Abstract
Many tumours harbour mutations in the p53 tumour-suppressor gene that result in the expression of a mutant p53 protein. This mutant p53 protein has, in most cases, lost wild-type transcriptional activity and can also acquire novel functions in promoting invasion and metastasis. One of the mechanisms underlying these novel functions involves the ability of the mutant p53 to interfere with other transcription factors, including the p53 family protein TAp63. To investigate whether simultaneous depletion of both p53 and TAp63 can recapitulate the effect of mutant p53 expression in vivo, we used a mouse model of pancreatic cancer in which the expression of mutant p53 resulted in the rapid appearance of primary tumours and metastases. As shown previously, loss of one allele of wild-type (WT) p53 accelerated tumour development. A change of one WT p53 allele into mutant p53 did not further accelerate tumour development, but did promote the formation of metastasis. By contrast, loss of TAp63 did not significantly accelerate tumour development or metastasis. However, simultaneous depletion of p53 and TAp63 led to both rapid tumour development and metastatic potential, although the incidence of metastases remained lower than that seen in mutant p53-expressing tumours. TAp63/p53-null cells derived from these mice also showed an enhanced ability to scatter and invade in tissue culture as was observed in mutant p53 cells. These data suggest that depletion of TAp63 in a p53-null tumour can promote metastasis and recapitulate—to some extent—the consequences of mutant p53 expression.
Keywords: p63, mutant p53, pancreatic tumours, PDAC, metastasis
INTRODUCTION
p63 belongs to a family of transcription factors that includes p53 and p73, and has been implicated in a multitude of processes that include epithelial stratification, cell death, aging, tumourigenesis and metastasis.1–3 Like its family members, p63 can be transcribed from two promoters, resulting in the expression of different isoforms that either contain the full N-terminal transactivation domain (TA) or lack this region (ΔN). However, despite lacking this TA, the ΔNp63 isoforms retain clear transcriptional regulatory function.4,5 Alternative splicing in the C-terminus of p63 can potentially generate more variants that are annotated as α, β, γ, giving rise to multiple isoforms.6 Although the detailed function of each of these C-terminal isoforms is unknown, recent work on TA and ΔN-specific knockout mice has demonstrated specific roles for the N-terminal isoforms.7 Loss of ΔNp63 results in lethality shortly after birth due to severe skin defects as a result of abnormal differentiation of epithelial cells.4 By contrast, TAp63 knockout animals are viable, but develop obesity, insulin resistance and glucose intolerance, and can generate metastasizing tumours.8,9
The role of TAp63 as a tumour suppressor and invasion/ metastasis inhibitor has been explored in more detail in the last few years. Although there is clear evidence that TAp63 can function to inhibit tumour development,9,10 mutations in TP63 are rarely found in tumours. However, recent data suggest that TAp63 can be inhibited by mutant forms of p53 that are frequently expressed in cancers, leading to enhanced invasion.11,12 Trp53 is frequently mutated (up to 50–80%) in many types of human cancer. The majority of these mutations are single amino-acid substitutions in the DNA-binding domain that result in the expression of a mutant p53 protein. This mutant p53 protein can exert dominant negative regulation over any remaining wild-type p53, but it can also gain novel functions in a variety of processes including invasion/metastasis, proliferation and prevention of regulated cell death. These activities of mutant p53 have been demonstrated in both cell culture and in various mouse models.13 Some of these acquired functions have been attributed to the ability of mutant p53 to bind and interfere with TAp63 function, leading to a decrease in the expression of metastasis-inhibiting target genes of TAp63, including Sharp-1, Cyclin-G2 and Dicer,11,14 or a change in microRNA expression.15–19 These results suggest that tumours expressing mutant p53 simultaneously lose wild-type p53 and TAp63 activity. However, mutant p53 can also interact with and regulate a variety of other transcription factors, such as p73, SREBPs, NF-Y or ETS-120–23—activities that have been shown to enhance cholesterol synthesis, induce proliferation, subvert senescence and decrease cell death. The relative contribution of these activities of the mutant p53, compared with its ability to inhibit TAp63, in controlling invasion and metastasis is not clear.
In order to address the contribution of inhibition of TAp63 function to the metastatic capacity of tumours depleted of p53 in vivo, we used a mouse pancreatic ductal adenocarcinoma (PDAC) model. As shown previously, primary tumours arising in pancreatic cells following the deletion of one p53 allele showed an accelerated rate of development. Cre-mediated expression of mutant p53 in place of one p53 allele caused the formation of primary tumours at a similar rate to that seen following the deletion of one p53 allele. Importantly, metastases were only detected in the mutant p53-expressing tumours, indicative of the gain of function of mutant p53 in driving metastasis. While the loss of TAp63 did not accelerate tumorigenesis, deletion of TAp63 in concert with the depletion of p53 resulted in the development of tumours with metastatic capacity.
RESULTS
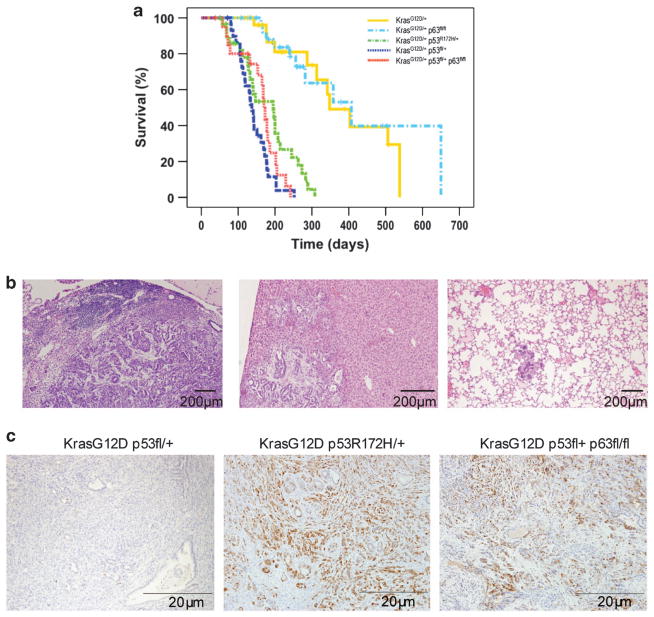
Mice with p53 mutations and mice with a loss of TAp63 and combined p53 depletion both develop sarcomas and carcinomas that are prone to metastasize.9,10,24,25 As mutant p53 has been shown to inhibit TAp63, we set out to explore the relative contribution of p53 and TAp63 loss, compared with mutant p53, to the promotion of a metastatic phenotype in vivo. For this purpose we used a mouse model of PDAC, where endogenous mutant p53 (LSL-Trp53R172H/ +) expression combined with the mutant KrasG12D in the pancreas resulted in decreased survival (compared with mutant KrasG12D expression alone) and clear evidence of metastasis in more than half (14/24) of the animals26,27 (Figure 1 and Table 1). Consistent with our previous findings, loss of one allele of p53 resulted in a similar decrease in the median survival from 220 days to 135 days, but did not induce metastatic spread (Figure 1a and Table 1). As mutant p53 expression substitutes for the expression of one p53 wild-type allele, the fact that p53 heterozygous mice and mutant p53 mice have a similar lifespan could indicate that p53 R172H did not retain WT p53 activity and that mutant p53 expression did not further accelerate primary tumour formation. To assess the impact of loss of TAp63 in this model, we crossed TAp63fl/fl mice with Pdx1-Cre, LSL (lox-stop-lox)-KrasG12D/ + mice to obtain a pancreas-specific loss of TAp63 expression (Figure 1 and Table 1). In this context, loss of TAp63 did not did not significantly reduce the median survival compared with KrasG12D/ + expression alone (220 days versus 248 days) (Figure 1a and Table 1) or significantly enhance metastatic potential of these more slowly developing tumours (1/11 mice with metastases compared to 2/10). Next, we crossed TAp63fl/fl mice with Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ + mice to generate tumours depleted for p53 and null for TAp63. While these mice showed a similar median survival as mice with deletion of one p53 allele (165 days versus 135 days) the pancreatic tumours from these mice were able to metastasize to the liver, lungs and the diaphragm, albeit at a significantly lower incidence (5/18, χ2 P =0.049) than that seen in the mutant p53-expressing tumours (Figure 1b and Table 1), although this was statistically significantly increased compared with Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +mice (χ2 P =0.0155). The increased incidence of metastasis in TAp63 −/− p53 +/ − tumours compared with the TAp63 −/− p53 +/ + tumours (5/18 compared with 2/11) is likely to be an underestimate of the true extent of this difference, as the lifespan (and so the time to develop metastases) is much shorter in the p53 +/− mice. In conclusion, our results are in agreement with the previous studies demonstrating that loss of TAp63 can drive a metastatic phenotype in a p53 heterozygous background.9
Figure 1.
Survival, metastasis and pERK1/2 staining of pancreatic tumours in different p63/(mutant) p53 mouse models. (a) The overall survival of Pdx1-Cre, LSL-KrasG12D/ + (yellow), Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ + (blue), Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/+, TAp63fl/fl(red, n =21), Pdx1-Cre, LSL-KrasG12D/ +, TAp63fl/fl(cyan) and Pdx1-Cre, LSL-KrasG12D/ +, Trp53172H/ + (green) mice was plotted in a Kaplan–Meier survival curve. Mice that succumbed to the disease before developing pancreatic cancer are shown as crossmarks on the survival curves. Significant differences in survival were detected between Pdx1-Cre, LSL-KrasG12D/ + mice (yellow) and Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ + mice (blue) (P =0.000), between Pdx1-Cre, LSL-KrasG12D/ + (yellow) and Pdx1-Cre, LSL-KrasG12D/ +, Trp53R172H/ + (green) (P =0.000), and between Pdx1-Cre, LSL-KrasG12D/ + (yellow) and Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl (red) (P =0.000) in a log-rank test. Significant differences in survival were also detected between Pdx1-Cre, LSL-KrasG12D/ +, TAp63fl/fl mice (cyan) and Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ + mice (blue) (P =0.000), between Pdx1-Cre, LSL-KrasG12D/ +, TAp63fl/fl (cyan) and Pdx1-Cre, LSL-KrasG12D/ +, LSL-Trp53R172H/ + (green) (P =0.000), and between Pdx1-Cre, LSL-KrasG12D/ +, TAp63fl/fl (cyan) and Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/+, TAp63fl/fl (red) (P =0.000) in a log-rank test. There was no significant difference in survival between Pdx1-Cre, LSL-KrasG12D/ + (yellow) and Pdx1-Cre, LSL-KrasG12D/ +, TAp63fl/fl (cyan) mice (P =0.697), or between Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ + (blue) and Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl (red) mice (P =0.235). (b) Metastases in Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl mice in the lymph nodes (left), liver (middle) and lung (right). (c) pERK1/2 staining in the pancreatic tumours of Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl and Pdx1-Cre, LSL-KrasG12D/ +, LSL-Trp53172H/ + mice.
Table 1.
Tumour growth and metastasis in genetically modified mice
| Genotype | Mean survival (overall) | Median survival (overall) | Mice with metastasis/total number of mice |
|---|---|---|---|
| Pdx1-Cre, LSL-KrasG12D/ + | 282 | 220 | 1/11 |
| Pdx1-Cre, LSL-KrasG12D/ +, TAp63fl/fl | 271 | 248 | 2/10 |
| Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ + | 138 | 135 | 0/27 |
| Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl | 146 | 165 | 5/18a,b |
| Pdx1-Cre, LSL-KrasG12D/ +, LSL-Trp53172H/+ | 159 | 140 | 14/24 |
Survival and metastasis of pancreatic tumours in different p63/(mutant) p53 mouse models. See Figure 1, Table 2 and Methods for further details.
The incidence of metastases in Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl compared with Pdx1-Cre, LSL-KrasG12D/+, LXL-Trp53172H/+ tested significantly lower in a χ2 test (P = 0.0489).
The incidence of metastases in Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl compared to Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/+ tested significantly higher in a χ2 test with Yate’s correction for low numbers (P = 0.0155)
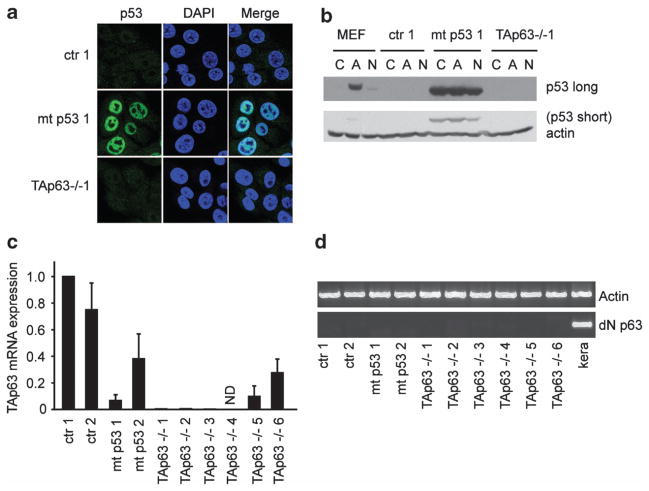
Although some of our studies were performed following deletion of only one p53 allele, no p53 could be detected in cell lines established from the pancreatic tumours of Pdx1-Cre, LSL-KrasG12D/+, Trp53fl/+, or Pdx1-Cre, LSL-KrasG12D/+, Trp53fl/ + and TAp63fl/fl mice (Figure 2), which could indicate that the wild-type p53 allele was lost during tumour formation. These data suggest that loss of TAp63 and p53 results in rapidly developing, metastatic tumours similar to those seen following the expression of mutant p53. However, loss of p53 and TAp63 does not fully phenocopy the expression of mutant p53 with respect to incidence of metastases, suggesting that mutant p53 exhibits activities additional to the inhibition of TAp63 in vivo to promote invasion.
Figure 2.
Cell lines established from the pancreatic tumours have lost wild-type p53 expression, have low TAp63 expression and no detectable ΔN p63 expression. (a) PDAC cell lines from Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ + (ctr), Pdx1-Cre, LSL-KrasG12D/ +, LSL-Trp53172H/ + (mt p53) or Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl (TAp63 −/−) were grown on coverslips and stained for p53 (green) or 4′,6-diamidino-2-phenylindole (blue). (b) The same cell lines as in (a) were incubated in 20 nM adryamycin (A) or 10 nM nutlin (N) or a control (C) for 24 h. A mouse embryonic fibroblast cell line was used as a positive control for wild-type p53 expression. Cells were lysed and p53 expression was determined using western blot. Actin was used as the loading control. (c) TAp63 expression in all PDAC cell lines was determined by qRT–PCR. Error bars indicate the standard error of the mean of three independent experiments. (d) ΔN p63 expression in all PDAC cell lines was determined by RT–PCR and the expression is shown using an agarose gel. As a positive control for ΔN p63 expression, mouse keratinocytes (kera) were used. Actin mRNA expression was used as a reference gene.
We previously detected a correlation between mutant p53 expression and increased ERK1/2 phosphorylation in pancreatic tumours, so we examined the steady-state protein levels of phospho-ERK1/2 staining in a selected group of the pancreatic tumours from each of our cohorts. While tumours derived from p53fl/ + mice showed low phospho-ERK1/2 staining, high levels of staining were detected in mutant p53-expressing tumours (Figure 1c and described in Muller et al.28). Interestingly, the phospho-ERK1/2 staining in the TAp63 −/−, p53 +/− tumours was increased compared with that in the control tumours (Figure 1c). Although many of these primary tumours with increased phospho-ERK1/2 did not generate overt or detectable metastases at the time of harvest, these data show a correlation between ERK activation and the acquisition of metastatic potential.
To more clearly examine the effect of TAp63 loss and p53 depletion on the invasive behaviour of the tumour cells, we generated cell lines from the pancreatic tumours and examined them for p53 and p63 expression (Figure 2). We obtained six PDAC cell lines from the Pdx1-Cre, LSL-KrasG12D/+, Trp53fl/ + and TAp63fl/fl mice (p53-null, TAp63-null), and compared these with cell lines derived from the pancreatic tumours of Pdx1-Cre, LSL-KrasG12D/ + and Trp53fl/+ mice (p53-null, wild-type TAp63), and from the pancreatic tumours of Pdx1-Cre, LSL-KrasG12D/+ and LSL-Trp53172H/+ mice (mutant p53, wild-type TAp63) (Figures 2 and 3 and Table 2). Notably, p53 expression could only be detected in the cell lines derived from the mutant p53 mice using immunofluorescence (Figure 2a) or western blot (Figure 2b), even in the presence of adriamycin and nutlin that can stabilize wild-type p53 in mouse embryonic fibroblasts (Figure 2b). These results therefore supported the suggestion that the cell lines established from tumours of the control and TAp63−/−, p53+/− mice have lost the second p53 allele during the process of tumourigenesis.
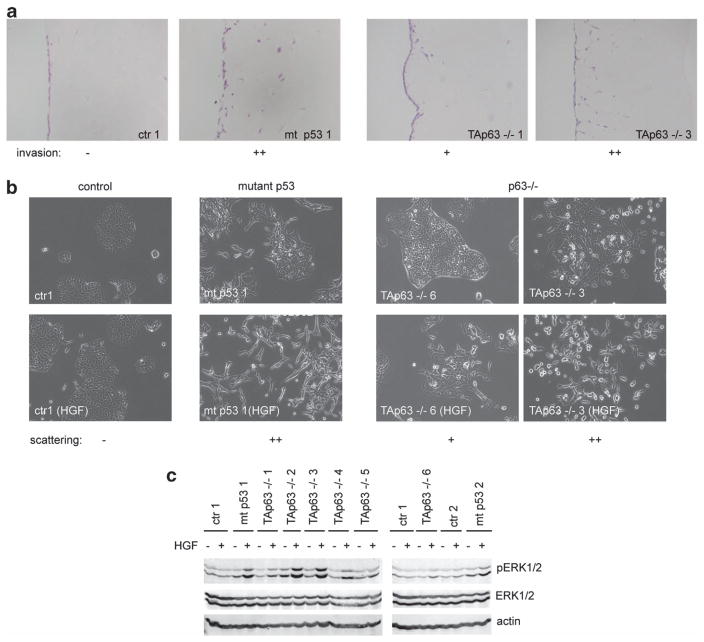
Figure 3.
Invasion, scattering and ERK1/2 activation of PDAC cell lines. (a) Invasion of PDAC cell lines from Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ + (ctr), Pdx1-Cre, LSL-KrasG12D/ +, LSL-Trp53172H/ + (mt p53) or Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/+, TAp63fl/fl (TAp63 −/−) was monitored in organotypic assays. Examples of no invasion (−), moderate invasion (+) or strong invasion (+ +) are shown, as indicated below the pictures. (b) Scattering of PDAC cell lines from Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ + (ctr), Pdx1-Cre, LSL-KrasG12D/ +, LSL-Trp53172H/ + (mutant p53) or Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl was monitored in response to HGF. Examples of no scattering (−), moderate scattering (+) or strong scattering (+ +) are shown, as indicated below the pictures. (c) ERK1/2 phosphorylation of PDAC cell lines from Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ + (ctr), Pdx1-Cre, LSL-KrasG12D/ +, LSL-Trp53172H/ + (mutant p53) or Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl was monitored in response to HGF using western blot analysis with pERK1/2-specific antibodies. ERK1/2 and actin expression were used as the loading controls.
Table 2.
Scattering, invasion and pERK1/2 staining of pancreatic tumours in different p63/(mutant) p53 mouse models
| Cell line | Genotype | Scattering (HGF) | Organotypic invasion | pERK1/2 (HGF) |
|---|---|---|---|---|
| Ctr 1 | Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ + | − | − | − |
| Ctr 2 | Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ + | − | +/ − | +/ − |
| TAp63−/− 1 | Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl | − | + | + + |
| TAp63−/− 2 | Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl | + + | + + | + + |
| TAp63−/− 3 | Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl | + + | + + | + + |
| TAp63−/− 4 | Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl | + + | + + | + |
| TAp63−/− 5 | Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl | − | +/ − | + |
| TAp63−/− 6 | Pdx1-Cre, LSL-KrasG12D/ +, Trp53fl/ +, TAp63fl/fl | + | +/ − | +/ − |
| Mtp53 1 | Pdx1-Cre, LSL-KrasG12D/ +, LSL-Trp53172H/ +, TAp63fl/fl | + + | + | + + |
| Mtp53 2 | Pdx1-Cre, LSL-KrasG12D/ +, LSL-Trp53172H/ +, TAp63fl/fl | + + | + + | + + |
The available antibodies for p63 were not reliable in allowing for the identification of p63 protein expression in the lysates of the primary tumours or in the pancreatic cell lines. We therefore used TA-specific oligos to detect mRNA expression. We were able to confirm that five of the cell lines from the TAp63 −/− mice had no or hardly detectable TAp63 mRNA expression, whereas one cell line (TAp63 −/− 6) showed a 5-fold reduction in TAp63 expression (Figure 2c), most likely reflecting incomplete deletion of TAp63. Remarkably, of the two mutant p53 cell lines, one (mt p53 1) had hardly detectable TAp63 expression whereas the other (mt p53 2) had a 3-fold reduction in TAp63 expression (Figure 2c). Notably, we could not detect another isoform of p63, ΔN p63 that has been shown to have tumour-promoting1 as well as anti-migratory functions,16 indicating that loss of TAp63 or mutant p53 expression does not coincide with an upregulation of ΔN p63 in these pancreatic tumours (Figure 2d)
Next we examined the cell lines for their invasive capacity in organotypic assays, scattering and HGF-mediated ERK1/2 phosphorylation. As expected, the two mutant p53 cell lines (mt p53 1 and mt p53 2) invaded the wells in organotypic plug assays (Table 2 and Figure 3a), showed clear scattering in response to HGF (Table 2 and Figure 3b), and efficiently induced ERK1/2 phosphorylation in response to HGF (Table 2 and Figure 3c). In comparison, the control cells (p53-null, TAp63 +/ +) did not scatter, hardly invaded and only mildly increased ERK1/2 phosphorylation after HGF exposure (Table 2 and Figures 2a–c). Of the six p53-null, TAp63 −/− cell lines, three (TAp63−/− 2, TAp63 −/− 3 and TAp63 −/− 4) were highly comparable to mutant p53-expressing cell lines with respect to efficient invasion in organotypic assays, and strong scattering in response to HGF (Table 2 and Figure 3). The other three p53-null, TAp63 −/− cell lines (TAp63−/− 1, TAp63 −/− 5 and TAp63 −/− 6) scattered more weakly and only invaded moderately into organotypic assays, although in each case these responses were greater than those seen in the control (p53-null, TAp63 +/ +) cells. Remarkably, all p53-null, TAp63 −/− lines showed an enhanced pERK1/2 induction after HGF exposure compared with the control cells (Table 2 and Figure 2), although not all of them induced ERK1/2 phosphorylation to the same extent as the mutant p53-expressing cells. These results correlated with our analysis of pERK1/2 staining in the primary tumours (Figure 1). In summary, the results of these cell lines support the in vivo data that loss of p63 expression in the context of p53 depletion can evoke a similar, albeit weaker response as that seen in mutant p53-expressing mice.
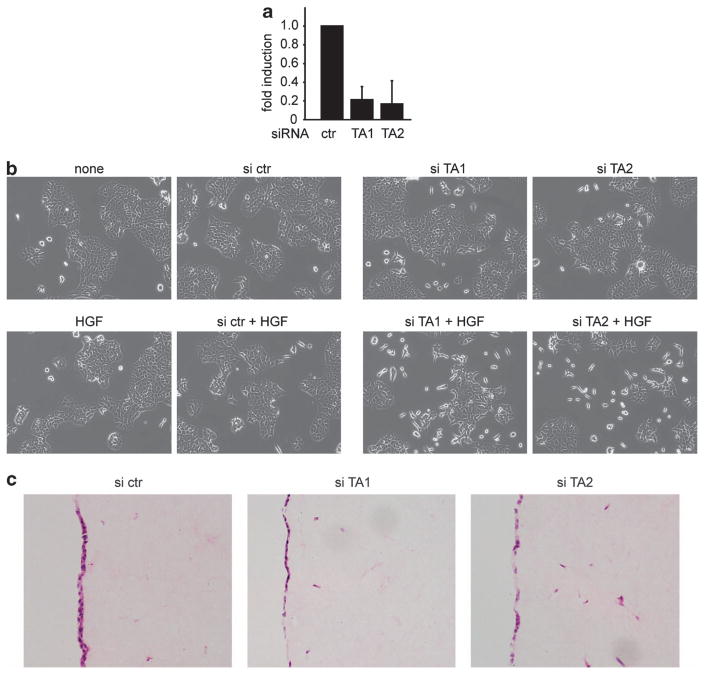
To demonstrate that the phenotypes we observed are not cell line specific, we used one of the control cell lines (ctr 1; p53-null, wild-type TAp63) and depleted-TAp63 expression using two different TA-specific siRNA oligos, verifying the knockdown using qRT–PCR (Figure 4a). A 5-fold reduction of TAp63 expression coincided with enhanced scattering in response to HGF (Figures 4a and b). The analysis of invasion using organotypic assays is complicated by the transient nature of the siRNA knockdown. Nevertheless, invasive cells were detected only after siRNA targeting of TAp63, but not in non-targeting control siRNA treated cells (Figure 4c). These results again support a role for the loss of TAp63 in driving an invasive phenotype in the context of loss of p53 expression.
Figure 4.
Loss of TAp63 expression enhances invasion and scattering of ctr PDAC cells. (a) qRT–PCR analysis of TAp63 mRNA expression in PDAC ctr cells that were transfected with two different siRNAs targeting TAp63. (b) Scattering of PDAC ctr cells that were transfected with two different siRNAs targeting TAp63 in response to HGF. (c) Organotypic invasion of PDAC ctr cells that were transfected with two different siRNAs targeting TAp63.
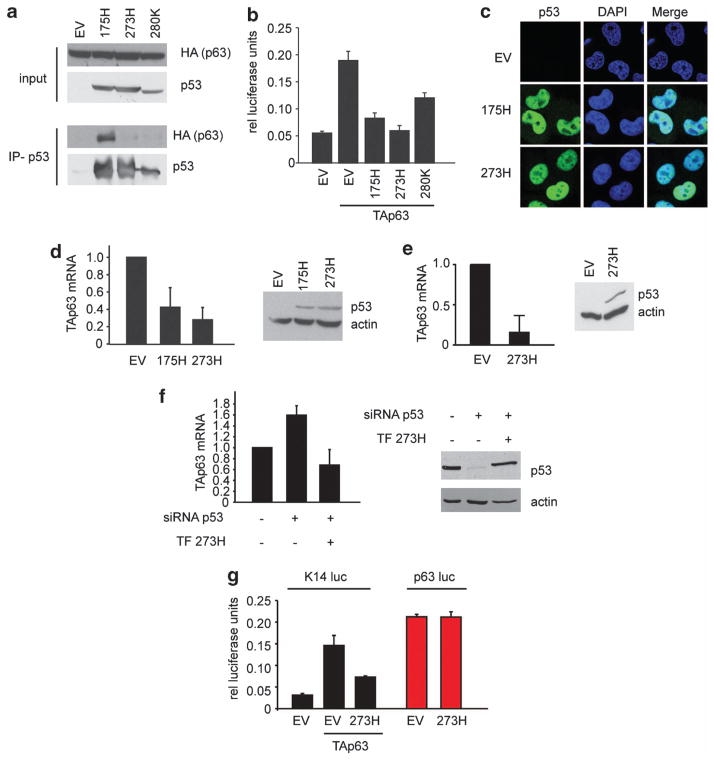
Cells expressing mutant p53 were invasive and metastatic without the genetic deletion of TAp63. Previous models have suggested that this reflects the ability of mutant p53 to bind and inactivate TAp63 function, possibly through the formation of mutant p53/TAp63 aggregates.22,29,30 Indeed, in agreement with others we could demonstrate that mutant p53 175H, a mutation that results in the conformational change of p53, could strongly bind to TAp63 (Figure 5a), whereas other p53 mutants such as 273H and 280K that have been described as DNA contact mutants rather than conformational mutants, showed much weaker interaction with TAp63 (Figure 5a).22,29 Despite this difference in interaction efficiency, mutant p53 273H and 280K could inhibit TAp63-mediated transcription to a similar extent as mutant p53 175H (Figure 5b). Notably, we could not detect any signs of aggregation under our culturing conditions in either H1299 cells or in the pancreatic tumour cells (Figure 5c and Figure 2a).
Figure 5.
Mutant p53 decreases TAp63 mRNA expression. (a) H1299 cells were transfected with an empty vector control (EV) or mutant p53 (175H, 273H or 280K) in combination with HA TAp63α. p53 was immunoprecipitated and the expression of TAp63 (HA) or p53 was determined in both the input and immunoprecipitation. (b) H1299 cells were transfected with an empty vector control (EV) or mutant p53 (175H, 273H or 280K) in combination with HA TAp63α, K14 luciferase and thymidine kinase renilla. Luciferase expression was determined and displayed as relative luciferase units corrected for TK renilla expression. Error bars indicate the standard deviation. (c) H1299 cells expressing an EV, 175H or 273H were grown on cover slips and were stained for p53 expression (green) or 4′,6-diamidino-2-phenylindole (blue). (d) TAp63 mRNA expression in PDAC ctr 1 cells that were transfected with control (EV) or human mutant p53 (175H or 273H) constructs (left panel). Expression of the constructs was validated by immunoblot analysis with p53 antibodies. Actin was used as the loading control. (e) TAp63 mRNA expression in H1299 cells that stably expressed a control (EV) or human mutant p53 (273H) (left panel). Expression of mutant p53 was validated by immunoblot analysis with p53 antibodies. Actin was used as the loading control. (f) TAp63 mRNA expression in MDA MB231 cells that were transfected with control or p53 siRNA in combination with a control construct or a mutant p53 (273H) construct that was insensitive to p53 siRNA targeting. Knockdown and expression of mutant p53 was validated by immunoblot analysis with p53 antibodies. Actin was used as the loading control. (g) H1299 cells were transfected with a control (EV) or mutant p53 (273H) in combination with the K14 luciferase (and TAp63) or a p63 promoter luciferase contruct. Luciferase expression was determined and displayed as relative luciferase units corrected for thymidine kinase renilla expression.
Interestingly, in our studies we observed that mutant p53 PDAC cell lines had very low TAp63 mRNA levels (Figure 2c), raising the possibility that mutant p53 could regulate TAp63 at the levels of both mRNA expression and protein function. To explore this more closely, we overexpressed human p53 R175H and R273H in one of the PDAC control lines (ctr 1), and detected a substantial decrease in TAp63 mRNA expression (Figure 5d). These results were not confined to the PDAC cells, as the overexpression of p53 R273H in human H1299 cells (human non-small-cell lung carcinoma that lacks endogenous WT p53 expression) similarly decreased TAp63 levels (Figure 5e). To confirm this effect in cells expressing endogenous mutant p53, we turned to MDA MB 231 breast cancer cells that express p53 R280K. After knockdown of this endogenous mutant p53, we detected an increased expression of TAp63 that was rescued by a simultaneous overexpression of an siRNA-insensitive p53 R273H construct (Figure 5f). To gain insight into the mechanism underlying this regulation, we examined the effect of mutant p53 on transcription from the TAp63 promoter, in a luciferase reporter. Interestingly, although mutant p53 could inhibit the ability of TAp63 to activate the expression from the K14 promoter (as expected, as p63 directly regulates the K14 gene31), we were unable to detect an effect of mutant p53 on the activity of the TAp63 promoter (Figure 5g), indicating that mutant p53 does not directly repress TAp63 transcription through binding to the promoter region. Together, these data demonstrate that mutant p53 not only inhibits TAp63 by interfering in its transcriptional functions, but also actively participates in reducing TAp63 mRNA expression.
DISCUSSION
In a previous mouse model, TAp63 and p53 were eliminated throughout the animal, resulting in the spontaneous formation of lymphomas, sarcomas and carcinomas that could readily metastasize.9 This tumour profile and metastasis phenotype was remarkably similar to the tumours found in mice expressing mutant p53.24,25 The notion that loss of TAp63 (and p53 depletion) can result in a similar outcome to mutant p53 expression is supported by a large number of studies showing that mutant p53 can inhibit TAp63, and that cells that have lost TAp63 expression display a similar invasive and metastatic phenotype to cells expressing mutant p53.11,12,14,15,17,22,29,32,33 However, numerous recent publications have described TAp63-independent activities of mutant p53, raising the question of what the contribution of TAp63 inhibition is to the mutant p53 metastasis phenotype. In order to assess the contribution of TAp63 to metastasis specifically, we used a metastatic mouse PDAC model to directly compare TAp63 loss to mutant p53 expression in the same genetic background of an activating Kras mutation and the loss of one wild-type p53 allele. Loss of TAp63 did not enhance the growth of the primary tumour and did not significantly increase the metastatic burden compared with the KrasG12D/ + mice. Mutant p53 promoted the formation of metastasis in 14 out of 24 animals, while 5 out of 18 animals in the TAp63 −/− p53 +/ − group developed metastasis. These data suggest that, at least in pancreatic tumours, loss of TAp63 expression in combination with a depletion of p53 has a similar, but less potent, effect to mutant p53 expression on metastasis, and that mutant p53 can induce TAp63-independent pathways to promote metastasis.
We have shown that by inhibiting TAp63, mutant p53 can promote Rab coupling protein-dependent recycling of integrin α5β1 and receptor tyrosine kinases to promote invasion and scattering.12,28 However, we also noticed that other cells that endogenously express mutant p53, but have very low expression levels of TAp63, could still scatter in a mutant p53-dependent but, importantly, TAp63-independent manner in response to HGF (hepatocyte growth factor),28 supporting the hypothesis that a decrease in TAp63 activity is not sufficient to explain the complete mutant p53 migration phenotype. Besides interfering with TAp63 function, mutant p53 proteins have been shown to interact and regulate a variety of other transcription factors, including p73, SP1, NF-Y, ETS-1, ETS-2 and SREBPs,34 each of which may contribute to a pro-oncogenic and pro-metastatic activity that is independent of TAp63.
Previous work demonstrated that mutant p53 can interact with all p53 family members and interfere with the transcriptional activity of these proteins.22,29 Here we noted that the pancreatic cell lines established from the mutant p53 tumours had a decreased or no detectable TAp63 expression, suggesting an additional mechanism by which mutant p53 can inhibit TAp63. However, we were unable to demonstrate an effect of mutant p53 on the activity of the TAp63 promoter, suggesting that mutant p53 does not act by directly regulating the transcription of TAp63. It is possible that mutant p53 regulates TAp63 mRNA stability or translation, and in this context it is interesting to note that mutant p53 can regulate the expression of many microRNAs;15,16,18,19 small RNA molecules that can promote the degradation of mRNA or block translation. Since p63 family members have been described as targets of multiple microRNAs, including miR-21, miR30b/c, miR-203 and miR-302,35–39 it will be interesting to determine whether any of the mutant p53-induced microRNAs can target TAp63.
The vast majority of all human PDACs contain activating Kras mutations,40 which signal through RAF and MEK to activate ERK1/2. ERK1/2 activation has been implicated in a variety of processes including proliferation, EMT, invasion, survival and differentiation.41,42 Many of the pancreatic tumours also harbour p53 mutations,43 which have been shown to strongly cooperate with activated Ras to drive malignancy.44 Interestingly, we have found that expression of mutant p53 in the pancreatic tumours resulted in an increase in ERK1/2 phosphorylation, implying that even though the constitutive active Kras in our model must have been activating this signalling pathway, mutant p53 could further ‘hyper’activate ERK1/2.28 Similar to mutant p53 tumours, TAp63 −/− p53-null tumours also displayed high phospho-ERK1/2 staining. Previously, we have demonstrated that in cells that do not express any p53, both TAp63 depletion and mutant p53 expression could drive integrin/RCP-dependent recycling of MET to enhance ERK1/2 signalling. The data presented here provide further evidence that loss of TAp63 in a p53-depleted background, similar to mutant p53-expressing tumours, activate the ERK1/2 signalling pathway. Furthermore, these data suggest that both mutant p53 and depletion of p63/p53 ‘hyper’activate ERK1/2 over and above the levels that are normally induced by Kras and that this increase helps to promote metastasis.
In summary, we conclude that loss of TAp63 and p53 depletion is less potent in driving metastasis than the expression of mutant p53, suggesting that mutant p53 also impinges on alternative pathways to promote metastasis. As mutations in p53 are one of the most frequently occurring events in malignancy, it will be important to further characterize all the pathways that the mutant p53 activates to drive invasion and metastasis.
MATERIALS AND METHODS
Mice
The Pdx1-Cre, LSL-KrasG12D, Trp53fl/ +, TAp63fl/fl and LSL-Trp53172H/ + mouse strains have been described in refs. 25,45–48 and experiments were conducted as described in Morton et al.27 Briefly, animals were kept in conventional animal facilities, monitored daily and experiments were carried out in compliance with UK Home Office guidelines. Mice genotypes were confirmed by PCR analysis (Transnetyx, Cordova, TN, USA). Tumour and metastatic burden was assessed by means of gross pathology and histology.
Cell lines, constructs, siRNA and transfection
MDA MB231 and H1299 cells were obtained from ATCC and cultured in Dulbeco modified eagles serum (DMEM, Invitrogen, Paisley, UK) supplemented with 10% fetal bovine serum, 1% glutamine (Invitrogen) and 1% pen/strep at 37 °C and 5% CO2. Mouse embryonic fibroblasts were cultured in DMEM supplemented with 10% fetal bovine serum, 1% glutamine and 1% pen/strep at 37 °C and 5% CO2. PDAC cell lines were established as described previously28 and maintained in DMEM, including 10% fetal bovine serum, 1% glutamine and 1% pen/strep at 37 °C and 5% CO2. Stable H1299 cells and constructs were established as previously described.12 The GNL 273H p53 construct was generated by mutagenesis, using the following oligos: forward, 5′-ACACTGGAAGACTCCAGTGGGAACC TACTGGGACGGAACAGCTTT-3′, reverse, 5′-TCAAAGCTGTTCCGTCCCAGTAG GTTCCCACTGGAGTCTTCCAGT-3′. The p63 luciferase reporter construct was a gift from M Dobbelstein and has been described in Waltermann et al.49
For overexpression and knockdown in PDAC cells and H1299 cells, we used lipofectamine (Invitrogen) or hiperfect (Qiagen, Crawley, UK), respectively, according to the manufacturer’s protocol. For knockdown experiments the following siRNAs were used: siTA1: 5′-AGAUUGAGAUUAG CAUGG A(TT)-3′, siTA2: 5′-CAGCCUAUAUGCUCAGUAC(TT)-3′, non-targeting sictr (Dharmacon; D-001810–10–20). Sip53: 5′-GACUCCAGUGGUAAUC UAC(TT)-3′.
For knockdown and overexpression in MDA MB231 cells, nucleofection (AMAXA, Lonza, Slough, UK, program X-013 and solution V) was used in which 30 pmol siRNA was combined with a 2-μg construct to transfect 5 × 106 cells.
Immunohistochemistry and immunofluorescence
Tissues were formalin-fixed, embedded in paraffin and stained for pERK1/2 (cell signalling) as described before.28
For immunofluorescence, cells were grown overnight on coverslips, washed in PBS and fixed in 4% paraformaldehyde for 10 min at 4 °C. Cells were blocked and permeabilized in 0.5% triton X-100 in PBS supplemented with 1% BSA (bovine serum albumin, Sigma-Aldrich, Gillingham, UK) for 1 h. Cells were next incubated in p53 FL393 antibody (1:150, Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h and washed three times with PBS–BSA, followed by incubation with a secondary rabbit alexa 488 antibody (1:250, Invitrogen). Finally, cells were washed three times with PBS and mounted on glass coverslips in the mounting medium containing 4′,6-diamidino-2-phenylindole (VECTASHIELD, Vector Laboratories, Peterborough, UK).
Immunoblot analysis and immunoprecipitation
For the detection of phosphorylated ERK1/2, cells were harvested directly in the sample buffer and sheared with an insulin needle. Lysates were subject to immunoblot analyses and activated ERK1/2 was detected with phosphospecific antibodies (1:1000, Cell Signaling, Danvers, MA, USA). For all other immublot analysis cells were lysed in NP-40 lysis buffer, containing 150 mM NaCl, 50 mM Tris-HCL pH 8.0 and 1% NP-40. Antibodies to detect the following proteins were used in western blotting: p53 1801 (1:5000), p53 DO-1 (1:5000), Actin (1:5000, Merck Millipore, Darmstadt, Germany), ERK1/2 (1:2000, Cell Signaling). For immunoprecipitation, cells were lysed in NP-40 and 10% of the lysates was taken as input. The rest of the lysate was incubated with protein G dynabeads (Invitrogen) and p53 antibody overnight at 4 °C and washed five times thoroughly with lysis buffer using a vortex.
qRT–PCR analysis
For qRT–PCR analysis RNA was isolated using Trizol (Invitrogen) according to the manufacturer’s protocol. cDNA was synthesized from 2 μg total RNA using a first strand synthesis kit (Invitrogen), according to the manufacturer’s instructions. In each qRT–PCR reaction 5 μl of cDNA material (diluted 40 ×) was combined with 10 μl Sybr green mastermix (Fermentas, York, UK), 2 μl oligo mixture (forward and reverse oligo at 10 μM) and 3 μl of H2O with the following PCR conditions: 94 °C annealing 2 min, 40 cycles of 30 s at 94 °C, 30 s at 60 °C and 1 min at 72 °C, followed by a 10-min 72 °C incubation. After finishing these cycles, a melting curve was generated to check for specificity. mRNA expression of mouse actin or human UBC (ubiquitin ligase C) was used as a reference. Oligos: mouse TAp63 forward: 5′-G TGGATGAACCTTCCGAAAA-3′; mouse ΔNp63 forward: 5′-CAAAACCCT GGAGCAGAAA-3′; mouse TAp63 and ΔNp63 reverse: 5′-GAGGAGCCGTTCT GAATCTG-3′ (as published by Laurikkala et al.50), mouse actin forward: 5′-AGAGAGGTATCCTGACCCTG-3′; actin reverse: 5′-GGCCATCTCCTGCTCGA AGT-3′, human TAp63 forward: 5′-ATTTTCTGGAACAGCCTATATG -3′; human TAp63 reverse: 5′-CTG TGG CCA CAT GGG GTC-3′; human UBC forward: 5′-GATGGTCGTACCCTGTCTGAC-3′; human UBC reverse: 5′-GGTCTTGCCAGT GAGTGTCTT-3′. As ΔN p63 expression could not be detected in PDAC cells using qRT–PCR, 15 μl of the qRT–PCR product was run on a 2% agarose gel containing ethidium bromide and visualized with a Geldoc imaging system (Bio-Rad Laboratories, Hemel Hempstead, UK).
Scattering
For scattering experiments, ~25 000 cells were plated in a six-well dish and allowed to settle for 16 h, followed by incubation in 30 ng/ml HGF (Sigma-Aldrich Company) for 40–48 h. Phase-contrast images were taken using an Olympus CKX41 microscope (Olympus, East Grinstead, UK) and RNA was harvested for RT–PCR analysis to verify knockdown after scattering.
Organotypic assays
Organotypic assays were performed as previously described.28,51 Briefly, collagen from rat tail tendons was isolated and combined with human fibroblasts to form a matrix. On this matrix 5 × 104cells were allowed to settle overnight after which the matrix and the cells were placed on a grid in an air–liquid interface. After 9 days on the grid the matrix and the cells were harvested in 4% paraformaldehyde and stained for hematoxylin and eosin using standard procedures.
Luciferase assays
For luciferase assays, cells were transfected with K14 luciferase or p63 luciferase in combination with thymidine kinase renilla. Luciferase and renilla expression were measured using dual luciferase, stop&glo substrate kit (Promega, Southhampton, UK) in a luminometer (Turner Biosystems, Sunnyvale, CA, USA) according to the manufacturer’s instructions.
Acknowledgments
Work in the labs of KHV and OJS was funded by CRUK. Work by PAJM and KHV was sponsored by the AICR. Part of the work was funded by AIRC (#5471) (2011-IG11955), AIRC 5xmille (#9979), Telethon Grant GGPO9133, Min. Salute (RF) to GM. We thank Dr N Rath and Dr M Olsen for providing the cDNA of mouse keratinocytes and Dr M Dobbelstein for providing the p63 promoter luciferase construct.
ABBREVIATIONS
- DMEM
Dulbecco modified eagles medium
- FBS
fetal bovine serum
- HGF
hepatocyte growth factor
- PDAC
pancreatic ductal adenocarcinoma
- RCP
Rab coupling protein
- WT
wild type
- TAp63
transactivating domain-containing p63
- ΔN
amino-deleted p63
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Contributor Information
KH Vousden, Email: k.vousden@beatson.gla.ac.uk.
PAJ Muller, Email: PM292@le.ac.uk.
References
- 1.Bergholz J, Xiao ZX. Role of p63 in development, tumorigenesis and cancer progression. Cancer Microenviron. 2012;5:311–322. doi: 10.1007/s12307-012-0116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine AJ, Tomasini R, McKeon FD, Mak TW, Melino G. The p53 family: guardians of maternal reproduction. Nat Rev Mol Cell Biol. 2011;12:259–265. doi: 10.1038/nrm3086. [DOI] [PubMed] [Google Scholar]
- 3.Su X, Chakravarti D, Flores ER. p63 steps into the limelight: crucial roles in the suppression of tumorigenesis and metastasis. Nat Rev Cancer. 2012;13:136–143. doi: 10.1038/nrc3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romano RA, Smalley K, Magraw C, Serna VA, Kurita T, Raghavan S, et al. DeltaNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development. 2012;139:772–782. doi: 10.1242/dev.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, et al. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci USA. 2007;104:3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mangiulli M, Valletti A, Caratozzolo MF, Tullo A, Sbisa E, Pesole G, et al. Identification and functional characterization of two new transcriptional variants of the human p63 gene. Nucleic Acids Res. 2009;37:6092–6104. doi: 10.1093/nar/gkp674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011;18:1487–1499. doi: 10.1038/cdd.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su X, Gi YJ, Chakravarti D, Chan IL, Zhang A, Xia X, et al. TAp63 Is a Master Transcriptional Regulator of Lipid and Glucose Metabolism. Cell Metab. 2012;16:511–525. doi: 10.1016/j.cmet.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin Y, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–991. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo X, Keyes WM, Papazoglu C, Zuber J, Li W, Lowe SW, et al. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol. 2009;11:1451–1457. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 12.Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 13.Lozano G. The oncogenic roles of p53 mutants in mouse models. Current Opin Genet Dev. 2007;17:66–70. doi: 10.1016/j.gde.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Girardini JE, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E, et al. A Pin1/ mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell. 2011;20:79–91. doi: 10.1016/j.ccr.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Neilsen PM, Noll JE, Mattiske S, Bracken CP, Gregory PA, Schulz RB, et al. Mutant p53 drives invasion in breast tumors through up-regulation of miR-155. Oncogene. 2012;32:2992–3000. doi: 10.1038/onc.2012.305. [DOI] [PubMed] [Google Scholar]
- 16.Tucci P, Agostino M, Grespi F, Marker EK, Terrinoni A, Vousden KH, et al. Loss of p63 and its miR-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc Natl Acad Sci USA. 2012;109:15312–15317. doi: 10.1073/pnas.1110977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neilsen PM, Noll JE, Suetani RJ, Schulz RB, Al-Ejeh F, Evdokiou A, et al. Mutant p53 uses p63 as a molecular chaperone to alter gene expression and induce a pro-invasive secretome. Oncotarget. 2011;2:1203–1217. doi: 10.18632/oncotarget.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong P, Karaayvaz M, Jia N, Kaneuchi M, Hamada J, Watari H, et al. Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis. Oncogene. 2012;32:1203–1217. doi: 10.1038/onc.2012.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Cheng B, Miao L, Mei Y, Wu M. Mutant p53-R273H gains new function in sustained activation of EGFR signaling via suppressing miR-27a expression. Cell Death Dis. 2013;4:e574. doi: 10.1038/cddis.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, et al. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sampath J, Sun D, Kidd VJ, Grenet J, Gandhi A, Shapiro LH, et al. Mutant p53 cooperates with ETS and selectively up-regulates human MDR1 not MRP1. J Biol Chem. 2001;276:39359–39367. doi: 10.1074/jbc.M103429200. [DOI] [PubMed] [Google Scholar]
- 24.Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 27.Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci USA. 2010;107:246–51. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller PA, Trinidad AG, Timpson P, Morton JP, Zanivan S, van den Berghe PV, et al. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene. 2012;32:1252–1265. doi: 10.1038/onc.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strano S, Fontemaggi G, Costanzo A, Rizzo MG, Monti O, Baccarini A, et al. Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J Biol Chem. 2002;277:18817–18826. doi: 10.1074/jbc.M201405200. [DOI] [PubMed] [Google Scholar]
- 30.Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol. 2011;7:285–295. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
- 31.Romano RA, Birkaya B, Sinha S. A functional enhancer of keratin14 is a direct transcriptional target of deltaNp63. J Invest Dermatol. 2007;127:1175–1186. doi: 10.1038/sj.jid.5700652. [DOI] [PubMed] [Google Scholar]
- 32.Liu K, Ling S, Lin WC. TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73. Mol Cell Biol. 2011;31:4464–4481. doi: 10.1128/MCB.05574-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noll JE, Jeffery J, Al-Ejeh F, Kumar R, Khanna KK, Callen DF, et al. Mutant p53 drives multinucleation and invasion through a process that is suppressed by ANKRD11. Oncogene. 2012;31:2836–2848. doi: 10.1038/onc.2011.456. [DOI] [PubMed] [Google Scholar]
- 34.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 35.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 36.Quintavalle C, Donnarumma E, Iaboni M, Roscigno G, Garofalo M, Romano G, et al. Effect of miR-21 and miR-30b/c on TRAIL-induced apoptosis in glioma cells. Oncogene. doi: 10.1038/onc.2012.410. (epub ahead of print 10 September 2012) [DOI] [PubMed] [Google Scholar]
- 37.Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, Aberdam D, Knight RA, Melino G, et al. miR-203 represses ’stemness’ by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–95. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 38.Melar-New M, Laimins LA. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J Virol. 2010;84:5212–5221. doi: 10.1128/JVI.00078-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheel AH, Beyer U, Agami R, Dobbelstein M. Immunofluorescence-based screening identifies germ cell associated microRNA 302 as an antagonist to p63 expression. Cell Cycle. 2009;8:1426–1432. doi: 10.4161/cc.8.9.8324. [DOI] [PubMed] [Google Scholar]
- 40.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 41.Neuzillet C, Hammel P, Tijeras-Raballand A, Couvelard A, Raymond E. Targeting the Ras-ERK pathway in pancreatic adenocarcinoma. Cancer Metastasis Rev. 2012;32:147–162. doi: 10.1007/s10555-012-9396-2. [DOI] [PubMed] [Google Scholar]
- 42.Collisson EA, Trejo CL, Silva JM, Gu S, Korkola JE, Heiser LM, et al. A central role for RAF-->MEK-->ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2012;2:685–693. doi: 10.1158/2159-8290.CD-11-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scarpa A, Capelli P, Mukai K, Zamboni G, Oda T, Iacono C, et al. Pancreatic adenocarcinomas frequently show p53 gene mutations. Am J Pathol. 1993;142:1534–1543. [PMC free article] [PubMed] [Google Scholar]
- 44.Buganim Y, Solomon H, Rais Y, Kistner D, Nachmany I, Brait M, et al. p53 Regulates the Ras circuit to inhibit the expression of a cancer-related gene signature by various molecular pathways. Cancer Res. 2010;70:2274–2284. doi: 10.1158/0008-5472.CAN-09-2661. [DOI] [PubMed] [Google Scholar]
- 45.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 46.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 48.Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009;5:64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waltermann A, Kartasheva NN, Dobbelstein M. Differential regulation of p63 and p73 expression. Oncogene. 2003;22:5686–5693. doi: 10.1038/sj.onc.1206859. [DOI] [PubMed] [Google Scholar]
- 50.Laurikkala J, Mikkola ML, James M, Tummers M, Mills AA, Thesleff I. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development. 2006;133:1553–1563. doi: 10.1242/dev.02325. [DOI] [PubMed] [Google Scholar]
- 51.Edward M, Gillan C, Micha D, Tammi RH. Tumour regulation of fibroblast hyaluronan expression: a mechanism to facilitate tumour growth and invasion. Carcinogenesis. 2005;26:1215–1223. doi: 10.1093/carcin/bgi064. [DOI] [PubMed] [Google Scholar]