Abstract
Understanding early determinants of type 2 diabetes is essential for refining disease prevention strategies. Proteomic technology may provide a useful approach to identify novel protein patterns potentially related to pathophysiological changes that lead up to diabetes. In this study, we sought to identify protein signals that are associated with diabetes incidence in a middle-aged population. Serum samples from 519 participants in a nested case–control selection (167 cases and 352 age-, sex- and BMI-matched normoglycemic control subjects, median follow-up 14.0 years) within the Whitehall-II cohort were analyzed by linear matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS). Nine protein peaks were found to be associated with incident diabetes. Rate ratios for high peak intensity ranged between 0.4 (95% CI, 0.2–0.8) and 4.0 (95% CI, 1.7–9.2) and were robust to adjustment for main potential confounders, including obesity, lipids and C-reactive protein. The proteins associated with these peaks may reflect diabetes pathogenesis. Our study exemplifies the utility of an approach that combines proteomic and epidemiological data.
Keywords: MALDI-TOF, Type 2 diabetes, Proteomics, Biomarker, Whitehall-II study, Random Forests
Introduction
Type 2 diabetes (T2DM) is a complex metabolic disorder, primarily characterized by abnormal glucose regulation [1]. In addition, several mechanisms not directly related to glucose regulation such as low-grade inflammation and adipocyte metabolism have been implicated in the pathogenesis of the disease [2, 3]. These processes involve upregulation of pro-inflammatory pathways and altered expression of cytokines and adipokines [4, 5]. Several biomarkers have been suggested to predict T2DM in early and progressive stages [6, 7]; however, it remains unclear whether currently known biomarkers accurately reflect all involved pathways and whether the associations between these markers and diabetes pathogenesis are causal. Indeed, it is conceivable that other metabolic systems could be causally involved or secondarily affected by the early changes leading to diabetes.
Current mass spectrometric methods enable the identification of not only novel protein patterns but also post-translationally modified proteins and in turn novel signal transduction pathways and their deregulation under pathological conditions as it can be expected that levels of many blood proteins are altered as a consequence of multiple early molecular pathophysiological changes. However, it should be noted that initial optimism over the possibilities of mass spectrometry as a tool for biomarker discovery has been tempered by the realization that confounding factors, including technical imprecisions and sample processing, represent important challenges to omics-based biomarker research [8–10].
Despite these limitations, we examine here the utility of a proteomic approach in an epidemiological setting. Specifically, we aimed to assess the value of MALDI-TOF-MS as a method for identifying protein peak signals that might serve as new markers of future diabetes.
Materials and methods
Study population
Data are from a nested case–control selection within the Whitehall-II study, an occupational cohort of 10,308 participants aged 35–55 years recruited from 20 civil service departments in London, UK, in 1985–1988 [11, 12]. Phase 3 (1991–94) was the first medical examination where glycemic status was determined by a 75-g oral glucose tolerance test (OGTT) and serves as the baseline for the current study. Further waves of data collection were carried out at 2.5-year intervals (phases 4–8), with OGTTs performed at phase 5 (1997–1999) and phase 7 (2002–2004). The case–control selection was performed based on blood sample availability. Cases (n = 167) are individuals with incident type 2 diabetes during a median follow-up period of 14.0 (IQR, 11.3–14.4) years and without type 2 diabetes at baseline. Controls (n = 352) had normal glucose tolerance at baseline and throughout follow-up and were frequency matched for age (5-year bands), sex and body mass index (BMI, 5 kg/m2 bands).
We used a complete case approach, excluding participants with missing information on any of the following covariates at baseline: age, sex, height, weight, smoking habits (never, ex, current), systolic blood pressure, total cholesterol, high density lipoprotein (HDL) cholesterol, triglycerides and high-sensitive C-reactive protein (CRP). Also, participants with prevalent or incident coronary heart disease, self-reported long-standing inflammatory illness or recent inflammatory symptoms, anti-inflammatory medication and non-white ethnicity were excluded. Baseline characteristics of the study population are summarized in Table 1. The study was approved by the University College London Medical School Committee on the Ethics of Human Research and conducted according to the Declaration of Helsinki. Written informed consent was obtained at baseline and renewed at each contact.
Table 1.
Baseline characteristics of study population
| Controls (n = 195) | Cases (n = 85) | |
|---|---|---|
| Male (%) | 73.3 (66.5;79.4) | 72.9 (62.2;82.0) |
| Age (years) | 50.7 (6.3) | 51.2 (5.8) |
| BMI (kg/m2) | 26.0 (3.5) | 26.7 (3.8) |
| Height (cm) | 172.9 (9.1) | 172.2 (8.6) |
| Diastolic blood pressure (mmHg) | 79.5 (9.3) | 83.3 (9.9) |
| Systolic blood pressure (mmHg) | 119.4 (11.8) | 126.5 (14.8) |
| Total cholesterol (mmol/l) | 6.6 (1.2) | 6.6 (1.2) |
| HDL cholesterol (mmol/l) | 1.5 (0.4) | 1.3 (0.3) |
| LDL cholesterol (mmol/l) | 4.5 (1.1) | 4.5 (1.1) |
| Triglycerides (mmol/l) | 1.4 (0.9) | 1.7 (0.9) |
| CRP (mg/l) | 0.8 (0.5;1.7) | 1.0 (0.6;2.2) |
| Smoking habits | ||
| Never-smoker (%) | 50.3 (43.0;57.5) | 44.7 (33.9;55.9) |
| Ex-smoker (%) | 35.9 (29.2;43.1) | 41.2 (30.6;52.4) |
| Current smoker (%) | 13.8 (9.3;19.5) | 14.1 (7.5;23.4) |
| Fasting plasma glucose (mmol/l) | 5.1 (0.4) | 5.4 (0.5) |
| 2-hour plasma glucose (mmol/l) | 5.0 (1.1) | 6.6 (1.9) |
| Anti-hypertensive treatment (%) | 6.2 (3.2;10.5) | 12.9 (6.6;22.0) |
| Lipid-lowering treatment (%) | 0.5 (0.0;2.8) | 2.4 (0.3;8.2) |
Data are means (SD), medians (interquartile range) or proportions (95% CI)
Measurements
Diabetes incidence was defined according to WHO criteria [13] and ascertained throughout follow-up based on a 75-g OGTT in combination with self-reports of diabetes or the use of glucose-lowering medication.
At baseline (phase 3), height, weight and blood pressure were measured according to a standard protocol. Venous blood samples were collected after an overnight fast in the morning or in the afternoon after no more than a light fat-free breakfast eaten before 08.00 h. After the initial venous blood samples were taken, the participants underwent a standard 2-h 75-g OGTT. Plasma glucose was analyzed both in the fasting and 2-h samples. In addition, total cholesterol, HDL and LDL cholesterol, triglycerides and CRP were determined in the fasting samples. Information on family history of diabetes (first-degree relative) and smoking habits (never, ex and current) was collected using a self-administered questionnaire. Missing values on family history of diabetes were set to ‘none’. Data collection and methods in the Whitehall-II study have been described in greater detail elsewhere [14, 15].
Serum sample extraction was performed using Dynabeads RPC 18 magnetic beads (Invitrogen, Carlsbad, CA, USA) as it was described previously [16]. Twelve pmol of horse myoglobin (used as internal standard, m/z = 16.952.55 Da) was added to each sample extract prior to MALDI-TOF-MS analysis. Two microliters of sample extract (containing internal standard) was spotted onto a MTP Ground Steel 384 target (Bruker Daltonics) with a droplet (0.5 μl) of a saturated solution of sinapinic acid dissolved in a 1:1 solution of acetonitrile/0.1% trifluoroacetic acid. Data acquisition was performed by linear MALDI-TOF-MS in positive ion mode on an Ultraflex III (Bruker Daltonics, Bremen, Germany) in the 1–20 kDa range.
Data processing
Prior to statistical analysis, mass spectra were calibrated using internal standards and subsequently processed using the open source software LIMPIC [17]. In this algorithm, calibrated signals were normalized using the intensity of internal calibrants. Since the mass spectrum may be affected by a low signal-to-noise ratio, its quality was enhanced by smoothing and baseline subtraction. Next, the detection of protein peaks in the single spectra was performed by finding all the local maxima and eliminating those with intensity lower than a non-uniform threshold, proportional to the noise level (signal-to-noise ratio threshold set to 3). Peak clustering was performed in order to identify in each spectrum the protein peaks corresponding to the same m/z (mass-to-charge ratio) class. The clustering procedure assumes that peaks within a given distance limit can be associated with the same class. Peaks are classified as protein or noise peaks on the basis of their consistency across the spectra. Qualification criteria for spectra compliance were a total ion count per spectrum of 1.5 × 104 and ion counts of transthyretin and myoglobin of at least 200. Only peaks with a frequency of 5% or more in the study sample were considered in the present study.
Statistical analysis
Given the relative high number of protein peaks to the number of participants in our study, we performed an initial screening of the peaks employing Random Forests (RF) analysis with incident diabetes as outcome [18]. In brief, the RF algorithm constructs an ensemble of classification trees (10.000 trees in this study) from several bootstrap samples of the original data and votes over trees in order to increase prediction [19]. Bootstrapping is sampling with replacement, and in each bootstrap, about one-third of the study participants are left out of the construction of a particular tree. This so-called out-of-bag sample is used as test data for calculating the error rate of the derived classification tree. For each protein peak included in the analysis, the RF algorithm computes an estimate of the increase in error rate of the classification tree had that peak not been used, a procedure named permutation test. The permutation test was used to rank the protein peaks, and the 20 highest ranking peaks were selected for further analysis.
For each of the 20 peaks, we created a binary variable indicating whether or not peak intensity was elevated. The threshold for elevated peak intensity was selected to maximize the difference in observed diabetes incidence between the two resulting subsets of participants and was determined by a single classification tree with one split.
Subsequently, univariate rate ratios for incident diabetes for the subset of participants with elevated peak intensity were assessed in Poisson regression analysis with individual follow-up time as offset. A level of significance of 5% was adjusted for multiple testing (n = 20) using the method by Benjamini et al. [20]. For each significant peak, we assessed the attenuating effect on the associations of including different levels of confounders in a staged approach, Model 1: univariate associations; Model 2: adjusting for age and sex; Model 3: further adjusting for family history of diabetes, BMI, smoking habits and systolic blood pressure; Model 4: further adjusting for total and HDL cholesterol, triglycerides and CRP.
Results
MALDI-TOF spectra from 304 (59%) of the 519 study participants met the qualification criteria for spectra compliance. Failure in meeting the qualification criteria for spectra compliance was due to technical reasons not related to case/control status of the participants. There were no differences in characteristics between participants with and without eligible spectra (Appendix Table 4). Our study population was comprised of the 280 individuals (92%) (85 cases and 195 controls) with complete data on the covariates. Study participants were middle aged and mainly male (73%) (Table 1).
A total of 364 different peaks were detected, but only 72 had a frequency of 5% or more in the study population and were considered in the present analysis. For the 20 highest ranking peaks from the RF analysis, nine were significantly associated with incident diabetes. Rate ratios for participants with peak intensity above the split threshold ranged between 0.4 (95% CI, 0.2–0.8) and 4.0 (95% CI, 1.7–9.2). The estimated rate ratios were robust toward confounder adjustment (Table 2). In three of the peaks (m/z, 3,774, 3,816 and 9,352) did adjustment for 2-h plasma glucose have a substantial attenuating effect on the rate ratios with incident diabetes.
Table 2.
Rate ratios for incident diabetes for participants with elevated peak intensity
| Peak presence
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| m/z | Controls (%) | Cases (%) | Elevated peak intensity (%) | Model 1 | Model 2 | Model 3 | Model 4 | Model 5 | Model 6 |
| 3,816 | 3 | 11 | 3 | 4.1 (1.8;9.5) | 4.1 (1.8;9.6) | 4.9 (2.1;11.7) | 4.4 (1.9;10.6) | 4.5 (1.9;10.8) | 1.9 (0.7;4.9) |
| 4,154 | 29 | 31 | 4 | 3.0 (1.5;5.9) | 3.0 (1.5;5.9) | 3.4 (1.7;6.8) | 3.8 (1.9;7.8) | 2.8 (1.3;5.8) | 3.0 (1.5;6.3) |
| 9,352 | 34 | 36 | 6 | 2.9 (1.5;5.4) | 3.0 (1.6;5.8) | 3.0 (1.5;5.8) | 3.0 (1.4;6.1) | 2.8 (1.3;5.8) | 2.4 (1.1;4.9) |
| 8,913 | 100 | 99 | 5 | 2.5 (1.2;5.3) | 2.6 (1.3;5.5) | 2.6 (1.2;5.4) | 2.4 (1.1;5.3) | 2.3 (1.0;5.1) | 2.1 (1.0;4.7) |
| 3,956 | 6 | 13 | 6 | 2.5 (1.3;4.8) | 2.6 (1.3;5.2) | 3.0 (1.5;6.2) | 2.7 (1.3;5.6) | 2.8 (1.4;5.8) | 2.1 (1.0;4.5) |
| 3,774 | 4 | 11 | 5 | 2.4 (1.1;4.9) | 2.4 (1.2;5.2) | 3.2 (1.5;7.0) | 3.1 (1.4;6.7) | 2.9 (1.3;6.3) | 1.8 (0.8;4.2) |
| 6,880 | 10 | 20 | 10 | 2.2 (1.3;3.8) | 2.2 (1.3;3.8) | 2.0 (1.1;3.5) | 1.8 (1.0;3.2) | 1.7 (0.9;3.0) | 1.7 (0.9;3.0) |
| 4,711 | 44 | 61 | 45 | 1.9 (1.2;2.9) | 1.9 (1.2;2.9) | 1.8 (1.1;2.7) | 1.8 (1.2;2.8) | 1.8 (1.2;2.8) | 1.6 (1.0;2.5) |
| 8,763 | 97 | 91 | 94 | 0.4 (0.2;0.8) | 0.4 (0.2;0.8) | 0.4 (0.2;0.7) | 0.4 (0.2;0.8) | 0.4 (0.2;0.9) | 0.4 (0.2;0.9) |
Model 1: unadjusted
Model 2: adjusted for age and sex
Model 3: further adjusted for family history of diabetes, BMI, smoking habits and systolic blood pressure
Model 4: further adjusted for total and HDL cholesterol, triglycerides and CRP
Model 5: Model 4 and further adjusted for fasting plasma glucose
Model 6: Model 4 and further adjusted for 2-h plasma glucose
Figure 1 provides an illustrative example of averaged spectra at the 4,154 Da peak for individuals with and without incident diabetes.
Fig. 1.
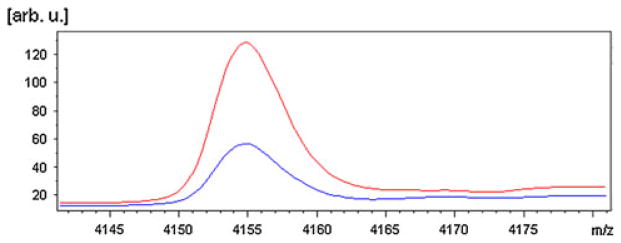
Comparison of average spectra at the 4,154 Da peak for individuals with incident diabetes (red) and without (blue) (color figure online)
Discussion
This study explored an approach in which data from mass spectrometric profiling by MALDI-TOF were combined with epidemiological data. We identified several peak thresholds in protein peak signal intensities that were robustly associated with incident diabetes.
Previously, a number of studies have employed mass spectrometry as a tool for biomarker discovery [21–24]. A recent FDA approval of a diagnostic tool issued from SELDI proteomic investigations illustrates the application of mass spectrometric methods for these purposes [25]. However, the cross-sectional nature and the small sample size of most of these biomarker studies have precluded information on the association between biomarkers and future development of disease as well as the ability to adjust for potential confounders. Therefore, a major strength of the present study is the use of a well-characterized prospective cohort where participants have been followed longitudinally for decades. A recent study by Wang et al. [26] did collect mass spectrometric data in the framework of an epidemiological study, but focused on individual amino acids as predictors of future development of diabetes.
Potential limitations of this study involve the exclusion of a large number of mass spectra, which failed to meet the qualification criteria. Although we are convinced that the reasons for this exclusion were entirely due to technical issues as the proportion lost was virtually identical among cases and controls and as the groups did not differ in baseline characteristics (Appendix Table 4), we cannot entirely exclude bias related to these exclusions. Subsequent studies should focus on reducing the number of excluded spectra. Also, general concerns raised in relation to mass spectrometric efforts represent additional important limitations to this study. Criticism has often focused on the poor reproducibility of mass spectrometric studies owing to factors impacting pre-analytical and analytical bias [27–30]. Although strict operating procedures were implemented for sample collection, it cannot be ruled out that bias has been introduced as a result of sample handling or storage. Samples were stored for different lengths of time, which may impact the observed changes in mass peaks.
Furthermore, the use of peak intensities from MALDI profiling in our statistical analyses may be problematic. The relationship between mass peak intensity and protein abundance is not fully understood, partly due to the varying ionization properties of different molecules [31]. Complicating matters further, the ionization process may cause molecules to fragment into several smaller molecules, resulting in a complex mass peak pattern that may be not readily interpreted [32]. Also, although mass spectrometry has been widely used as a biomarker discovery tool, few candidate biomarkers have been qualified or verified and even fewer validated [33].
However, the relatively high number of samples included in the present study should contribute to reducing the impact of these potential biases. Furthermore, peak intensities were normalized to an internal standard added in equal amounts to each sample, aiming to limit the effect of interferences with other analytes on the processed signal intensities [29].
We used a two-step approach for statistical analysis using the RF method for the initial screening of peaks. RF is a nonparametric approach; thus, we could model the relationship between variables without the need to specify a particular model. In particular, it does not require the specification of a particular linear or nonlinear relationship between predictor and response variables. Also, it provides a variable ranking method for selecting the most predictive peaks for incident diabetes that takes into account peak interaction. RF has shown excellent performance in accuracy among current classification algorithms [34–36] and has been used in several microarray and proteomics studies [37, 38].
The number of significant peaks found in our study was greater than the number of peaks that could be expected to result from chance. Furthermore, most rate ratios for incident diabetes were very robust toward confounder adjustment, providing further support for our findings although validation is an essential first step prior to application of this approach to larger cohorts.
Finally, the identities and functions of the molecules corresponding to the specific peaks detected here remain to be established. However, using the Expert Protein Analysis System (ExPASy) TagIdent tool [39], we are suggesting plausible candidates based on the m/z values (Table 3).
Table 3.
Suggested protein identities based on mass values of detected peaks
| Detected peak (m/z) | Suggested protein |
|---|---|
| 3,816 | CGRP-1 |
| 4,154 | PP |
| 9,352 | MCP-4 |
| 8,913 | apoC-II |
| 3,956 | (N/A) |
| 3,774 | GLP-2 |
| 6,880 | Lkn-1 |
| 4,711 | (N/A) |
| 8,763 | apoC-III |
CGRP-1 calcitonin gene-related peptide 1; PP pancreatic polypeptide; MCP-4 monocyte chemotactic protein; ApoC-II apolipoprotein C-II; GLP-2 glucagon-like peptide 2; Lkn-1 leukotactin-1
The search was performed with a ±0.7% tolerance in m/z value, allowing for experimental error or possible protein modifications, and an isoelectric point (pI) interval from 3 to 10.
Calcitonin gene-related peptide 1 (CGRP-1) is within the specified mass range of the peak detected here at 3,816 Da. In skeletal muscle in vitro and in rat models, CGRP-1 has been shown to affect glucose regulation through inhibition of insulin-stimulated glycogen synthesis [40, 41]. Similarly, pancreatic prohormone (PP) is in the specified range of the peak at 4,154 Da. The physiological role for pancreatic hormone is not well described, although it may act as a regulator of pancreatic functions. Indeed, PP deficiency following pancreatic resection may lead to hyperglycemia due to impaired hepatic insulin action [42].
The peak detected at 9,352 Da may represent monocyte chemotactic protein 4 (MCP-4). In a recent study of patients with symptomatic carotid atherosclerosis, MCP-4 was suggested to have pro-atherogenic effects as an inflammatory mediator of platelet and monocyte activation [43]; these effects may accompany incident diabetes in our study sample.
The peak at 8,913 Da may be apolipoprotein C-II (apoC-II). Béliard et al. [44] found increased total ApoC-II concentrations in type 2 diabetic patients compared to healthy control subjects. The peak detected at 3,774 Da may be an alternative representation of CGRP-1 (also suggested for the peak detected at 3,816 Da) or glucagon-like peptide 2 (GLP-2). GLP-2 may indirectly increase glucose levels through stimulation of glucagon secretion [45, 46].
Leukotactin-1 may represent the peak at 6,880 Da. Lkn-1 is involved in inflammatory diseases and essential in the development of atherosclerosis as a potent chemoattractant for leukocytes and by enhancing adhesion molecules on endothelial cells and leukocytes [47, 48]. As for MCP-4, it is not unlikely that diabetes in our sample occurs in concert with early pro-atherogenic events.
Finally, the peak at 8,763 Da may represent apolipoprotein C-III (apoC-III). Increased levels of apoC-III have been found in type 2 diabetic patients [44], and one study reported high apoC-III levels to be a major diabetogenic factor [49].
For the peaks detected at 3,956 and 4,711 Da, no plausible identities have been suggested using the Expasy TagIdent tool. However, using MALDI-TOF on serum samples, one study found identified a peak at 3,956 Da as internal fragment of inter-α-trypsin inhibitor heavy chain 4 (ITIH4), a marker for ovarian cancer [50].
In addition to the already established mechanisms, the suggested proteins may play previously unknown roles in glucose metabolism. Besides the proteins suggested here, there were a number of uncharacterized proteins within the specified range. These proteins may also represent candidate identities for the peaks detected in this study. Also, as previously mentioned, it should be pointed out that the peaks detected here may in fact not be intact proteins, but rather fragments of larger proteins. Upcoming analyses will entail LC/MS of eluates in order to confirm identities of the peaks.
Our data suggest that the combination of a relatively simple mass spectrometric method with epidemiological data may provide a good starting point for assessing further the value of proteomic profile information in predicting future diabetes. The normalized peak intensities provide a semi-quantitative measure of serum protein concentration; therefore, these efforts may help determine the possible biological relevance of a given peak intensity in relation to the split thresholds.
Using a mass spectrometry-based protein profiling platform, we detected several protein peaks significantly associated with incident diabetes in a population free of diabetes at baseline. The present study can be viewed as a proof-of-concept warranting further studies, where an increase in sample size may allow further detection of proteins possibly involved in the early pathophysiology of type 2 diabetes.
Acknowledgments
The Whitehall II study is supported by the Medical Research Council, the British Heart Foundation, the US National Institutes of Health (R01HL36310, R01AG013196, R01AG034454).
Appendix
See Table 4
Table 4.
Baseline characteristics of participants with and without MALDI-TOF spectra
| Without (n = 230) | With (n = 329) | p value for difference* | |
|---|---|---|---|
| Males (%) | 74.3 (68.2;79.9) | 70.8 (65.6;75.7) | 0.358 |
| Age (years) | 50.5 (6.3) | 51.0 (6.1) | 0.420 |
| BMI (kg/m2) | 26.9 (4.6) | 26.4 (3.8) | 0.111 |
| Height (cm) | 173.6 (8.8) | 172.6 (9.2) | 0.268 |
| Diastolic blood pressure (mmHg) | 82.2 (9.8) | 80.7 (9.7) | 0.079 |
| Systolic blood pressure (mmHg) | 123.8 (13.7) | 121.7 (13.5) | 0.077 |
| Total cholesterol (mmol/l) | 6.7 (1.1) | 6.6 (1.2) | 0.437 |
| HDL cholesterol (mmol/l) | 1.4 (0.4) | 1.4 (0.4) | 0.745 |
| LDL cholesterol (mmol/l) | 4.5 (1.0) | 4.5 (1.1) | 0.820 |
| Triglycerides (mmol/l) | 1.8 (1.4) | 1.6 (1.0) | 0.008 |
| CRP (mg/l) | 0.9 (0.5;1.6) | 0.9 (0.5;2) | 0.400 |
| Smoking habits (%) | |||
| Never-smoker (%) | 54.8 (48.1;61.3) | 45.3 (39.8;50.8) | 0.027 |
| Ex-smoker (%) | 33.9 (27.8;40.4) | 34.0 (28.9;39.4) | 0.975 |
| Current smoker (%) | 6.1 (3.4;10.0) | 13.4 (9.9;17.5) | 0.004 |
| Fasting plasma glucose (mmol/l) | 5.2 (0.5) | 5.2 (0.5) | 0.991 |
| 2-h plasma glucose (mmol/l) | 5.8 (1.7) | 5.4 (1.6) | 0.020 |
| Anti-hypertensive treatment (%) | 7.4 (4.4;11.6) | 8.5 (5.7;12.1) | 0.631 |
| Lipid-lowering treatment (%) | 0.0 (0.0;1.6) | 0.9 (0.2;2.6) | 0.074 |
Data are means (SD), medians (interquartile range) or proportions (95% CI).
After controlling for multiple testing, there were no significant differences in characteristics between the two groups
Footnotes
Conflict of interest TMJ, DRW and DV are employed by the Steno Diabetes Center A/S, a research hospital working in the Danish National Health Service and owned by Novo Nordisk A/S. TMJ, DRW and DV hold shares in Novo Nordisk Inc. Remaining authors declare no duality of interest.
Contributor Information
Troels Mygind Jensen, Email: tmyj@steno.dk, Steno Diabetes Center A/S, Gentofte, Denmark.
Daniel R. Witte, Steno Diabetes Center A/S, Gentofte, Denmark
Damiana Pieragostino, Department of Biomedical Sciences, University “G. d’Annunzio” of Chieti-Pescara, Chieti, Italy.
James N. McGuire, Hagedorn Research Institute, Gentofte, Denmark
Ellis D. Schjerning, Hagedorn Research Institute, Gentofte, Denmark
Chiara Nardi, IRCCS-Santa Lucia Foundation, Rome, Italy.
Andrea Urbani, IRCCS-Santa Lucia Foundation, Rome, ItalyDepartment of Internal Medicine, University “Tor Vergata”, Rome, Italy.
Mika Kivimäki, Department of Epidemiology & Public Health, University College London, London, UK.
Eric J. Brunner, Department of Epidemiology & Public Health, University College London, London, UK
Adam G. Tabàk, Department of Epidemiology & Public Health, University College London, London, UKSemmelweis University Faculty of Medicine, 1st Department of Medicine, Budapest, Hungary
Dorte Vistisen, Steno Diabetes Center A/S, Gentofte, Denmark.
References
- 1.Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: principles of pathogenesis and therapy. Lancet. 2005;365:1333–1346. doi: 10.1016/S0140-6736(05)61032-X. [DOI] [PubMed] [Google Scholar]
- 2.Duncan BB, Schmidt MI, Pankow JS, Ballantyne CM, Couper D, Vigo A, Hoogeveen R, Folsom AR, Heiss G. Low-grade systemic inflammation and the development of type 2 diabetes: the atherosclerosis risk in communities study. Diabetes. 2003;52:1799–1805. doi: 10.2337/diabetes.52.7.1799. [DOI] [PubMed] [Google Scholar]
- 3.Cusi K. The role of adipose tissue and lipotoxicity in the pathogenesis of type 2 diabetes. Curr Diab Rep. 2010;10:306–315. doi: 10.1007/s11892-010-0122-6. [DOI] [PubMed] [Google Scholar]
- 4.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kolb H, Mandrup-Poulsen T. An immune origin of type 2 diabetes? Diabetologia. 2005;48:1038–1050. doi: 10.1007/s00125-005-1764-9. [DOI] [PubMed] [Google Scholar]
- 6.Kolberg JA, Jorgensen T, Gerwien RW, Hamren S, McKenna MP, Moler E, Rowe MW, Urdea MS, Xu XM, Hansen T, Pedersen O, Borch-Johnsen K. Development of a type 2 diabetes risk model from a panel of serum biomarkers from the Inter99 cohort. Diabetes Care. 2009;32:1207–1212. doi: 10.2337/dc08-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salomaa V, Havulinna A, Saarela O, Zeller T, Jousilahti P, Jula A, Muenzel T, Aromaa A, Evans A, Kuulasmaa K, Blankenberg S. Thirty-one novel biomarkers as predictors for clinically incident diabetes. PLoS One. 2010;5:e10100. doi: 10.1371/journal.pone.0010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albrethsen J. The first decade of MALDI protein profiling: a lesson in translational biomarker research. J Proteomics. 2011;74:765–773. doi: 10.1016/j.jprot.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 9.Albrethsen J. Reproducibility in protein profiling by MALDI-TOF mass spectrometry. Clin Chem. 2007;53:852–858. doi: 10.1373/clinchem.2006.082644. [DOI] [PubMed] [Google Scholar]
- 10.Ransohoff DF. Lessons from controversy: ovarian cancer screening and serum proteomics. J Natl Cancer Inst. 2005;97:315–319. doi: 10.1093/jnci/dji054. [DOI] [PubMed] [Google Scholar]
- 11.Marmot M, Brunner E. Cohort profile: the Whitehall II study. Int J Epidemiol. 2005;34:251–256. doi: 10.1093/ije/dyh372. [DOI] [PubMed] [Google Scholar]
- 12.Herder C, Brunner EJ, Rathmann W, Strassburger K, Tabák AG, Schloot NC, Witte DR. Elevated levels of the anti-inflammatory interleukin-1 receptor antagonist precede the onset of type 2 diabetes: the Whitehall II study. Diabetes Care. 2009;32:421–423. doi: 10.2337/dc08-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization. Report of a WHO Consultation, Part 1: Diagnosis and classification of diabetes mellitus. Geneva: World Health Organisation; 1999. Definition, diagnosis and classification of diabetes mellitus and its complications. [Google Scholar]
- 14.Brunner EJ, Marmot MG, Nanchahal K, Shipley MJ, Stansfeld SA, Juneja M, Alberti KG. Social inequality in coronary risk: central obesity and the metabolic syndrome. Evidence from the Whitehall II study. Diabetologia. 1997;40:1341–1349. doi: 10.1007/s001250050830. [DOI] [PubMed] [Google Scholar]
- 15.Tabák AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimäki M, Witte DR. Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet. 2009;373:2215–2221. doi: 10.1016/S0140-6736(09)60619-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen HG, Overgaard J, Lajer M, Hubalek F, Højrup P, Pedersen L, Tarnow L, Rossing P, Pociot F, McGuire JN. Finding diabetic nephropathy biomarkers in the plasma peptidome by high-throughput magnetic bead processing and MALDI-TOF-MS analysis. Proteomics Clin Appl. 2010;4:697–705. doi: 10.1002/prca.200900169. [DOI] [PubMed] [Google Scholar]
- 17.Mantini D, Petrucci F, Pieragostino D, Del-Boccio P, Di-Nicola M, Di-Ilio C, Federici G, Sacchetta P, Comani S, Urbani A. LIMPIC: a computational method for the separation of protein MALDI-TOF-MS signals from noise. BMC Bioinformatics. 2007;8:101. doi: 10.1186/1471-2105-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 19.Breiman L, Friedman JH, Olshen R, Stone CJ. Classification and regression trees. Wadsworth; California: 1984. [Google Scholar]
- 20.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 21.Suhre K, Meisinger C, Doring A, Altmaier E, Belcredi P, Gieger C, Chang D, Milburn MV, Gall WE, Weinberger KM, Mewes HW, Hrabe de Angelis M, Wichmann HE, Kronenberg F, Adamski J, Illig T. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One. 2010;5:e13953. doi: 10.1371/journal.pone.0013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huffman KM, Shah SH, Stevens RD, Bain JR, Muehlbauer M, Slentz CA, Tanner CJ, Kuchibhatla M, Houmard JA, Newgard CB, Kraus WE. Relationships between circulating metabolic intermediates and insulin action in overweight to obese, inactive men and women. Diabetes Care. 2009;32:1678–1683. doi: 10.2337/dc08-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis GD, Asnani A, Gerszten RE. Application of metabolomics to cardiovascular biomarker and pathway discovery. J Am Coll Cardiol. 2008;52:117–123. doi: 10.1016/j.jacc.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sundsten T, Ostenson CG, Bergsten P. Serum protein patterns in newly diagnosed type 2 diabetes mellitus–influence of diabetic environment and family history of diabetes. Diabetes Metab Res Rev. 2008;24:148–154. doi: 10.1002/dmrr.789. [DOI] [PubMed] [Google Scholar]
- 25.Fung ET. A recipe for proteomics diagnostic test development: the OVA1 test, from biomarker discovery to FDA clearance. Clin Chem. 2010;56:327–329. doi: 10.1373/clinchem.2009.140855. [DOI] [PubMed] [Google Scholar]
- 26.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O’Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu J, Coombes KR, Morris JS, Baggerly KA. The importance of experimental design in proteomic mass spectrometry experiments: some cautionary tales. Brief Funct Genomic Proteomic. 2005;3:322–331. doi: 10.1093/bfgp/3.4.322. [DOI] [PubMed] [Google Scholar]
- 28.De-Bock M, de-Seny D, Meuwis MA, Chapelle JP, Louis E, Malaise M, Merville MP, Fillet M. Challenges for biomarker discovery in body fluids using SELDI-TOF-MS. J Biomed Biotechnol. 2010:Art no 906082. doi: 10.1155/2010/906082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villar-Garea A, Griese M, Imhof A. Biomarker discovery from body fluids using mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci. 2007;849:105–114. doi: 10.1016/j.jchromb.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 30.Pieragostino D, Petrucci F, Del-Boccio P, Mantini D, Lugaresi A, Tiberio S, Onofrj M, Gambi D, Sacchetta P, Di-Ilio C, Federici G, Urbani A. Pre-analytical factors in clinical proteomics investigations: impact of ex vivo protein modifications for multiple sclerosis biomarker discovery. J Proteomics. 2010;73:579–592. doi: 10.1016/j.jprot.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 31.Szajli E, Fehér T, Medzihradszky KF. Investigating the quantitative nature of MALDI-TOF MS. Mol Cell Proteomics. 2008;7:2410–2418. doi: 10.1074/mcp.M800108-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Ekblad L, Baldetorp B, Fernö M, Olsson H, Bratt C. In-source decay causes artifacts in SELDI-TOF MS spectra. J Proteome Res. 2007;6:1609–1614. doi: 10.1021/pr060633y. [DOI] [PubMed] [Google Scholar]
- 33.Parker CE, Pearson TW, Anderson NL, Borchers CH. Mass-spectrometry-based clinical proteomics–a review and prospective. Analyst. 2010;135:1830–1838. doi: 10.1039/c0an00105h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez-Recio O, Forni S. Genome-wide prediction of discrete traits using bayesian regressions and machine learning. Genet Select Evol. 2011;43:7. doi: 10.1186/1297-9686-43-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu B, Abbott T, Fishman D, McMurray W, Mor G, Stone K, Ward D, Williams K, Zhao H. Comparison of statistical methods for classification of ovarian cancer using mass spectrometry data. Bioinformatics. 2003;19:1636–1643. doi: 10.1093/bioinformatics/btg210. [DOI] [PubMed] [Google Scholar]
- 36.Ziegler A, DeStefano AL, Konig IR. Data mining, neural nets, trees—problems 2 and 3 of genetic analysis workshop 15. Genet Epidemiol. 2007;31:S51–S60. doi: 10.1002/gepi.20280. [DOI] [PubMed] [Google Scholar]
- 37.Díaz-Uriarte R, Alvarez de Andrés Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan Y, Murphy TB, Byrne JC, Brennan L, Fitzpatrick JM, Watson RW. Applying random forests to identify biomarker panels in serum 2D-DIGE data for the detection and staging of prostate cancer. J Proteome Res. 2011;10:1361–1373. doi: 10.1021/pr1011069. [DOI] [PubMed] [Google Scholar]
- 39.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leighton B, Foot EA. The role of the sensory peptide calcitonin-gene-related peptide(s) in skeletal muscle carbohydrate metabolism: effects of capsaicin and resiniferatoxin. Biochem J. 1995;307:707–712. doi: 10.1042/bj3070707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muff R, Born W, Fischer JA. Calcitonin, calcitonin gene-related peptide, adrenomedullin and amylin: homologous peptides, separate receptors and overlapping biological actions. Eur J Endocrinol. 1995;133:17–20. doi: 10.1530/eje.0.1330017. [DOI] [PubMed] [Google Scholar]
- 42.Slezak LA, Andersen DK. Pancreatic resection: effects on glucose metabolism. World J Surg. 2001;25:452–460. doi: 10.1007/s002680020337. [DOI] [PubMed] [Google Scholar]
- 43.Breland UM, Michelsen AE, Skjelland M, Folkersen L, Krohg-Sørensen K, Russell D, Ueland T, Yndestad A, Paulsson-Berne G, Damås JK, Oie E, Hansson GK, Halvorsen B, Aukrust P. Raised MCP-4 levels in symptomatic carotid atherosclerosis: an inflammatory link between platelet and monocyte activation. Cardiovasc Res. 2010;86:265–273. doi: 10.1093/cvr/cvq044. [DOI] [PubMed] [Google Scholar]
- 44.Béliard S, Nogueira JP, Maraninchi M, Lairon D, Nicolay A, Giral P, Portugal H, Vialettes B, Valéro R. Parallel increase of plasma apoproteins C-II and C-III in Type 2 diabetic patients. Diabet Med. 2009;26:736–739. doi: 10.1111/j.1464-5491.2009.02757.x. [DOI] [PubMed] [Google Scholar]
- 45.Sørensen LB, Flint A, Raben A, Hartmann B, Holst JJ, Astrup A. No effect of physiological concentrations of glucagon-like peptide-2 on appetite and energy intake in normal weight subjects. Int J Obes Relat Metab Disord. 2003;27:450–456. doi: 10.1038/sj.ijo.0802247. [DOI] [PubMed] [Google Scholar]
- 46.Meier JJ, Nauck MA, Pott A, Heinze K, Goetze O, Bulut K, Schmidt WE, Gallwitz B, Holst JJ. Glucagon-like peptide 2 stimulates glucagon secretion, enhances lipid absorption, and inhibits gastric acid secretion in humans. Gastroenterology. 2006;130:44–54. doi: 10.1053/j.gastro.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 47.Yu R, Kim CS, Kawada T, Kwon TW, Lim TH, Kim YW, Kwon BS. Involvement of leukotactin-1, a novel CC chemokine, in human atherosclerosis. Atherosclerosis. 2004;174:35–42. doi: 10.1016/j.atherosclerosis.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 48.Reape TJ, Groot PH. Chemokines and atherosclerosis. Atherosclerosis. 1999;147:213–225. doi: 10.1016/s0021-9150(99)00346-9. [DOI] [PubMed] [Google Scholar]
- 49.Onat A, Hergenç G, Ayhan E, Uğur M, Kaya H, Tuncer M, Can G. Serum apolipoprotein C-III in high-density lipoprotein: a key diabetogenic risk factor in Turks. Diabet Med. 2009;26:981–988. doi: 10.1111/j.1464-5491.2009.02814.x. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z, Yip C, Ying Y, Wang J, Meng XY, Lomas L, Yip TT, Fung ET. Mass spectrometric analysis of protein markers for ovarian cancer. Clin Chem. 2004;50:1939–1942. doi: 10.1373/clinchem.2004.036871. [DOI] [PubMed] [Google Scholar]


