age intervals (21 total activity traits).
Our study was motivated by three basic goals: (1) the primary purpose was to see if we would uncover unique epistatic interactions not previously found in the epistasis scan for the average activity over the entire 3 week period. We expected to find additional epistatic interactions affecting the activity traits in restricted age intervals, and were interested to know how pervasive these might be. (2) We also wanted to see to what extent we might find epistatic QTL combinations affecting traits in more than one interval (epistatic pleiotropy), and as was true for singlelocus QTL effects (Leamy et al. 2010), whether these combinations would affect the activity traits primarily in the early or in the late age intervals. (3) Finally, it was of interest to assess the total impact of epistasis to compare its magnitude with that of single-locus effects on these traits throughout the age intervals.
Materials and methods
The population and traits
All mice used in this study were from an F2 population generated from an intercross of two inbred strains, C57L/J and C3H/HeJ. These strains were chosen for crossing because they previously showed considerable divergence in wheel-running activity traits (Lightfoot et al. 2004). A total of 310 F2 mice were produced and when they were from 35 to 62 days of age, each was measured daily for three separate activity traits for several weeks. These traits included total daily distance in km, total daily exercise time in min, and average daily running speed in m/min (distance/duration). Measurements in all mice were taken with a running wheel interfaced with a computer that counted the total wheel revolutions and recorded the time each mouse spent exercising (Lightfoot et al. 2004). Throughout this study, we followed guidelines approved by the UNC Charlotte IACUC, the American Physiological Society, and the American College of Sports Medicine.
To adjust for potential age effects on the activity traits and to standardize the testing period, we first age-matched the available activity data to the extent possible to a 63 day old start age and a total testing period of 3 weeks. Using these criteria, our final sample consisted of data from 297 mice that were 63 days of age and 13 mice that were 60 days of age at the start of testing. Since all 310 mice had been exposed to running wheels for an average of 13 days (range 1–28 days) before the start age, however, we tested for potential effects of this prior exposure to the running wheel. Multivariate analyses of variance (MANOVAs) showed that this factor was not statistically significant for any of the activity traits throughout the age intervals, so no adjustments to these data were required. We also tested for potential effects of body weight (at sacrifice) differences on the activity traits in these mice, and MANOVAs also showed no significant effects for this variable. Effects of sex, litter size, and rearing blocks reached significance in MANOVAs, however, so all activity traits were adjusted for these factors as described in Leamy et al. (2010).
To analyze within-individual variation in the three activity traits, we calculated their mean values for each of the seven successive three-day intervals over the 21 day testing period. These distributions proved to be normal (P > 0.05) and less variable than those for the single-day values, and were presumed to increase the statistical power to detect epistasis influencing the activity traits. The seven 3 day intervals for each of the three activity traits also provided snapshots in time as the mice aged throughout the 9–12 week period and allowed us to assess the genetic changes that occurred during this period. Thus, we generated a total of 21 separate activity traits for the analysis. These distance, duration, and speed values over each of the seven 3 day intervals are designated DT1–DT7, DR1– DR7, and SP1–SP7, respectively.
DNA was collected for all mice after sacrifice and stored for eventual genotyping that was accomplished with the use of 129 single-nucleotide polymorphisms (SNPs). These SNPs were chosen for their polymorphism between the C57L/J and C3H/HeJ progenitor strains and to provide coverage of all 20 chromosomes in the genome (the average intermarker interval was approximately 14 cm). Occasional SNPs could not be resolved, reducing the effective sample size in some instances slightly below 310.
Epistasis analysis
We conducted separate two-way genome scans to test for epistasis affecting each of the 21 (adjusted) physical activity traits in the F2 generation mice. To accomplish this, we first assigned additive (Xa) and dominance genotypic index values (Xd) for C3H/HeJ homozygotes, heterozygotes, and C57L/J homozygotes at the site of each SNP marker, and also imputed index values at every 2 cm location between flanking markers (see Leamy et al. 2008). We then conducted regression analyses for all pairs of locations for each of the 190 possible pairs of 20 chromosomes. In these regressions, the activity trait was the dependent variable, and the independent variables were the additive and dominance genotypic index values from each of the two chromosomes as well all four of their pairwise products (XaXa, XaXd, XdXa, XdXd). The collective test of the four epistatic components (aa, ad, da, dd) estimated by regression on these four index score interactions was used as an overall indication of epistasis.
We plotted the pairs of positions for all probabilities of 1% or less generated in these tests and assumed that those pairs exhibiting the lowest probability within areas of plotted probabilities were potential QTL combinations exhibiting epistasis. For all epistatic combinations whose associated probabilities reached the 0.1% level, we also used the regression analyses to provide estimates of the additive by additive (aa), additive by dominance (ad), dominance by additive (da), and dominance by dominance (dd) genotypic epistatic terms (Leamy et al. 2008). Tests for the individual significance of each of these four genotypic epistasis terms were done via individual t tests using the conventional 5% significance level.
Evaluation of epistasis probabilities
With the hundreds of epistasis tests conducted for each activity trait, we expected many to reach significance because of chance alone. We therefore adjusted for this multiple comparisons problem by first using the method of Li and Ji (2005) to estimate the effective number of independent tests (markers) on each chromosome. Because of linkage disequilibrium between associated loci on each chromosome, this effective number is less than the actual number of markers on each chromosome (Cheverud 2001). The calculated effective number of markers on each chromosome varied from 3 to 5. We then calculated the sum of the crossproducts of these effective numbers of markers for all 190 pairs of chromosomes to estimate the total number of independent epistasis tests. This sum was 3,032, suggesting that for any trait we might expect about 30 tests to be significant at the 1% level, and 3 at the 0.1% level because of chance alone. Epistasis therefore was indicated if the number of putative instances of epistasis significantly exceeded these values (using chi-square tests each with 1 df).
These values for the number of significance epistatis tests expected by chance also were useful in two other ways. First, we used them to assess the probability of false positive instances of epistasis. This was done by estimating the false discovery rate, or FDR (Storey and Tibshirani 2003), simply by the number of epistatic tests expected to be significant by chance alone divided by the total number actually found to be significant. Second, we used the expected numbers of independent tests of epistasis, 3,032, to test individual instances of epistasis. For this purpose, we calculated the 0.05 Bonferroni threshold level of significance as 0.05/3,032 = 1.65 × 10−5. Thus, the probability associated with any individual epistatic pairwise QTL combination was considered significant if it was less than this threshold value (Leamy et al. 2008). The 5% Bonferroni value is considered a very conservative genome- wide threshold level (Holland 1998), however, so we also considered any QTL by QTL interactions with probabilities less than 0.001 to be suggestive of epistasis. This actually is a conservative measure itself since division of 0.05 by the product of the highest effective number of independent markers (5) on two chromosomes yields a probability of 0.05/(5 × 5) = 0.002 that is higher than 0.001. And for chromosome pairs with fewer effective numbers of markers (such as those with 3), the actual suggestive epistatic probability would be even higher: 0.05/(3 × 3) = 0.0055.
Results
A number of pairwise QTL combinations epistatically affecting the activity traits reached the 0.1% (suggestive) probability level although, only three of these also reached significance at the 5% genomewise threshold level of 1.65 × 10−5. Details of the locations and effects for all suggestive epistatic combinations are given in Online Resources 1–3, respectively, for the distance, duration, and speed traits, and Table 1 provides a summary of the specific numbers of combinations and chromosomes involved. Across all seven intervals, the total number of suggestive epistatic QTL combinations found was 221, including 71 for the distance traits, 89 for the duration traits, and 61 for the speed traits. While for any individual trait only about 3 epistatic combinations were expected at the 0.1% probability level by chance alone, chi-square tests showed that the number of QTL pairs affecting the traits significantly (P < 0.05) exceeded this value in 18 of the 21 total instances. Epistatic effects were most frequent for the duration traits and least frequent for the speed traits where their numbers reached significance only for SPD1, SPD3, SPD5 and SPD6. This trend also is reflected in the false discovery rates, which tend to be lowest for the duration traits and highest for the speed traits. QTLs on all 20 chromosomes participated in epistatic interactions, with chromosome 3 being particularly involved for the distance and duration traits as was chromosome 12 for the speed traits.
Table 1.
Summary statistics for QTL pairs exhibiting epistasis for the activity traits throughout the 7 intervals
| Interval | Number of pairs |
False discovery rate |
Number of chromosomes |
Most frequent chromosomes (occurrences) |
|---|---|---|---|---|
| Distance | ||||
| 1 | 15** | 0.20 | 14 | 3, 12, 13 (4) |
| 2 | 10** | 0.30 | 14 | 4, 5, 9, 13, 18, 19 (2 Each) |
| 3 | 11** | 0.28 | 13 | 1, 3 (3) |
| 4 | 8* | 0.38 | 11 | 3, 6 (3 Each) |
| 5 | 7* | 0.38 | 11 | 2, 5, 7 (2 Each) |
| 6 | 9** | 0.34 | 11 | 5 (4) |
| 7 | 10** | 0.30 | 15 | 1, 3, 4, 5, 6, (2 Each) |
| All | 71** | 0.30 | 20 | 3 (16), 5 (11), 1, 12 (10) |
| Duration | ||||
| 1 | 14** | 0.22 | 15 | 3 (4) |
| 2 | 13** | 0.23 | 19 | 7 (3) |
| 3 | 17** | 0.18 | 16 | 17 (5) |
| 4 | 11** | 0.28 | 15 | 3 (4) |
| 5 | 16** | 0.19 | 17 | 3, 12 (4 Each) |
| 6 | 11** | 0.28 | 12 | 10 (4) |
| 7 | 7* | 0.43 | 9 | 5, 10 (3 Each) |
| All | 89** | 0.24 | 20 | 3 (20), 10 (18), 12 (15) |
| Speed | ||||
| 1 | 11** | 0.28 | 16 | 9 (3) |
| 2 | 7 | 0.43 | 11 | 7, 9, X (2 Each) |
| 3 | 10** | 0.30 | 13 | 11 (3) |
| 4 | 7 | 0.43 | 8 | 12, 15 (4 Each) |
| 5 | 10** | 0.30 | 12 | 1 (4) |
| 6 | 9** | 0.34 | 11 | X (3) |
| 7 | 7 | 0.43 | 11 | 12, 15, 18 (2 Each) |
| All | 61** | 0.37 | 20 | 12 (13), 1, 15, 18 (10 Each) |
Shown are the number of QTL pairs reaching significance (P < 0.001), false discovery rates (=3.043/number of significant pairs), the number of different chromosomes involved in epistasis, and the chromosomes most frequently involved in epistasis (with the number of occurrences).
P < 0.05;
P < 0.01
We searched the suggestive epistatic interactions affecting each of the three activity traits throughout the seven intervals to discover any that appeared to replicate the 30 interactions found in our previous study using 3 week averages (Leamy et al. 2008). For ease in comparison, we show the locations of these replicate epistatic interactions from both sets of results in Table 2. As may be seen, 18 epistatic interactions (7 for the distance traits, 6 for the duration traits, and 5 for the speed traits) found for the interval traits appear to be the same as those discovered previously, leaving 12 interactions not replicated in this study. Most (14) of the replicate epistatic interactions affect traits in more than one interval, including one interaction involving QTLs on chromosomes 12 and 15 affecting speed in 5 different intervals. All epistatic interactions other than these 18 replicates affecting the activity traits are new to this study.
Table 2.
Colocalization of epistatic effects on activity traits averaged over a 3 week period with those calculated at each of 7 different intervals
| Three week averages | Intervals | |||||||
|---|---|---|---|---|---|---|---|---|
| CH1 | Loc1 | CH2 | Loc2 | CH1 | Loc1 | CH2 | Loc2 | Traits affected |
| Distance | ||||||||
| 1 | 99 | 7 | 32 | 1 | 96–98 | 7 | 30–38 | DT5, DT7 |
| 1 | 109 | 8 | 76 | |||||
| 2 | 61 | 3 | 46 | 2 | 52 | 3 | 40–42 | DT4, DT5 |
| 3 | 74 | 9 | 69 | 3 | 72–74 | 9 | 68–70 | DT6, DT7 |
| 3 | 62 | 10 | 34 | 3 | 60 | 10 | 30–34 | DT1, DT3, DT4 |
| 4 | 54 | X | 94 | 4 | 44 | X | 94 | DT7 |
| 6 | 80 | 15 | 4 | |||||
| 9 | 25 | 19 | 37 | 9 | 18–24 | 19 | 28–30 | DT6, DT7 |
| 11 | 26 | 12 | 55 | 11 | 22–24 | 12 | 48–52 | DT1, DT3 |
| 13 | 17 | 18 | 56 | |||||
| Duration | ||||||||
| 1 | 25 | 4 | 64 | 1 | 20–22 | 4 | 56–58 | DR6, DR7 |
| 1 | 107 | 8 | 74 | |||||
| 2 | 109 | 5 | 85 | 2 | 94–98 | 5 | 82 | DR3, DR6 |
| 3 | 68 | 10 | 41 | 3 | 62–66 | 10 | 30 | DR1, DR4 |
| 4 | 110 | 14 | 32 | 4 | 102 | 14 | 32 | DR5 |
| 5 | 19 | 11 | 54 | 5 | 10–16 | 11 | 46–60 | DR2, DR7 |
| 5 | 35 | 19 | 55 | |||||
| 6 | 74 | 11 | 94 | |||||
| 6 | 84 | 15 | 6 | |||||
| 10 | 0 | 17 | 53 | |||||
| 11 | 28 | 12 | 53 | 11 | 24–26 | 12 | 48 | DR1, DR3, DR5 |
| 12 | 17 | 14 | 15 | |||||
| Speed | ||||||||
| 2 | 71 | 7 | 38 | |||||
| 4 | 92 | 11 | 18 | 4 | 84 | 11 | 14 | SP3 |
| 8 | 88 | X | 32 | |||||
| 10 | 80 | 11 | 56 | 10 | 82–84 | 11 | 52–56 | SP3–SP5 |
| 10 | 48 | 19 | 15 | 10 | 52 | 19 | 4 | SP7 |
| 12 | 25 | 15 | 8 | 12 | 14–22 | 15 | 0–4 | SP3–SP7 |
| 13 | 15 | 18 | 58 | 13 | 14 | 18 | 50 | SP6 |
| 15 | 10 | 19 | 55 | |||||
Shown are locations (Loc1 and Loc2) in cM from the most proximal SNP marker for QTLs on each pair of chromosomes (CH1 and CH2). The interval traits affected by the replicate epistatic interactions also are given
To assess the extent of epistatic pleiotropy among the epistatic combinations, we tallied all instances of two or more QTL pairs that colocalized to the same or similar positions (no > 12 cM apart) on the same pair of chromosomes. We found 43 epistatic pleiotropic QTL pairs affecting two or more traits, reducing the total number of different epistatic combinations affecting the activity traits from 221 to 161. Thus, epistatic pleiotropy was common, occurring among about 27% (43/161) of the epistatic combinations. The specific numbers of instances of epistatic pleiotropy for the distance (16) duration (18) and speed traits (9) parallel the trend already seen for the total number of significant epistatic QTL pairs where duration was most affected and speed was least affected. Of the 43 total instances of QTL pairs affecting multiple traits, 34 affected two traits, nine affected three traits and one affected five traits.
Inspection of the data in the Online Resources suggests a tendency for the QTL pairs exhibiting epistatic pleiotropy to affect traits either in the early (1–4) or in the late (4–7) intervals (counting the 4th transitional interval as either early or late). This is particularly noticeable for the distance traits, where this is seen for all 16 QTL pairs, 7 affecting distance only in the early age intervals and the other 9 affecting distance only in the later age intervals. This trend also occurs for the speed traits where 7 of 9 pairs affect speed only in the early or late intervals, but not for duration where 11 of the 18 total cases of epistatic pleiotropy can be identified as affecting speed only in the early or late intervals. Nonetheless, over all traits, this trend holds in 34 of 43 total instances.
Another trend among those interactions exhibiting epistatic pleiotropy is that their epistatic components generally are quite similar in magnitude and sign especially for traits in adjacent age intervals. For example, the two interactions of QTLs on chromosomes 3 and 12 affecting distance in intervals 1 and 2 (common QTL G in Online Resource 1) both exhibited positive, statistically significant ad and dd effects. Epistatic components generated in pleiotropic interactions affecting traits in more disparate age intervals, however, tend to show greater differences. For example, the common QTLs designated E in Online Resource 2 show significant aa and ad effects for duration in interval 2 but significant ad and da effects for duration in interval 6. Even where these sorts of differences occur, however, the signs of the components always are the same.
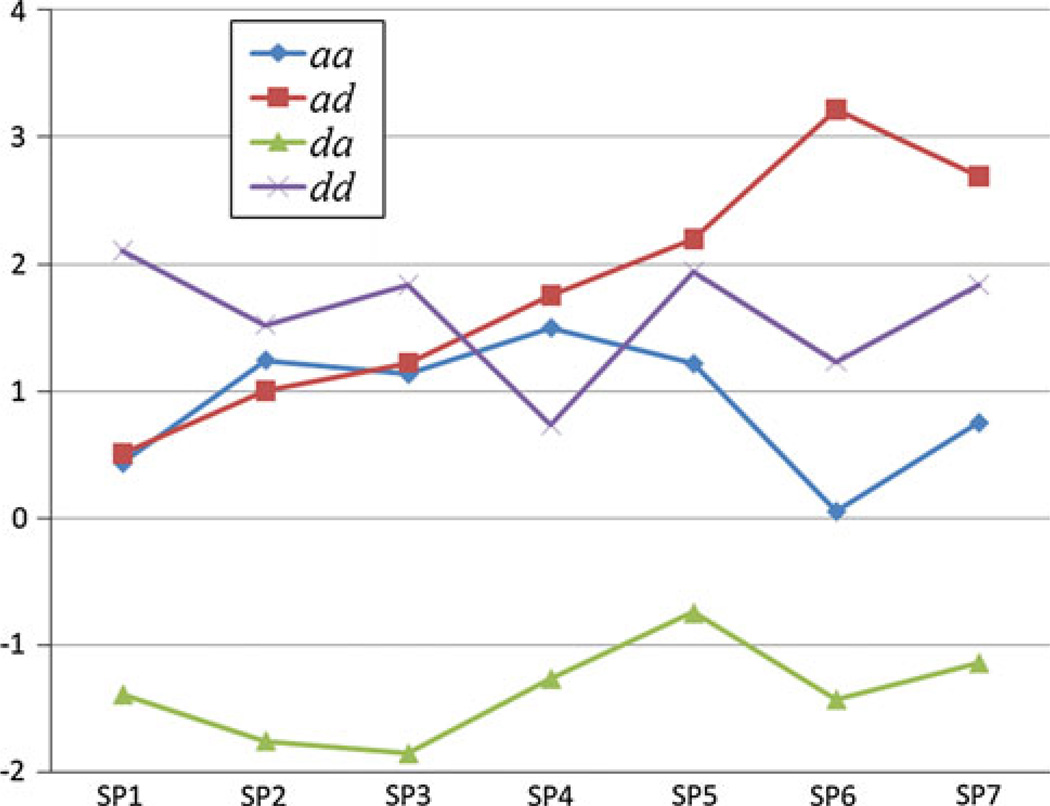
To illustrate how the magnitude of the epistatic components can change over the seven intervals, we plotted the values for all four components generated from the interaction of QTLs on chromosomes 9 and 13 affecting speed (Fig. 1). This particular interaction exhibited suggestive epistatic effects on speed in intervals 3–7 (genomewise significance for SP6), although, for completeness we also calculated the components for SP1 and SP2 as well. This figure clearly shows that the aa, da, and dd components are basically similar across the seven intervals (P > 0.05 in regression analyses) whereas values for the additive by dominance (ad) component steadily increase throughout the intervals (b = +0.43, P < 0.01). The peak ad value occurs in interval 6, and probably accounts for the genome wise significance for SP6 exhibited by this interaction.
Fig. 1.
Patterns of epistatic components affecting speed over all 7 intervals. Illustrated are the values for the four epistatic components (aa, ad, da, and dd) generated from the epistatic effects of QTLs on chromosomes 9 and 13 affecting speed throughout the 7 intervals
To assess the overall impact of epistasis on the activity traits, we calculated the absolute (standardized), significant, epistatic components generated from each of the epistatic interactions as well as their percentage contribution to the total phenotypic variance of each trait. We present the means of these components calculated over all seven intervals (Table 3) since ANOVAs showed they did not significantly differ among the intervals. Over all three traits, the aa values average the lowest (0.35–0.37), the aa/da values are higher (0.53–0.66), and the dd values average the highest (0.79–0.96). The dd values for distance are especially impressive, averaging nearly one standard deviation. The percentage contributions of epistasis to the total phenotypic variation for each of the activity traits beyond that contributed by single effects of QTLs were obtained from multiple regression analyses of the significant epistatic components. This was accomplished by subtracting the R2 values from the full (single and epistatic QTL effects included) and reduced (single QTL effects only) regression models, where this difference expresses the amount of variation contributed by the epistatic interactions to the total phenotypic variation (explained plus unexplained by regression) captured in the models. The mean percentages (Table 3) across all age intervals suggest that the average impact of epistasis is highest (35%) for duration, but is somewhat lower in magnitude for both distance (27%) and speed (22%).
Table 3.
Epistatic components generated from the QTL-QTL interactions affecting the activity traits
| Interval | % | aa | ad | da | dd | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | STD | N | Mean | STD | N | Mean | STD | N | Mean | STD | N | ||
| Distance | 26.7 | 0.35 | 0.08 | 23 | 0.66 | 0.16 | 38 | 0.59 | 0.18 | 38 | 0.96 | 0.25 | 31 |
| Duration | 34.9 | 0.37 | 0.11 | 33 | 0.60 | 0.18 | 54 | 0.53 | 0.15 | 42 | 0.79 | 0.18 | 40 |
| Speed | 22.3 | 0.35 | 0.08 | 22 | 0.64 | 0.15 | 28 | 0.55 | 0.14 | 37 | 0.86 | 0.22 | 34 |
Shown are means, standard deviations (STD) and sample sizes (N) for the significant standardized, absolute epistatic components generated from the QTL pairs affecting distance, duration, and speed averaged over all 7 intervals. The mean percentage (%) impact of the significant components on the total phenotypic variability across all intervals for each of the traits also is given
Discussion
Evidence for epistasis
We undertook this study to assess the potential role of epistasis in the genetic architecture of within-individual physical activity, and in fact discovered an abundance of epistatic interactions affecting all three activity traits throughout the seven age intervals. Our evidence for epistasis was based primarily on the numbers of these interactions reaching the 0.001 probability levels beyond those expected by chance alone. Thus, 3 significant epistatic interactions were expected at the 0.001 level by chance for each trait in any given age interval, but the actual numbers found varied from seven to as high as 15 (Table 1). While the FDR values suggested that as many as a third of these might be false positives, this still leaves a sizeable number of interactions that should represent true instances of epistasis affecting the activity traits. The QTLQTL interactions that reached the 5% genome wise threshold level of significance offer additional evidence for epistasis affecting the activity traits. There were a total of only three such interactions, but given the large number of epistasis tests, this is not particularly surprising. In fact in our previous investigation (Leamy et al. 2008), no epistatic interactions affecting the physical activity traits averaged over the entire 3 week period reached this level of significance.
Concordance of epistatic interactions
A large number of epistatic interactions affecting the physical activity traits in specific intervals were new; that is, they were not seen in our previous scan for epistastic effects on these traits averaged over the entire 3 week period. At the 0.001 probability level, we discovered a total of 161 epistatic interactions (counting common QTL combinations as one each), only 18 of which appeared to be replicated from our previous study (Leamy et al. 2008). This suggests that the remaining 143 interactions affecting the interval activity traits are new. Assuming some unknown fraction of these are false positives, this still leaves many interactions that must represent true instances of epistasis uniquely affecting these activity traits throughout the age intervals.
There was little concordance of the epistatic QTL combinations with the single-effect QTLs previously found affecting the activity traits throughout the age intervals (Leamy et al. 2010). Thus, none of the epistatic interactions we discovered at the 0.1% significance level (Online Resources 1–3) involved any pair of QTLs previously found for these specific traits. And only a few interactions contained one QTL that colocalized with any previously found. Based on locations, two QTLs for distance, five QTLs for duration, and five QTLs for distance we earlier identified may be involved in several of the epistatic interactions affecting these traits at specific ages. This suggests that if epistasis tests for the interval traits had been performed only on the QTLs we previously found, we would have grossly underestimated the extent of epistasis affecting these traits that has been revealed in the full genome scan.
Epistatic patterns among the age intervals
At the 0.001 suggestive probability level, most of the epistatic interactions affected the activity traits in a single age interval. Specifically, this was true in 37 of 53 cases for distance, 46 of 64 cases for duration, and 35 of 44 cases for speed, or 118 out of the total of 161 (Online Resources 1–3). This suggests that novel epistatic interactions influenced activity of the mice in this population at specific times during the course of the 3 week testing period. Ordinarily we should not expect to detect these sorts of interactions in epistasis scans of traits averaged over an entire testing period, although, five of our interactions replicated from the original study influenced activity in only one interval (Table 2). Perhaps this occurred because correlations of the trait values in each interval with their 3 week averages all are relatively high, varying from 0.73 to 0.84 over all activity traits.
Although, most epistatic interactions affected single-interval traits, it will be recalled that we did discover 43 interactions affecting the activity traits in more than one interval. Since a given trait such as distance measured in two or more intervals can be regarded as separate traits, we considered these examples of epistatic pleiotropy (Wolf et al. 2005). It was interesting that the majority of these 43 epistatic interactions appeared to affect the activity traits (especially distance and speed) primarily in the early or in the late age intervals. We previously found this same trend for single-locus effects of QTLs in this mouse population, although, mainly for duration (Leamy et al. 2010).
It seems reasonable to expect epistatic pleiotropic effects on traits especially in adjacent age intervals. Thus, we previously showed that the autocorrelation pattern of distance, duration, and speed throughout the age intervals was generally monotonic, with decreasing associations for each of these traits with increasingly distant age intervals (Leamy et al. 2010). This may explain why so few epistatic interactions, at least at the 0.001 probability level, affected traits in more than two age intervals.
The high correlations among traits especially in adjacent intervals also presumably explain the similarity in the magnitude of the epistatic components generated by the epistatic QTL pairs exhibiting pleiotropy. Even in those few interactions jointly affecting traits in non-adjacent intervals, the significant components still were reasonably similar. Wolf et al. (2005) have shown that in an orthogonal epistasis model such as we have used here, epistatic pleiotropic interactions must show significance for the same epistatic component(s) in order to contribute to the covariation of two or more traits. Thus, a common QTL interaction exhibiting significant aa epistasis on one trait but dd epistasis on another trait would generate no covariation between the traits because these epistatic components are independent. Wolf et al. (2005) also showed that common epistatic components with the same sign will generate a positive covariance whereas those that differ in sign will generate a negative covariance. Given the patterns of the significant components exhibited by the epistatic pleiotropic interactions described above, therefore, we would expect epistasis to generate positive covariances between the activity traits, with larger values especially for traits in adjacent age intervals. This is precisely the pattern shown by the correlations of the activity traits among the intervals (Leamy et al. 2010).
Even if the magnitude and sign of these coefficients generated by common epistatic interactions are similar for traits especially in adjacent intervals, however, this does not mean they cannot exhibit significant changes over a period of time. The example we illustrated with the epistatic interaction of QTLs on chromosomes 9 and 13 affecting speed throughout the intervals clearly showed a significant increase in one component (ad) over time. As another example, the dd component for a common epistatic interaction of QTLs on chromosomes 1 and 18 exhibited a suggestive trend with values of −0.83, −1.01, and −1.11 for SP5–SP7, respectively (Online Resource 3). Although, this interaction did not exhibit suggestive or significant effects on speed in the first four intervals, we calculated the appropriate dd values for comparative purposes, and they were −0.12, −0.49, −0.79, and −0.83. This trend of decrease in the dd values is both statistically significant (b = −0.15, P < 0.01) and remarkably consistent throughout the intervals. No doubt similar trends can be found for other epistatic interactions.
Impact of epistasis
The magnitude of the standardized epistatic components generated by the interactions affecting the activity traits was quite high. Over all three traits throughout the age intervals, the means for the aa, ad, da, and dd components were 0.36, 0.63, 0.58, and 0.86. These values compare quite closely with the estimates of 0.35, 0.59, 0.57 and 0.85 previously made in the epistatic analysis of these traits averaged over the entire 3 week period (Leamy et al. 2008). However, they are considerably higher than the means of the single-locus additive and dominance effects for these traits that varied only from 0.22 to 0.32 standard deviations (Leamy et al. 2010). It is likely that the dominance epistatic components especially are inflated because of sampling variation in this population, especially since estimates of epistatic components from other populations of larger size have consistently been smaller, typically around 0.3–0.4 standard deviations (Cheverud et al. 2001; Peripato et al. 2004; Wolf et al. 2005; Leamy et al. 2008). Nonetheless, the magnitude of these epistatic components suggests that epistasis may be more important than singlelocus effects on these activity traits.
Another indication of the impact of epistasis was seen by its large contribution to the phenotypic variation in the traits. These contributions averaged 22–35% across all seven intervals for each of the three traits, more than estimates for single-locus effects that averaged just over 15% for all three traits (Leamy et al. 2010). Some of these interactions are expected to be false positives, but it also is true that other interactions reaching non-suggestive probability levels (such as 1%) will also include some fraction of true positives. So although, we cannot know the true extent of epistasis affecting these activity traits, it seems reasonable to conclude that it is considerable.
Conclusions
We have located a large number of epistatic interactions of QTLs that influence the distance, duration, and speed run by mice in our F2 population over each of seven three-day intervals. A few of these interactions apparently are the same as found in our original study using the average of these physical activity traits measured daily over the entire 21 days period (Leamy et al. 2008), but the majority are novel and appear to affect the activity traits (especially distance and speed) in specific age intervals. The overall impact of epistasis as measured both by the magnitude of the epistatic components and by the contribution of epistasis to the overall phenotypic variability of the traits was also quite impressive. It therefore seems reasonable to conclude that epistasis is as important, if not more so, than single-locus effects of genes on activity traits at specific ages, and thus, clearly is an important component of the genetic architecture of physical activity.
Supplementary Material
Acknowledgments
We should like to thank two anonymous reviewers for excellent revision suggestions on an earlier version of this paper. This work was supported in part by grants from the National Institutes of Health (NIDDK DK61635 to J. Timothy Lightfoot, NIAMS AR050085 to J. Timothy Lightfoot and Larry J. Leamy, and NIDDK DK076050 to Daniel Pomp).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10709-011-9586-9) contains supplementary material, which is available to authorized users.
Contributor Information
Larry J. Leamy, Email: ljleamy@uncc.edu, Department of Biology, University of North Carolina, Charlotte, NC 28223, USA.
Daniel Pomp, Department of Genetics, University of North Carolina, Chapel Hill, NC 27599, USA; Department of Nutrition, University of North Carolina, Chapel Hill, NC 27599, USA; Department of Cell and Molecular Physiology, University of North Carolina, Chapel Hill, NC 27599, USA; Carolina Center for Genome Science, University of North Carolina, Chapel Hill, NC 27599, USA.
J. Timothy Lightfoot, Sydney and JL Huffines Institute for Sports Medicine and Human Performance, Texas A&M University, College Station, TX 77845, USA; Department of Health and Kinesiology, Texas A&M University, College Station, TX 77845, USA.
References
- Cheverud JM. A simple correction for multiple comparisons in interval mapping genome scans. Heredity. 2001;2:52–58. doi: 10.1046/j.1365-2540.2001.00901.x. [DOI] [PubMed] [Google Scholar]
- Cheverud J, Vaughn T, Pletscher L, Peripato A, Adams E, Erickson C, King-Ellison K. Genetic architecture of adiposity in the cross of large (LG/J) and small (SM/J) inbred mice. Mamm Genome. 2001;12:3–12. doi: 10.1007/s003350010218. [DOI] [PubMed] [Google Scholar]
- Holland JB. EPISTACY: a SAS program for detecting two-locus epistatic interactions using genetic marker information. J Hered. 1998;89:374–375. [Google Scholar]
- Houle-Leroy P, Garland T, Jr, Swallow JG, Guderley H. Effects of voluntary activity and genetic selection on muscle metabolic capacities in house mice M. domesticus. J Appl Physiol. 2000;89:1608–1615. doi: 10.1152/jappl.2000.89.4.1608. [DOI] [PubMed] [Google Scholar]
- Kas MJH, de Mooij-van Malsen JR, de Krom M, van Gassen KLI, van Lith HA, Olivier B, Oppelaar H, Hendriks J, de Wit M, Groot Koerkamp MJA, Holstege FCP, van Oost BA, de Graan PNE. High-resolution genetic mapping of mammalian motor activity levels in mice. Genes Brain Behav. 2009;8:13–22. doi: 10.1111/j.1601-183X.2008.00435.x. [DOI] [PubMed] [Google Scholar]
- Leamy LJ, Pomp D, Lightfoot JT. An epistatic genetic basis for physical activity traits in mice. J Hered. 2008;99:639–646. doi: 10.1093/jhered/esn045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy LJ, Pomp D, Lightfoot JT. A search for quantitative trait loci controlling within-individual variation of physical activity traits in mice. BMC Genet. 2010;11:83. doi: 10.1186/1471-2156-11-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman I, Harrison BC, Freeman K, Hewett TE, Allen DL, Robbins J, Leinwand LA. Genetic variability in forced and voluntary endurance exercise performance in seven inbred mouse strains. J Appl Physiol. 2002;92:2245–2255. doi: 10.1152/japplphysiol.01045.2001. [DOI] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Lightfoot JT, Turner MJ, Daves M, Vordermark A, Kleeberger SR. Genetic influence on daily wheel running activity level. Physiol Genomics. 2004;19:270–276. doi: 10.1152/physiolgenomics.00125.2004. [DOI] [PubMed] [Google Scholar]
- Lightfoot JT, Turner M, Kleinfehn A, Jedlick A, Oshimura T, Marzec J, Gladwell W, Leamy L, Kleeberger S. Quantitative trait loci (QTL) associated with maximum exercise endurance in mice. J Appl Physiol. 2007;103:105–110. doi: 10.1152/japplphysiol.01328.2006. [DOI] [PubMed] [Google Scholar]
- Lightfoot JT, Turner MJ, Pomp D, Kleeberger SR, Leamy LJ. Quantitative trait loci for physical activity traits in mice. Physiol Genomics. 2008;32:401–408. doi: 10.1152/physiolgenomics.00241.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrenberg DL, Wang S, Hannon RM, Garland T, Jr, Pomp D. QTL underlying voluntary exerercise in mice: interactions with the “mini-muscle” locus and sex. J Hered. 2010;101:42–53. doi: 10.1093/jhered/esp066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peripato AC, de Brito RA, Matioli SR, Pletscher LS, Vaughn TT, Cheverud JM. Epistasis affecting litter size in mice. J Evol Biol. 2004;17:593–602. doi: 10.1111/j.1420-9101.2004.00702.x. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genome-wide studies. PNAS. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Leamy LJ, Routman EJ, Cheverud JM. Epistatic pleiotropy and the genetic architecture of covariation within early and late-developing skull trait complexes in mice. Genetics. 2005;171:683–694. doi: 10.1534/genetics.104.038885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HeS, Vitaterna MH, Laposky AD, Shimomura K, Turek FW. Genetic analysis of daily physical activity using a mouse chromosome substitution strain. Physiol Genomics. 2009;39:47–55. doi: 10.1152/physiolgenomics.00066.2009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.