Abstract
Spinocerebellar ataxia 13 (SCA13) is an autosomal dominant disease resulting from mutations in KCNC3 (Kv3.3), a voltage-gated potassium channel. The KCNC3R420H mutation was first identified as causative for SCA13 in a four-generation Filipino kindred with over 20 affected individuals. Electrophysiological analyses in oocytes previously showed that this mutation did not lead to a functional channel and displayed a dominant negative phenotype. In an effort to identify the molecular basis of this allelic form of SCA13, we first determined that human KCNC3WT and KCNC3R420H display disparate posttranslational modifications, and the mutant protein has reduced complex glycan adducts. Immunohistochemical analyses demonstrated that KCNC3R420H was not properly trafficking to the plasma membrane and surface biotinylation demonstrated that KCNC3R420H exhibited only 24% as much surface expression as KCNC3WT. KCNC3R420H trafficked through the ER but was retained in the Golgi. KCNC3R420H expression results in altered Golgi and cellular morphology. Electron microscopy of KCNC3R420H localization further supports retention in the Golgi. These results are specific to the KCNC3R420H allele and provide new insight into the molecular basis of disease manifestation in SCA13.
Keywords: Spinocerebellar ataxia, dominant inheritance, SCA13, KCNC3, voltage-gated potassium channel, Golgi, protein trafficking
Introduction
The spinocerebellar ataxias (SCAs), a heterogeneous group of neurologic diseases bound by the shared phenotypes of ataxia and motor incoordination, are characterized both by their mode of inheritance and, when known, the particular causative mutation. We previously demonstrated that two different point mutations in the Shaw subtype KCNC3 (Kv3.3) voltage-gated potassium channel located at 19q13.33 segregate within a four generation Filipino kindred [KCNC3R420H (c.1259G>A)] and a three generation French pedigree [KCNC3F448L (c.1344C>A)] causing SCA13 (Subramony et al., 2013; Waters et al., 2006). The KCNC/Kv3 family contains four related genes (KCNC1/Kv3.1, KCNC2/Kv3.2, KCNC3/Kv3.3, and KCNC4/Kv3.4), that can assemble into both homotetrameric and heterotetrameric channels (Chang et al., 2007; Rudy and McBain, 2001). This family of channels has large voltage-dependent K+ currents evident upon membrane depolarization at −20mV, values more positive than any other known mammalian voltage-gated K+ channels (Rudy and McBain, 2001).
KCNC3R420H causes a late-onset neurodegenerative phenotype similar to other SCA subtypes, while patients with KCNC3F448L present with early-onset ataxia accompanied by delayed motor milestones, cognitive delay, and epilepsy. Analyses of the electrophysiological profile for KCNC3R420H in a Xenopus oocyte heterologous system was consistent with a non-functional channel, whereas the KCNC3F448L mutation altered the gating of KCNC3 thus prolonging the open state (Waters et al., 2006) with these biophysical profiles for these mutants duplicated in the zebra fish KCNC3 genes (Mock et al., 2010). Autosomal dominant inheritance in the Filipino family is characterized by cerebellar signs including gait and limb ataxia, brisk tendon reflexes and mild cognitive impairment (Subramony et al., 2013), as well as abnormal sound localization cueing (Middlebrooks et al., 2013). The most pronounced gross pathological feature in both the Filipino and French kindred was severe, isolated cerebellar atrophy revealed by magnetic resonance (MR) imaging (Subramony et al., 2013; Waters et al., 2006). This pathological hallmark in SCA13, as well as other SCAs is in sharp contrast to the complete lack of alterations in brain anatomy and motor function in any single mouse gene ablation models (Espinosa et al., 2001; McMahon et al., 2004; Zagha et al., 2010) or studies in the zebrafish (Issa et al., 2011; Mock et al., 2010).
We therefore sought to understand the underlying mechanisms that might explain the dominant inheritance of the KCNC3R420H allele in SCA13 patients, given that this channel mutation is electrophysiologically non-functional (Mock et al., 2010; Waters et al., 2006). Our studies reveal that KCNC3R420H displays altered post-translational modifications and aberrant intracellular trafficking with associated changes in Golgi and cellular morphology and dominant phenotypes in the Drosophila wing and eye.
Materials and Methods
Cell Culture, Transient and Stable Transfection
COS-1 cells (ATCC, CRL 1650) were cultured in Dulbecco's Modified Eagle's Medium (DMEM, Omega) with 10% FBS (Omega), 4 mM glutamine, and antibiotic/antimycotic (ABAM) (Gibco) at 37°C in humidified air with 5% CO2. Human neuroblastoma cells, SH-SY5Y (ATCC, CRL 2266) were cultured similarly in 1:1 Eagle's Minimum Essential Medium: F12 Medium (ATCC, 30-2003). The human KCNC3WT cDNA was kindly provided by Dr. James L. Rae (Mayo Foundation) (Rae and Shepard, 2000) and subcloned into pcDNA1 (Invitrogen). KCNC3WT and individual mutants (KCNC3R420H or KCNC3F448L) were generated by Quikchange Mutagenesis (Stratagene) and used for transient transfection in COS-1 cells. For co-transfection, equimolar amounts (1:1) of each plasmid were used. cDNAs for KCNC3WT, KCNC3R420H or KCNC3F448L were subcloned into a pEF-IRES-puro vector for stable selection in SH-SY5Y cells using puromycin and individual colonies selected for clonal expression. Transfections were performed using Lipofectamine LTX (Invitrogen) as per manufacturer's instructions.
Immunoblotting
Total cellular protein (30 μg, Bicinchoninic Acid (BCA) assay (Pierce)) was separated by SDS-PAGE (4-12% gradient, Invitrogen) and electrotransferred to nitrocellulose membrane (Millipore). The membranes were then blocked for 1 h with 5% nonfat dry milk in TBST [10 mM Tris-HCl (pH 7.5), 200 mM NaCl, 0.1% (v/v) Tween 20] at room temperature. The membranes were incubated overnight at 4°C with primary antibodies to KCNC3 (Alomone, APC-102, 1:200) or β1 integrin (Chemicon 1952-20, 1:500) washed three times with TBST, incubated with a peroxidase-conjugated secondary antibody for 1 h, washed again three times, and subjected to ECL (Perkin Elmer). To address glycosylation from total cellular protein isolated from SH-SY5Y cells stably expressing KCNC3WT and KCNC3R420H, protein isolates were treated with either PNGase F or Endo H per manufacturer's instructions (New England Biolabs) followed by immunoblot analysis.
Immunohistochemistry
For immunofluorescence microscopy, transiently transfected COS-1 cells were permeabilized with Dulbecco's PBS containing 0.1% Triton X-100 for 20 min and blocked with 1% bovine serum albumin and 5% goat serum (Sigma) for 1 h. PBS washed cells were incubated with antibodies to KCNC3 (Alomone, APC-102, 1:500), cadherin (Abcam, ab6528, 1:500), calnexin (Abcam, ab2798, 1:200), GM130 (BD Bioscience, 1:200), 58K/FTCD (Abcam, ab27043,1:50), β-tubulin (Sigma, T 4026, 1:200) followed by corresponding secondary antibodies conjugated with Alexa Fluor 488 or with Alexa Fluor 568 fluorescent dyes (Molecular Probes). Antigen retrieval was performed in 10 mM sodium citrate and control reactions were devoid of primary antibodies or stained with blocking antibodies. Samples were imaged with a Leica TCS/SP spectral confocal scanner (Leica Microsystems, Mannheim, Germany) in dual emission mode to distinguish autofluorescence from specific staining.
Membrane Protein Surface Biotinylation
To study surface localization of membrane proteins, transiently transfected COS-1 cells were subjected to surface biotinylation as per manufacturer's protocols (Cell Surface Protein Isolation Kit, Pierce). Isolated protein was quantified by BCA assay and then subjected to immunoblot analysis for KCNC3 and β1 integrin as described above. Densitometry on the resulting immunoblots was performed using ImageJ software (v1.48) to determine the proportion of unmodified to glycosylated proteins for both mutant and WT KCNC3.
Acceptor Photobleaching Fluorescence Resonance Energy Transfer (apFRET)
COS-1 cells were grown on 1 cm coverslips and fixed and mounted for processing. Cells transiently transfected with KCNC3WT or KCNC3R420H were labeled with fluorescently tagged antibodies to KCNC3 (rhodamine, acceptor fluorophore) and GM130 or 58K/FTCD (FITC, donor fluorophore). After 6 h, cells were rinsed in PBS and covered with a thin layer of 50% PEG solution (Sigma) with gentle rocking for 90 s. The PEG solution was removed by repeated rinses with media and the cells were allowed to recover for 4 h. Pre-bleach images were obtained by laser excitation at 488nm (FITC) or 543 nm (rhodamine). Acceptor bleaching (acceptor) was performed within a region of interest by repeated pulses at 543 nm. A post-bleach FITC (donor) image excited with 543 nm is also detected. apFRET efficiency was calculated by pixel density as the difference between maximal and absorbed output of the donor (post-bleach minus pre-bleach) divided by the maximal (post-bleach) output of the donor. The data analysis resulted from n=9 for R420H, n=8 for WT and n=6 F448L.
Electron Microscopy
COS-1 cells transiently transfected with KCNC3WT and KCNC3R420H were trypsinized, pelleted, fixed in 1% osmium tetroxide in H2O for 1 h, and then dehydrated with subsequent immersion in 30-100% acetone and embedded in Eponate (Ted Pella). After overnight polymerization at 56°C, samples were stained with methylene blue, counterstained with 5% uranyl acetate in methanol and in Reynold’s lead citrate for 10 min each and viewed with a Jeol 100CX transmission electron microscope at 80kV. Digital images were collected with a XR40 Digital Camera (Advance Microscopy Techniques).
Generation of transgenic flies and Drosophila genetics
The cDNAs encoding KCNC3WT and KCNC3R420H were first subjected to Quikchange Mutagenesis (Stratagene) to optimize the Kozak sequence for appropriate translation in flies. The 3.2 kb cDNAs were sub-cloned into the fly expression vector pUAST (Brand and Perrimon, 1993) via EcoRI (5’) and NotI (3’) ligation. The resulting constructs were sequenced to verify the integrity of the DNAs and injected into yw embryos by standard procedures (Rainbow Transgenics, Inc.). The driver strains da-Gal4 (ubiquitous) and gmr-Gal4 (all eye cells), and the reporter UAS-LacZ were obtained from the Bloomington Drosophila Stock Center. Fly stocks were maintained on standard Drosophila medium at 25°C. For experiments, homozygous females for the Gal4 strains were crossed with UAS males to generate progeny expressing KCNC3 in the desired tissue. All crosses were initially placed at 25°C for two days and the progeny was raised at 28°C to maximize expression.
Drosophila imaging
For imaging KCNC3 eyes, targeted eye expression was achieved by crossing gmr-Gal4 females with KCNC3WT, KCNC3R420H and LacZ flies. Freshly frozen eyes from 1-2 day-old females were imaged as Z-stacks with a Leica Z16 APO using a 2× Plan-Apo objective and single in-focus images produced with the Montage Multifocus module of the Leica Application Software. For scanning electron microscopy (SEM), flies were serially dehydrated in ethanol, air dried in hexamethyldisilazane (Electron Microscope Sciences), and metal coated for observation in a Hitachi S4000 SEM. The retinal sections were generated by fixing the fly heads in 3% glutaraldehyde, washed in phosphate buffer, post-fixed in 1% OsO4, dehydrated in ethanol and propylene oxide, embed in Spurr, and subsequently mounted. Blocks were then cut at 1 μm, stained with toluidine-blue (Fernandez-Funez et al., 2000) and imaged with a plan-apo 63× NA: 1.4 (oil) objective using AxioVision (Zeiss) in an Axio-Observer Z1 microscope (Zeiss). Wings from da-Gal4 females crossed with KCNC3WT, KCNC3R420H and LacZ flies were collected and kept for two days in a glycerol/ethanol mix. Wings were mounted on slides in the glycerol/ethanol mix, and imaged with a plan-neo 5× NA: 0.16 and a plan-apo 20× NA: 0.7 objectives using AxioVision as above.
Drosophila Immunoblot Analysis
For immunoblot analysis of fly protein, da-Gal4 females were crossed with KCNC3WT, KCNC3R420H and LacZ flies. One fly per genotype of 1-2 day-old females was used for this analysis. Single flies were homogenized in 30 μl of RIPA buffer containing complete protease inhibitors (Roche) and centrifuged for 1 min at 1,000 rpm. Then, 25 μL of supernatant was mixed with loading buffer and resolved by SDS-PAGE, transferred to nitrocellulose membrane, immunoblotted, and the signal was revealed by chemiluminescence (Pierce). Anti-KCNC3 (1:4000, Alomone labs) was used to detect KCNC3 with α-tubulin (1:250k, Sigma) as a loading control.
Statistical Analysis
Quantitative results are given as means ± SEM. Statistical significance was determined using Student’s t-test. Values of p=0.05 were considered significant.
Results
KCNC3R420H displays altered levels of glycosylation and cellular localization
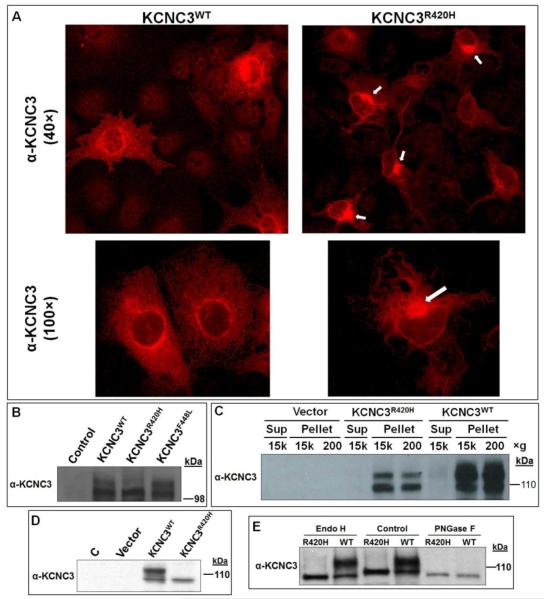
Given the lack of an animal model that displays symptoms analogous to the ataxic phenotype observed in SCA13 patients, specifically the later onset, slowly progressive form observed in the Filipino kindred, we assessed both biochemical and cell biological parameters for KCNC3R420H that might explain the fully penetrant autosomal dominant inheritance. We utilized transient transfection of the human KCNC3 constructs in COS-1 cells because of their highly efficient transfection rate. Immunohistochemistry by confocal microscopy of COS-1 cells transiently transfected with KCNC3WT or KCNC3R420H reproducibly demonstrated an altered pattern of intracellular localization for KCNC3R420H (Fig. 1A). Expression of KCNC3WT demonstrated classical membrane protein localization with punctate staining of the plasma membrane and distinct perinuclear localization, presumably due to ER and Golgi trafficking of newly synthesized protein. In contrast, KCNC3R420H showed intense staining in a single perinuclear area that extended out into the cytoplasm with limited staining associated with plasma membrane localization (Fig. 1A).
Figure 1. Localization and differential protein modification of KCNC3WT, KCNC3F448L, and KCNC3R420H in COS-1 and SH-SY5Y cells.
(A) OS-1 cells transiently transfected with either KCNC3WT or KCNC3R420H were analyzed by confocal fluorescence microscopy using a KCNC3 antibody. KCNC3WT shows perinuclear distribution and punctate staining of plasma membrane whereas KCNC3R420H displays primarily perinuclear staining (white arrows) and very limited fluorescence associated with the plasma membrane. Top and bottom panels were obtained at 40× and 100× magnification, respectively. (B) Immunoblot analysis of KCNC3 in COS-1 cells transiently transfected with human KCNC3WT, KCNC3F448L, or KCNC3R420H expression vectors illustrates a significant reduction of a slower migrating (upper) band consistent with altered posttranslational modifications. (C) Immunoblot analysis of total protein from COS-1 cells transiently transfected with KCNC3WT or KCNC3R420H subjected to differential centrifugation verifies that there was no fractional loss of the KCNC3R420H protein within our isolation protocol and illustrates the diminished upper band. (D) Immunoblot analysis of KCNC3 in stably transfected human neuroblastoma SH-SY5Y cells illustrating the complete absence of the upper band. (E) Immunoblot analysis of KCNC3 in stably transfected SH-SY5Y cells was used to evaluate the treatment of total protein with Endo H or PNGase F. The upper band was resistant to Endo H whereas this species disappeared when total cell extracts were subjected to PNGase F treatment, consistent with the removal of a complex glycan. Both enzymes however led to cleavage of the lower band (control lanes) generating a faster migrating band presumably due to release of a secondary high mannose modification as further illustrated in Figure S1. The removal of a high mannose moiety is consistent with the known activity of Endo H.
Protein glycosylation and proper protein folding are associated with the transit of membrane proteins through the endoplasmic reticulum (ER) and the Golgi apparatus. The level of expression for KCNC3WT, KCNC3R420H, and KCNC3R448L were analyzed by immunoblot analysis (Fig. 1B). KCNC3WT and KCNC3R448L displayed comparable banding patterns, one at the expected size as well as a slower migrating band. KCNC3R420H, however, showed a diminution of the upper band potentially associated with altered posttranslational modification. KCNC3F448L was utilized here and in subsequent experiments since biophysical experiments had implied its normal trafficking to the plasma membrane and as an assurance that observations with KCNC3R420H were specific to this mutation. As an integral membrane protein, differential centrifugation on the protein isolates was employed to verify that there was no fractional loss of the KCNC3R420H protein within our isolation protocol (Fig. 1C). As in Figure 1B, the KCNC3R420H protein showed a markedly reduced level of the presumptively post-translationally modified protein. Based on densitometry, the proportion of upper band is ~50% less for the mutant protein. A dramatic reduction in the upper band for the mutant protein is consistent with observations using different vectors and cell lines (Figs. 1B-D and S1). These representative immunoblots also demonstrate that COS-1 cells do not express endogenous KCNC3 protein.
To verify altered levels of posttranslational modification of KCNC3R420H in a human neuronal cell line, we generated SH-SY5Y stably transfected cells with the empty vector and constructs expressing KCNC3WT and KCNC3R420H proteins. As in the COS-1 cells, SH-SY5Y cells did not express detectable levels of endogenous KCNC3 protein, whereas the cells stably transfected with KCNC3WT showed a similar banding pattern to that observed in COS-1 cells (Fig. 1D). The stable expression of KCNC3R420H displayed only the lower band, with no detectable upper band. The slower migrating bands detected by the KCNC3 antibody may be the result of various posttranslational modifications. Cartwright et al. (Cartwright et al., 2007) previously showed that Kv3.1-3.4 channels have two highly conserved N-glycosylation sites that are likely modified in rat brain by N-linked oligosaccharides, one of which is a complex glycan. We therefore utilized endoglycosidase H (Endo H), which cleaves high mannose and some hybrid oligosaccharides of N-linked glycoproteins, and peptide-N-glycosidase F (PNGase F), which cleaves between the innermost GlcNAc and asparagine residues of high mannose, hybrid, and complex oligosaccharides to investigate the glycosylation pattern. Treatment of cellular extracts from the stably expressing SH-SY5Y cells with Endo H did not affect the upper band but resulted in a faster migrating lower band of both KCNC3WT and KCNC3R420H (Fig. 1E). This small change in migration has been observed in multiple experiments (Fig. S1). In contrast, digestion of cellular extracts with PNGase F resulted in the complete disappearance of the slower migrating upper band and an identical drop in the lower band (Figs. 1E and S1). These results are consistent with modification of KCNC3WT with both complex oligosaccharides and high mannose, whereas KCNC3R420H is only modified by high mannose given that both Endo H and PNGase F create a slightly faster migrating lower band and the complete absence of the upper band in human neuroblastoma cells.
KCNC3R420H channel displays limited trafficking to the plasma membrane
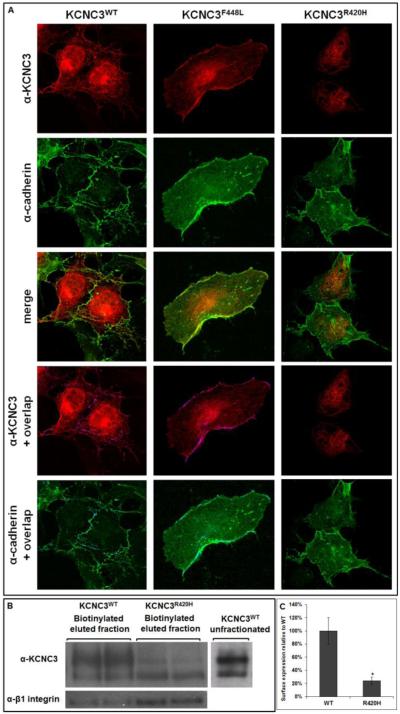
To visualize the cellular distribution of the KCNC3WT, KCNC3F448L, and KCNC3R420H channel proteins, we investigated the presence of KCNC3 relative to the plasma membrane protein cadherin. Cadherin co-staining with either KCNC3WT or KCNC3F448L showed overlapping surface targeting based on the merged image. The overlapping yellow pixels from the merged image were also visualized as a purple overlay on the KCNC3 image or blue on cadherin, emphasizing the co-registered fluorescence between KCNC3 and cadherin. KCNC3R420H, in contrast, did not appear to overlap with cadherin at the plasma membrane, again demonstrating that the majority of the mutant protein is intracellular (Fig. 2A).
Figure 2. KCNC3R420H exhibits limited cell surface expression.
(A) COS-1 cells transiently transfected with KCNC3WT, KCNC3F448L, or KCNC3R420H were analyzed by confocal fluorescence microscopy using antibodies to KCNC3 and cadherin as a plasma membrane marker. The top two rows illustrate the images for each antibody in cells expressing KCNC3WT, KCNC3F448L, or KCNC3R420H. Merged images are provided in the middle row. The bottom two rows illustrate either KCNC3 or cadherin along with overlapping fluorescence illustrated as purple or blue, respectively. KCNC3WT or KCNC3F448L showed co-registration with cadherin labeling at the plasma membrane as expected for normal trafficking of an integral membrane protein. (B) COS-1 cells transiently transfected with either KCNC3WT or KCNC3R420H were subjected to surface biotinylation and analyzed by immunoblot to either KCNC3 or β1-integrin as a positive control for surface expression. (C) Corresponding densitometry demonstrates a 76% reduction in surface expression relative to KCNC3WT. Data are presented as mean ± SEM (n=4) and * denotes p = 0.05. The lack of any obvious overlap with cadherin staining for KCNC3R420H is consistent with surface biotinylation results demonstrating limited plasma membrane trafficking for this mutant.
To further address the impact of altered glycosylation and aberrant trafficking of the mutant channel, surface biotinylation studies were performed. These experiments revealed that the relative level of KCNC3R420H transported to and inserted into the plasma membrane was substantially reduced as compared to KCNC3WT and KCNC3F448L (Fig. 2B). Densitometry of biotinylated protein normalized to KCNC3WT, with the plasma membrane protein β1-integrin as a loading control, demonstrated 76% less surface expression of KCNC3R420H (Fig. 2C).
KCNC3R420H channel is retained in the Golgi based on immunohistochemistry
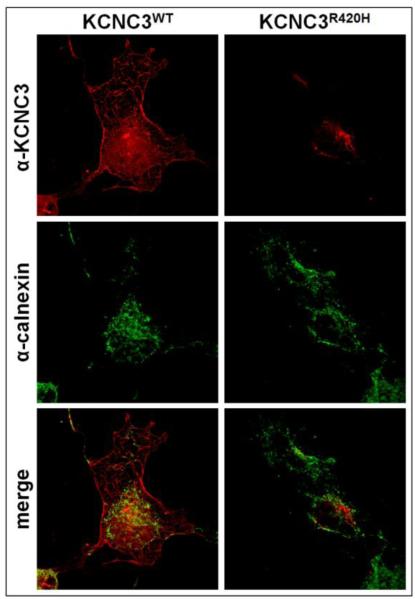
Integral membrane proteins are normally synthesized and trafficked through the ER and Golgi. We sought to determine which stage of this process might explain the intracellular retention of KCNC3R420H. We first compared the transient expression of KCNC3 to the integral ER membrane chaperone calnexin, which plays a central role in recognizing glycosylated proteins and aiding in protein folding (Moremen et al., 2012). As expected, KCNC3WT exhibits overlap with calnexin in the ER, while the majority of KCNC3WT is detected at the plasma membrane (Fig. 3). In contrast, KCNC3R420H does not co-register with calnexin in the ER, supporting its aberrant trafficking (Fig. 3).
Figure 3. KCNC3R420H does not co-register with a marker for endoplasmic reticulum (ER).
COS-1 cells transiently transfected with KCNC3WT or KCNC3R420H were analyzed by confocal fluorescence microscopy using antibodies to KCNC3 and calnexin as a marker for ER. KCNC3WT co-registered with calnexin, whereas KCNC3R420H showed no overlap with calnexin.
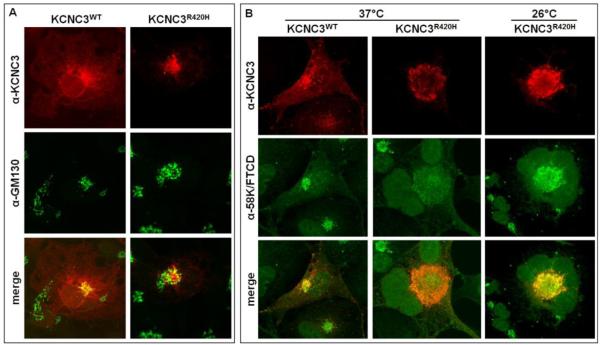
We next investigated KCNC3R420H trafficking by studying its localization in the Golgi apparatus. Transiently expressed KCNC3WT overlaps with the GM130 (Golgi matrix protein of 130 kDa), a protein associated with the cis-Golgi (Chang et al., 2012) (Fig. 4A). Immunohistochemical analysis of KCNC3R420H in comparison to GM130 indicates a partial overlap with the Golgi apparatus (Fig. 4A). To further delineate the Golgi-specific localization of KCNC3R420H, we also compared localization of KCNC3 and 58K Golgi protein/formiminotransferase cyclodeaminase (FTCD), an enzyme that couples histidine degradation to folate metabolism and also participates in the interaction of the Golgi complex with the vimentin intermediate filament (IF) cytoskeleton (Gao and Sztul, 2001; Hennig et al., 1998; Mao et al., 2004). As with GM130, KCNC3WT overlapped with the 58K/FTCD Golgi marker, with the majority of the protein localized to the plasma membrane at 37°C (Fig. 4B). Results on the localization of KCNC3R420H demonstrate a striking overlap with the 58K/FTCD Golgi protein with limited to no expression elsewhere at 37°C (Fig 4B). Previous studies on the cystic fibrosis transmembrane conductance regulator (CFTR) demonstrated that a mutant form of the protein (ΔF508) exhibiting aberrant trafficking was rescued with proper trafficking to the plasma membrane by growth at 26°C (Wilke et al., 2012). We therefore replicated the experiment with KCNC3R420H at 26°C, however, unlike the results reported for CFTR, the aberrant trafficking of the KCNC3 mutant was not rescued by this growth condition (Fig. 4B).
Figure 4. KCNC3R420H localization overlaps with Golgi markers.
(A) COS-1 cells transiently transfected with KCNC3WT or KCNC3R420H were analyzed by confocal fluorescence microscopy using antibodies to KCNC3 and GM130 (Golgi matrix protein of 130 kDa), a marker of the cis-Golgi. KCNC3WT displayed strong co-registration with the GM130 whereas KCNC3R420H labeling showed a partial overlap. (B) COS-1 cells transiently transfected with KCNC3WT or KCNC3R420H were grown at 37°C and analyzed by confocal fluorescence microscopy using antibodies to KCNC3 and the 58K Golgi protein/formiminotransferase cyclodeaminase (58K/FTCD) (first two columns). KCNC3WT co-registered with 58K/FTCD also indicative of steady-state trafficking through the Golgi whereas all KCNC3R420H staining completely overlapped with 58K/FTCD demonstrating almost total retention in the Golgi. KCNC3R420H was also analyzed by growth of transiently transfected COS-1 cells at 26°C to address the effects on rescue of plasma membrane trafficking (third column), with no effect.
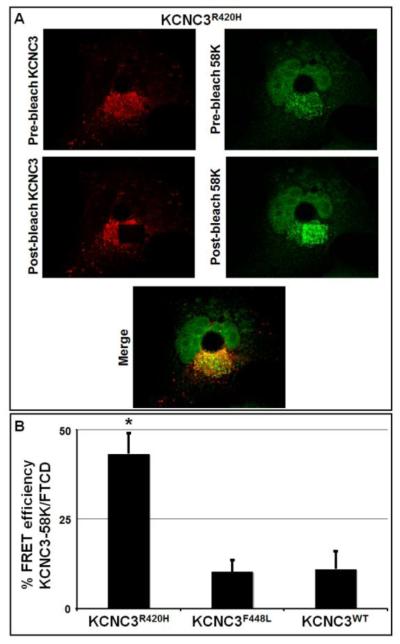
Fluorescence resonant energy transfer (FRET) demonstrates Golgi localization of KCNC3R420H
To extend the immunohistochemical results, FRET was used to demonstrate protein co-localization (<7 nm). Given the fluorescent co-registration of KCNC3R420H with Golgi-specific proteins with KCNC3R420H, acceptor photobleaching fluorescence resonance energy transfer (apFRET) (Day and Davidson, 2012; Day and Schaufele, 2005) was utilized to demonstrate co-localization of KCNC3 with specific regions of the Golgi apparatus. Rhodamine-tagged KCNC3 and FITC-tagged GM130 were used for initial apFRET analysis. Interestingly, both KCNC3WT and KCNC3R420H showed comparable apFRET efficiency, implying that KCNC3R420H was not retained in the cis-Golgi (data not shown). However, a negative FRET result could be due to the positioning of the respective fluorophores and is thus not conclusive. apFRET was further utilized with the FITC-tagged Golgi marker 58K/FTCD in comparison to KCNC3. A representative set of images used for apFRET analysis is provided (Fig. 5A). The apFRET measurements using KCNC3F448L and KCNC3WT in conjunction with 58K/FTCD demonstrated minimal FRET efficiency, exemplifying the steady-state levels of these proteins in the Golgi due to normal trafficking (Fig. 5B). In contrast, KCNC3R420H displays a 4-fold greater apFRET efficiency than KCNC3WT or KCNC3F448L confirming co-localization with 58K/FTCD and retention of KCNC3R420H in the Golgi (Fig. 5B).
Figure 5. KCNC3R420H co-localizes with the Golgi marker 58K/FTCD by acceptor photobleaching FRET (apFRET).
COS-1 cells were transiently transfected with KCNC3WT, KCNC3F448L, or KCNC3R420H. KCNC3 protein was visualized using an antibody conjugated with rhodamine as the acceptor fluorophore, while the 58K/FTCD protein was visualized similarly with FITC. Cells were then analyzed by apFRET. (A) Representative pre-bleach and post-bleach images for KCNC3R420H transfected cells are provided. Images were obtained by laser excitation at 488 nm (FITC) or 543 nm (rhodamine). Acceptor bleaching was performed within a region of interest by repeated pulses at 543 nm. (B) Quantitative results are presented as % FRET efficiency as described in the Materials and Methods. Data are presented as mean ± SEM (KCNC3R420H, n=9; KCNC3WT, n=8; KCNC3F448L, n=6) and * denotes a p = 0.05 of KCNC3R420H level as compared to KCNC3WT.
KCNC3R420H causes subcellular perturbations and alterations in cellular morphology
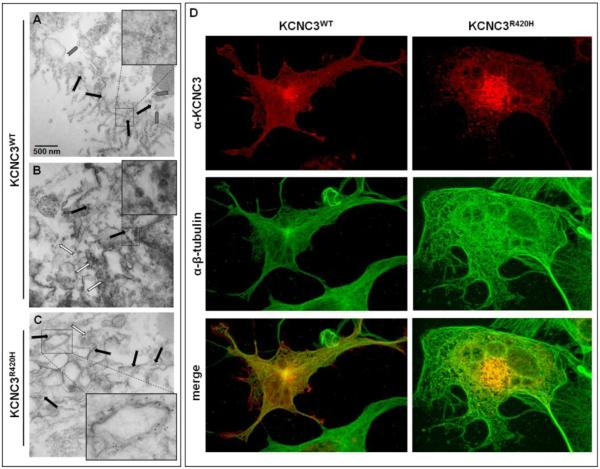
Having demonstrated the retention of KCNC3R420H in the Golgi by various methods, we next sought to further visualize the intracellular localization of this mutant using immunogold transmission electron microscopy (TEM). A high concentration of gold particles (black arrows) is apparent at the phospholipid bilayer and nearby vesicles (grey arrows) in cells expressing KCNC3WT (Fig. 6A), with minimal intracellular expression or association with clathrin-ordered protein (COP) vesicles (white arrows) (Fig. 6B). These results are consistent with the normal trafficking of a plasma membrane protein. Cells expressing KCNC3R420H display a high concentration of intracellular gold particles associated with a swollen and dysmorphic Golgi structure along with a reduction in the number of COP vesicles (Fig. 6C). Given the observed alterations in Golgi structure, we investigated potential perturbations in gross cytoskeletal structure by β-tubulin immunohistochemistry in cells expressing KCNC3. Cells expressing KCNC3R420H displayed altered cellular morphology relative to cells expressing KCNC3WT (Fig. 6D), consistent with many of the other representative images of KCNC3R420H-expressing cells (Figs. 1, 2, and 4).
Figure 6. KCNC3R420H causes alterations in the Golgi apparatus and cellular morphology.
COS-1 cells were transiently transfected with KCNC3WT or KCNC3R420H and visualized by immunogold staining of KCNC3 followed by TEM. (A) Black arrows indicate immunogold-stained KCNC3WT near the cell surface with gray arrows denoting KCNC3WT potentially in secretory vesicles. The inset provides an enlarged area containing gold particles. (B) White arrows denote COP (clathrin-ordered protein) vesicles near the Golgi apparatus in the KCNC3WT-expressing cells. Black arrows show the limited number of intracellular immunogold-stained KCNC3WT consistent with steady-state trafficking. The inset provides an enlarged area highlighting the COP vesicles. (C) White arrows denote COP vesicles whereas black arrows indicate immunogold-stained KCNC3R420H. KCNC3R420H concentrates on intracellular membranes associated with what appears to be a dysmorphic Golgi apparatus. The inset provides an example of a swollen Golgi cisternae containing a high level of KCNC3R420H associated with the membrane. (D) COS-1 cells expressing KCNC3WT or KCNC3R420H were examined by confocal fluorescence imaging with antibodies to KCNC3 and β-tubulin. KCNC3WT-expressing cells display an ordered tubulin network while KCNC3R420H cells show altered morphology.
Effect of co-transfection of KCNC3WT and KCNC3R420H
It has been previously reported that co-expression of KCNC3WT and KCNC3R420H results in channels displaying a ~90% reduction in normalized peak current amplitudes at +60 mV (Minassian et al., 2012; Waters et al., 2006). However, these studies did not address channel formation or membrane localization. To address co-expression analogous to previous studies, we repeated our immunohistochemical studies using transient transfection at a 1:1 ratio of KCNC3WT:KCNC3R420H. Combined expression of these proteins resulted in little to no surface expression of KCNC3 when compared to staining for the surface protein cadherin (Fig. 7). Having demonstrated the retention of KCNC3R420H alone in the Golgi (Figs. 4, 5, and 6A-C), we also sought to determine if the KCNC3R420H protein could cause retention of KCNC3WT in the Golgi. As shown in Figure 7, the majority of the KCNC3 in the co-transfected cells co-registers with the Golgi marker 58K/FTCD by immunohistochemical analysis, implying that either the mutant protein sequesters KCNC3WT through direct interaction or KCNC3R420H disrupts a trafficking pathway specific to KCNC3. This latter argument is based on the normal trafficking of cadherin (Figs. 2 and 7, top) and β-integrin (Fig. 2B) in spite of KCNC3R420H expression. This experiment is limited in that the two proteins cannot be identified separately, given only a single amino acid difference. However as with other co-transfection experiments, such as in the generation of iPS cells or cell lines for the production of virus, the co-expression from multiple plasmids in the same cell is well demonstrated. In addition, the images (Fig. 7) also illustrate an altered cellular morphology in cells expressing a SCA13 patient-relevant 1:1 ratio of mutant to KCNC3WT protein.
Figure 7. KCNC3WT and KCNC3R420H co-expression leads to Golgi retention and limited cell-surface expression for both proteins.
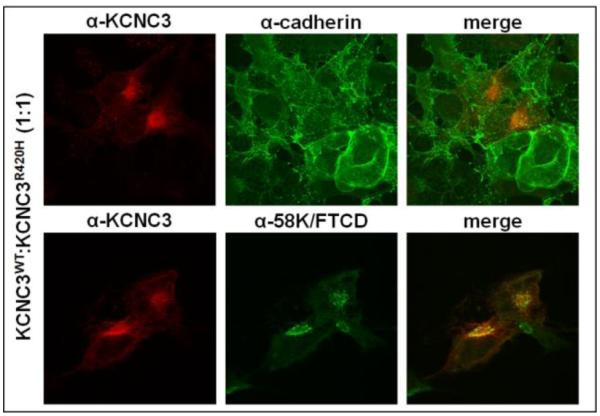
COS-1 cells transiently transfected with KCNC3WT and KCNC3R420H at a 1:1 ratio were examined by confocal fluorescence imaging with antibodies to KCNC3 and either cadherin or 58K/FTCD for plasma membrane localization or Golgi retention, respectively. Co-expression of KCNC3WT and KCNC3R420H results in limited co-registration of KCNC3 with cadherin and strong overlap between KCNC3 and 58K/FTCD.
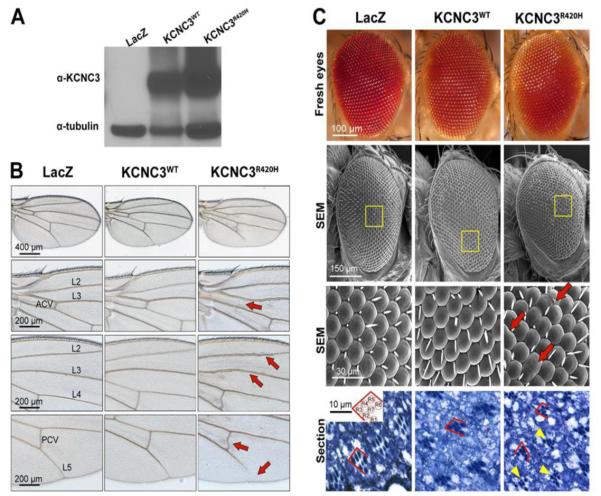
KCNC3R420H perturbs wing and eye development in Drosophila melanogaster
As discussed, no animal model-based studies have demonstrated morphological changes resulting from KCNC3R420H expression. Having established cellular alterations resulting from expression of the mutant protein, we sought to determine whether these disruptions affect cells in the complex cellular environment of intact tissues and organs. For this, we generated transgenic Drosophila melanogaster carrying human KCNC3WT or KCNC3R420H under the control of the UAS/Gal4 expression system (Brand and Perrimon, 1993) by crossing with flies carrying the ubiquitous daughterless-Gal4 (da-Gal4) driver. To examine protein expression levels, protein homogenates from whole flies expressing LacZ, KCNC3WT and KCNC3R420H were assayed by immunoblot analysis for KCNC3 protein. In control flies expressing LacZ, we detected no signal with the mammalian KCNC3 antibody. Flies expressing either KCNC3WT or KCNC3R420H produced specific immunoreactivity at the expected molecular weight with comparable expression levels (Fig. 8A).
Figure 8. KCNC3R420H perturbs Drosophila eye and wing morphology.
(A) Total protein from a single fly ubiquitously expressing LacZ or human KCNC3WT or KCNC3R420H as a consequence of crosses with the da-Gal4 driver (Materials and Methods) was subjected to immunoblot using antibodies against human KCNC3 and tubulin as a loading control. (B) Wings from da-Gal4 females crossed with LacZ and KCNC3WT flies show identical wing morphology with perfectly patterned veins. KCNC3R420H-expressing flies show abnormal veins, including loss of the anterior cross vein (ACV), thickening of longitudinal veins L2 and L3, a distorted posterior cross vein (PCV) and partial loss of L5 (red arrows). (C) Images from fresh eyes (top), micrographs from scanning electron microscope (SEM) (middle two rows), and histological sections (bottom) from flies expressing constructs targeted with the eye-specific gmr-Gal4 driver are provided. Higher resolution SEM (third row) are derived from the yellow boxed regions (second row). Eyes from flies expressing LacZ or KCNC3WT are perfectly organized. Expression of KCNC3R420H resulted in fused ommatidia (red arrows). Toluidine-blue-stained eye sections (bottom row) illustrate the arrangement of photoreceptor cells within each ommatidium. The inset shows the normal geometric organization of photoreceptors in all ommatidia. Eyes from LacZ and KCNC3WT flies display normal photoreceptor polarity (red lines). KCNC3R420H eyes contain aberrant differentiation of photoreceptors (yellow arrowheads) and abnormal cell polarity (red lines).
The Drosophila wing consists of a simple epithelium folded onto itself with characteristic size and shape. The wing also contains robust pattern elements, including the sensory elements in the wing margin and the veins inside the wing. Expression of LacZ or KCNC3WT under the control of da-Gal4 resulted in wings with normal size and shape and normal veins (Fig. 8B). However, expression of KCNC3R420H resulted in abnormal wing vein development, including the loss of the anterior cross vein (ACV), thickening of longitudinal veins L2 and L3, a distorted posterior cross vein (PCV) and partial loss of L5 (Fig. 8B, red arrows). These alterations in vein patterning resemble changes observed in signaling pathways implicated in wing growth and patterning (Price et al., 1997).
To address the consequences of targeted KCNC3R420H expression, we used the gmr-Gal4 driver to achieve specific expression in all differentiating eye cells, including photoreceptors and other support cells. The Drosophila eye consists of 800 ommatidia neatly organized in a lattice that is easily disorganized by perturbations in genes and pathways involved in the specification of photoreceptors and the recruitment of accessory cells. Images from fresh eyes illustrate no gross changes in eye morphology (Fig. 8C, top row). Flies expressing LacZ or KCNC3WT produced eyes with normal ommatidia and lattice structure (Fig. 8C). In contrast, expression of KCNC3R420H resulted in small perturbations of the eye, including multiple fused ommatidia and disruption of the normal lattice organization (Fig. 8C, SEM). Each ommatidium contains eight precisely organized photoreceptor cells, although only seven are visible due to the stacking of R7 and R8 (Fig. 8C, inset, bottom row). Sectioning these eyes to uncover the arrangement of photoreceptor cells within each ommatidium revealed that expression of LacZ or KCNC3WT did not perturb the differentiation or organization of the photoreceptor cells (Fig. 8C, bottom row). Expression of KCNC3R420H , however, caused aberrant differentiation of photoreceptor cells (Fig. 8C, bottom row, yellow arrowheads) and abnormal cell polarity (red lines). These phenotypes are consistent with the alterations in wing patterning, suggesting that KCNC3R420H affects key signaling pathways in both tissues.
Discussion
SCA13 is an autosomal dominant disease and, as such, expression of the KCNC3R420H allele is causative in the presence of the KCNC3WT allele. Studies in Xenopus oocytes and zebrafish (Issa et al., 2011; Minassian et al., 2012; Mock et al., 2010; Zhao et al., 2013) have implicated dominant negative effects of KCNC3R420H based on electrophysiological studies demonstrating the lack of channel activity in heterologous systems. However, evidence for the underlying molecular basis of disease is lacking. Zhao et al. (Zhao et al., 2013) recently attempted to address this by illustrating both a reduction in protein half-life and cell surface expression for KCNC3R420H overexpressed in CHO cells. These phenomena were attributed to increased protein degradation. It is, however, counterintuitive that reduced protein levels of KCNC3R420H would be causative in a dominantly inherited disease. Our results, however, demonstrate that KCNC3R420H displays aberrant glycosylation and trafficking with a significant reduction in surface expression in conjunction with altered Golgi and cellular morphology. Furthermore, expression of the human KCNC3R420H during the life cycle of Drosophila results in developmental abnormalities of both wing and eye.
To address biochemical or cellular alterations associated with KCNC3R420H expression, we demonstrated an altered pattern of glycosylation similar to the previous characterization of KCNC3WT glycan modifications (Cartwright et al., 2007). Enzymatic digestion of KCNC3WT by PNGase F removes both complex carbohydrate and high mannose modifications, whereas Endo H treatment of cellular extracts only removes high mannose modifications, implying that KCNC3WT exists with both types of carbohydrate moieties. In contrast, KCNC3R420H lacks the complex carbohydrate modifications but may possess high mannose chains based on a small increase in protein mobility following treatment with either Endo H or PNGase F.
Given these results, we also studied the intracellular localization and trafficking of both KCNC3WT and KCNC3R420H and were able to demonstrate that KCNC3R420H was retained in an intracellular compartment with a limited presence at the plasma membrane. The markedly reduced trafficking of KCNC3R420H to the plasma membrane was confirmed by surface biotinylation and the lack of co-registration with cadherin at the membrane. Further characterization of trafficking suggests that KCNC3R420H traverses the endoplasmic reticulum. Immunohistochemical results indicate that KCNC3R420H co-registers with both the cis-Golgi marker GM130 and 58K/FTCD whereas apFRET data specifically demonstrated co-localization with the 58K/FTCD Golgi protein. Bloom and Brashear's characterization of 58K/FTCD described it as a peripheral membrane protein exposed to the cytoplasmic side of the Golgi, directly interacting with the microtubule network (Bloom and Brashear, 1989). Gao and Sztul further demonstrated that 58K/FTCD specifically interacts with the Golgi membrane and vimentin filaments proximal to the Golgi (Gao and Sztul, 2001). Therefore, our immunohistochemistry and apFRET data indicate that KCNC3R420H is retained in the Golgi membranes, unlike cadherin and β-integrin which traffic normally to the plasma membrane even in cells expressing KCNC3R420H. The retention of KCNC3R420H in the Golgi also appears to affect the integrity of the Golgi apparatus based on EM and cellular morphology as illustrated by β-tubulin co-staining.
In simulating the natural existence of both a wild-type and mutant allele in SCA13 patients, co-expression of KCNC3WT and KCNC3R420H appears to inhibit surface expression of KCNC3WT with Golgi retention of both proteins comparable to KCNC3R420H alone. At the cellular level, this may explain how KCNC3R420H contributes to the autosomal dominant inheritance causative in SCA13 patients. The morphological alterations induced by expression of KCNC3R420H in the wing and eye during Drosophila development also provide insights into phenotypic impacts in a complex cellular environment. Our current efforts are therefore focused on the development of a mouse model that directly mimics autosomal dominant inheritance in SCA13.
In summary, we have demonstrated that a mutation that is causative for SCA13, KCNC3R420H, exhibits altered glycosylation coupled with aberrant trafficking with a significant fraction of the protein retained in the Golgi. The cellular consequences of this retention are altered Golgi and cellular morphology. Most relevant is the observation that co-expression of KCNC3WT and KCNC3R420H, analogous to a heterozygote genotype, results in co-retention of both proteins. The latter result provides a rationale for disease manifestation in SCA13.
Supplementary Material
Highlights.
KCNC3R420H displays aberrant glycosylation KCNC3R420H is retained in the Golgi by immunohistochemistry and FRET Coexpression of KCNC3R420H and KCNC3WT causes retention of both proteins in the Golgi KCNC3R420H expression results in Golgi and cell dysmorphology
Acknowledgments
The authors would like to thank Daniela Arbelaez and Praneetha Muthumani for their technical assistance in completing this work. Support provided, in part, by NIH NINDS K23 NS054715 (MFW), the McKnight Brain Institute, and the Rosalyn Heilman Research Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data
Supplemental data include one figure.
References
- Bloom GS, Brashear TA. A novel 58-kDa protein associates with the Golgi apparatus and microtubules. J Biol Chem. 1989;264:16083–92. [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Carlson KM, et al. Emerging pathogenic pathways in the spinocerebellar ataxias. Curr Opin Genet Dev. 2009;19:247–53. doi: 10.1016/j.gde.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright TA, et al. Complex oligosaccharides are N-linked to Kv3 voltage-gated K+ channels in rat brain. Biochim Biophys Acta. 2007;1770:666–71. doi: 10.1016/j.bbagen.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Chang SH, et al. GOLGA2/GM130, cis-Golgi matrix protein, is a novel target of anticancer gene therapy. Mol Ther. 2012;20:2052–63. doi: 10.1038/mt.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SY, et al. Distribution of Kv3.3 potassium channel subunits in distinct neuronal populations of mouse brain. J Comp Neurol. 2007;502:953–72. doi: 10.1002/cne.21353. [DOI] [PubMed] [Google Scholar]
- Day RN, Davidson MW. Fluorescent proteins for FRET microscopy: monitoring protein interactions in living cells. Bioessays. 2012;34:341–50. doi: 10.1002/bies.201100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RN, Schaufele F. Imaging molecular interactions in living cells. Mol Endocrinol. 2005;19:1675–86. doi: 10.1210/me.2005-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa F, et al. Alcohol hypersensitivity, increased locomotion, and spontaneous myoclonus in mice lacking the potassium channels Kv3.1 and Kv3.3. J Neurosci. 2001;21:6657–65. doi: 10.1523/JNEUROSCI.21-17-06657.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Funez P, et al. Identification of genes that modify ataxin-1-induced neurodegeneration. Nature. 2000;408:101–6. doi: 10.1038/35040584. [DOI] [PubMed] [Google Scholar]
- Fowler PW, Sansom MS. The pore of voltage-gated potassium ion channels is strained when closed. Nat Commun. 2013;4:1872. doi: 10.1038/ncomms2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Sztul E. A novel interaction of the Golgi complex with the vimentin intermediate filament cytoskeleton. J Cell Biol. 2001;152:877–94. doi: 10.1083/jcb.152.5.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman-Wohl DS, et al. Kv3.3b: a novel Shaw type potassium channel expressed in terminally differentiated cerebellar Purkinje cells and deep cerebellar nuclei. J Neurosci. 1994;14:511–22. doi: 10.1523/JNEUROSCI.14-02-00511.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding AE. The clinical features and classification of the late onset autosomal dominant cerebellar ataxias. A study of 11 families, including descendants of the 'the Drew family of Walworth'. Brain. 1982;105:1–28. doi: 10.1093/brain/105.1.1. [DOI] [PubMed] [Google Scholar]
- Hennig D, et al. A formiminotransferase cyclodeaminase isoform is localized to the Golgi complex and can mediate interaction of trans-Golgi network-derived vesicles with microtubules. J Biol Chem. 1998;273:19602–11. doi: 10.1074/jbc.273.31.19602. [DOI] [PubMed] [Google Scholar]
- Issa FA, et al. Spinocerebellar ataxia type 13 mutant potassium channel alters neuronal excitability and causes locomotor deficits in zebrafish. J Neurosci. 2011;31:6831–41. doi: 10.1523/JNEUROSCI.6572-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joho RH, Hurlock EC. The role of Kv3-type potassium channels in cerebellar physiology and behavior. Cerebellum. 2009;8:323–33. doi: 10.1007/s12311-009-0098-4. [DOI] [PubMed] [Google Scholar]
- Long SB, et al. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–82. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- Mao Y, et al. Structure of the bifunctional and Golgi-associated formiminotransferase cyclodeaminase octamer. EMBO J. 2004;23:2963–71. doi: 10.1038/sj.emboj.7600327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A, et al. Allele-dependent changes of olivocerebellar circuit properties in the absence of the voltage-gated potassium channels Kv3.1 and Kv3.3. Eur J Neurosci. 2004;19:3317–27. doi: 10.1111/j.0953-816X.2004.03385.x. [DOI] [PubMed] [Google Scholar]
- Middlebrooks JC, et al. Mutation in the kv3.3 voltage-gated potassium channel causing spinocerebellar ataxia 13 disrupts sound-localization mechanisms. PLoS One. 2013;8:e76749. doi: 10.1371/journal.pone.0076749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minassian NA, et al. Altered Kv3.3 channel gating in early-onset spinocerebellar ataxia type 13. J Physiol. 2012;590:1599–614. doi: 10.1113/jphysiol.2012.228205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock AF, et al. Functional effects of spinocerebellar ataxia type 13 mutations are conserved in zebrafish Kv3.3 channels. BMC Neurosci. 2010;11:99. doi: 10.1186/1471-2202-11-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen KW, et al. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13:448–62. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JV, et al. Dominant enhancers of Egfr in Drosophila melanogaster: genetic links between the Notch and Egfr signaling pathways. Genetics. 1997;147:1139–53. doi: 10.1093/genetics/147.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae JL, Shepard AR. Kv3.3 potassium channels in lens epithelium and corneal endothelium. Exp Eye Res. 2000;70:339–48. doi: 10.1006/exer.1999.0796. [DOI] [PubMed] [Google Scholar]
- Reeber SL, et al. Architecture and development of olivocerebellar circuit topography. Front Neural Circuits. 2012;6:115. doi: 10.3389/fncir.2012.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy B, McBain CJ. Kv3 channels: voltage-gated K+ channels designed for high-frequency repetitive firing. Trends Neurosci. 2001;24:517–26. doi: 10.1016/s0166-2236(00)01892-0. [DOI] [PubMed] [Google Scholar]
- Subramony SH, et al. Comprehensive phenotype of the p.Arg420his allelic form of spinocerebellar ataxia type 13. Cerebellum. 2013;12:932–6. doi: 10.1007/s12311-013-0507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MF, et al. Mutations in voltage-gated potassium channel KCNC3 cause degenerative and developmental central nervous system phenotypes. Nat Genet. 2006;38:447–51. doi: 10.1038/ng1758. [DOI] [PubMed] [Google Scholar]
- Waters MF, Pulst SM. Sca13. Cerebellum. 2008;7:165–9. doi: 10.1007/s12311-008-0039-7. [DOI] [PubMed] [Google Scholar]
- Weiser M, et al. Differential expression of Shaw-related K+ channels in the rat central nervous system. J Neurosci. 1994;14:949–72. doi: 10.1523/JNEUROSCI.14-03-00949.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke M, et al. Rescue of murine F508del CFTR activity in native intestine by low temperature and proteasome inhibitors. PLoS One. 2012;7:e52070. doi: 10.1371/journal.pone.0052070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagha E, et al. Dendritic Kv3.3 potassium channels in cerebellar purkinje cells regulate generation and spatial dynamics of dendritic Ca2+ spikes. J Neurophysiol. 2010;103:3516–25. doi: 10.1152/jn.00982.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, et al. Spinocerebellar ataxia-13 Kv3.3 potassium channels: arginine-to-histidine mutations affect both functional and protein expression on the cell surface. Biochem J. 2013;454:259–65. doi: 10.1042/BJ20130034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.