Abstract
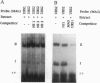
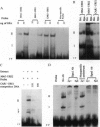
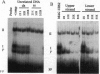
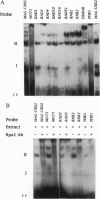
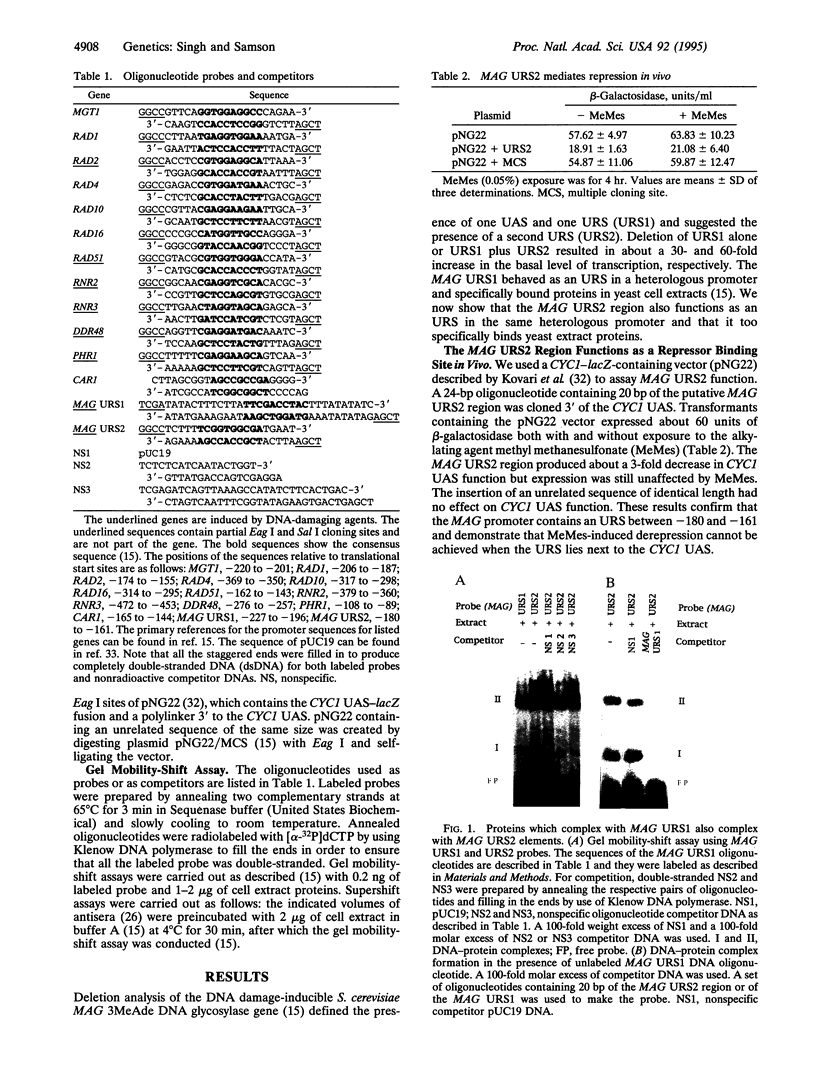
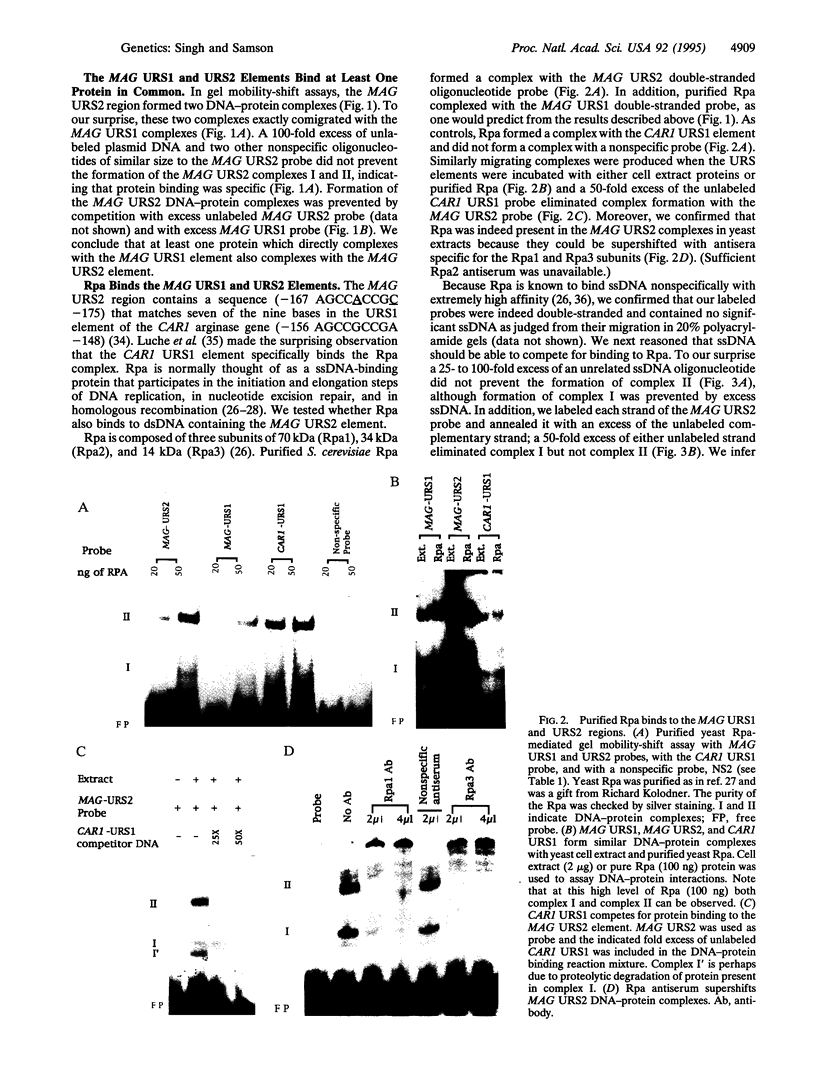
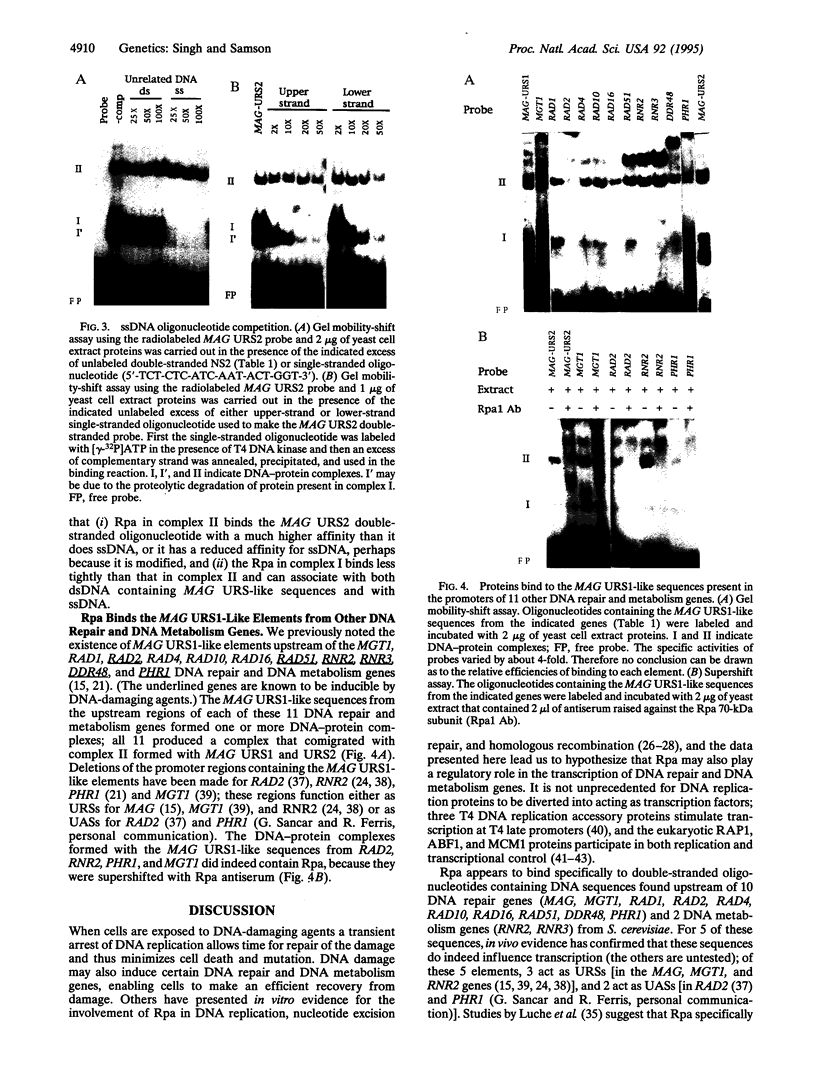
Saccharomyces cerevisiae responds to DNA damage by arresting cell cycle progression (thereby preventing the replication and segregation of damaged chromosomes) and by inducing the expression of numerous genes, some of which are involved in DNA repair, DNA replication, and DNA metabolism. Induction of the S. cerevisiae 3-methyladenine DNA glycosylase repair gene (MAG) by DNA-damaging agents requires one upstream activating sequence (UAS) and two upstream repressing sequences (URS1 and URS2) in the MAG promoter. Sequences similar to the MAG URS elements are present in at least 11 other S. cerevisiae DNA repair and metabolism genes. Replication protein A (Rpa) is known as a single-stranded-DNA-binding protein that is involved in the initiation and elongation steps of DNA replication, nucleotide excision repair, and homologous recombination. We now show that the MAG URS1 and URS2 elements form similar double-stranded, sequence-specific, DNA-protein complexes and that both complexes contain Rpa. Moreover, Rpa appears to bind the MAG URS1-like elements found upstream of 11 other DNA repair and DNA metabolism genes. These results lead us to hypothesize that Rpa may be involved in the regulation of a number of DNA repair and DNA metabolism genes.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alani E., Thresher R., Griffith J. D., Kolodner R. D. Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J Mol Biol. 1992 Sep 5;227(1):54–71. doi: 10.1016/0022-2836(92)90681-9. [DOI] [PubMed] [Google Scholar]
- Barker D. G., White J. H., Johnston L. H. The nucleotide sequence of the DNA ligase gene (CDC9) from Saccharomyces cerevisiae: a gene which is cell-cycle regulated and induced in response to DNA damage. Nucleic Acids Res. 1985 Dec 9;13(23):8323–8337. doi: 10.1093/nar/13.23.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basile G., Aker M., Mortimer R. K. Nucleotide sequence and transcriptional regulation of the yeast recombinational repair gene RAD51. Mol Cell Biol. 1992 Jul;12(7):3235–3246. doi: 10.1128/mcb.12.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill S. J., Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991 Sep;5(9):1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Kimmerly W. J., Rine J., Kornberg R. D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jan;8(1):210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carty M. P., Zernik-Kobak M., McGrath S., Dixon K. UV light-induced DNA synthesis arrest in HeLa cells is associated with changes in phosphorylation of human single-stranded DNA-binding protein. EMBO J. 1994 May 1;13(9):2114–2123. doi: 10.1002/j.1460-2075.1994.tb06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Derfler B., Maskati A., Samson L. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7961–7965. doi: 10.1073/pnas.86.20.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Derfler B., Samson L. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 1990 Dec;9(13):4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Samson L. Induction of S.cerevisiae MAG 3-methyladenine DNA glycosylase transcript levels in response to DNA damage. Nucleic Acids Res. 1991 Dec 11;19(23):6427–6432. doi: 10.1093/nar/19.23.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole G. M., Mortimer R. K. Failure to induce a DNA repair gene, RAD54, in Saccharomyces cerevisiae does not affect DNA repair or recombination phenotypes. Mol Cell Biol. 1989 Aug;9(8):3314–3322. doi: 10.1128/mcb.9.8.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley D., Kenny M. K., Munn M., Rupp W. D., Lane D. P., Wood R. D. Requirement for the replication protein SSB in human DNA excision repair. Nature. 1991 Feb 7;349(6309):538–541. doi: 10.1038/349538a0. [DOI] [PubMed] [Google Scholar]
- Demple B., Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- Demple B. Regulation of bacterial oxidative stress genes. Annu Rev Genet. 1991;25:315–337. doi: 10.1146/annurev.ge.25.120191.001531. [DOI] [PubMed] [Google Scholar]
- Din S., Brill S. J., Fairman M. P., Stillman B. Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes Dev. 1990 Jun;4(6):968–977. doi: 10.1101/gad.4.6.968. [DOI] [PubMed] [Google Scholar]
- Dutta A., Ruppert J. M., Aster J. C., Winchester E. Inhibition of DNA replication factor RPA by p53. Nature. 1993 Sep 2;365(6441):79–82. doi: 10.1038/365079a0. [DOI] [PubMed] [Google Scholar]
- Eisenberg S., Civalier C., Tye B. K. Specific interaction between a Saccharomyces cerevisiae protein and a DNA element associated with certain autonomously replicating sequences. Proc Natl Acad Sci U S A. 1988 Feb;85(3):743–746. doi: 10.1073/pnas.85.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. DNA damage induction of ribonucleotide reductase. Mol Cell Biol. 1989 Nov;9(11):4932–4940. doi: 10.1128/mcb.9.11.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Zhou Z., Allen J. B., Navas T. A. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays. 1993 May;15(5):333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- Erdile L. F., Heyer W. D., Kolodner R., Kelly T. J. Characterization of a cDNA encoding the 70-kDa single-stranded DNA-binding subunit of human replication protein A and the role of the protein in DNA replication. J Biol Chem. 1991 Jun 25;266(18):12090–12098. [PubMed] [Google Scholar]
- He Z., Brinton B. T., Greenblatt J., Hassell J. A., Ingles C. J. The transactivator proteins VP16 and GAL4 bind replication factor A. Cell. 1993 Jun 18;73(6):1223–1232. doi: 10.1016/0092-8674(93)90650-f. [DOI] [PubMed] [Google Scholar]
- Herendeen D. R., Kassavetis G. A., Barry J., Alberts B. M., Geiduschek E. P. Enhancement of bacteriophage T4 late transcription by components of the T4 DNA replication apparatus. Science. 1989 Sep 1;245(4921):952–958. doi: 10.1126/science.2672335. [DOI] [PubMed] [Google Scholar]
- Hurd H. K., Roberts J. W. Upstream regulatory sequences of the yeast RNR2 gene include a repression sequence and an activation site that binds the RAP1 protein. Mol Cell Biol. 1989 Dec;9(12):5359–5372. doi: 10.1128/mcb.9.12.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. S., Prakash L. Transcript levels of the Saccharomyces cerevisiae DNA repair gene RAD18 increase in UV irradiated cells and during meiosis but not during the mitotic cell cycle. Nucleic Acids Res. 1991 Feb 25;19(4):893–898. doi: 10.1093/nar/19.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan M. B., Zhan Q., el-Deiry W. S., Carrier F., Jacks T., Walsh W. V., Plunkett B. S., Vogelstein B., Fornace A. J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992 Nov 13;71(4):587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- Kovari L., Sumrada R., Kovari I., Cooper T. G. Multiple positive and negative cis-acting elements mediate induced arginase (CAR1) gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Oct;10(10):5087–5097. doi: 10.1128/mcb.10.10.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larner J. M., Lee H., Hamlin J. L. Radiation effects on DNA synthesis in a defined chromosomal replicon. Mol Cell Biol. 1994 Mar;14(3):1901–1908. doi: 10.1128/mcb.14.3.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Botchan M. R. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell. 1993 Jun 18;73(6):1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Sedgwick B., Sekiguchi M., Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- Liu V. F., Weaver D. T. The ionizing radiation-induced replication protein A phosphorylation response differs between ataxia telangiectasia and normal human cells. Mol Cell Biol. 1993 Dec;13(12):7222–7231. doi: 10.1128/mcb.13.12.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche R. M., Smart W. C., Marion T., Tillman M., Sumrada R. A., Cooper T. G. Saccharomyces cerevisiae BUF protein binds to sequences participating in DNA replication in addition to those mediating transcriptional repression (URS1) and activation. Mol Cell Biol. 1993 Sep;13(9):5749–5761. doi: 10.1128/mcb.13.9.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura K., Prakash S. Nucleotide sequence, transcript mapping, and regulation of the RAD2 gene of Saccharomyces cerevisiae. J Bacteriol. 1986 Jun;166(3):914–923. doi: 10.1128/jb.166.3.914-923.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura K., Prakash S. Transcript levels of the Saccharomyes cerevisiae DNA repair gene RAD23 increase in response to UV light and in meiosis but remain constant in the mitotic cell cycle. Nucleic Acids Res. 1990 Aug 25;18(16):4737–4742. doi: 10.1093/nar/18.16.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine G. T., Sinha P., Tye B. K. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics. 1984 Mar;106(3):365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian J., Kraus B., Sancar G. B. Expression of the yeast PHR1 gene is induced by DNA-damaging agents. Mol Cell Biol. 1990 Sep;10(9):4630–4637. doi: 10.1128/mcb.10.9.4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siede W., Friedberg E. C. Regulation of the yeast RAD2 gene: DNA damage-dependent induction correlates with protein binding to regulatory sequences and their deletion influences survival. Mol Gen Genet. 1992 Mar;232(2):247–256. doi: 10.1007/BF00280003. [DOI] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Ubiquitous upstream repression sequences control activation of the inducible arginase gene in yeast. Proc Natl Acad Sci U S A. 1987 Jun;84(12):3997–4001. doi: 10.1073/pnas.84.12.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treger J. M., Heichman K. A., McEntee K. Expression of the yeast UB14 gene increases in response to DNA-damaging agents and in meiosis. Mol Cell Biol. 1988 Mar;8(3):1132–1136. doi: 10.1128/mcb.8.3.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Samson L. The Saccharomyces cerevisiae MGT1 DNA repair methyltransferase gene: its promoter and entire coding sequence, regulation and in vivo biological functions. Nucleic Acids Res. 1992 Jul 25;20(14):3599–3606. doi: 10.1093/nar/20.14.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Singh K. K., Chen B., Samson L. A common element involved in transcriptional regulation of two DNA alkylation repair genes (MAG and MGT1) of Saccharomyces cerevisiae. Mol Cell Biol. 1993 Dec;13(12):7213–7221. doi: 10.1128/mcb.13.12.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagle K., McEntee K. The DNA damage-inducible gene DIN1 of Saccharomyces cerevisiae encodes a regulatory subunit of ribonucleotide reductase and is identical to RNR3. Mol Cell Biol. 1990 Oct;10(10):5553–5557. doi: 10.1128/mcb.10.10.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zhan Q., Carrier F., Fornace A. J., Jr Induction of cellular p53 activity by DNA-damaging agents and growth arrest. Mol Cell Biol. 1993 Jul;13(7):4242–4250. doi: 10.1128/mcb.13.7.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]