Abstract
In the mammalian cortex, neurons form extremely complicated networks and exchange information at synapses. Changes in synaptic strength, as well as addition/removal of synapses, occur in an experience-dependent manner, providing the structural foundation of neuronal plasticity. As postsynaptic components of the most excitatory synapses in the cortex, dendritic spines are considered to be a good proxy of synapses. Taking advantages of mouse genetics and fluorescent labeling techniques, individual neurons and their synaptic structures can be labeled in the intact brain. Here we introduce a transcranial imaging protocol using two-photon laser scanning microscopy to follow fluorescently labeled postsynaptic dendritic spines over time in vivo. This protocol utilizes a thinned-skull preparation, which keeps the skull intact and avoids inflammatory effects caused by exposure of the meninges and the cortex. Therefore, images can be acquired immediately after surgery is performed. The experimental procedure can be performed repetitively over various time intervals ranging from hours to years. The application of this preparation can also be expanded to investigate different cortical regions and layers, as well as other cell types, under physiological and pathological conditions.
Keywords: Neuroscience, Issue 87, dendritic spine, mouse cortex, in vivo, two-photon microscopy, thinned-skull, imaging
Introduction
The mammalian cortex participates in many brain functions, from sensory perception and movement control to abstract information processing and cognition. Various cortical functions build upon different neural circuits, which are made up of different types of neurons communicating and exchanging information at individual synapses. The structure and function of synapses are consistently being modified in response to experiences and pathologies. In the mature brain, synaptic plasticity takes the form of both strength changes and addition/removal of synapses, playing important roles in formation and maintenance of a functional neural circuitry. Dendritic spines are the postsynaptic components of the majority of excitatory synapses in the mammalian brain. The constant turnover and morphological changes of spines are believed to serve as a good indicator of modifications in synaptic connections1-7.
Two-photon laser scanning microscopy offers deep penetration through thick, opaque preparations and low phototoxicity, which makes it suitable for live imaging in the intact brain8. In combination with fluorescent labeling, two-photon imaging provides a powerful tool to peek into the living brain and follow structural reorganization at individual synapses with high spatial and temporal resolution. Various methods have been used to prepare mice for live imaging9-13. Here, we describe a thinned-skull preparation of in vivo two-photon imaging to investigate the structural plasticity of postsynaptic dendritic spines in the mouse cortex. Using this approach, our recent studies have depicted a dynamic picture of dendritic spine changes in response to motor skill learning With increasing availability of transgenic animals with fluorescently labeled neuronal subsets and rapid development of in vivo labeling techniques, similar procedures described here can also be applied to investigate other cell types and cortical regions, combined with other manipulations, as well as used in disease models16-23.
Protocol
Approval needs to be obtained from home institutions before commencement of the surgery and imaging study. Experiments described in this manuscript were performed in accordance with the guidelines and regulations from the University of California, Santa Cruz Institutional Animal Care and Use Committee.
1. Surgery
Autoclave all surgical instruments and sterilize the workspace with 70% alcohol thoroughly before surgery.
Anesthetize the mouse by intraperitoneal (IP) injection of KX anesthetic solution (200 mg/kg ketamine and 20 mg/kg xylazine) according to the mouse’s body weight. Note: KX dosage may be adjusted according to the strain, age, and health status of the mice. Veterinary consultation to determine the optimal dose is also recommended.
Perform a toe-pinch test periodically by pressing the mouse’s toes and checking for a reflexive response to monitor the anesthesia status. Make sure that the mouse is fully anesthetized before starting the surgery. Note: During prolonged imaging sessions, check the mouse’s anesthesia status periodically, administrate additional KX if necessary.
Place the mouse on a heating pad to maintain the body temperature during the surgery.
Shave the head of the mouse using a razor blade to expose the scalp.
Sterilize the shaved area by wiping the skin with alternating alcohol pads and betadine. Remove any residual hair clippings.
Gently apply eye ointment to lubricate both eyes, therefore preventing permanent damage caused by dehydration during the experiment.
Make a straight incision along the midline of the scalp, and move the skin laterally towards the edges of the skull.
Remove the connective tissue attached to the skull.
2. Thinned-skull Preparation
Identify the imaging region based on stereotaxic coordinates. Note: Try to avoid large blood vessels, which block light penetration and blur the imaged structures. Vasculature is best observed when the skull is moist with sterile saline.
Use the high-speed micro drill to thin a circular region of skull (0.5-1.0 mm diameter). Move the drill parallel to the skull surface, rather than holding it against the skull and pressing down. Drill until both the outer compact bone layer and the middle spongy bone layer are removed. Note: To prevent damage caused by overheating, avoid prolonged contact between the drill bit and the skull.
Continue thinning the inner compact bone layer with a microsurgical blade in the center of the drill-thinned region. Hold the microsurgical blade at an angle of approximately 45° to scrape the skull without pressing down against the skull until an evenly thinned small region (200-300 μm diameter) with a thickness of approximately 20-30 μm is obtained. Due to the auto-fluorescence of the skull, the thickness can be measured by scanning the distance between upper and lower surface of the skull under the two-photon microscope. Note: although a skull thickness of less than 20 μm provides good imaging quality, it is not recommended because it makes the thinning process in subsequent re-imaging sessions difficult. To avoid over-thinning, the thickness of the skull needs to be checked periodically during the surgery (see Discussion).
3. Immobilization
Carefully remove bone debris.
Place a small drop of cyanoacrylate glue on each edge of the center opening of the sterile head plate, which has been autoclaved (see Figure 1). Hold the head plate tightly against the skull with the thinned region located in the center of the opening. Note: If the glue contaminates the thinned region by accident, remove it carefully with the microsurgical blade.
Gently pull the skin from the sides of the skull to the edges of the center opening of the head plate.
Wait approximately 10 min until the head plate is well attached to the skull.
Place the head plate on two lateral blocks of the holding plate and then tighten the screws over the edges of the head plate to immobilize the mouse on the holding plate (see Figure 1). Note: Make sure that there is no skin or whiskers between the head plate and the blocks before tightening the screws. A good immobilized preparation should show no observable movement of the skull under the dissecting microscope when the back of the animal is gently patted.
Rinse the exposed skull with saline to remove unpolymerized glue. Note: It is important to remove any remaining glue, as unpolymerized glue blurs images and damages microscope objectives.
4. Imaging
Take a photo of the vasculature of the exposed skull with the thinned region as Map 1, which is used to relocate the imaged region in subsequent imaging sessions (see Figure 2).
Place the mouse under the imaging microscope. Locate the thinned area under a 10X air objective using epifluorescence and move the thinnest area to the center of the view.
Add a drop of saline on the top of the skull and switch to the 60X objective, which has been rinsed with sterile water. Select a region where individual dendritic spines are clearly visualized along dendrites. Identify and label the corresponding region on Map 1 by comparing vasculature between the epifluorescent view and the vasculature photo.
Tune the two-photon laser wavelength according to the fluorophores. For instance, 920 nm for YFP; 890 nm for GFP; 1,000 nm for DsRed and tdTomato24.
Acquire image stacks with 2 μm steps along the z-axis using the 60X objective. This image stack covers an approximately 200 μm x 200 μm area (512 x 512 pixels) and is used as Map 2 for relocation during subsequent imaging sessions (see Figure 2).
Acquire nine image stacks within Map 2 using 3X digital zoom. Each image stack covers an approximate area of 70 μm x 70 μm (512 x 512 pixels), with 0.7 μm steps along the z-axis. Note: The intensity of the laser should be below 40 mW when measured at samples to minimize phototoxicity.
5. Recovery
Following imaging, gently detach the head plate from the skull.
Thoroughly clean the skull and the skin to remove all the remaining glue. Note: Any remaining glue on the skin will cause irritations and slow down skin healing while any remaining glue on the skull will cause erosion of the skull and angiogenesis in the newly grown bone layer, making subsequent relocation and re-imaging difficult.
Rinse the skull and the skin with saline several times.
Suture the scalp with sterile surgical suture.
Keep the animal on a heating pad in a separate cage. Administer buprenorphine analgesic (0.1 mg/kg) subcutaneously to mitigate post-operative pain. Return the animal to the home cage after full recovery. Monitor the animal closely (check at least once daily) until the incision is healed and sutures are removed. Administer subsequent injections of buprenorphine analgesic if needed.
6. Reimaging
Repeat Steps 1.1-1.8.
Repeat Steps 3.1-3.6
Locate previously imaged region by comparing the vasculature pattern to Map 1 and the dendritic branch pattern to Map 2.
If reimaging is performed within 1 week, repeat Step 2.3 to remove the thin layer of newly grown bone on top of the thinned region using the microsurgical blade. If reimaging is performed after more than 1 week, repeat Steps both 2.2 and 2.3. Note: The newly grown bone layer consists of a less condensed structure compared to the original compact bone layer, leading to reduced image quality. Therefore, to acquire the same quality, it is necessary to thin the skull slightly more than the previous imaging session.
Adjust the position and orientation to obtain image stacks that match previously taken image stacks under the two-photon microscope.
Acquire images as in Step 4.6.
Repeat Steps 5.1-5.5 after imaging.
Representative Results
In YFP-H line mice25, yellow fluorescent protein expresses in a subset of layer V pyramidal neurons, which project their apical dendrites to the superficial layers in the cortex. Through the thinned-skull preparation, the fluorescently labeled dendritic segments can be repetitively imaged under two-photon microscope over various imaging intervals, ranging from hours to months. Here we show an example of a four-time imaging of the same dendrites over 8 days in the motor cortex of a 1 month old mouse, where individual spines as well as filopodia can be clearly visualized along the dendrite. Usually, the depth of image stacks is approximately 100-200 μm from the pial surface. Various analyses can be performed based on these images. For instance, the spine formation, elimination and turnover can be quantified by comparing images from different sessions. Spine density can be calculated by dividing the number of spines by the length of the dendritic segment. Changes of spine motility and morphology can also be analyzed.
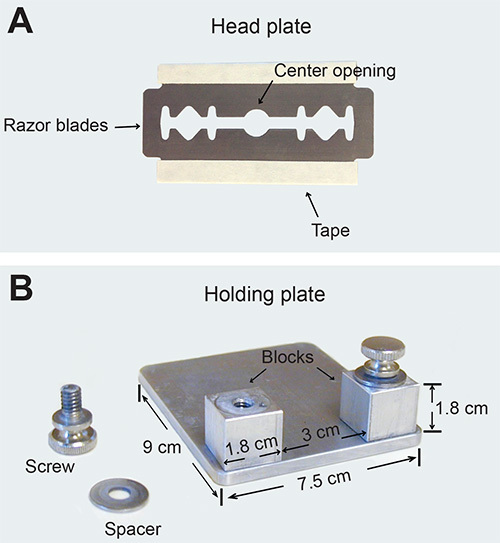 Figure 1. Custom-made immobilization plates of thinned-skull preparation for two-photon in vivo imaging. (A) A photograph of the head plate, which is made of two or three razor blades glued together, with sharp edges covered by tapes. (B) A photograph of the holding plate, which consists of 1 stainless steel plate, 2 stainless steel blocks, 2 screws, and 2 spacers.
Figure 1. Custom-made immobilization plates of thinned-skull preparation for two-photon in vivo imaging. (A) A photograph of the head plate, which is made of two or three razor blades glued together, with sharp edges covered by tapes. (B) A photograph of the holding plate, which consists of 1 stainless steel plate, 2 stainless steel blocks, 2 screws, and 2 spacers.
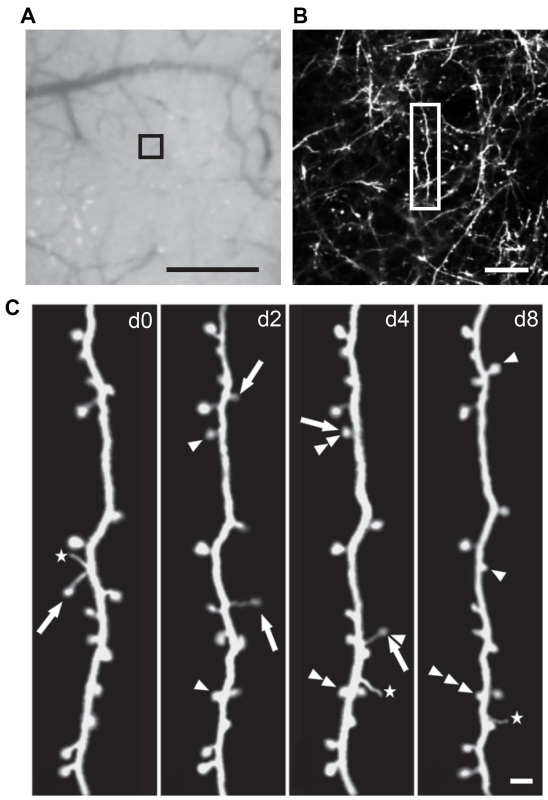 Figure 2. Transcranial two-photon imaging through a thinned-skull preparation in the mouse motor cortex,showing dynamics of dendritic spines over eight days. (A) A CCD image of the vasculature pattern with the thinned skull area (Map 1). The black box indicates the region where two-photon in vivo images were acquired. (B) A low-magnification maximum projection of dendritic branches in the motor cortex of a 1 month old mouse (Map 2). (C) Repetitive images of the same dendritic segment reveal newly formed spines (arrowheads), eliminated spines (arrows), and filopodia (stars) on day 0, 2, 4, and 8. The left panel is a higher-magnification view of the dendritic segment shown in the boxed region in (B). Scale bars: 500 μm (A), 20 μm (B), and 2 μm (C).
Figure 2. Transcranial two-photon imaging through a thinned-skull preparation in the mouse motor cortex,showing dynamics of dendritic spines over eight days. (A) A CCD image of the vasculature pattern with the thinned skull area (Map 1). The black box indicates the region where two-photon in vivo images were acquired. (B) A low-magnification maximum projection of dendritic branches in the motor cortex of a 1 month old mouse (Map 2). (C) Repetitive images of the same dendritic segment reveal newly formed spines (arrowheads), eliminated spines (arrows), and filopodia (stars) on day 0, 2, 4, and 8. The left panel is a higher-magnification view of the dendritic segment shown in the boxed region in (B). Scale bars: 500 μm (A), 20 μm (B), and 2 μm (C).
Discussion
To obtain a successful thinned-skull preparation, several steps in this protocol are crucial. 1) The thickness of the skull. The cranial bone has a sandwich structure, with two layers of high-density compact bone and a middle layer of low-density spongy bone. While the high-speed micro drill is suitable for removing the outer layers of compact bone and spongy bone, the microsurgical blade is ideal for thinning the inner layer of compact bone. As the thickness and stiffness of the skull increases during development, imaging of adult mice requires more bone to be removed in order to obtain images of good quality. The thinned area exhibits a transparent solid appearance and provides good imaging quality when the thickness of the skull is approximately 20-30 μm. The skull thickness should be checked periodically during the thinning process as over-thinning makes the thinning process in subsequent reimaging sessions difficult. 2) The size of the thinned region. It is important to establish a stable thinned-skull configuration, where a cone-like cavity is achieved. The architecture of the thinned region with a proper size prevents causing damage to the cortex and helps thinning in subsequent re-imaging sessions. It is recommended to make the bottom area (approximately 200-300 μm in diameter) of the thinned region smaller than the top opening (approximately 0.5-1.0 mm in diameter). 3) The stability of the images. A well-immobilized preparation helps to improve the image quality by reducing respiratory-induced movement artifacts. It is important to keep the skull dry and devoid of connective tissue and bone debris before the head plate is attached on to the skull. Extra glue can also be applied to fill the remaining gaps between the head plate and the skull. In addition, targeting an area away from big vasculature also minimizes the movements caused by blood pumping.
Investigation of the changes in the brain of living animals with high spatial and temporal resolution using two-photon microscopy requires generation of an optical window. In addition to thinned-skull preparation, fluorescently labeled structures can also be visualized by removing the skull and replacing it with a cover glass (usually 3-5 mm in diameter) that provides a clear imaging window10,13. While both thinned-skull and open-skull imaging protocols are applied broadly for in vivo imaging studies, each method has its own advantages and is suitable for different experimental designs. For instance, the open-skull preparation is useful for studies that investigate relatively large brain areas (e.g., 5 mm diameter) and require many imaging sessions with short intervals (i.e., days). Once the cranial window is successfully implanted, no additional surgery is required. However, a post-surgery period (usually approximately 1 month) is required before the first imaging session to ameliorate potential inflammatory responses caused by surgery. Re-imaging becomes impossible when the cranial window is blocked by regrowth of the bone and/or thickening of the meninges. In contrast, animals with a thinned-skull preparation can be imaged immediately after the initial surgery, making it more suitable for chronic imaging of younger animals (e.g., 2 weeks old). It is suitable for investigation with long imaging intervals (i.e. months to years), as the bone regrowth and dura thickening is not problematic. However, re-thinning of the skull is usually required for subsequent re-imaging sessions (even with 2 day intervals) due to the bone regrowth. Moreover, the newly grown bone layer consists of a less condensed structure compared to the original compact bone, greatly reducing the transparency of the thinned-skull region. Therefore, to acquire the same quality of images, it is necessary to completely remove the newly grown bone layer. And such attempts usually lead to the final thickness of the skull in the subsequent imaging thinner than the previous preparation. Since the skull thickness had been prepared to 20-30 μm in the initial imaging session, leaving limited room for re-thinning, the total imaging times of thinned-skull preparation is usually less than 5 times. Recently, a new experimental approach named polished and reinforced thinned-skull (PoRTS) has been developed to combine both thinned- and open-skull preparations11,26,27.
So far, the majority of in vivo imaging in the cortex has been performed using transgenic mouse lines expressing EGFP or YFP under neuronal specific thy1 promoter. These transgenic mice express fluorescent proteins sparsely in a subset, but mixed population of cortical neurons25. Since many lines of evidence suggest that experience-dependent structural plasticity happens in selective cell types of well-defined circuits, it is important to label and image in a cell-type specific manner. To date, many approaches have been applied to achieve this goal. For example, pyramidal neurons in different cortical layers can be targeted by performing in utero electroporation at defined embryonic stages28,29. Similarly, injection of viral vectors engineered to express a fluorescent protein can also be used to label cells in different brain regions30. In addition, many lines of transgenic mice have been generated to drive the expression of Cre recombinase under cell type specific promoters31,32. Injection of viral vectors such as adeno-associated virus carrying fluorescent reporter gene following a floxed stop codon into these mice offers the possibility to target a specified class of neurons in a restricted region33. By combining these new labeling approaches with our thinned-skull preparation, changes in synaptic connections of cortical neurons can be investigated by two-photon in vivo imaging in a cell type- and/or circuit-specific manner.
With the increasing availability of transgenic animals with fluorescently labeled cell populations and rapid development of in vivo labeling techniques, similar procedures described here can also be applied to investigate other cell types (glial cells) and vasculature in the living brain. Combined with behavioral manipulations and disease models, two-photon in vivo imaging will greatly expand our understandings of the molecular, cellular, and circuit mechanisms underlying brain functions.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
We thank James Perna for the graphic illustration. This work was supported by grants from the National Institute of Mental Health to Y.Z.
References
- Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nature reviews. Neuroscience. 2009;10:647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- Fu M, Zuo Y. Experience-dependent structural plasticity in the cortex. Trends in neurosciences. 2011;34:177–187. doi: 10.1016/j.tins.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Zuo Y. Spine plasticity in the motor cortex. Current opinion in neurobiology. 2011;21:169–174. doi: 10.1016/j.conb.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms KJ, Dunaevsky A. Dendritic spine plasticity: looking beyond development. Brain research. 2007;1184:65–71. doi: 10.1016/j.brainres.2006.02.094. [DOI] [PubMed] [Google Scholar]
- Segal M. Dendritic spines and long-term plasticity. Nature reviews. Neuroscience. 2005;6:277–284. doi: 10.1038/nrn1649. [DOI] [PubMed] [Google Scholar]
- Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Current opinion in neurobiology. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Alvarez VA, Sabatini BL. Anatomical and physiological plasticity of dendritic spines. Annual review of neuroscience. 2007;30:79–97. doi: 10.1146/annurev.neuro.30.051606.094222. [DOI] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Yang G, Pan F, Parkhurst CN, Grutzendler J, Gan WB. Thinned-skull cranial window technique for long-term imaging of the cortex in live mice. Nature protocols. 2010;5:201–208. doi: 10.1038/nprot.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nature protocols. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew PJ, et al. Chronic optical access through a polished and reinforced thinned skull. Nature methods. 2010;7:981–984. doi: 10.1038/nmeth.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szu JI, et al. Thinned-skull cortical window technique for in vivo optical coherence tomography imaging. J Vis Exp. 2012. [DOI] [PMC free article] [PubMed]
- Mostany R, Portera-Cailliau C. A craniotomy surgery procedure for chronic brain imaging. J Vis Exp. 2008. [DOI] [PMC free article] [PubMed]
- Xu T, et al. Rapid formation and selective stabilization of synapses for enduring motor memories. Nature. 2009;462:915–919. doi: 10.1038/nature08389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Yu X, Lu J, Zuo Y. Repetitive motor learning induces coordinated formation of clustered dendritic spines in vivo. Nature. 2012;483:92–95. doi: 10.1038/nature10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos D, et al. ATP mediates rapid microglial response to local brain injury in vivo. Nature neuroscience. 2005;8:752–758. doi: 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- Tsai J, Grutzendler J, Duff K, Gan WB. Fibrillar amyloid deposition leads to local synaptic abnormalities and breakage of neuronal branches. Nature neuroscience. 2004;7:1181–1183. doi: 10.1038/nn1335. [DOI] [PubMed] [Google Scholar]
- Pan F, Aldridge GM, Greenough WT, Gan WB. Dendritic spine instability and insensitivity to modulation by sensory experience in a mouse model of fragile X syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17768–17773. doi: 10.1073/pnas.1012496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Condello C, Schain A, Harb R, Grutzendler J. CX3CR1 in microglia regulates brain amyloid deposition through selective protofibrillar amyloid-beta phagocytosis. J Neurosci. 2010;30:17091–17101. doi: 10.1523/JNEUROSCI.4403-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay ME, Zettel ML, Ison JR, Allen PD, Majewska AK. Effects of aging and sensory loss on glial cells in mouse visual and auditory cortices. Glia. 2012;60:541–558. doi: 10.1002/glia.22287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam CK, Yoo T, Hiner B, Liu Z, Grutzendler J. Embolus extravasation is an alternative mechanism for cerebral microvascular recanalization. Nature. 2010;465:478–482. doi: 10.1038/nature09001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly EA, Majewska AK. Chronic imaging of mouse visual cortex using a thinned-skull preparation. J Vis Exp. 2010. [DOI] [PMC free article] [PubMed]
- Marker DF, Tremblay ME, Lu SM, Majewska AK, Gelbard HA. A thin-skull window technique for chronic two-photon in vivo imaging of murine microglia in models of neuroinflammation. J Vis Exp. 2010. [DOI] [PMC free article] [PubMed]
- Svoboda K, Yasuda R. Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron. 2006;50:823–839. doi: 10.1016/j.neuron.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Feng G, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Shih AY, Mateo C, Drew PJ, Tsai PS, Kleinfeld D. A polished and reinforced thinned-skull window for long-term imaging of the mouse brain. J Vis Exp. 2012. [DOI] [PMC free article] [PubMed]
- Zhang L, et al. Imaging glioma initiation in vivo through a polished and reinforced thin-skull cranial window. J Vis Exp. 2012. [DOI] [PMC free article] [PubMed]
- Pacary E, et al. Visualization and genetic manipulation of dendrites and spines in the mouse cerebral cortex and hippocampus using in utero electroporation. J Vis Exp. 2012. [DOI] [PMC free article] [PubMed]
- Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Developmental biology. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- Lowery RL, Majewska AK. Intracranial injection of adeno-associated viral vectors. J Vis Exp. 2010. [DOI] [PMC free article] [PubMed]
- Taniguchi H, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zariwala HA, et al. A Cre-dependent GCaMP3 reporter mouse for neuronal imaging in vivo. J Neurosci. 2012;32:3131–3141. doi: 10.1523/JNEUROSCI.4469-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman SJ, Huang ZJ. High-resolution labeling and functional manipulation of specific neuron types in mouse brain by Cre-activated viral gene expression. PloS one. 2008;3 doi: 10.1371/journal.pone.0002005. [DOI] [PMC free article] [PubMed] [Google Scholar]


