Abstract
The cytochrome P450c17α gene (CYP17) encodes a key biosynthesis enzyme of estrogen, which is critical in regulating adipogenesis and adipocyte development in humans. We therefore hypothesized that CYP17 is a candidate gene for predicting obesity. In order to test this hypothesis, we performed a family-based association test to investigate the relationship between the CYP17 gene and obesity phenotypes in a large sample comprising 1873 subjects from 405 Caucasian nuclear families of European origin recruited by the Osteoporosis Research Center of Creighton University, USA. Both single SNPs and haplotypes were tested for associations with obesity-related phenotypes, including body mass index (BMI) and fat mass. We identifiedthree SNPs to be significantly associated with BMI, including rs3740397, rs6163, and rs619824. We further characterized the linkage disequilibrium structure for CYP17 and found that the whole CYP17 gene was located in a single-linkage disequilibrium block. This block was observed to be significantly associated with BMI. A major haplotype in this block was significantly associated with both BMI and fat mass. In conclusion, we suggest that the CYP17 gene has an effect on obesity in the Caucasian population. Further independent studies will be needed to confirm our findings.
Keywords: CYP17, Body mass index, Obesity, Association study
INTRODUCTION
The prevalence of obesity is increasing throughout the world, making it a worldwide public health problem (Khan and Bowman, 1999). Obesity is characterized by excessive body fat that adversely affects health and is a major risk factor for type 2 diabetes mellitus, coronary heart disease, hypertension, excess mortality, and some forms of cancer (Kopelman, 2000). It is estimated that the obese population in developed countries has more than doubled over the last decade (James, 2004).
Many studies have shown that estrogen plays an important role in obesity. Some of them indicate that menopause-induced estrogen deficiency is associated with increased abdominal obesity (Tchernof et al., 1998) and higher total cholesterol in plasma (Maynar et al., 2001). Estrogen replacement therapy can prevent menopause-induced gains in adipose tissue mass (Gambacciani et al., 2001). Mattiasson et al. (2002) found decreased intra-abdominal and intra-pelvic fat compartments in postmenopausal women after one year of estrogen treatment. Puder et al. (2006) found that serum estradiol level was inversely associated with central fat accumulation in women with regular menstrual cycles. The cytochrome P450c17α gene (CYP17) plays an important role in the biosynthesis of estrogen. The CYP17 gene mediates the early steps of endogenous estrogen biosynthesis by converting pregnenolone and progesterone to androgen and estrogen precursors. However, no study has explored the relationship between the CYP17 gene and obesity.
In this study, we aimed to test for associations between polymorphisms in CYP17 and obesity susceptibility in a large Caucasian sample of 1873 subjects from 405 nuclear families.
MATERIAL AND METHODS
Subjects
The study was approved by the Creighton University Institutional Review Board in USA. All the study subjects were Caucasians of European origin and signed informed-consent documents before they entered the study in the Osteoporosis Research Center of Creighton University to search for genes underlying common human complex traits, including obesity and osteoporosis, etc. The detailed design and recruitment procedures were published previously (Deng et al., 2002). A total of 405 nuclear families were recruited with 1873 subjects, including 740 parents, 744 daughters and 389 sons. Among these, 341 families were composed of both parents and at least one offspring. In the remaining 64 families, there were at least two children with either one or no parent. The average family size was 4.62 ± 1.78 (mean ± SD, standard deviation), ranging from 3 to 12, and there were 1512 sibling pairs in total. For each subject, information on age, gender, medical history, and family history was acquired.
Phenotype measurement
Obesity phenotypes included body mass index (BMI) and body fat mass. BMI was calculated as the ratio of total body weight divided by the square of height (in units of kg/m2). Weight was measured in light indoor clothing, using a calibrated balance beam scale, and height was measured using a calibrated stadiometer. Fat mass was measured by a Hologic 4500 dual-energy X-ray absorptiometry (DXA) scanner (Hologic Corporation, Waltham, MA, USA). The measurement precision of BMI as reflected by the coefficient 0.2%. The coefficient of variation for fat mass was 1.2%. for artifacts and confounding features. Subjects with significant excluded from the analysis.
SNP selection and genotyping
For each subject, genomic DNA was extracted from whole blood using a commercial isolation kit (Gentra Systems, Minneapolis, MN, USA) following the procedure detailed in the kit. SNPs were selected mainly according to public databases such as dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) and SNPper (http://snpper.chip.org/bio/snpper-enter). We selected 7 SNPs in CYP17 based on the following criteria: 1) validation status, especially in Caucasians; 2) an average density of 1 SNP per 4 kb; 3) degree of heterozygosity, i.e., minor allele frequencies >0.05; 4) functional relevance and importance, and 5) reported to dbSNP by various sources. Seven SNPs were successfully genotyped using the high-throughput BeadArray SNP genotyping technology from Illumina Inc. (San Diego, CA, USA). The average rate of missing genotype data reported by Illumina is ~0.05%.
Statistical analyses
We used PedCheck (O’Connell and Weeks, 1998) to check Mendelian consistency of SNP genotype data within families before analysis. Any inconsistent genotypes were removed. The error checking option embedded in Merlin (Abecasis et al., 2002) was run to identify and disregard the genotypes flanking excessive recombinants, further reducing genotyping errors. Hardy-Weinberg equilibrium for each SNP was tested in parents using the PEDSTATS procedure implemented in Merlin. Haplotype analyses were performed by the Haploview software (Barrett et al., 2005) to generate graphical representation of linkage disequilibrium (LD) structure as measured by D′. The Box-Cox transformation was used for converting the raw phenotypic data to normal distribution by Minitab (State College, PA, USA).
For single- and multiple-marker haplotype transmission, the program FBAT was used (http://www.biostat.harvard.edu/fbat/fbat.htm). This program is based on the original transmission disequilibrium test, in which alleles transmitted to affected offspring are compared with the expected distribution of alleles among offspring (Ewens and Spielman, 1995). All FBAT analyses assumed an additive model. The statistical analyses were performed on the transformed data using age and gender as covariates that significantly affect the obesity phenotypes. Permutated P values were obtained through 10,000 permutations to account for the multiple haplotypes within each block by the haplotype version of FBAT (HBAT) (Horvath et al., 2004).
RESULTS
The basic characteristics of the studied samples are presented in Table 1. Among all the 1873 subjects, 262 females (125 mothers, 137 daughters) and 193 males (119 fathers, 74 sons) were obese (BMI ≥ 30 kg/m2).
Table 1.
Descriptive characteristics of the study subjects from 405 nuclear families.
| Mother (N = 380) | Father (N = 360) | Daughter (N = 744) | Son (N = 389) | |
|---|---|---|---|---|
| Age (years) | 62.4 ± 10.4 | 62.8 ± 10.7 | 37.7 ± 10.3 | 36.0 ± 10.9 |
| Height (m) | 1.62 ± 0.06 | 1.77 ± 0.07 | 1.65 ± 0.06 | 1.80 ± 0.07 |
| Weight (kg) | 73.81 ± 15.21 | 90.30 ± 15.26 | 70.40 ± 16.42 | 87.60 ± 15.94 |
| BMI (kg/m2) | 28.04 ± 5.57 | 28.85 ± 4.42 | 25.77 ± 5.86 | 27.02 ± 4.39 |
| Fat mass (kg) | 28.70 ± 9.87 | 24.73 ± 7.90 | 24.44 ± 10.45 | 20.13 ± 8.11 |
Data are reported as means ± SD of the raw phenotypic values without adjustment for covariates; total sample size, N = 1873.
We performed single-SNP association analyses. The results are summarized in Table 2. For BMI, significant associations were 0.016), rs6163 (P = 0.018) and rs619824 (P = 0.039). For fat mass, only one SNP showed a marginal association signal (rs619824: P = 0.058).
Table 2.
Single-SNP association results for CYP17 gene polymorphisms with body mass index (BMI) and fat mass.
| No. | SNP ID | Physical positiona | Allelesb | MAF | Location | BMI P value | Fat mass P value |
|---|---|---|---|---|---|---|---|
| 1 | rs11191416 | 104594906 | A/C | 0.094 | Promoter | 0.320 | 0.237 |
| 2 | rs6163 | 104586914 | C/A | 0.397 | Exon1 | 0.018 | 0.079 |
| 3 | rs3740397 | 104582665 | C/G | 0.396 | Intron5 | 0.016 | 0.083 |
| 4 | rs10883783 | 104581142 | T/A | 0.301 | Intron7 | 0.064 | 0.299 |
| 5 | rs4919685 | 104577352 | C/A | 0.301 | 3′ UTR | 0.064 | 0.299 |
| 6 | rs4919682 | 104574320 | G/A | 0.287 | 3′ UTR | 0.080 | 0.310 |
| 7 | rs619824 | 104571278 | C/A | 0.442 | 3′ UTR | 0.039 | 0.058 |
Positions are based on the NCBI web site (http://www.ncbi.nlm.nih.gov/SNP/).
The second allele is the minor allele. MAF = minor allele frequency. Results with P values <0.05 are shown in bold.
We further characterized the LD structure for CYP17 and conducted haplotype association analyses. The 7 SNPs we tested spanned a ~23-kb genetic region within or around the CYP17 gene, with average marker distance of 1 SNP per 3.9 kb. These 7 SNPs included 1 promoter marker, 1 exonic marker, 2 intronic markers, and 3 3′-untranslated region markers. Only 1 haplotype block was identified in the wholeCYP17 gene (Figure 1). Table 3 summarizes the haplotype association results. After 10,000 permutations, the whole block of CYP17 was significantly associated with BMI (permutation P value = 0.019). For the single haplotype, we also found a major haplotype, ACCTCGC, with the frequency of 0.571, to be significantly associated both with BMI (P = 0.013) and fat mass (P = 0.021).
Figure 1.
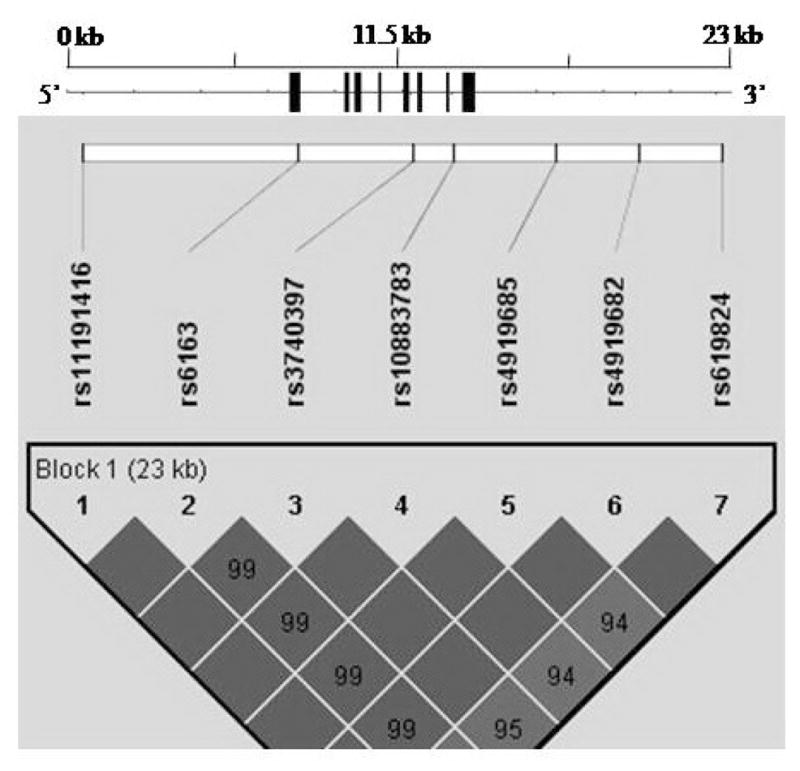
Gene structure and linkage disequilibrium pattern for the CYP17 gene.
Table 3.
Haplotype association results for CYP17 gene polymorphisms with body mass index (BMI) and fat mass.
| Block | Haplotype | Haplotype frequencya | BMI P valueb | Fat mass P value |
|---|---|---|---|---|
| Block1 | ACCTCGC | 0.571 | 0.013 | 0.021 |
| AAGAAAA | 0.269 | 0.050 | 0.160 | |
| CAGTCGA | 0.091 | 0.331 | 0.242 | |
| ACCTCGA | 0.054 | 0.484 | 0.844 | |
| 0.019 | 0.086 |
We only listed the common haplotypes with estimated frequencies of more than 5%.
Results with P values <0.05 are shown in bold. Permutation P value of the whole block is shown in bold italics.
DISCUSSION
In this study, we explored the relationship between the estrogen-related gene CYP17 and obesity-related phenotypes in a large Caucasian sample using family-based association analysis. We identified 3 SNPs and a haplotype in CYP17 to be significantly associated with BMI, suggesting that the CYP17 gene may have potential influence on variation of BMI and the pathogenesis of obesity.
In the pathway of estrogen biosynthesis, CYP17 catalyzes the conversion of 17-hydroxypregnenolone to dehydroprenenolone (DHEA). DHEA was marketed as a weight-loss pill for years. Some studies found that DHEA could enhance muscle strength and muscle mass. Morales et al. (1998) treated 16 older people for six months using 100 mg oral DHEA daily and observed that DHEA decreased fat mass and increased both knee muscle strength and lumbar back strength. Villareal and Holloszy (2006) found that DHEA replacement had the beneficial effect of enhancing the increases in muscle mass and strength induced by heavy resistance exercise in elderly individuals. Moreover, many studies proved that DHEA could help reduce excessive fat storage (Hansen et al., 1997; Han et al., 1998; Villareal and Holloszy, 2004). A possible mechanism is that DHEA reduces abdominal fat by activating the peroxisome proliferator activated receptor, which can up-regulate the transcription of fatty acid transport proteins that facilitate fatty acid entry into cells, as well as the expression of enzymes involved in the β-oxidation of fatty acids (Villareal and Holloszy, 2004). The close relationship between DHEA and obesity partially supported our observed association between CYP17 and obesity.
The transcription of CYP17 is regulated by cyclic AMP (cAMP) and steroidogenic factor-1 (SF-1) (Lund et al., 1997; Dammer et al., 2007). SNP rs6163, located in exon 1, is close to the major cAMP response elements (−243 to −225 bp) and an SF-1 binding site (−80 to −40 bp) (Demeter et al., 1996; Bischof et al., 1998). Previous studies found that the mutation or deletion of exon 1 induced the declined activities of cytochrome P450c17α (Yanase et al., 1989; Laflamme et al., 1996). Ahlgren et al. (1992) found exon 4 and exon 6 mutations in a cytochrome P450c17α-deficient patient. SNP rs3740397, located in intron 5, is very close to exon 6. The SNPs of CYP17 are all located in one block and thus in strong LD. Therefore, rs6163 and rs3740397 may be related to the regulation of gene expression of CYP17 and the activity of cytochrome P450c17α.
In summary, we are the first to suggest the potential effect of one key estrogen biosynthesis gene, CYP17, on BMI variation, which might have important implications for understanding the mechanism involved in the pathology of obesity. Our findings could be a starting point for follow-up molecular functional studies.
Acknowledgments
Research supported by the National Natural Science Foundation of China (#81000363, #31000554), the Fundamental Research Funds for the Central Universities, the Ph.D. Programs Foundation of Ministry of Education of China (#20100201120058), the Shanghai Leading Academic Discipline Project (#S30501), and the startup fund from Xi’an Jiaotong University. The study was also funded by grants from the NIH (#R01 AR050496, #R21 AG027110, #R01 AG026564, #P50 AR055081, #R01 AR057049-01A1, and #R21 AA015973).
Footnotes
Conflict of interest
All the authors declare that they have no conflicts of interest.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Ahlgren R, Yanase T, Simpson ER, Winter JS, et al. Compound heterozygous mutations (Arg 239→stop, Pro 342→Thr) in the CYP17 (P45017 alpha) gene lead to ambiguous external genitalia in a male patient with partial combined 17 alpha-hydroxylase/17,20-lyase deficiency. J Clin Endocrinol Metab. 1992;74:667–672. doi: 10.1210/jcem.74.3.1740503. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bischof LJ, Kagawa N, Waterman MR. The bovine CYP17 promoter contains a transcriptional regulatory element cooperatively bound by tale homeodomain proteins. Endocr Res. 1998;24:489–495. doi: 10.3109/07435809809032637. [DOI] [PubMed] [Google Scholar]
- Dammer EB, Leon A, Sewer MB. Coregulator exchange and sphingosine-sensitive cooperativity of steroidogenic factor-1, general control nonderepressed 5, p54, and p160 coactivators regulate cyclic adenosine 3′,5′-monophosphate-dependent cytochrome P450c17 transcription rate. Mol Endocrinol. 2007;21:415–438. doi: 10.1210/me.2006-0361. [DOI] [PubMed] [Google Scholar]
- Demeter AM, Michael MD, Kilgore MW, Simpson ER. 17alpha-Hydroxylase gene expression in the bovine ovary: mechanisms regulating expression differ from those in adrenal cells. J Steroid Biochem Mol Biol. 1996;59:21–29. doi: 10.1016/s0960-0760(96)00088-x. [DOI] [PubMed] [Google Scholar]
- Deng HW, Deng H, Liu YJ, Liu YZ, et al. A genomewide linkage scan for quantitative-trait loci for obesity phenotypes. Am J Hum Genet. 2002;70:1138–1151. doi: 10.1086/339934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens WJ, Spielman RS. The transmission/disequilibrium test: history, subdivision, and admixture. Am J Hum Genet. 1995;57:455–464. [PMC free article] [PubMed] [Google Scholar]
- Gambacciani M, Ciaponi M, Cappagli B, De Simone L, et al. Prospective evaluation of body weight and body fat distribution in early postmenopausal women with and without hormonal replacement therapy. Maturitas. 2001;39:125–132. doi: 10.1016/s0378-5122(01)00194-3. [DOI] [PubMed] [Google Scholar]
- Han DH, Hansen PA, Chen MM, Holloszy JO. DHEA treatment reduces fat accumulation and protects against insulin resistance in male rats. J Gerontol A Biol Sci Med Sci. 1998;53:B19–B24. doi: 10.1093/gerona/53a.1.b19. [DOI] [PubMed] [Google Scholar]
- Hansen PA, Han DH, Nolte LA, Chen M, et al. DHEA protects against visceral obesity and muscle insulin resistance in rats fed a high-fat diet. Am J Physiol. 1997;273:R1704–R1708. doi: 10.1152/ajpregu.1997.273.5.R1704. [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Lake SL, Silverman EK, et al. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- James PT. Obesity: the worldwide epidemic. Clin Dermatol. 2004;22:276–280. doi: 10.1016/j.clindermatol.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Khan LK, Bowman BA. Obesity: a major global public health problem. Annu Rev Nutr. 1999;19:xiii–xvii. doi: 10.1146/annurev.nutr.19.1.0. [DOI] [PubMed] [Google Scholar]
- Kopelman PG. Obesity as a medical problem. Nature. 2000;404:635–643. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- Laflamme N, Leblanc JF, Mailloux J, Faure N, et al. Mutation R96W in cytochrome P450c17 gene causes combined 17 alpha-hydroxylase/17-20-lyase deficiency in two French Canadian patients. J Clin Endocrinol Metab. 1996;81:264–268. doi: 10.1210/jcem.81.1.8550762. [DOI] [PubMed] [Google Scholar]
- Lund J, Bakke M, Mellgren G, Morohashi K, et al. Transcriptional regulation of the bovine CYP17 gene by cAMP. Steroids. 1997;62:43–45. doi: 10.1016/s0039-128x(96)00157-2. [DOI] [PubMed] [Google Scholar]
- Mattiasson I, Rendell M, Tornquist C, Jeppsson S, et al. Effects of estrogen replacement therapy on abdominal fat compartments as related to glucose and lipid metabolism in early postmenopausal women. Horm Metab Res. 2002;34:583–588. doi: 10.1055/s-2002-35420. [DOI] [PubMed] [Google Scholar]
- Maynar M, Mahedero G, Maynar I, Maynar JI, et al. Menopause-induced changes in lipid fractions and total fatty acids in plasma. Endocr Res. 2001;27:357–365. doi: 10.1081/erc-100106013. [DOI] [PubMed] [Google Scholar]
- Morales AJ, Haubrich RH, Hwang JY, Asakura H, et al. The effect of six months treatment with a 100 mg daily dose of dehydroepiandrosterone (DHEA) on circulating sex steroids, body composition and muscle strength in age-advanced men and women. Clin Endocrinol. 1998;49:421–432. doi: 10.1046/j.1365-2265.1998.00507.x. [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puder JJ, Monaco SE, Sen GS, Wang J, et al. Estrogen and exercise may be related to body fat distribution and leptin in young women. Fertil Steril. 2006;86:694–699. doi: 10.1016/j.fertnstert.2006.02.085. [DOI] [PubMed] [Google Scholar]
- Tchernof A, Calles-Escandon J, Sites CK, Poehlman ET. Menopause, central body fatness, and insulin resistance: effects of hormone-replacement therapy. Coron Artery Dis. 1998;9:503–511. doi: 10.1097/00019501-199809080-00006. [DOI] [PubMed] [Google Scholar]
- Villareal DT, Holloszy JO. Effect of DHEA on abdominal fat and insulin action in elderly women and men: a randomized controlled trial. JAMA. 2004;292:2243–2248. doi: 10.1001/jama.292.18.2243. [DOI] [PubMed] [Google Scholar]
- Villareal DT, Holloszy JO. DHEA enhances effects of weight training on muscle mass and strength in elderly women and men. Am J Physiol Endocrinol Metab. 2006;291:E1003–E1008. doi: 10.1152/ajpendo.00100.2006. [DOI] [PubMed] [Google Scholar]
- Yanase T, Kagimoto M, Suzuki S, Hashiba K, et al. Deletion of a phenylalanine in the N-terminal region of human cytochrome P-450(17 alpha) results in partial combined 17 alpha-hydroxylase/17,20-lyase deficiency. J Biol Chem. 1989;264:18076–18082. [PubMed] [Google Scholar]


