Abstract
The Niemann-Pick C1 (NPC1) protein has a central role in regulating the efflux of lipoprotein-derived cholesterol from late endosomes/lysosomes and transport to other cellular compartments. Since the NPC1 protein has been shown to regulate the transport of cholesterol to cellular compartments enriched with the ubiquitous cholesterol-binding and transport protein caveolin-1, the present study was performed to determine whether the NPC1 and caveolin-1 proteins interact and function to modulate efflux of low density lipoprotein (LDL)-derived cholesterol from endocytic compartments. To perform these studies, normal human fibroblasts were grown in media with lipoprotein-deficient serum (LPDS) or media with LPDS supplemented with purified human LDL. The results indicated reciprocal co-immunoprecipitation and partial co-localization of the NPC1 and caveolin-1 proteins that was decreased when fibroblasts were grown in media with LDL. Consistent with interaction of the NPC1 and caveolin-1 proteins, a highly conserved caveolin-binding motif was identified within a cytoplasmic loop located adjacent to the sterol-sensing domain (SSD) of the NPC1 protein. To examine the functional relevance of this interaction, fibroblasts were transfected with caveolin-1 siRNA and found to accumulate increased amounts of LDL-derived cholesterol within late endosomes/ lysosomes. Together, this report presents novel results demonstrating that the NPC1 and caveolin-1 proteins interact to modulate efflux of LDL-derived cholesterol from late endocytic compartments.
Introduction
A number of studies have indicated that most tissues with the exception of liver obtain cholesterol primarily through endogenous biosynthesis and to a lesser extent through receptor-mediated endocytosis of remnant lipoproteins (Dietschy et al. 1993, Osono et al. 1995, Turley et al. 1981). The endocytosis of lipoproteins, particularly cholesterol-enriched low-density lipoprotein (LDL) and the subsequent transport of LDL-derived cholesterol through cellular compartments, serves an important role in maintaining intracellular cholesterol homeostasis. Early studies performed using human fibroblasts demonstrated that LDL particles bind to both the LDL receptor and LDL receptor-related protein (LRP) present on the cell surface to facilitate the internalization of these particles into endocytic compartments (Brown and Goldstein 1975, Brown et al. 1976). It is within early endosomes, and to a lesser extent late endosomes/lysosomes, that the bulk of cholesteryl ester contained within the hydrophobic core of these particles is hydrolyzed by an acidic lipase to generate LDL-derived cholesterol (Goldstein et al. 1975, Sugii et al. 2003).
The transport of LDL-derived cholesterol from endocytic compartments is primarily regulated by both the Niemann-Pick C2 (NPC2) and Niemann-Pick C1 (NPC1) proteins, which have been shown to function in a non-redundant and cooperative manner (Infante et al. 2008b, Sleat et al. 2004). The soluble NPC2 protein is contained within the lumen of endocytic compartments, where it binds and facilitates the transport of cholesterol to the limiting membrane (Cheruku et al. 2006, Friedland et al. 2003, Ko et al. 2003). In contrast, the membrane-bound NPC1 protein is associated with a unique late-endosome, commonly referred to as the NPC1 compartment, that transiently interacts with cholesterol-enriched endocytic compartments (Garver et al. 2000, Neufeld et al. 1999). Although the human NPC1 cDNA opening reading frame predicts a protein of 1,278 amino acids with an estimated molecular weight of 142 kDa, the NPC1 protein migrates as a doublet corresponding to 170 and 190 kDa due to variable asparagine-linked glycosylation (Watari et al. 1999a, Watari et al. 1999b). Studies indicate that both the 170 and 190 kDa NPC1 proteins are capable of interacting with endocytic compartments but that the 170 kDa NPC1 protein is 50% less efficient in facilitating the efflux of cholesterol from these compartments (Watari et al. 1999b). The NPC1 protein is also capable of binding cholesterol through two distinct domains, referred to as the amino or N-terminal domain (NTD) and the sterol-sensing domain (SSD) (Infante et al. 2008a, Liu et al. 2009, Ohgami et al. 2004). A series of eloquent studies have demonstrated that the NPC2 protein transfers cholesterol to the NPC1-NTD positioned within the endocytic lumen, followed by the NPC1 protein inserting the hydrophobic isooctyl side chain of cholesterol into the endocytic membrane (Infante et al. 2008a, Infante et al. 2008b, Kwon et al. 2009).
In general, studies performed using either NPC1 or NPC2 human fibroblasts have demonstrated that these proteins regulate the transport of LDL-derived cholesterol to different cellular compartments, including the Golgi appartus, plasma membrane, and endoplasmic reticulum (Blanchette-Mackie et al. 1988, Coxey et al. 1993, Underwood et al. 1998, Wojtanik & Liscum 2003). More recent studies have indicated that a distinct region of the Golgi apparatus, the trans-Golgi network (TGN), preferentially receives cholesterol derived from the NPC1 compartment and distributes this cholesterol to the endoplasmic reticulum and plasma membrane (Garver et al. 2002, Urano et al. 2008). It should be noted that the TGN and TGN-derived vesicles are enriched with caveolin-1, which is known to be a ubiquitous cholesterol-binding and transport protein (Dupree et al. 1993, Li & Papadopoulos 1998, Murata et al. 1995). Consistent with this result, a number of studies indicate that caveolin-1 is actively involved in facilitating the transport of cholesterol between cellular compartments (Fielding & Fielding 1996, Smart et al. 1996, Uittenbogaard et al. 2002, Uittenbogaard et al. 1998). Moreover, studies have shown that caveolin-1 partially co-localizes with the NPC1 compartment and that expression of caveolin-1 is significantly increased in both human fibroblasts and mouse tissues as a result of decreased NPC1 protein function, thereby suggesting that the NPC1 protein and caveolin-1 may interact and function in an undefined manner (Garver et al. 1997a, Garver et al. 2000, Garver et al. 1997b, Higgins et al. 1999).
The present study was performed to determine whether the NPC1 and caveolin-1 proteins interact and function to modulate the efflux of LDL-derived cholesterol from endocytic compartments. To perform these studies, normal human fibroblasts were grown in media containing lipoprotein-deficient serum (LPDS), which represented the control or basal culture condition, in addition to media containing LPDS supplemented with purified human LDL. In brief, the results indicated i) an inverse association between relative amounts of the NPC1 and caveolin-1 proteins when fibroblasts were grown in media with LDL, ii) reciprocal co-immunoprecipitation of the NPC1 and caveolin-1 proteins was decreased when fibroblasts were grown in media with LDL, iii) partial co-localization of the NPC1 and caveolin-1 proteins was decreased when fibroblasts were grown in media with LDL, iv) identification of a conserved caveolin-binding motif located adjacent to the SSD of the NPC1 protein, and v) an increased amount of LDL-derived cholesterol within late endosomes/lysosomes resulting from caveolin-1 siRNA knockdown. Together, this report presents novel results demonstrating that the NPC1 and caveolin-1 proteins interact to modulate efflux of LDL-derived cholesterol from endocytic compartments.
Materials and Methods
Materials
DMEM, PBS, trypsin-EDTA, and 100 U/ml penicillin/streptomycin (P/S) were purchased from Invitrogen Corporation (Carlsbad, CA). Fetal bovine serum (FBS) and lipoprotein-deficient serum (LPDS) were purchased from Cocalico Laboratories (Reamstown, PA). Complete protease inhibitor cocktail tablets were purchased from Roche Diagnostic (Indianapolis, IN). Human low-density lipoprotein was purchased from Ray-Biotech (Norcross, GA). The RNeasy Mini Kit was purchased from Qiagen (Valencia, CA). The TaqMan Gene Expression Assay including PCR primers (NPC1, Hs00264835_m1 and caveol in-1, Hs00184697_ml) were purchased from Applied Biosystems (Foster City, CA). The Niemann-Pick C1 (NPC1) antibody was generated in rabbits against amino acids 1254–1273 (NKAKSCATEERYGTERER) of the human NPC1 protein and purchased from Invitrogen Corporation (Carlsbad, CA). The caveolin-1 antibodies (clones C060 and 2297) were purchased from BD Biosciences (San Jose, CA). The lysosome-associated membrane protein-1 (LAMP-1) antibody (clone H4A3) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Stealth caveolin-1 siRNA duplex oligomers, Stealth non-specific siRNA Negative Control duplex oligomers, Lipofectamine 2000, and Opti-MEM I Reduced Serum Medium were purchased from Invitrogen Corporation (Carlsbad, CA). The peroxidase, Cy2, and Cy3-conjugated goat secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). The West Pico SuperSignal Substrate, Bicinchoninic Acid (BCA) protein assay, and Protein A-Sepharose beads were purchased from Pierce Chemical Company (Rockford, IL).
Cell culture and harvest
Normal human fibroblasts (CRL-2097) were purchased from the American Type Culture Collection (Manassas, VA) and cultured using basic media (DMEM, 10% FBS, and 1% P/S) in a humidified incubator at 37°C with 5% CO2. At ~ 30% confluence, the cells were rinsed with PBS and media was changed to DMEM, 5% LPDS, and 1% P/S to deplete cellular sterol pools and increase expression of the LDL receptor. When the cells reached ~ 60% confluence (24 h), media was changed to fresh DMEM, 5% LPDS, and 1% P/S or the same media supplemented with human LDL (50 µg/ml LDL). The cells were allowed to incubate (24 h) and were harvested for experimentation.
RNA preparation
Total RNA was extracted from fibroblasts using the RNeasy Mini Kit followed by treatment of the RNA with RNase-free DNase to remove residual contaminating DNA. The concentration of RNA was determined by absorbance at 260 nm, with the purity of RNA determined by the ratio of absorbance at 260 nm and 280 nm.
Quantitative RT-PCR
The relative amounts of target mRNA were determined using quantitative reverse transcription polymerase-chain reaction (qRT-PCR) analysis. Reverse transcription was performed using 2.5 µM random hexamers, 4.0 mM dNTP’s, 15 mM MgCl2, 50 U reverse transcriptase, and 100 U RNAase inhibitor to produce cDNA. Pre-developed commercially available primers and probes were used for detection of NPC1 and caveolin-1 mRNA. The qRT-PCR was performed with a TaqMan Gene Expression Assay containing PCR primers and TaqMan MGB probe (FAM dye-labeled) using an ABI-PRISM Sequence Detection System (Applied Biosystems, Foster City, CA). The quantification of PCR products was normalized to 18S rRNA (internal control).
Reciprocal co-immunoprecipitation
Fibroblasts were solubilized using Buffer A (PBS containing 1% v/v Triton X-100, 60 mM octylglucoside, and protease inhibitor cocktail), homogenized using needle aspiration, and centrifuged (100,000 × g at 4°C for 30 min) to remove insoluble material. The protein concentration of this solubilized homogenate was determined and 250 µg aliquots were diluted to 1.0 ml with Buffer A. The primary antibodies (rabbit antihuman NPC1 or mouse anti-human caveolin-1) were added to the solubilized and diluted homogenate (1:250 dilution) and incubated during nutation (4°C for 4 h). The Protein A-Sepharose beads were rinsed and blocked with Buffer A containing 10 mg/ml FAFA (4° C for 4 h) and collected by centrifugation (1,000 × g at 4°C for 1 min). The solubilized and diluted homogenate containing primary antibodies was added onto the Protein A-Sepharose beads and incubated during nutation (4°C for 16 h). The beads were collected by centrifugation (1,000 × g at 4°C for 5 min) and rinsed in a sequential manner with Buffer A containing 10 mg/ml FAFA and decreasing concentrations of NaCl (0.5 M, 0.4 M, 0.3 M, 0.2 M, and 0.1 M NaCl). Finally, the rinsed beads were mixed with SDS-PAGE sample buffer (50°C for 5 min to detect NPC1 and 100°C for 5 min to detect caveolin-1) in preparation for SDS-PAGE.
Caveolin-1 siRNA knockdown
Fibroblasts were transfected with caveolin-1 siRNA duplex oligomers to knockdown caveolin-1 gene expression. In brief, cells were plated using basic media without antibiotics (DMEM and 10% FBS) and grown to ~ 30% confluence. The cells were rinsed with PBS and media was changed to DMEM and 5% LPDS, followed by transfection (5 h) with either 2.0 nM Stealth caveolin-1 siRNA duplex oligomers or 2.0 nM nonspecific siRNA Negative Control duplex oligomers using Lipofectamine 2000. After transfection, the cells were rinsed with PBS and incubated in DMEM and 5% LPDS (48 h). The media was changed to DMEM and 5% LPDS or the same media supplemented with human LDL (50 µg/ml LDL) and incubated (24 h). Finally, the cells were rinsed with PBS and harvested for experimentation.
Immunoblot analysis
The relative amounts of NPC1 and caveolin-1 protein were determined using immunoblot analysis. Protein samples were separated using either 7% or 13% SDS-PAGE under reduced conditions and transferred to a nitrocellulose membrane. In brief, blocking buffer (10 mM sodium phosphate pH 7.4, 150 mM NaCl, 0.05% Tween 20, and 5% non-fat dry milk) was used to block non-specific sites on the nitrocellulose membrane (2 h). The membranes were incubated in blocking buffer containing rabbit anti-human NPC1 antibody (1:1,500 dilution) or mouse anti-human caveolin-1 antibody (1:750 dilution) (4°C for 16 h). The membranes were rinsed (3 × 5 min) and incubated with blocking buffer containing peroxidase-conjugated goat secondary antibody (1:5,000 dilution) (1.5 h). This particular combination of host specific primary and secondary antibodies prevented cross-reaction while performing immunoblot analysis to detect reciprocal co-immunoprecipiation of the NPC1 or caveolin-1 proteins. Finally, the membranes were rinsed (3 × 5 min) and bound secondary antibodies were identified using enhanced chemiluminescence.
Fluorescence labeling
The fibroblasts were fixed onto coverslips using PBS containing 4% paraformaldehyde (30 min). After fixation, the coverslips were rinsed with PBS (3 × 5 min) and placed in quenching buffer (PBS and 50 mM NH4Cl) for 15 min. The coverslips were then rinsed with PBS (3 × 5 min) and placed in Buffer B (PBS, 10% goat serum, and 0.05% w/v saponin) or Buffer C (PBS, 10% goat serum, and 0.05% w/v filipin) for 90 min. Following this, they were placed in Buffer B or C containing rabbit anti-human NPC1 antibody (1:250 dilution), mouse anti-human caveolin-1 antibody (1:250 dilution), or mouse anti-human LAMP-1 antibody (1:250 dilution) for 90 min. They were then rinsed with PBS (3 × 5 min) and placed in Buffer B or C containing Cy2 and Cy3-conjugated goat secondary antibody (1:500 dilution) for 90 min. Finally, the coverslips were rinsed with PBS (3 × 5 min) and mounted onto slides using Aqua Poly/Mount.
Deconvolution fluorescence microscopy
Deconvolution fluorescence microscopy is a relatively new technique that decreases the amount of unfocused and distorted fluorescence through computational processing, thereby promoting restoration of multiple focal planes into a high-resolution three-dimensional fluorescent image. The images were obtained using an Olympus IX-70 inverted microscope equipped with a 60X and 100X (NA 1.4) oil immersion objective (Olympus America, Melville, NY), Photometrics cooled CCD camera (Roper Scientific Instruments, Tucson, AZ), and DeltaVision RT restoration microscopy system software (Applied Precision, Issaquah, WA). The emission wavelengths used for obtaining fluorescent images were 350 nm for the DAPI filter, 528 nm for the Cy2 filter, and 617 nm for the Cy3 filter. For the different fluorescent probes, 10 sections were obtained using a distance of 0.2 µm (recommended step size for the NA of the objectives) set between focal planes. The data, subjected to 5 deconvolution iterations, was projected using Soft-WoRX software and processed using identical parameters with Adobe Photoshop software CS2 (Adobe Systems, Mountain View, CA). Finally, the percentage of co-localization between the NPC1 and caveolin-1 proteins, and the intensity of filipin-staining for cholesterol within LAMP-1 vesicles, was determined using the color range selection and area measurement/ intensity features with Adobe Photoshop software CS2.
Identification and sequence comparison of caveolin-binding motifs
The three potential caveolin-binding motifs, denoted by the principal consensus sequence jXjXXXXj and the less common consensus sequences jXXXXjXXj and jXjXXXXjXXj, where j represents one of three aromatic amino acids (Trp W, Phe F, or Tyr Y), were used to screen the human NPC1 amino acid sequence at the National Center for Biotechnology Information (NCBI). Moreover, sequence comparison of caveolin-binding motifs among orthologous NPC1 proteins was performed according to the following species and reference sequences: Human (GenBank: AAH63302.1), Chimpanzee (NCBI: XP_001155163.1), Mouse (NCBI: NP_032746.2), Pig (NCBI: NP_00999487.1), Dog (NCBI: NP_001003107.1), Hamster (GenBank: AAF31692.1), Cattle (Genbank: AAI51277.1), Horse (NCBI: XP_001490228.2) , Bi rd (NCBI: XP_419162.2), Rat (GenBank: EDL86695.1), and Cat (NCBI: NP_001009829.2).
Statistical analysis
For all experiments, quantitative data is represented as the mean ± standard deviation (SD) using five plates of human fibroblasts (qRT-PCR and immunoblot analysis) or ten fluorescent images (quantification of colocalization) of human fibroblasts grown using both culture conditions (LPDS or LDL). Significant differences (P ≤ 0.05) between two groups of data were determined using the two-tailed Student’s t-test assuming equal variance.
Results
Relative amounts of the NPC1 and caveolin-1 proteins
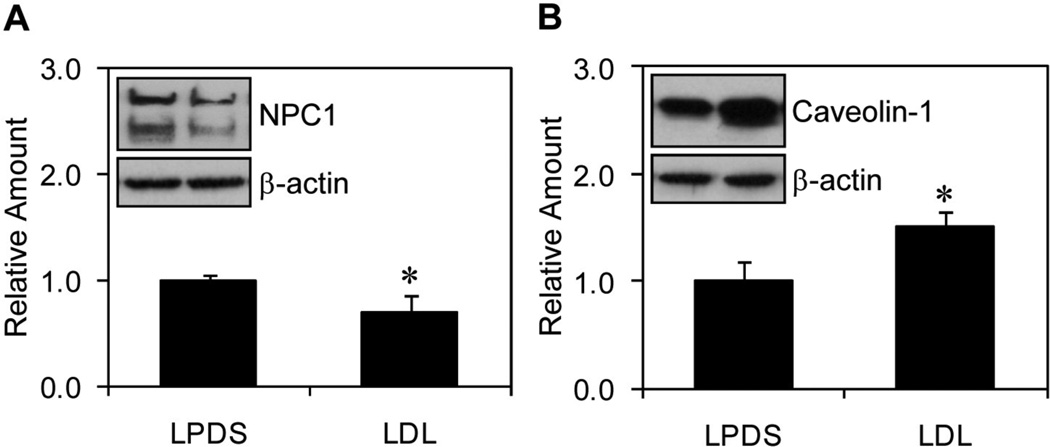
Fibroblasts were grown using the two culture conditions (LPDS and LDL) and processed to determine relative amounts of the NPC1 and caveolin-1 proteins. The results indicated a significant average decrease (31%) in the relative amounts of NPC1 protein for fibroblasts grown in media with LDL compared to fibroblasts grown in media with LPDS (Figure 1A). In contrast, the results indicated a significant average increase (52%) in the relative amounts of caveolin-1 protein for fibroblasts grown in media with LDL compared to fibroblasts grown in media with LPDS (Figure 1B). Similarly, consistent with transcriptional regulation of the NPC1 and caveolin-1 genes, the results indicated a significant average decrease (33%) in the relative amounts of NPC1 mRNA and a significant average increase (33%) in the relative amounts of caveolin-1 mRNA for fibroblasts grown in media with LDL compared to fibroblasts grown in media with LPDS (data not shown).
Figure 1.
Relative amounts of the NPC1 and caveolin-1 proteins. The average amounts of NPC1 protein (A) and caveolin-1 protein (B) for fibroblasts grown in media with LPDS were normalized to β-actin (internal control) and assigned a value of 1.0, while the average amounts of NPC1 protein and caveolin-1 protein for fibroblasts grown in media with LDL were expressed as fold change. A representative immunoblot showing the relative amounts of NPC1 and caveolin-1 protein for fibroblasts grown in LPDS (left lane) and LDL (right lane) are included as insets within respective graphs. Quantitative data is represented as the mean ± S.D. using five plates of fibroblasts grown using both culture conditions. *P ≤ 0.05 compared to fibroblasts grown in media with LPDS.
Reciprocal co-immunoprecipitation of the NPC1 and caveolin-1 proteins
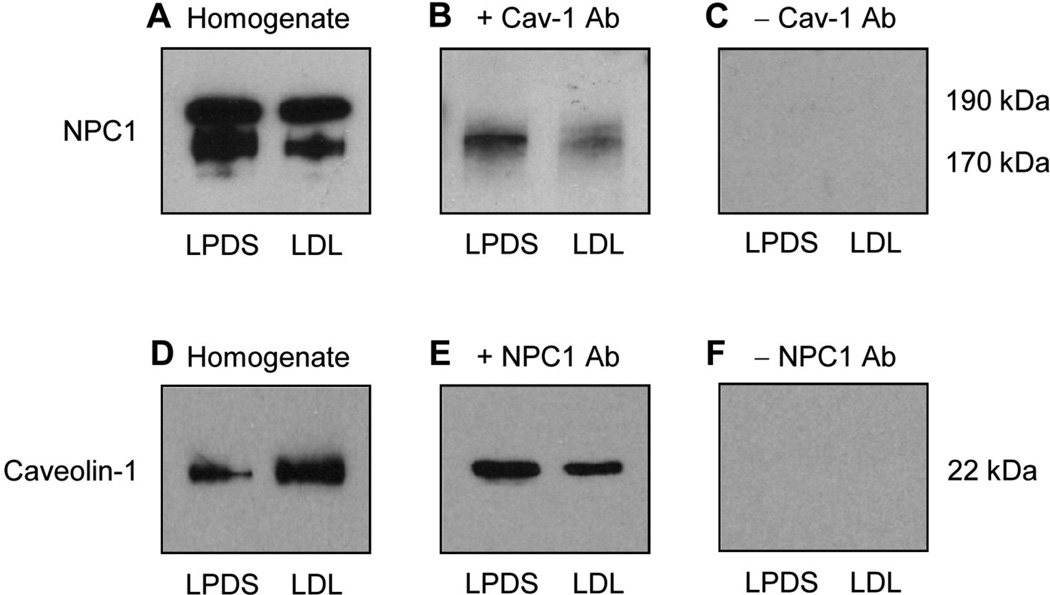
Fibroblasts were grown using the two culture conditions (LPDS and LDL) and processed to perform reciprocal co-immunoprecipitation of the NPC1 and caveolin-1 proteins. The results indicated that immunoprecipitation of caveolin-1 promoted co-immunoprecipitation of the lower molecular weight NPC1 protein (170 kDa) when fibroblasts were grown in media with LPDS or LDL (Figure 2A, B, C). Similarly, immunoprecipitation of the NPC1 protein promoted co-immunoprecipitation of caveolin-1 when fibroblasts were grown in media with LPDS or LDL (Figure 2D, E, F). However, it must be noted that co-immunoprecipitation of either the NPC1 or caveolin-1 protein was decreased for fibroblasts grown in media with LDL compared to fibroblasts grown in media with LPDS (Figure 2B, E).
Figure 2.
Reciprocal co-immunoprecipitation of the NPC1 and caveolin-1 proteins. An equivalent amount of solubilized and diluted homogenate protein from fibroblasts grown in media with LPDS or LDL was incubated in the presence or absence of rabbit anti-human NPC1 antibody or mouse antihuman caveolin-1 antibody and resulting antibody-antigen complexes were precipitated using Protein A-Sepharose beads. The beads were incubated with sample buffer and immunoblot analysis was performed using reciprocal antibodies (same caveolin-1 or NPC1 antibody, respectively). The solubilized and diluted homogenate protein (A, D) served as a positive control for identification of the co-immunoprecipitated proteins (B, E), while the solubilized and diluted homogenate protein incubated in the absence of NPC1 or caveolin-1 primary antibody (C, F) served as a negative control.
Cellular distribution of the NPC1 and caveolin-1 proteins
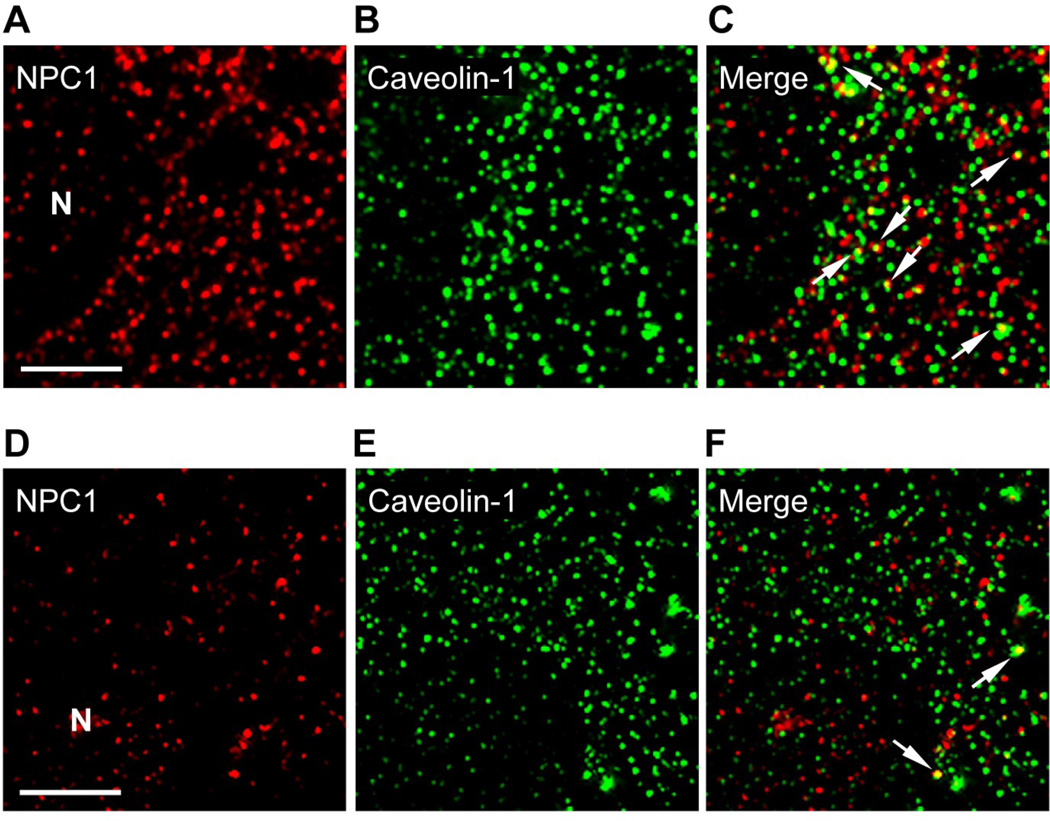
Fibroblasts were grown using the two culture conditions (LPDS and LDL) and processed to perform de-convolution fluorescence microscopy. The NPC1 protein, which is known to be primarily associated with a unique population of late endosomes, was represented by vesicles approximately 1.5 mm in diameter and distributed throughout the perinuclear region of the cytoplasm when fibroblasts were grown in media with LPDS or LDL (Figure 3A, D). In contrast, caveolin-1 was represented by a relatively increased number of similarly sized vesicles that were also distributed throughout the perinuclear region of the cytoplasm when fibroblasts were grown in media with LPDS or LDL (Figure 3B, E). The merged images indicated partial co-localization of these vesicles when fibroblasts were grown in media with LPDS or LDL (Figure 3C, F).
Figure 3.
Cellular distribution of the NPC1 and caveolin-1 proteins. A representative fibroblast grown in LPDS is provided in the top set of images (A, B, C), while a representative fibroblast grown in LDL is provided in the bottom set of images (D, E, F). Partial co-localization of the NPC1 and caveolin-1 proteins are indicated using white arrows. Fluorescent images were obtained using a deconvolution fluorescence microscope equipped with an Olympus 100× (NA 1.4) oil-immersion objective. All images were acquired and processed using identical conditions. Bar represents 15 mm. N, nucleus.
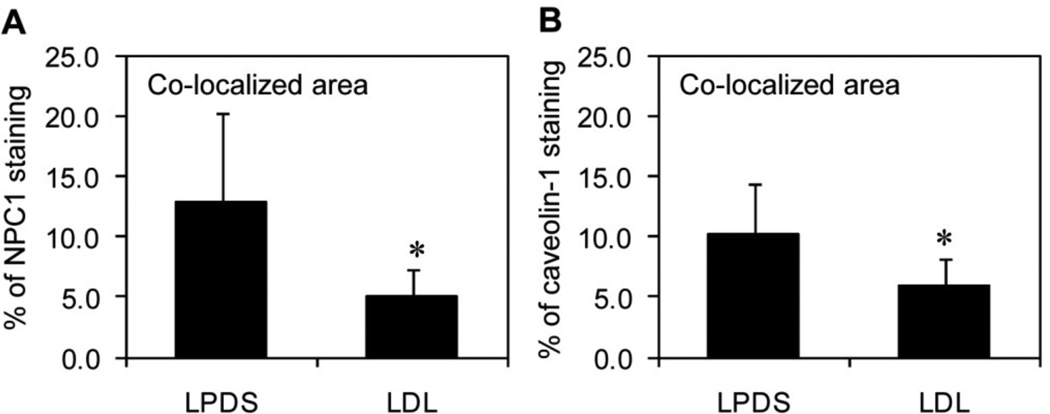
Percentage of co-localization between the NPC1 and caveolin-1 proteins
Fibroblasts were grown using the two culture conditions (LPDS and LDL) and processed to perform de-convolution fluorescence microscopy and determine the percentage of co-localization between the NPC1 and caveolin-1 proteins. The results indicated that, on average, caveolin-1 co-localized with 13% of the NPC1 protein when fibroblasts were grown in media with LPDS (Figure 4A). In contrast, the average amount of caveolin-1 that co-localized with the NPC1 protein was significantly decreased (62%) when fibroblasts were grown in media with LDL. Similarly, on average, the NPC1 protein co-localized with 10% of caveolin-1 when fibroblasts were grown in media with LPDS, but the average amount was significantly decreased (40%) when fibroblasts were grown in media with LDL (Figure 4B).
Figure 4.
Percentage of co-localization between the NPC1 and caveolin-1 proteins. The total areas representing NPC1 or caveolin-1 staining were independently identified using the color range selection feature within Adobe Photoshop software, followed by determining the overlapping area and percentage of co-localization for caveolin-1 staining (A) and NPC1 staining (B) from fibroblasts grown in media with LPDS or LDL. Fluorescent images were obtained using a deconvolution fluorescence microscope equipped with an Olympus 100× (NA 1.4) oil-immersion objective. Quantitative data is represented as the mean ± S.D. using ten images acquired in a random manner and processed using identical conditions. *P ≤ 0.05 compared to fibroblasts grown in media with LPDS.
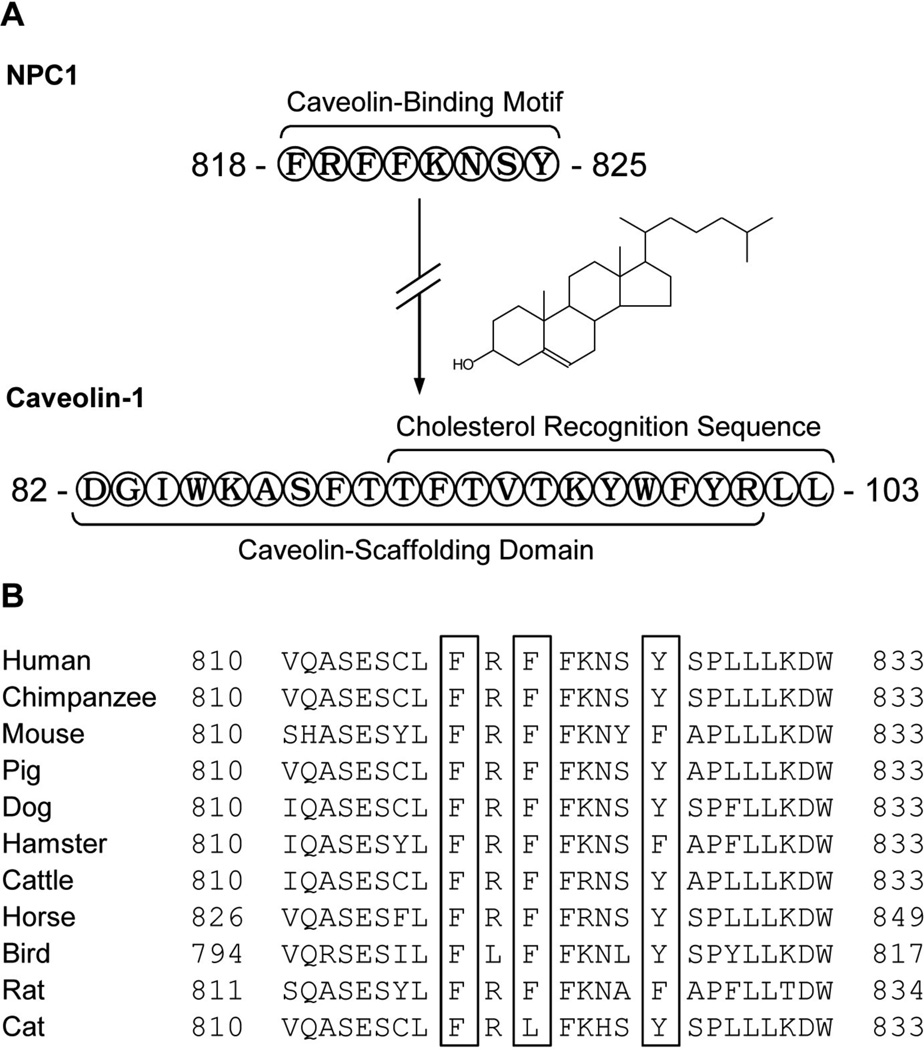
Identification and sequence comparison of caveolin-binding motifs associated with orthologous NPC1 proteins
The principal caveolin-binding motif, denoted by the consensus sequence jXjXXXXj, where j represents Trp (W), Phe (F), or Tyr (Y), but not the less common caveolin-binding motifs denoted by the consensus sequences jXXXXjXXj and jXjXXXXjXXj, was identified within the NPC1 protein amino acid sequence. The amino acid consensus sequence (818-FRFFKNSY-825) is encoded within cytoplasmic loop 4 adjacent to the sterol-sensing domain (amino acids 616–797) of the human NPC1 protein. Since the NPC1 protein caveolin-binding motif is positioned within the cytoplasm, it would theoretically permit interaction with the caveolin-scaffolding domain (82-DGIWKASFTTFTVTKYWFYR-101) of caveolin-1 also positioned within the cytoplasm (Figure 5A). A comparison of the amino acid sequences from orthologous NPC1 proteins indicated that the NPC1 protein caveolin-binding motif is highly conserved among several different species (Figure 5B). The one species (cat) that does not encode the common caveolin-binding motif consensus sequence, instead encodes what has been reported as a permissible caveolin-like binding motif (I, L, or V instead of W, F or Y in place of the second j) also capable of interaction with the caveolin-scaffolding domain.
Figure 5.
Identification and sequence comparison of caveolin-binding motifs associated with orthologous NPC1 proteins. A schematic representation of the topology for both the NPC1 and caveolin-1 proteins is provided (A). The human NPC1 protein contains a previously unidentified caveolin-binding motif (818-FRFFKNSY-825) located within cytoplasmic loop 4 adjacent to the sterol-sensing domain (amino acids 616–797). It is proposed that the NPC1 protein caveolin-binding motif interacts with the caveolin-1 protein through the caveolin-scaffolding domain (82-DGIWKASFTTFTVTKYWFYR-101). The caveolin-1 protein also possesses a cholesterol recognition sequence (91-TFTVTKYWFYRLL-103) located near the cytofacial membrane and cytoplasm boundary that partially overlaps with the caveolin-scaffolding domain. A comparison of the amino acid sequences between orthologous NPC1 proteins indicates that the NPC1 protein caveolin-binding motif (jXjXXXXj), where j mostly represents the amino acids Trp (W), Phe (F), or Tyr (Y), is conserved among several different species (B).
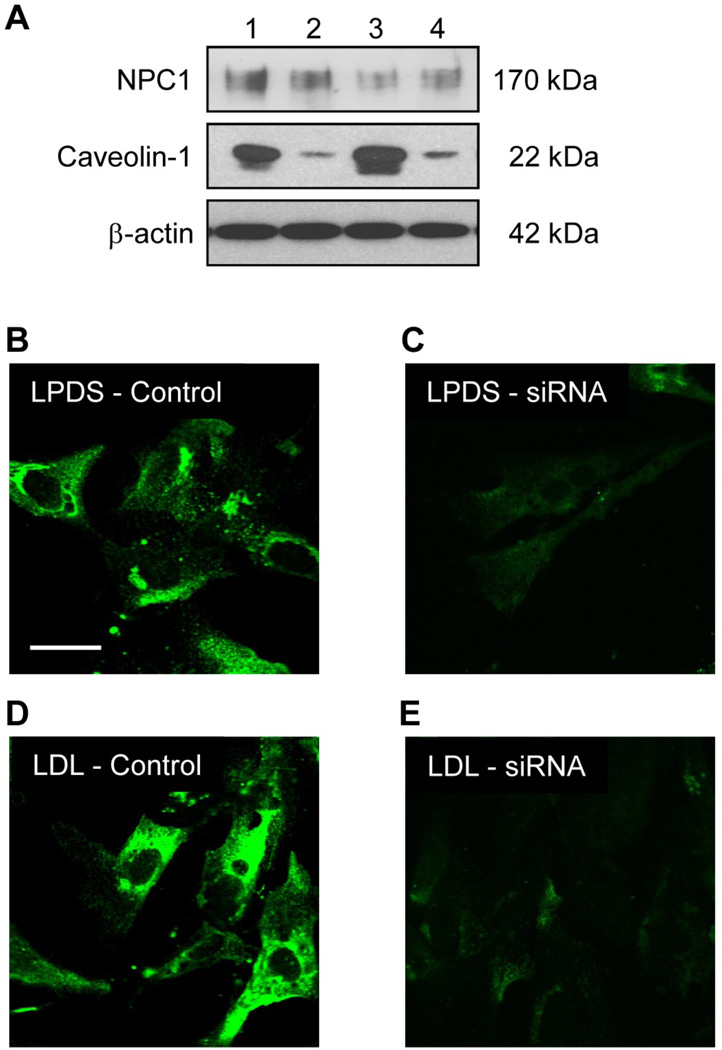
Relative amounts of the NPC1 and Caveolin-1 proteins resulting from caveolin-1 siRNA knockdown
Fibroblasts were transfected with either non-specific siRNA or caveolin-1 siRNA and grown using the two culture conditions (LPDS and LDL) to determine the relative average amounts of the NPC1 and caveolin-1 proteins. The results indicated no significant difference in the relative amounts of the NPC1 protein for fibroblasts transfected with caveolin-1 siRNA when fibroblasts were grown in media with LPDS or LDL (Figure 6A). In contrast, the results indicated a significant decrease in the relative average amounts of caveolin-1 protein for fibroblasts transfected with caveolin-1 siRNA when fibroblasts were grown in media with LPDS or LDL (89% and 92%, respectively), compared to fibroblasts transfected with the non-specific (control) siRNA (Figure 6A). Consistent with the significantly decreased average amounts of caveolin-1 protein determined using immunoblot analysis, the relative amounts of caveolin-1 protein were noticeably decreased for fibroblasts transfected with caveolin-1 siRNA when fibroblasts were grown in media with LPDS or LDL compared to fibroblasts transfected with the non-specific (control) siRNA upon examination using fluorescence microscopy (Figure 6B, C, D, F).
Figure 6.
Relative amounts of the NPC1 and caveolin-1 proteins resulting from caveolin-1 siRNA knockdown. The relative amounts of the NPC1 and caveolin-1 proteins for fibroblasts transfected with non-specific siRNA (control) or caveolin-1 siRNA (siRNA) and grown in media with LPDS or LDL was determined using immunoblot analysis, where 1 = LPDS Control, 2 = LPDS siRNA, 3 = LDL Control, and 4 = LDL siRNA (A). A field of representative fibroblasts transfected with non-specific siRNA (control) or caveolin-1 siRNA (siRNA) and grown in media with LPDS (B and C) or LDL (D and E) is provided. Fluorescent images were obtained using a deconvolution fluorescence microscope equipped with an Olympus 60× (NA 1.4) oil-immersion objective. All images were acquired and processed using identical conditions.
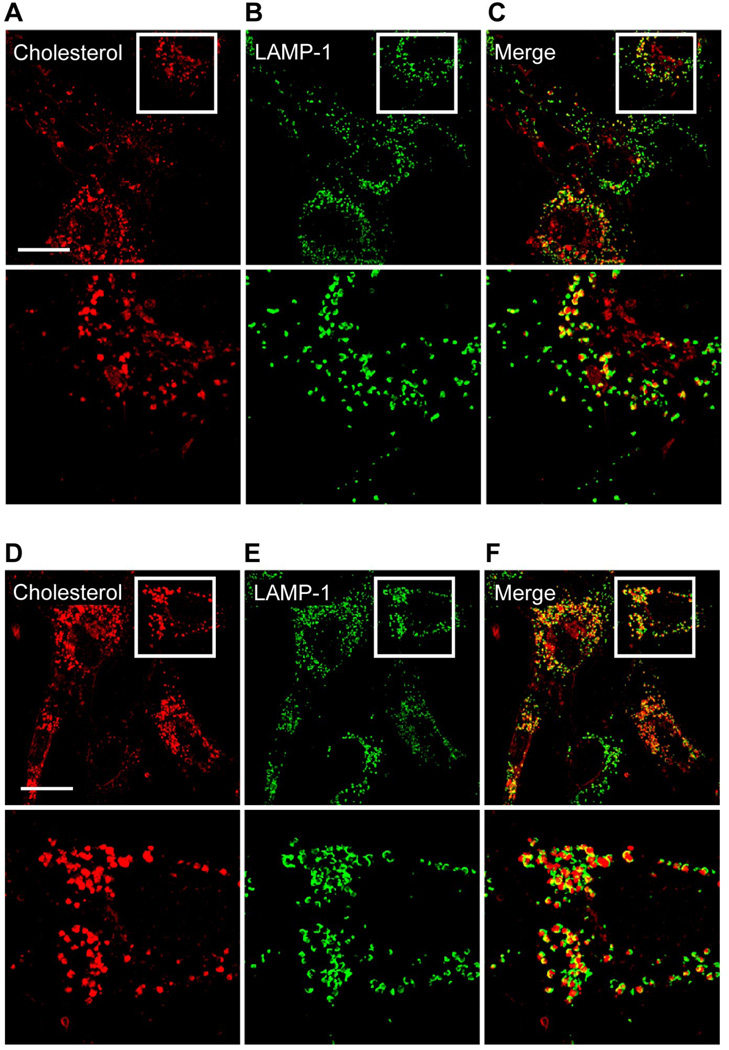
Cellular distribution of cholesterol in relation to late endosomes/lysosome resulting from caveolin-1 siRNA knockdown
Fibroblasts were transfected with non-specific siRNA or caveolin-1 siRNA and grown in media with LDL to determine the relative cellular distribution of cholesterol in relation to late endosomes/lysosomes using deconvolution fluorescence microscopy. The images indicated one primary cellular distribution for cholesterol which was localized within perinuclear cytoplasmic vesicles (Figure 7A, D). With respect to fibroblasts transfected with the non-specific siRNA (control), the images indicated that only a certain population of the LAMP-1 containing vesicles, represented by semi-rounded and hollowed vesicles, co-localized with cholesterol (Figure 7A, B, C). In contrast, fibroblasts transfected with caveolin-1 siRNA had a noticeably increased population of LAMP-1 containing vesicles that colocalized with cholesterol (Figure 7D, E, F). There was no noticeable difference between fibroblasts transfected with the non-specific siRNA (control) and caveolin-1 siRNA when grown in media with LPDS (data not shown), thereby suggesting an accumulation of cholesterol within the LAMP-1 containing vesicles was derived from LDL.
Figure 7.
Cellular distribution of cholesterol in relation to late endosomes/lysosomes resulting from caveolin-1 siRNA knockdown. Fibroblasts were processed to label cholesterol using filipin and the LAMP-1 protein (a marker protein for late endosomes/lysosomes). Fibroblasts transfected with a non-specific siRNA (control) and grown in media with LDL are provided in the top set of images (A, B, C), while fibroblasts transfected with caveolin-1 siRNA and grown in media with LDL are provided in the bottom set of images (D, E, F). A defined area (square white outline) within fibroblasts are enlarged and placed directly below the corresponding image to visualize the cellular distribution of cholesterol (A, D), the LAMP-1 protein (B, E), and merged images (C, F). Fluorescent images were obtained using a deconvolution fluorescence microscope equipped with an Olympus 60× (NA 1.4) oil-immersion objective. All images were acquired and processed using identical conditions. Bar represents 15 mm.
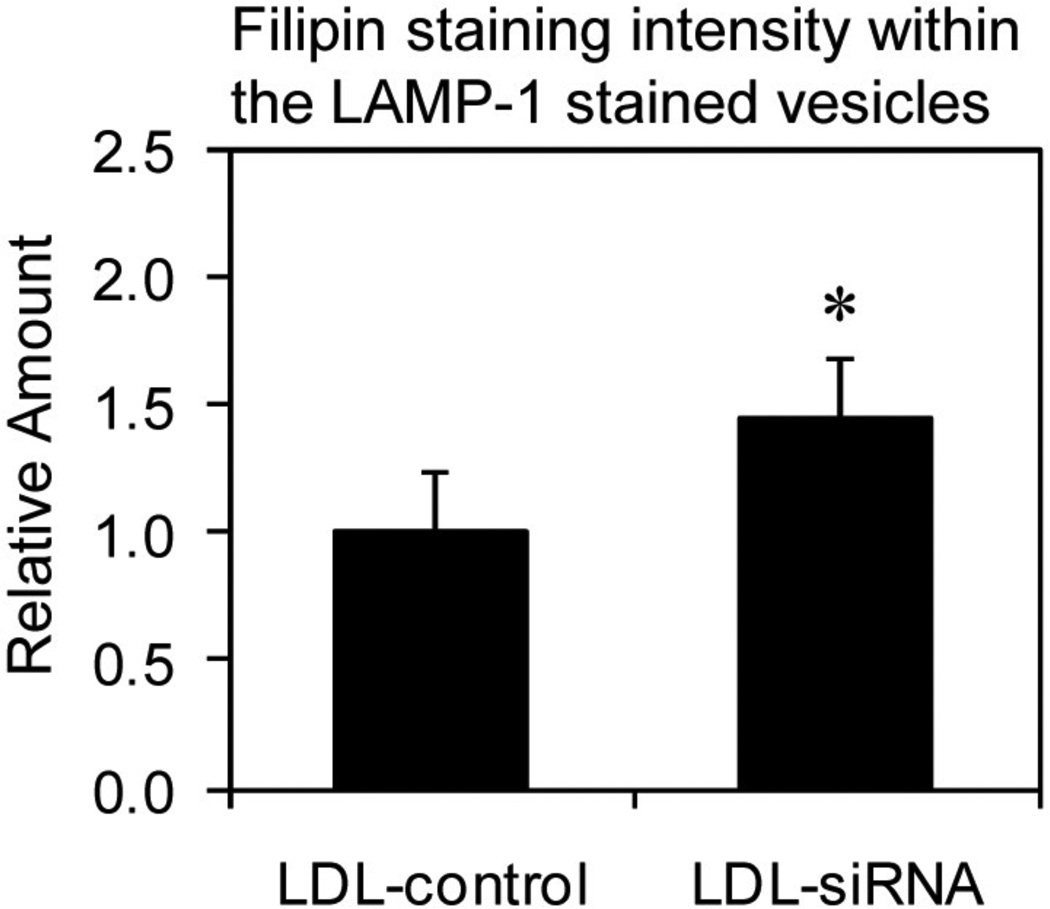
Relative amounts of cholesterol associated with late endosomes/lysosomes resulting from caveolin-1 siRNA knockdown
Fibroblasts were transfected with non-specific siRNA or caveolin-1 siRNA and grown in media with LDL to determine the relative amount of LDL-derived cholesterol associated with late endosomes/lysosomes using deconvolution fluorescence microscopy. The results indicated a significant average increase (44%) in the relative amounts of cholesterol localized within LAMP-1 containing vesicles, which are known to represent late endosomes/lysosomes, for fibroblasts transfected with caveolin-1 siRNA compared to fibroblasts transfected with non-specific siRNA (control) (Figure 8). These results again suggest an accumulation of cholesterol within the LAMP-1 containing vesicles was derived from LDL.
Figure 8.
Relative amounts of cholesterol associated with late endosomes/lysosomes resulting from caveolin-1 siRNA knockdown. The total area representing LAMP-1 containing vesicles was identified using the color range selection feature, followed by determining the intensity of cholesterol staining within this area to calculate the relative amounts of cholesterol associated with late endosomes/lysosomes. Fluorescent images were obtained using a deconvolution fluorescence microscope equipped with an Olympus 60× (NA 1.4) oil-immersion objective. Quantitative data is represented as the mean ± S.D. using ten fields acquired in random manner and processed using identical conditions for fibroblasts grown in media with LDL. *P ≤ 0.05 compared to fibroblasts transfected with non-specific siRNA (control) and grown in media with LDL.
Discussion
The present study was performed to determine whether the NPC1 and caveolin-1 proteins interact and function to modulate the efflux of LDL-derived cholesterol from endocytic compartments. To perform these studies, normal human fibroblasts were grown in media containing LPDS, which represented the control or basal culture condition, in addition to media containing LPDS supplemented with purified human LDL. In brief, the results indicated i) an inverse association between relative amounts of the NPC1 and caveolin-1 proteins when fibroblasts were grown in media with LDL; ii) that reciprocal co-immunoprecipitation of the NPC1 and caveolin-1 proteins was decreased when fibroblasts were grown in media with LDL; iii) that partial co-localization of the NPC1 and caveolin-1 proteins was decreased when fibroblasts were grown in media with LDL; iv) identification of a conserved caveolin-binding motif located adjacent to the SSD of the NPC1 protein, and v) an increased amount of LDL-derived cholesterol within late endosomes/lysosomes resulting from caveolin-1 siRNA knockdown. Together, this report presents novel results demonstrating that the NPC1 and caveolin-1 proteins interact to modulate efflux of LDL-derived cholesterol from late endocytic compartments.
The results indicated an inverse association between relative amounts of the NPC1 and caveolin-1 proteins when fibroblasts were grown in media with LDL. Specifically, compared to fibroblasts grown in media with LPDS, supplementation of media with LDL significantly decreased the relative average amounts of NPC1 protein, but significantly increased the relative average amounts of caveolin-1 protein. A logical explanation for the inverse expression of the NPC1 and caveolin-1 genes is supported by studies indicating that LDL-dependent feedback inhibition of the SREBP pathway promotes downregulation of the NPC1 gene and upregulation of the caveolin-1 gene (Bist et al. 1997, Garver et al. 2008, Hailstones et al. 1998). However, it should be noted that feedback inhibition of the SREBP pathway and subsequent transcriptional regulation of the NPC1 and caveolin-1 genes may only serve as a partial explanation. It is well accepted that decreased NPC1 protein function alters the efflux of cholesterol from endocytic compartments and impairs feedback inhibition of the SREBP pathway (Garver et al. 2007, Kruth et al. 1986, Liscum et al. 1989, Pentchev et al. 1986). As a result, decreased NPC1 protein function should decrease expression of the caveolin-1 gene. This is contrary to previous results indicating that NPC1 heterozygous (NPC1+/−) and NPC1 homozygous affected (NPC1−/−) human fibroblasts and mouse livers have significantly increased amounts of caveolin-1 protein, suggesting that the caveolin-1 protein may compensate for decreased NPC1 protein function (Garver et al. 1997a, Garver et al. 1997b Garver et al. 2002). Consistent with this hypothesis, the relative average amounts of the NPC2 protein, which is well known to interact and function in a coordinate manner with the NPC1 protein, is likewise increased in NPC1−/− mouse livers (Klein et al. 2006).
To determine whether the NPC1 and caveolin-1 proteins interact, studies were performed using reciprocal co-immunoprecipitation. The results indicated that immunoprecipitation of either the NPC1 or caveolin-1 proteins resulted in co-immunoprecipitation of the caveolin-1 and NPC1 proteins, respectively. Interestingly, of the two NPC1 proteins (170 and 190 kDa) which differ due to asparagine-linked glycosylation within the NTD, only the lower molecular weight NPC1 protein co-immunoprecipitated with caveolin-1. Previous studies have indicated that this lower molecular weight NPC1 protein is ~50% less efficient in facilitating efflux of LDL-derived cholesterol from endocytic compartments (Watari et al. 1999a, Watari et al. 1999b). Moreover, the results indicated that reciprocal co-immunoprecipitation of the NPC1 or caveolin-1 proteins was decreased when fibroblasts were grown in media with LDL, suggesting that the increased efflux of cholesterol from endocytic compartments partially inhibits interaction of the NPC1 and caveolin-1 proteins.
Consistent with interaction of the NPC1 and caveolin-1 proteins, the results indicated partial colocalization of the NPC1 and caveolin-1 proteins when examined using deconvolution fluorescence microscopy. This particular result confirms earlier reports describing partial co-localization of the NPC1 and caveolin-1 proteins using both human and mouse fibroblasts (Garver et al. 2000, Higgins et al. 1999). Moreover, consistent with the decreased interaction of the NPC1 and caveolin-1 proteins when fibroblasts are grown in media with LDL, quantification of high-resolution fluorescent images revealed significantly less co-localization when fibroblasts were grown using this condition. Interestingly, a recent study has indicated that caveolin-1 enriched vesicles that transiently interact with late endosomes/lysosomes are capable of forming caveolin-1 enriched subdomains that participate in the selective release of endosomal cargo (Pelkmans et al. 2004).
Studies have indicated that caveolin-1 is capable of specific and direct interaction with a number of proteins through a scaffolding domain (Human caveolin-1: 82-DGIWKASFTTFTVTKYWFYR-101) and that this domain partially overlaps with a cholesterol recognition sequence (Human caveolin-1: 91-TFTVTKYWFYRLL-103) that has stable and high-affinity for cholesterol (Li & Papadopoulos 1998, Murata et al. 1995). Both the caveolin-scaffolding domain and cholesterol recognition sequence are located at the cytofacial membrane and cytoplasm boundary near the amino-terminus of caveolin-1. This caveolin-scaffolding domain has been shown to interact with proteins that possess a caveolin-binding motif consensus sequence (jXjXXXXj), where j represents the amino acids Trp (W), Phe (F), or Tyr (Y) (Couet et al. 1997). Importantly, the human NPC1 protein was found to possess a previously unidentified caveolin-binding motif (818-FRFFKNSY-825) located within cytoplasmic loop 4 adjacent to the sterol-sensing domain (amino acids 616–797). A computer database search also revealed that the human NPC1 protein caveolin-binding motif is highly conserved among different species. This being the case, it is interesting to note that Patched, a membrane-bound protein with extensive sequence homology to the NPC1 protein, has also been shown to possess a caveolin-binding motif located within a cytoplasmic loop adjacent to the sterol-sensing domain and capable of interacting with caveolin-1 (Karpen et al. 2001).
To determine whether the caveolin-1 protein might have a role in modulating the efflux of LDL-derived cholesterol from endocytic compartments, fibroblasts were transfected with caveolin-1 siRNA to decrease expression of the caveolin-1 gene, and grown in media with LDL. The results clearly demonstrated a significant increase in the relative average amounts of LDL-derived cholesterol within LAMP-1 containing vesicles when compared to fibroblasts transfected with control siRNA and grown using similar conditions. However, compared to fibroblasts transfected with control siRNA, the total concentration of cellular cholesterol or cholesteryl ester was not significantly different for fibroblasts transfected with caveolin-1 siRNA, thereby suggesting that decreased amounts of caveolin-1 protein alters the distribution of cellular cholesterol (data not shown). This particular result is consistent with an earlier report indicating that a caveolin-1 dominant-negative mutant promotes altered intracellular cholesterol homeostasis characterized by an accumulation of cholesterol within late endosomes/lysosomes, although the concentration of cellular cholesterol remains unchanged (Pol et al. 2001, Roy et al. 1999). Moreover, a recent report using cells derived from the caveolin-1 deficient mouse indicated that caveolin-1 has a key role in maintaining intracellular cholesterol homeostasis, characterized by only a modest change in the concentration of cellular cholesterol and cholesteryl ester (Frank et al. 2006). This being the case, it is interesting to note that whole body cholesterol homeostasis, including plasma lipid levels, remain relatively normal for both the caveolin-1 deficient mouse and a patient diagnosed with a rare form of congenital lipodystrophy (Kim et al. 2008, Razani et al. 2002, Razani et al. 2001).
Conclusion
In summary, the present study determined that the NPC1 and caveolin-1 proteins interact and function to modulate efflux of LDL-derived cholesterol from endocytic compartments. A plausible model may be envisioned whereby the conserved NPC1 protein caveolin-binding motif allows interaction with the caveolin-1 scaffolding domain in the absence of LDL-derived cholesterol, but which is interrupted by the binding of cholesterol to the high-affinity caveolin-1 cholesterol recognition sequence which prompts dissociation of the NPC1 and caveolin-1 proteins and subsequent transport of cholesterol to other caveolin-1 cellular compartments, most likely the TGN. Clearly, additional studies will be necessary to further define how the NPC1 and caveolin-1 proteins modulate the efflux of LDL-derived cholesterol from endocytic compartments and maintain intracellular cholesterol homeostasis.
Acknowledgments
This work was supported in part by grants received from the National Institutes of Health (R21 DK071544), the Ara Parseghian Medical Research Foundation, and private donations for the investigation of childhood genetic and metabolic diseases. The authors would like to express their appreciation to Sarah Mount Patrick for expertise in performing deconvolution fluorescence microscopy and quantitation of fluorescence co-localization.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
Author Contributions
DJ and WSG conceived and performed these studies, while DJ, RAH, RAO and WSG interpreted results and prepared the manuscript.
References
- Bist A, Fielding PE, Fielding CJ. Two sterol regulatory element-like sequences mediate upregulation of caveolin gene transcription in response to low density lipoprotein free cholesterol. Proc Natl Acad Sci USA. 1997;94:10693–10698. doi: 10.1073/pnas.94.20.10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette-Mackie EJ, Dwyer NK, Amende LM, Kruth HS, Butler JD, Sokol J, Comly ME, Vanier MT, August JT, Brady RO, Pentchev PG. Type C Niemann-Pick disease: Low density lipoprotein uptake is associated with premature cholesterol accumulation in the Golgi complex and excessive cholesterol storage in lysosomes. Proc Natl Acad Sci USA. 1988;85:8022–8026. doi: 10.1073/pnas.85.21.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. Regulation of the activity of the low density lipoprotein receptor in human fibroblasts. Cell. 1975;6:307–316. doi: 10.1016/0092-8674(75)90182-8. [DOI] [PubMed] [Google Scholar]
- Brown MS, Ho YK, Goldstein JL. The lowdensity lipoprotein pathway in human fibroblasts: Relation between cell surface receptor binding and endocytosis of low-density lipoprotein. Ann NY Acad Sci. 1976;275:244–257. doi: 10.1111/j.1749-6632.1976.tb43358.x. [DOI] [PubMed] [Google Scholar]
- Cheruku SR, Xu Z, Dutia R, Lobel P, Storch J. Mechanism of cholesterol transfer from the Niemann-Pick type C2 protein to model membranes supports a role in lysosomal cholesterol transport. J Biol Chem. 2006;281:31594–31604. doi: 10.1074/jbc.M602765200. [DOI] [PubMed] [Google Scholar]
- Couet J, Li S, Okamoto T, Ikezu T, Lisanti MP. Identification of peptide and protein ligands for the caveolin-scaffolding domain. J Biol Chem. 1997;272:6525–6533. doi: 10.1074/jbc.272.10.6525. [DOI] [PubMed] [Google Scholar]
- Coxey RA, Pentchev PG, Campbell G, Blanchette-Mackie EJ. Differential accumulation of cholesterol in Golgi compartments of normal and Niemann-Pick type C fibroblasts incubated with LDL: A cytochemical freeze-fracture study. J Lipid Res. 1993;34:1165–1176. [PubMed] [Google Scholar]
- Dietschy JM, Turley SD, Spady DK. Role of liver in the maintenance of cholesterol and low density lipoprotein homeostasis in different animal species, including humans. J Lipid Res. 1993;34:1637–1659. [PubMed] [Google Scholar]
- Dupree P, Parton RG, Raposo G, Kurzchalia TV, Simons K. Caveolae and sorting in the trans-Golgi network of epithelial cells. EMBO J. 1993;12:1597–1605. doi: 10.1002/j.1460-2075.1993.tb05804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding PE, Fielding CJ. Intracellular transport of low density lipoprotein derived free cholesterol begins at clathrin-coated pits and terminates at cell surface caveolae. Biochemistry. 1996;35:14932–14938. doi: 10.1021/bi9613382. [DOI] [PubMed] [Google Scholar]
- Friedland N, Liou H-L, Lobel P, Stock AM. Structure of a cholesterol-binding protein deficient in Niemann-Pick type C2 disease. Proc Nat Acad Sci USA. 2003;100:2512–2517. doi: 10.1073/pnas.0437840100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver WS, Erickson RP, Wilson JM, Colton TL, Hossain GS, Kozloski MA, Heidenreich RA. Altered expression of caveolin-1 and increased cholesterol in detergent insoluble membrane fractions from liver in mice with Niemann-Pick disease type C. Biochim Biophys Acta. 1997a;1361:272–280. doi: 10.1016/s0925-4439(97)00047-1. [DOI] [PubMed] [Google Scholar]
- Garver WS, Heidenreich RA, Erickson RP, Thomas MA, Wilson JM. Localization of the murine Niemann-Pick C1 protein to two distinct intracellular compartments. J Lipid Res. 2000;41:673–687. [PubMed] [Google Scholar]
- Garver WS, Hsu SC, Erickson RP, Greer WL, Byers DM, Heidenreich RA. Increased expression of caveolin-1 in heterozygous Niemann-Pick type II human fibroblasts. Biochem Biophys Res Comm. 1997b;236:189–193. doi: 10.1006/bbrc.1997.6929. [DOI] [PubMed] [Google Scholar]
- Garver WS, Jelinek D, Oyarzo JN, Flynn J, Zuckerman M, Krishnan K, Chung BH, Heidenreich RA. Characterization of liver disease and lipid metabolism in the Niemann-Pick C1 mouse. J Cell Biochem. 2007;101:498–516. doi: 10.1002/jcb.21200. [DOI] [PubMed] [Google Scholar]
- Garver WS, Krishnan K, Gallagos JR, Michikawa M, Francis GA, Heidenreich RA. Niemann-Pick C1 protein regulates cholesterol transport to the trans-Golgi network and plasma membrane caveolae. J Lipid Res. 2002;43:579–589. [PubMed] [Google Scholar]
- Garver WS, Jelinek D, Francis GA, Murphy BD. The Niemann-Pick C1 gene is downregulated by feedback inhibition of the SREBP pathway in humans fibroblasts. J Lipid Res. 2008;49:1090–1102. doi: 10.1194/jlr.M700555-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Dana SE, Faust JR, Beaudet AL, Brown MS. Role of lysosomal acid lipase in the metabolism of plasma low density lipoprotein. Observations in cultured fibroblasts from a patient with cholesteryl ester storage disease. J Biol Chem. 1975;250:8487–8495. [PubMed] [Google Scholar]
- Hailstone D, Sleer LS, Parton RG, Stanley KK. Regulation of caveolin and caveolae by cholesterol in MDCK cells. J Lipid Res. 1998;39:369–379. [PubMed] [Google Scholar]
- Higgins ME, Davies JP, Chen FW, Ioannou YA. Niemann-Pick C1 is a late endosome-resident protein that transiently associates with lysosomes and the trans-Golgi network. Mol Gene Metab. 1999;68:1–13. doi: 10.1006/mgme.1999.2882. [DOI] [PubMed] [Google Scholar]
- Infante RE, Radhakrishnan A, Abi-Mosleh L, Kinch LN, Wang ML, Grishin NV, Goldstein JL, Brown MS. Purified NPC1 Protein II. Localization of sterol binding to a 240-amino acid soluble luminal loop. J Biol Chem. 2008a;283:1064–1075. doi: 10.1074/jbc.M707944200. [DOI] [PubMed] [Google Scholar]
- Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, Goldstein JL. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci USA. 2008b;105:15287–15292. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen HE, Bukowski JT, Hughes T, Gratton J, Sessa WC, Gailani MR. The sonic hedgehog receptor patched associates with caveolin-1 in cholesterol-rich microdomains of the plasma membrane. J Biol Chem. 2001;276:19503–19511. doi: 10.1074/jbc.M010832200. [DOI] [PubMed] [Google Scholar]
- Kim CA, Delepin M, Boutet M, El Mourabit H, Le Lay S, Meier M, Nemani M, Bridel E, Leite CC, Bertola DR, Semple RK, O'Rahilly S, Dugail I, Capeau J, Lathrop M, Magre J. Association of a homozygous nonsense caveolin mutation with Berardinelli-Seip congenital lipodystrophy. J Clin Endo Metab. 2008;93:1129–1134. doi: 10.1210/jc.2007-1328. [DOI] [PubMed] [Google Scholar]
- Klein A, Amigo L, Retamal MJ, Morales MG, Miquel JF, Rigotti A, Zanlungo S. NPC2 is expressed in human and murine liver and secreted into bile: Potential implications for body cholesterol homeostasis. Hepatology. 2006;43:126–133. doi: 10.1002/hep.20985. [DOI] [PubMed] [Google Scholar]
- Ko DC, Binkley J, Sidow A, Scott MP. The integrity of a cholesterol-binding pocket in Niemann-Pick C2 protein is necessary to control lysosome cholesterol levels. Proc Natl Acad Sci USA. 2003;100:2518–2525. doi: 10.1073/pnas.0530027100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruth HS, Comly ME, Butler JD, Vanier MT, Fink JK, Wenger DA, Patel S, Pentchev PG. Type C Niemann-Pick disease. Abnormal metabolism of low density lipoprotein in homozygous and heterozygous fibroblasts. J Biol Chem. 1986;261:16769–16774. [PubMed] [Google Scholar]
- Kwon HJ, Abi-Mosleh, Wang ML, Deisenhofer J, Goldstein JL, Brown MS, Infante RE. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137:1213–1224. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Papadopoulos V. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/ interaction amino acid sequence and consensus pattern. Endocrinology. 1998;139:4991–4997. doi: 10.1210/endo.139.12.6390. [DOI] [PubMed] [Google Scholar]
- Liscum L, Ruggiero RM, Faust JR. The intracellular transport of low density lipoprotein-derived cholesterol is defective in Niemann-Pick type C fibroblasts. J Cell Biol. 1989;108:1625–1636. doi: 10.1083/jcb.108.5.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Lu P, Chu JWK, Sharom FJ. Characterization of fluorescent sterol binding to purified human NPC1. J Biol Chem. 2009;284:1840–1852. doi: 10.1074/jbc.M803741200. [DOI] [PubMed] [Google Scholar]
- Murata M, Peranen J, Schreiner R, Wieland F, Kurzchalia TV, Simons K. VIP21/caveolin is a cholesterol-binding protein. Proc Natl Acad Sci USA. 1995;92:10339–10343. doi: 10.1073/pnas.92.22.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld EB, Wastney M, Patel S, Suresh S, Conney AM, Dwyer NK, Roff CF, Ohno K, Morris JA, Carstea ED, Incardona JP, Strauss JF, Vanier MT, Patterson MC, Brady RO, Pentchev PG, Blanchette-Mackie EJ. The Niemann-Pick C1 protein resides in a vesicular compartment linked to retrograde transport of multiple lysosomal cargo. J Biol Chem. 1999;274:9627–9635. doi: 10.1074/jbc.274.14.9627. [DOI] [PubMed] [Google Scholar]
- Ohgami N, Ko DC, Thomas M, Scott MP, Chang CCY, Chang TY. Binding between the Niemann-Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc Natl Acad Sci USA. 2004;101:12473–12478. doi: 10.1073/pnas.0405255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osono Y, Woollett LA, Herz J, Dietschy JM. Role of the low density lipoprotein receptor in the flux of cholesterol through the plasma and across the tissues of the mouse. J Clin Invest. 1995;95:1124–1132. doi: 10.1172/JCI117760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L, Burli T, Zerial M, Helenius A. Caveolin-stabilized membrane domains as multifunctional transport and sorting devices in endocytic membrane traffic. Cell. 2004;118:767–780. doi: 10.1016/j.cell.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Pentchev PG, Kruth HS, Comly ME, Butler JD, Vanier MT, Wenger DA, Patel S. Type C Niemann-Pick disease. A parallel loss of regulatory responses in both the uptake and esterification of low density lipoprotein-derived cholesterol in cultured fibroblasts. J Biol Chem. 1986;261:16775–16780. [PubMed] [Google Scholar]
- Pol A, Luetterforst R, Lindsay M, Heino S, Ikonen E, Parton RG. A caveolin dominant negative mutant associates with lipid bodies and induces intracellular cholesterol imbalance. J Cell Biol. 2001;152:1057–1070. doi: 10.1083/jcb.152.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B, Engelman JA, Wang XB, Schubert W, Zhang XL, Marks CB, Macaluso F, Russell RG, Li M, Pestell RG, Di Vizio D, Hou H, Kneitz B, Lagaud G, Christ GJ, Edelmann W, Lisanti MP. Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J Biol Chem. 2001;276:38121–38138. doi: 10.1074/jbc.M105408200. [DOI] [PubMed] [Google Scholar]
- Razani B, Combs TP, Wang XB, Frank PG, Park DS, Russell RG, Li M, Tang B, Jelicks LA, Scherer PE, Lisanti MP. Caveolin-1-deficient mice are lean, resistant to diet-induced obesity, and show hyper-triglyceridemia with adipocyte abnormalities. J Biol Chem. 2002;277:8635–8647. doi: 10.1074/jbc.M110970200. [DOI] [PubMed] [Google Scholar]
- Roy S, Luetterforst R, Harding A, Apolloni A, Etheridge M, Stang E, Rolls B, Hancock JF, Parton RG. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat Cell Biol. 1999;1:98–105. doi: 10.1038/10067. [DOI] [PubMed] [Google Scholar]
- Sleat DE, Wiseman JA, El-Banna M, Price SM, Verot L, Shen MM, Tint GS, Vanier MT, Walkley SU, Lobel P. Genetic evidence for non-redundant functional cooperativity between NPC1 and NPC2 in lipid transport. Proc Natl Acad Sci USA. 2004;101:5886–5891. doi: 10.1073/pnas.0308456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart EJ, Ying Y, Donzell WC, Anderson RGW. A role for caveolin in transport of cholesterol from endoplasmic reticulum to plasma membrane. J Biol Chem. 1996;271:29427–29435. doi: 10.1074/jbc.271.46.29427. [DOI] [PubMed] [Google Scholar]
- Sugii S, Reid PC, Ohgami N, Du H, Chang TY. Distinct endosomal compartments in early trafficking of low density lipoprotein-derived cholesterol. J Biol Chem. 2003;278:27180–27189. doi: 10.1074/jbc.M300542200. [DOI] [PubMed] [Google Scholar]
- Turley SD, Spady DK, Dietschy JM. Role of liver in the synthesis of cholesterol and the clearance of low density lipoproteins in the cynomolgus monkey. J Lipid Res. 1995;36:67–79. [PubMed] [Google Scholar]
- Uittenbogaard A, Everson WV, Matveev SV, Smart EJ. Cholesteryl ester is transported from caveolae to internal membranes as part of a caveolin-annexin II lipid-protein complex. J Biol Chem. 2002;277:4925–4931. doi: 10.1074/jbc.M109278200. [DOI] [PubMed] [Google Scholar]
- Uittenbogaard A, Ying Y, Smart EJ. Characterization of a cytosolic heat-shock protein-caveolin chaperone complex. Involvement in cholesterol trafficking. J Biol Chem. 1998;273:6525–6532. doi: 10.1074/jbc.273.11.6525. [DOI] [PubMed] [Google Scholar]
- Underwood KW, Jacobs NL, Howley A, Liscum L. Evidence for a cholesterol transport pathway from lysosomes to endoplasmic reticulum that is independent of the plasma membrane. J Biol Chem. 1998;273:4266–4274. doi: 10.1074/jbc.273.7.4266. [DOI] [PubMed] [Google Scholar]
- Urano Y, Watanabe H, Murphy SR, Shibuya Y, Geng Y, Peden AA, Chang CCY, Chang TY. Transport of LDL-derived cholesterol from the NPC1 compartment to the ER involves the trans-Golgi network and the SNARE protein complex. Proc Natl Acad Sci USA. 2008;105:16513–16518. doi: 10.1073/pnas.0807450105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watari H, Blanchette-Mackie EJ, Dwyer NK, Glick JM, Patel S, Neufeld EB, Brady RO, Pentchev PG, Strauss JF., III Niemann-Pick C1 protein: Obligatory roles for N-terminal domains and lysosomal targeting in cholesterol mobilization. Proc Natl Acad Sci USA. 1999a;96:805–810. doi: 10.1073/pnas.96.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watari H, Blanchette-Mackie EJ, Dwyer NK, Watari M, Neufeld EB, Patel S, Pentchev PG, Strauss JF., III Mutations in the leucine zipper motif and sterol-sensing domain inactivate the Niemann-Pick C1 glycoprotein. J Biol Chem. 1999b;274:21861–21866. doi: 10.1074/jbc.274.31.21861. [DOI] [PubMed] [Google Scholar]
- Wojtanik KM, Liscum L. The transport of low density lipoprotein-derived cholesterol to the plasma membrane is defective in NPC1 cells. J Biol Chem. 2003;278:14850–14856. doi: 10.1074/jbc.M300488200. [DOI] [PubMed] [Google Scholar]