Abstract
STAT3 can be transcriptionally activated by phosphorylation of its tyrosine 705 or serine 727 residue. In mouse embryonic stem cells (mESCs), leukemia inhibitory factor (LIF) signaling maintains pluripotency by inducing JAK-mediated phosphorylation of STAT3 Y705 (pY705). However, the function of phosphorylated S727 (pS727) in mESCs remains unclear. In this study, we examined the roles of STAT3 pY705 and pS727 in regulating mESC identities, using a small molecule-based system to post-translationally modulate the quantity of transgenic STAT3 in STAT3−/− mESCs. We demonstrated that pY705 is absolutely required for STAT3-mediated mESC self-renewal, while pS727 is dispensable, serving only to promote proliferation and optimal pluripotency. S727 phosphorylation is regulated directly by fibroblast growth factor/Erk signaling and crucial in the transition of mESCs from pluripotency to neuronal commitment. Loss of S727 phosphorylation resulted in significantly reduced neuronal differentiation potential, which could be recovered by a S727 phosphorylation mimic. Moreover, loss of pS727 sufficed LIF to reprogram epiblast stem cells to naïve pluripotency, suggesting a dynamic equilibrium of STAT3 pY705 and pS727 in the control of mESC fate.
Keywords: Embryonic stem cell, STAT3, Phosphorylation, Self-renewal, Differentiation
Introduction
The signal transducer and activator of transcription 3 (STAT3) functions in various cytokine-driven signaling pathways [1]. Cyto-plasmic STAT3 monomers dimerize when phosphorylated at tyrosine 705 (Y705) by Janus kinases (JAKs) associated with cytokine-stimulated receptors, and then translocate to the nucleus, where the homodimers activate target gene transcription. STAT3 can also be phosphorylated at serine 727 (S727) by members of the mitogen-activated protein kinases (MAPK) and c-Jun N-terminal kinase families [2]. While phosphorylated Y705 (pY705) is generally believed to be essential for STAT3’s transcriptional activity; the function of phosphorylated S727 (pS727) remains controversial, as this modification has been reported to have both down- and upregulatory effects on STAT3’s transcriptional activity [3-6].
In general, STAT3 activation is associated with proliferation, inhibition of apoptosis and cellular transformation [7, 8]. However, activation of STAT3 also suppresses tumor growth [9] and induces differentiation and apoptosis in some contexts [10-12]. The mechanisms underlying STAT3’s diverse and sometimes opposing roles are still largely unknown. It is assumed that STAT3 recruits specific coactivators and activates distinct gene expression programs based on the genetic background, type and developmental stage of the cell [13]. This hypothesis raises an interesting issue of whether STAT3 pY705 and pS727 play a role in this process, considering their significance in STAT3-mediated control of gene transcription.
STAT3 plays an indispensable role in leukemia inhibitory factor (LIF)-mediated mouse embryonic stem cell (mESC) self-renewal. LIF treatment results in the activation of the JAK/STAT3 pathway and subsequently the expression of pluripotency genes [14, 15]. Recently, we developed a new culture medium that sustains mESC self-renewal by circumventing the otherwise obligatory LIF/STAT3 pathway [16]. This medium contains N2B27 and two inhibitory small molecules (2i): PD0325901 (PD) and CHIR99021 (CHIR), which specifically block MAPK and glycogen synthase kinase 3 (GSK3) pathways, respectively. Using the N2B27 + 2i system, we were able to establish a STAT3−/− mESC line [16, 17], which allows fine manipulation of STAT3 signaling and therefore provides a unique and powerful tool for investigating how STAT3 functions in mESCs.
Phosphorylation of Y705 is believed to be the key event in the transcriptional activation of STAT3. Phosphorylation of S727, however, has not been functionally linked to STAT3-mediated mESC fate determination. In this study, we sought to define the exact role of STAT3 phosphorylation in mESC self-renewal and differentiation. By studying the function of various STAT3 mutants in STAT3−/− mESCs, we demonstrated unambiguously that Y705 and S727 differentially regulate mESC fates in a phosphorylation-dependent manner.
Materials and Methods
mESC Culture
46C ESCs, R1 ESCs, and STAT3−/− mESCs were cultured on plates precoated with 0.1% gelatin or mitotically inactive mouse embryonic fibroblasts. 46C ESCs and R1 ESCs were grown in Glasgow minimum essential medium (Sigma, St. Louis, MO, www.sigmaaldrich.com) supplemented with 10% fetal bovine serum (Hyclone, Waltham, MA, http://www.thermoscientific.com/hyclone), 2 mM L-glutamine (Gibco, Carlsbad, CA, http://www.lifetechnologies.com/gibco), 0.1 mM nonessential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), 0.1 mM β-mercaptoethanol (a formulation hereinafter referred to as “mESC medium”), and 1,000 units/ml murine LIF (Stemgent, Cambridge, MA, www.stemgent.com). STAT3−/− mESCs were grown in N2B27 medium supplemented with 3 μM CHIR99021 and 1 μM PD0325901 as previously described [18]. CHIR99021 and PD0325901 were synthesized in the Division of Signal Transduction Therapy, University of Dundee, UK. C-myc inhibitor 10058-F4 was purchased from EMD Millipore (Billerica, MA, www.millipore.com) and used at concentration of 50 μM.
Transgenesis of STAT3 Mutant Cell Lines
Various STAT3 mutant genes were generated using the Quik-Change Site-Directed Mutagenesis Kit according to manufacturer’s instructions (Stratagene, Santa Clara, CA, http://www.genomics.agilent.com) and fused with the destabilizing domain (DD) at the STAT3 N-terminus. The DD-STAT3 mutants were then cloned into a CAG-ires-puro vector between the XhoI and NotI sites (Supporting Information Fig. S1C). Plasmids were electroporated into STAT3−/− mESCs. Transfected STAT3−/− mESCs were plated onto drug-resistant DR4 feeder cells [19], and allowed to propagate for 2 days before puromycin (1 μg/ml, Invitrogen, Carlsbad, CA, http://www.lifetechnologies.com/invitrogen) selection was initiated. Positive clones were expanded and transgene expression was confirmed by western blotting. Mutant cell lines were routinely maintained in N2B27 + 2i medium. For c-myc over-expression, the coding region of c-myc was cloned and inserted into the PiggyBac expression vector, which was later transfected together with transposase vector into STAT3−/− + DD-STAT3-S727A cells.
Protein and RNA Analysis
For protein analysis, cell lysis buffer was prepared by adding 50 μl of β-mercaptoethanol to 950 μl of Laemmli Sample Buffer (BioRad, Hercules, CA, www.bio-rad.com) and 1,000 μl of water. Cells were washed with phosphate-buffered saline and lysed in the culture wells. Samples were collected and heated to 95 °C before being subjected to electrophoresis on BioRad Tris-HCl precast gels. Electrotransfer to PVDF membranes was performed and blots were probed with primary antibodies overnight at 4 °C, and then for 1 hour at room temperature with appropriate secondary antibodies. Pierce ECL Western Blotting Substrate (Rockford, IL, www.piercenet.com) was used to detect the signal. The primary antibodies used include STAT3 (1:2,000, BD Biosciences, San Jose, CA, http://www.bdbiosciences.com), P-STAT3-Y705 (1:2,000, Cell Signaling, Danvers, MA, http://www.cellsignal.com/), P-STAT3-S727 (1:2,000, BD Biosciences), c-myc (1:500, Calbiochem, Billerica, MA, www.millipore.com), α-Tubulin (1:4,000, Invitrogen), Oct4 (1:3,000, Santa Cruz, Dallas, TX, http://www.scbt.com/), Sox2 (1:1,000, Santa Cruz), Klf4 (1:1,000, R&D Systems, Minneapolis, MN, http://www.rndsystems.com/), and Nanog (1:1,000, R&D Systems). For RNA analysis, total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA, www.qiagen.com), and 1 μg of the corresponding RNA yield was used to synthesize cDNA (Quanti-Tect Transcription Kit, Qiagen). The quantitative real-time PCR (RT-PCR) mixtures were prepared using SYBR Green PCR Master Mix and run on an ABI7900HT Fast RT-PCR System (Applied Biosystems, Carlsbad, CA, http://www.lifetechnologies.com/applied-biosystems). RT-PCR was carried out using the Paq5000 DNA polymerase (Stratagene). Relative expression levels of pertinent genes were analyzed and normalized against gapdh.
Self-renewal Assays
To determine the self-renewal potential of STAT3 mutant cell lines, we plated cells in 12-well plates at a density of 1,000 cells per well and cultured them in N2B27 + 2i medium overnight. Different culture conditions were applied the next day. After 7 days, cells were washed, fixed and stained for alkaline phosphatase (AP) activity in accordance with manufacturer’s instructions (Sigma). Each treatment was performed in triplicate. For long-term tests of self-renewal, cells were plated at a density of 1.2 × 104 cells per square centimeter in 12-well plates and cultured for at least six passages before morphological observation.
Neural Differentiation and Immunostaining
To induce neural differentiation, we trypsinized, collected by centrifugation, and resuspended 3 × 106 to 5 × 106 ESCs in 10 ml of mESC medium with 1 μM of Shield 1 (S1, Clontech, Mountain View, CA, www.clontech.com). This single-cell suspension was deposited onto a 10 cm nonadhesive bacteriological petridish. After 2 days in this condition, cells aggregated in suspension and formed embryoid bodies (EBs). Medium was changed every other day and retinoic acid (RA, sigma) was added to a final concentration of 1 μM for days 4–8. On day 9, EBs were collected and trypsinized into a single-cell suspension, which was plated into two wells of a BD Matrigel Basement Membrane Matrix (BD Biosciences)-coated 12-well plate and cultured in the mESC1S1 condition. Immunostaining was performed according to a standard protocol, using primary antibodies including Nestin (1:200, Santa Cruz) and Tuj1 (1:1000, Covance, Princeton, NJ, www.covance.com). Alexa Flour fluorescent secondary antibodies (Invitrogen) were used at a dilution of 1:2,000 and nuclei were stained with DAPI. For RT-PCR analysis, STAT3−/− + DD-STAT3-WT and STAT3−/− + DD-STAT3-S727A cells were collected 0, 2, 4, 6, 8, 10, and 12 days after the onset of differentiation, for RNA isolation.
Cell Proliferation Assay
Cells were plated on feeders in 12-well plates at a starting density of 2 × 104 cells per well in mESC medium supplemented with 1,000 U/ml LIF and 1 μM of S1. After 24, 48, 72, or 96 hours, cells were harvested and counted under a microscope. The experiment was performed in quadruplicate and paired student’s t-tests were used for statistical analysis. Doubling times were calculated by the software available from www.doubling-time.com.
Kinase Inhibitor Assay
Cells were plated in 12-well plates at a density of 100,000 cells per well and starved in basal medium with S1 overnight. On the next morning, cells were pre-treated by different kinase inhibitors for 2 hours, and subsequently stimulated with serum, LIF or serum+LIF for 1 hour. Cell lysates were then collected for western blot analysis. Inhibitors used in the experiment include JNK inhibitor SP600125 (50 μM, Sigma), Erk 1/2 inhibitor PD0325901 (1 μM), mTOR inhibitor rapamycin (100 nM, Calbiochem), CDK inhibitor olomoucine (50 μM, Sigma), PI3K inhibitor LY294002 (50 μM, Calbiochem), PKC inhibitor Go6983 (1 μM, Calbiochem) and JAK inhibitor 1 (5 μM, Calbiochem).
Results
Establishment of an Inducible STAT3 Expression System in STAT3−/− mESCs
To evaluate the role of STAT3 Y705 and S727 phosphorylation in regulating mESC self-renewal, we introduced full-length STAT3 (STAT3-WT), STAT3β (an isoform lacking 55 amino acid residues in the C-terminal domain), STAT3-Y705F (a mutant in which tyrosine 705 is replaced by phenylalanine), and STAT3-S727A (a mutant in which serine 727 is replaced by alanine) transgenes into STAT3−/− mESCs. Initially, we used the CAG promoter to drive constitutive expression of the transgenes. However, the majority of STAT3−/− mESCs transfected with STAT3-Y705F vector died soon after the first passage, and the surviving colonies expressed the transgene only weakly (Supporting Information Fig. S1A), suggesting that STAT3-Y705F has a cytotoxic effect in mESCs. In addition, we found that the expression level of STAT3-WT in STAT3−/− mESCs greatly exceeded the endogenous level of STAT3 in wild-type 46C ESCs (Supporting Information Fig. S1B) and that STAT3-WT-expressing STAT3−/− mESCs were able to self-renew in the absence of LIF for several passages. These observations led us to surmise that the phenotypes exhibited by these cell lines might lack physiological relevance. Therefore, we sought to control the expression of the STAT3 mutants to obtain more meaningful data regarding their functions in ESCs.
We next modified the sequences encoding the various STAT3 transgenes (WT, Y705F, and S727A) to include an N-terminal destabilizing domain (DD) and introduced those into STAT3−/− mESCs (Fig. 1A; Supporting Information Fig. S1C). When expressed in cells, the DD-STAT3 fusion protein was rapidly and constitutively degraded in a proteasome-dependent fashion. Shield1 (S1), a stabilizing ligand, can bind specifically to DD and protect DD-tagged proteins from degradation [20]. This fast-acting regulation allows DD-tagged protein to accumulate within a matter of hours following the addition of S1 and does not disturb gene transcriptional control. Using this system, we established STAT3−/− + DD-STAT3-Y705F mESC lines that could be maintained in the 2i condition as robustly as other STAT3 transgenic lines (Fig. 1B). Expression levels were even among different STAT3 mutants, and responded to S1 treatment dose-dependently (Fig. 1C). Immunofluorescence staining confirmed a stable level of DD-STAT3-Y705F expression in the presence of S1; in its absence, no DD-STAT3-Y705F leak expression was apparent (Supporting Information Fig. S1D). DD-STAT3-WT was phosphorylated at both S727 and Y705 in LIF-stimulated STAT3−/− 1 DD-STAT3-WT mESCs, whereas DD-STAT3-Y705F and DD-STAT3-S727A were phosphorylated only at S727 and Y705 sites, respectively, in cells expressing either of the corresponding transgenes, confirming site-specific loss of phosphorylation potential (Supporting Information Fig. S1E).
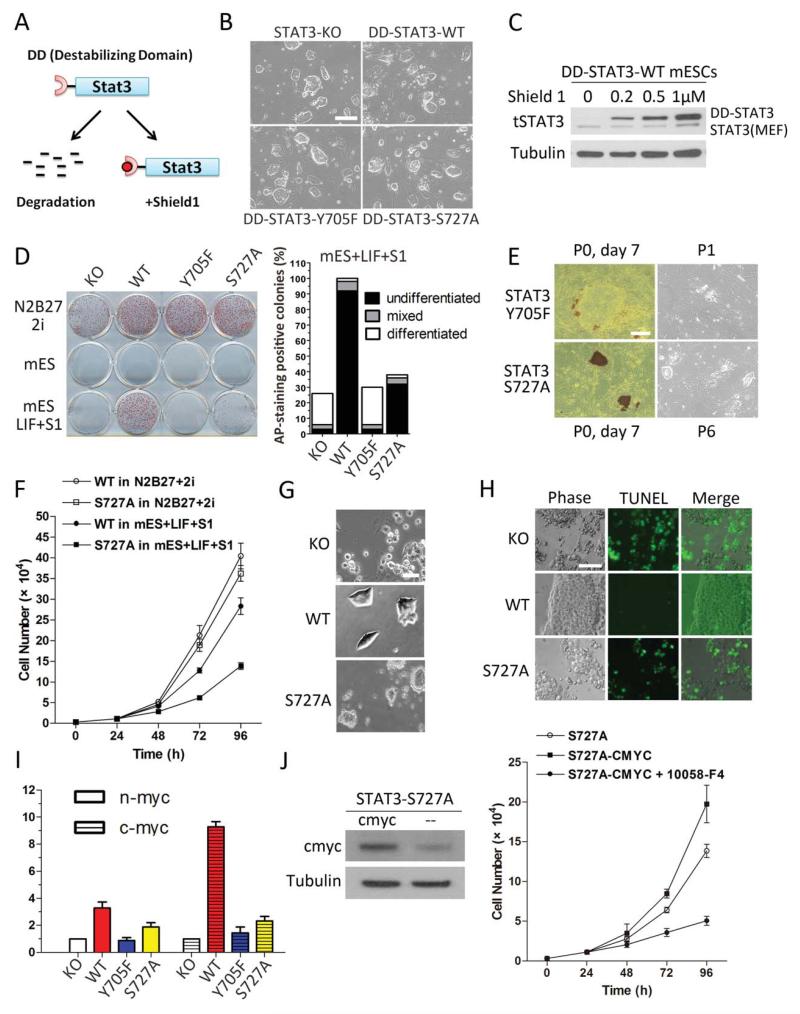
Figure 1.
Diverse functions of STAT3 at different phosphorylation sites revealed using transgenic STAT3 in STAT3−/− mouse embryonic stem cells (mESCs). (A): Schematic diagram showing the principle of destabilizing domain (DD)-STAT3 expression system. (B): STAT3−/− + DD-STAT3 transgenic mESCs in N2B27 + 2i medium display typical mESC morphology. (C): Dose-dependent modulation of DD-STAT3 protein level by Shield 1 (S1). Cells were cultured in N2B27 + 2i medium and treated with various concentrations of S1 overnight. DD-STAT3-WT expression in STAT3−/− mESCs is depicted as an example. The two bands in the STAT3 blot represent STAT3 from mouse embryonic fibroblast feeder cells (lower band) and DD-STAT3 fusion protein (upper band). (D): Comparison of self-renewal potential of different STAT3−/− + DD-STAT3 cells under various culture conditions: N2B27 + 2i, mESC, and mESC+LIF+S1. We counted the numbers of differentiated, undifferentiated and mixed colonies these mESCs formed after 7 days in the mESC+LIF+S1 culture condition. (E): Alkaline phosphatase staining and phase contrast images showing the short and long-term self-renewal potential of STAT3−/− + DD-STAT3-Y705F and STAT3−/− + DD-STAT3-S727A mESCs under the mESC+LIF+S1 condition. (F): Growth curves of STAT3−/− + DD-STAT3-WT and STAT3−/− + DD-STAT3-S727A mESCs in mESC+LIF+S1. Their growth curves in N2B27 + 2i are used as positive controls. (G): Representative images showing different STAT3−/− + DD-STAT3 mESCs plated at a density of 1 × 104 cells per square centimeter on gelatin-coated plates and cultured in mESC+LIF+S1 overnight. (H): TUNEL-FITC staining of STAT3−/− + DD-STAT3 mESCs in mESC+LIF+S1. (I): Myc transcription level in STAT3−/− + DD-STAT3 mESCs in mESC+S1 after 1 hour of leukemia inhibitory factor (LIF) stimulation. (J): Improved proliferation of STAT3−/− + DD-STAT3-S727A mESCs with forced expression of c-myc: left: western blot showing overexpression of c-myc; right: growth curve of cells in mESC+LIF+S1 condition, 10058-F4: c-myc inhibitor (50 μM) (B, E, G, H) Scale bars = 50 μm. Abbreviations: DD, destabilizing domain; KO, knock out; LIF, leukemia inhibitory factor; mESC, mouse embryonic stem cell; WT, wild type.
STAT3 Functions Diversely in mESCs via Different Phosphorylation Sites
To investigate the roles of STAT3 S727 and Y705 phosphorylation in mESC fate regulation, we performed functional rescue assays. Immediately after overnight culture in the N2B27 + 2i condition, the DD-transgenic STAT3−/− mESCs were switched to conventional mESC medium supplemented with LIF and S1. As expected, STAT3−/− mESCs started to differentiate 3-4 days after the switch to LIF+S1, and gradually died out beginning on day 6 (Supporting Information Fig. S2A). In contrast, STAT3−/− + DD-STAT3-WT and STAT3−/− + DD-STAT3-S727A mESCs formed colonies that tested positive for AP activity when stained 7 days after the switch to LIF+S1 (Fig. 1D). STAT3−/− + DD-STAT3-Y705F mESCs failed to form AP-positive colonies in LIF+S1 (Fig. 1D), consistent with previous reports that pY705 is indispensable for STAT3-mediated self-renewal in mESCs [14]. Highly differentiated, morphologically flat colonies consisting of squamous epithelial-type cells predominated among STAT3−/− + DD-STAT3-Y705F mESCs. These cells could not be expanded beyond one passage in the mESC+LIF+S1 condition (Fig. 1E).
A considerable proportion of the STAT3−/− + DD-STAT3-S727A mESCs died within 2 days of being switched to the LIF1S1 condition. However, surviving colonies were uniformly positive for AP activity, and could be maintained continuously while retaining an undifferentiated morphology resembling that of the STAT3−/− + DD-STAT3-WT colonies (Fig. 1D, 1E). Previous reports have shown that STAT3 S727 phosphorylation is associated with cell survival and mitogenicity in various tumor and primary cells [21-23]. Consistent with this, we observed that STAT3−/− + DD-STAT3-S727A mESCs in LIF+S1 proliferated more slowly than did STAT3−/− + DD-STAT3-WT mESCs, with doubling times of 19.71 and 16.87 hours, respectively (Fig. 1F). During passaging, STAT3−/− + DD-STAT3-S727A mESCs underwent extensive cell death. In a substrate attachment assay, both STAT3−/− and STAT3−/− + DD-STAT3-S727A mESCs adhered poorly to gelatin and gave strong apoptotic signals as detected by TUNEL-FITC staining (Fig. 1G, 1H), indicating that reduced cell adhesion might be the reason for STAT3−/− + DD-STAT3-S727A mESC death during passaging in LIF+S1. STAT3−/− + DD-STAT3-WT mESCs attached strongly to gelatin substrate and formed compact colonies, abilities that are hallmarks of the wild-type phenotype and which indicate that the DD-STAT3-WT transgene efficiently rescued the deficiency of the STAT3−/− mESCs. Together, these results support the notion that pS727 plays an important role in mESC survival.
Myc is a key transcription factor involved in cell proliferation, cell growth and apoptosis. In mESCs, myc serves as a downstream target of LIF/ STAT3 signaling to promote pluripotency [24, 25], but does so mostly by stimulating proliferation in a manner independent of the core pluripotency factor network [26]. The impaired proliferation yet efficient self-renewal exhibited by STAT3−/− + DD-STAT3-S727A mESCs prompted us to investigate whether differential expression of myc plays a role in the associated phenotype. Following LIF stimulation of STAT3−/− + DD-STAT3-WT mESCs, c-myc transcripts were increased 10-fold and n-myc transcripts, 3.5-fold (Fig. 1I). In contrast, no changes in c-myc or n-myc expression were detected in STAT3−/− or STAT3−/− + DD-STAT3-Y705F mESCs. C-myc and n-myc expression in STAT3−/− + DD-STAT3-S727A mESCs increased only slightly, suggesting compromised myc transactivation. Consistently, we observed that forced expression of c-myc in STAT3−/− + DD-STAT3-S727A mESCs led to increased proliferation rate, which could be abrogated by treatment with c-myc inhibitor 10058-F4 (Fig. 1J).
pY705, But Not pS727, Is Absolutely Required for STAT3-Mediated mESC Self-Renewal
We analyzed the expression of core pluripotency factors in different transgenic STAT3−/− mESCs to further clarify the minimal requirement for self-renewal. Oct4 expression was the same among the different transgenic cells cultured in LIF+S1, yet Sox2 and Nanog expression was greatly suppressed in STAT3−/− + DD-STAT3-Y705F mESCs (Fig. 2A), which also exhibited significant downregulation of Klf4, a direct STAT3 downstream target. STAT3−/− + DD-STAT3-S727A mESCs showed expression of all core pluripotency factors, but to a much lesser degree than did the STAT3−/− + DD-STAT3-WT mESCs. The low transcription rate of known STAT3 downstream target genes (socs3, jun, gbx2, fos, klf5, and pim3) in STAT3−/− + DD-STAT3-S727A cells also indicates that this STAT3 mutant possesses impaired transcriptional activity (Fig. 2B). These data are consistent with the results of the self-renewal assays (Fig. 1D, 1E) and suggest that pS727 confers optimal mESC self-renewal by enhancing STAT3 transcriptional activity, but is not essential for minimal pluripotency.
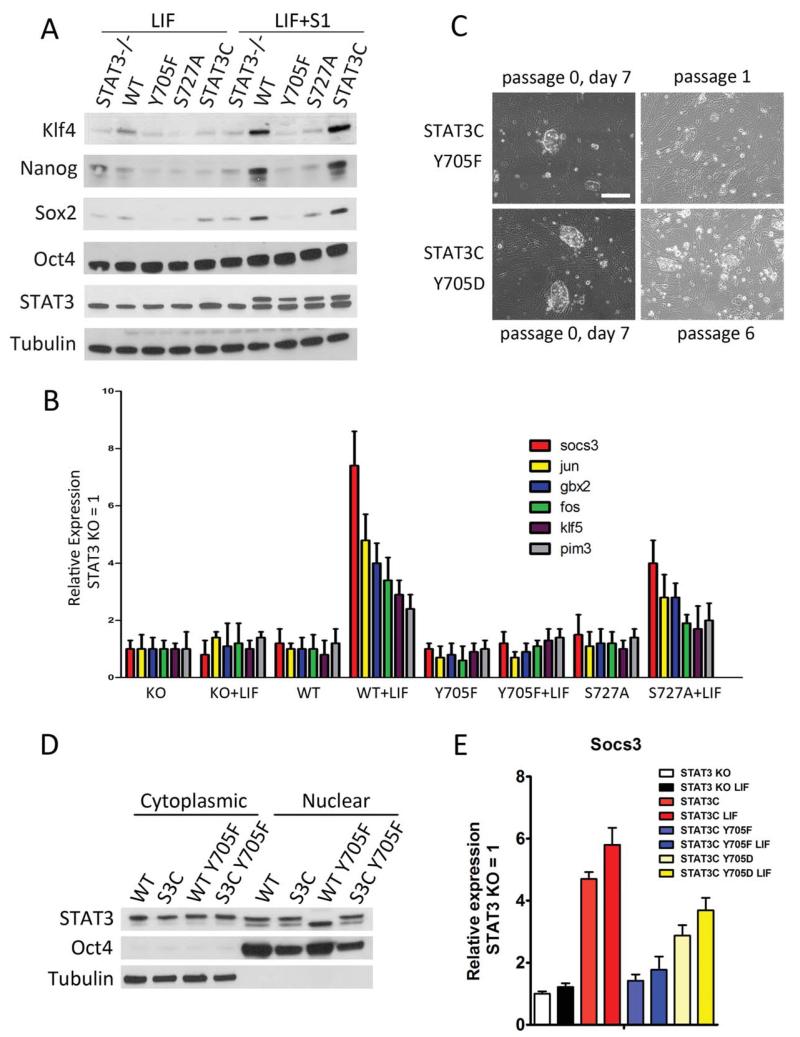
Figure 2.
pY705, but not pS727, is absolutely required for STAT3-mediated mouse embryonic stem cell (mESC) self-renewal. (A): Western blot analysis showing differential expression of core pluripotency factors among STAT3−/− mESCs expressing the indicated versions of destabilizing domain (DD)-STAT3. (B): Differential transcription of STAT3 target genes (socs3, jun, gbx2, fos, klf5, and pim3) among STAT3−/− mESCs expressing the indicated versions of DD-STAT3 after 1 hour of leukemia inhibitory factor (LIF) stimulation. (C): Phase contrast images showing the short and long-term self-renewal potential of STAT3−/− + STAT3C-Y705F and STAT3−/− + STAT3C-Y705D mESCs in mESC+LIF+S1. Scale bar = 50 μm (D). Western blot analysis indicating the presence of STAT3C-Y705F protein in the nucleus following LIF stimulation. (E): Quantitative real-time polymerase chain reaction analysis of socs3 expression in STAT3−/− + DD-STAT3 mESCs after 1 hour of LIF stimulation. Abbreviations: KO, knock out; LIF, leukemia inhibitory factor; WT, wild type.
Because pY705 mediates both the dimerization and transcriptional activation of STAT3, we investigated its necessity in each event. Introduction of two cysteines at residues 662 and 664 within the C-terminal loop of the SH2 domain in STAT3 renders it constitutively transcriptionally active (STAT3C) [27]. We introduced a DD-STAT3C transgene into STAT3−/− mESCs and examined the resultant phenotype. In the presence of S1, STAT3−/− + DD-STAT3C cells remained in an undifferentiated state over many passages even in the absence of LIF, and exhibited strong activation of all pluripotency genes (Fig. 2A; Supporting Information Fig. S2B). We found that STAT3−/− + DD-STAT3C-Y705F mESCs could be maintained initially in the presence of S1, but died soon after being passaged, indicating a defect in self-renewal capacity (Fig. 2C). On the contrary, a transgene encoding a phosphomimetic mutation (Y705D) supported self-renewal in STAT3−/− mESCs (Fig. 2C). To examine the subcellular localization of DD-STAT3C-Y705F, we performed nuclear fractionation and found that DD-STAT3C-Y705F, unlike DD-STAT3-Y705F, accumulated in the nucleus in the LIF+S1 condition (Fig. 2D). However, it failed to activate gene transcription, as shown by the quantity of Socs3 transcripts (Fig. 2E). These results establish that STAT3 pY705 mediates target gene transcription and is absolutely required for STAT3-mediated mESC self-renewal.
STAT3 S727 Phosphorylation Promotes Neuronal Differentiation of mESCs
In mESCs, LIF/STAT3 signaling maintains long-term self-renewal by suppressing spontaneous differentiation. STAT3 also plays important roles in neurogenesis [28], neural stem cell fate determination [29], and neural regeneration after injury [30]. We thus examined the ability of the transgenic STAT3−/− mESCs to undergo differentiation by method of embryoid body (EB) formation. STAT3−/− and STAT3−/− + DD-STAT3-Y705F mESCs produced massive numbers of neural precursor cells and efficiently differentiated into neurons afterwards. STAT3−/− + DD-STAT3-WT mESCs initially showed a delay in neural progenitor production, but by day 18 had given rise to about the same number of neurons that STAT3−/− mESCs had by this day (Fig. 3A). These observations establish that loss of STAT3 pY705 results in early exit from the pluripotent state but appears to exert no effect on neuronal fate determination. Strikingly, STAT3−/− + DD-STAT3-S727A mESCs gave rise to far fewer neural progenitor cells than did the other transgenic STAT3−/− mESC lines. These STAT3−/− + DD-STAT3-S727A mESC-derived neural progenitors exhibited low expression of the neural stem cell marker Nestin and produced Tuj1-positive neurons only sporadically (Fig. 3A). We also compared the exact time of induction of neural precursor genes in STAT3−/− + DD-STAT3-WT and STAT3−/− + DD-STAT3-S727A mESCs as they underwent differentiation (Fig. 3B). Starting from day 6 (2 days after addition of retinoic acid, the differentiation-inducing agent), sox1 and nestin mRNA expression was observed in STAT3−/− + DD-STAT3-WT cells, indicating efficient initiation of neural differentiation. On the other hand, in STAT3−/− + DD-STAT3-S727A cells, sox1 and nestin expression was not only delayed (occurring at day 8 for sox1 and day 10 for nestin), but also weakened. Additionally, the Oct4 expression level in STAT3−/− + DD-STAT3-S727A mESCs also suggested impeded differentiation as pluripotent cells were still present at day 12. To further establish a correlation between STAT3 pS727 and mESC neural differentiation, we used a transgene encoding DD-STAT3-S727D, which contains a phosphomimetic mutation at S727. STAT3−/− + DD-STAT3-S727D mESCs underwent neuronal differentiation more efficiently than did STAT3−/− + DD-STAT3-S727A mESCs (Fig. 3C, 3D).
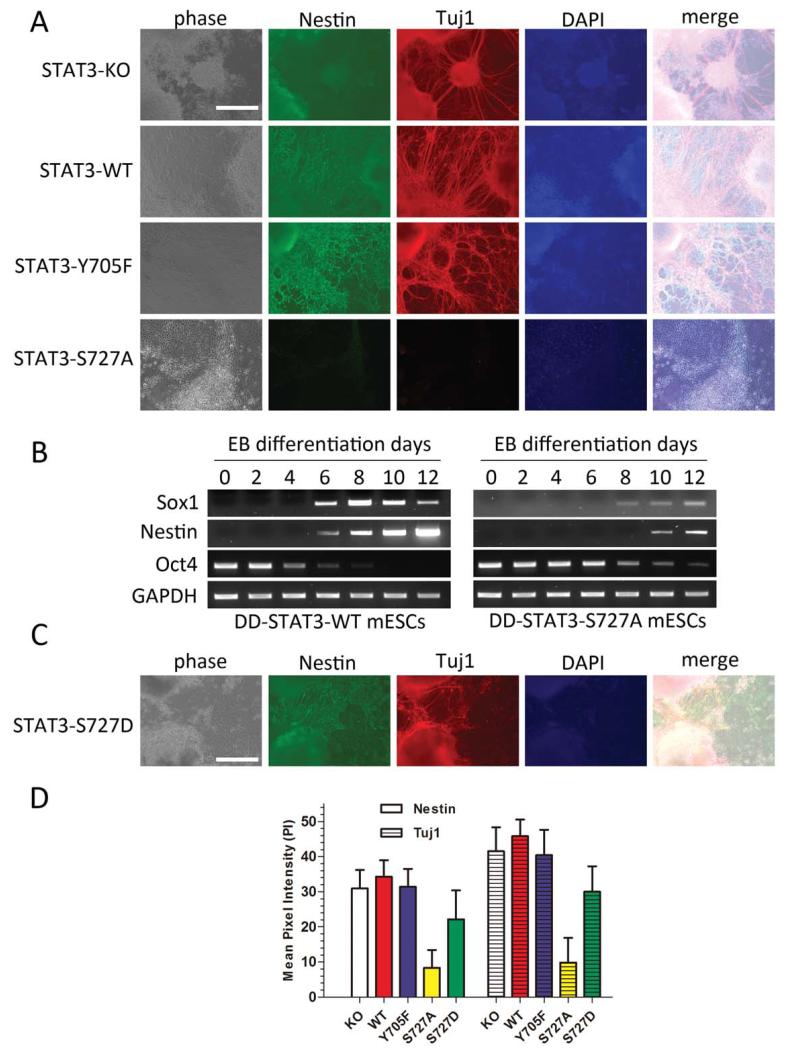
Figure 3.
STAT3 S727 phosphorylation promotes neuronal differentiation of mouse embryonic stem cell (mESCs). (A): Immunofluorescence staining of different STAT3−/− + destabilizing domain (DD)-STAT3 transgenic mESCs with neural precursor cell marker Nestin (green) and neuron marker Tuj1 (red). Nuclei were stained with DAPI (blue). Cells were fixed and stained with above antibodies on day 18 of differentiation. (B): Real-time polymerase chain reaction analysis of induction of neural precursor genes (sox1 and nestin) in EB differentiation of STAT3−/− + DD-STAT3-WT and STAT3−/− + DD-STAT3-S727A cells. Oct4 is a marker for undifferentiated mESCs remaining in the population. (C): Immunofluorescence staining of STAT3−/− + DD-STAT3-S727D mESCs. (D): Quantitative comparison of the pixel intensity among STAT3−/− mESCs expressing various versions of DD-STAT3. The measurements were averaged on at least two different sets of images. (A, B): Scale bars 5 50 μm. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; DD, destabilizing domain; KO, knock out; mESC, mouse embryonic stem cell; WT, wild type.
Loss of STAT3 S727 Phosphorylation Enables LIF-Induced mEpiSC Reprogramming
Since STAT3−/− + DD-STAT3-S727A mESCs were resistant to neuronal differentiation after LIF withdrawal, we hypothesized that pS727 might play an important role in the mouse epiblast stem cell (mEpiSC) stage, a necessary transition state for mESCs undergoing differentiation. We cultured STAT3−/− + DD-STAT3 transgenic mESCs in the fibroblast growth factor (FGF)2/Activin A condition to convert them to mEpiSCs (Fig. 4A). The cells formed large, flat colonies and expressed a high level of EpiSC-specific marker fgf5, a low level of ESC-specific marker rex1, and a reduced level of the pluripotency marker Oct4 (Fig. 4B). We then carried out a serum-free neural induction protocol by exposing monolayer mEpiSCs to N2B27 medium [31] supplemented with S1. Consistent with the observation in Figure 3A, in STAT3−/−, STAT3−/− + DD-STAT3-Y705F, and STAT3−/− + DD-STAT3-WT mEpiSCs, neural-like cell populations began to emerge on day 6 and were predominant on day 11, whereas STAT3−/− + DD-STAT3-S727A mEpiSCs remained largely undifferentiated (Fig. 4C, bottom panel).
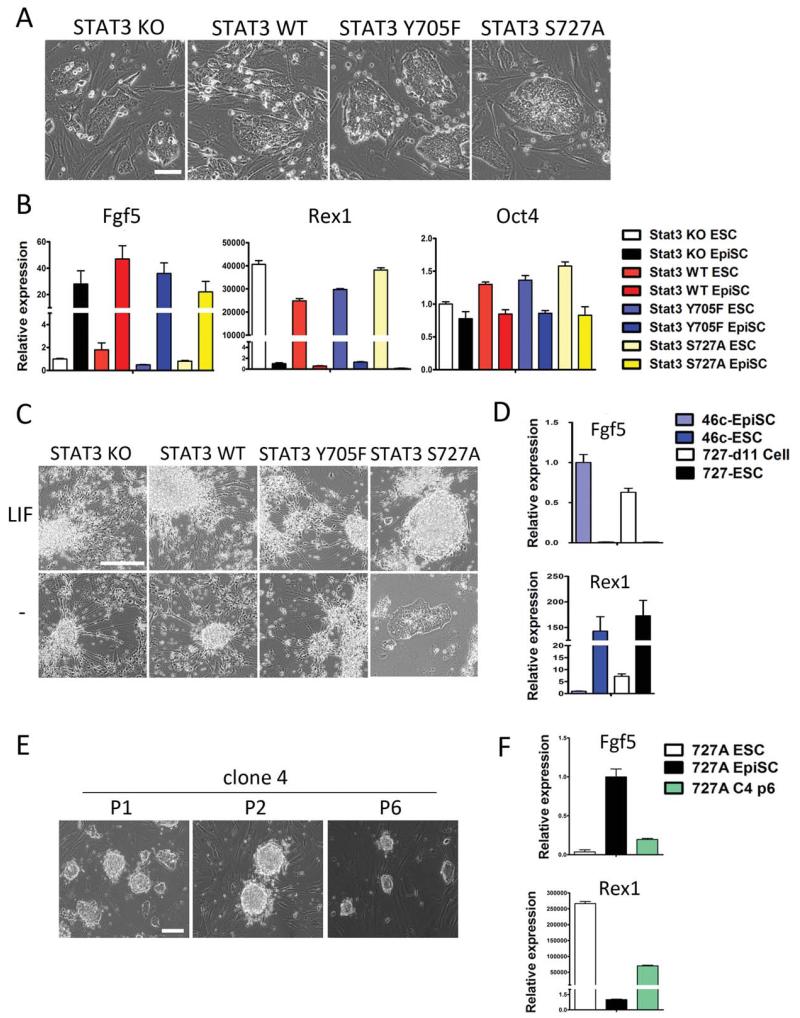
Figure 4.
Loss of STAT3 S727 phosphorylation enables leukemia inhibitory factor (LIF)-induced mouse epiblast stem cell (mEpiSC) reprogramming. (A): Micrographs of STAT3−/− mEpiSCs (p12) expressing indicated versions of destabilizing domain (DD)-STAT3, cultured in mESC+FGF+Activin A. (B): Gene expression profile of STAT3−/− mEpiSCs expressing indicated versions of DD-STAT3. fgf5: EpiSC-specific; Rex1: embryonic stem cell (ESC)-specific; Oct4: shared. (C): Morphology of STAT3−/− + DD-STAT3 transgenic mEpiSCs in N2B27+S1 medium on day 11 with or without administration of LIF. (D): Relative expression of fgf5 and Rex1 in STAT3−/− + STAT3-S727A cells after being subject to LIF+N2B27+S1 reprogramming condition for 11 days. 46C-mEpiSCs, 46C-mouse ESCs (mESCs), and STAT3−/− + STAT3-S727A mESCs were used as controls. (E): mESC-like clones picked from reprogrammed STAT3−/− + DD-STAT3-S727A d11 cells. (F): fgf5 and Rex1 expression in a representative clone of STAT3 −/− + STAT3-S727A d11 mESC-like cells (p6). (A, C, E) Scale bars = 50 μm. Abbreviations: EpiSC, epiblast stem cell; ESC, embryonic stem cell; KO, knock out; LIF, leukemia inhibitory factor; WT, wild type.
It has been reported that JAK/STAT3 activation can reprogram mEpiSCs to a state of naïve pluripotency [32, 33]. We tested this by culturing STAT3−/− + DD-STAT3 transgenic mEpiSCs under LIF+N2B27. Interestingly, in this condition, STAT3−/−, STAT3−/− + DD-STAT3-Y705F, and STAT3−/− + DD-STAT3-WT mEpiSCs still differentiated into neural progenitor cells, presumably due to the strong neural-inductive effect of N2B27. Only STAT3−/− + DD-STAT3-S727A mEpiSCs remained undifferentiated (Fig. 4C, top panel) and gradually regained a mESC identity as indicated by the increased expression of Rex1 and decreased expression of fgf5 by day 11 (Fig. 4D). We isolated several colonies from these cells and continued to culture them in this condition. On passage 6, these colonies developed the small, rounded morphology typical of mESC colonies and exhibited a mESC-like gene expression pattern (Fig. 4E, 4F). These results suggest that LIF-induced Y705 phosphorylation enables reprogramming of mEpiSCs into a naïve state of pluripotency when S727 phosphorylation is absent.
PD0325901 Inhibition of Serum-Induced S727 Phosphorylation Prevents mESC Differentiation
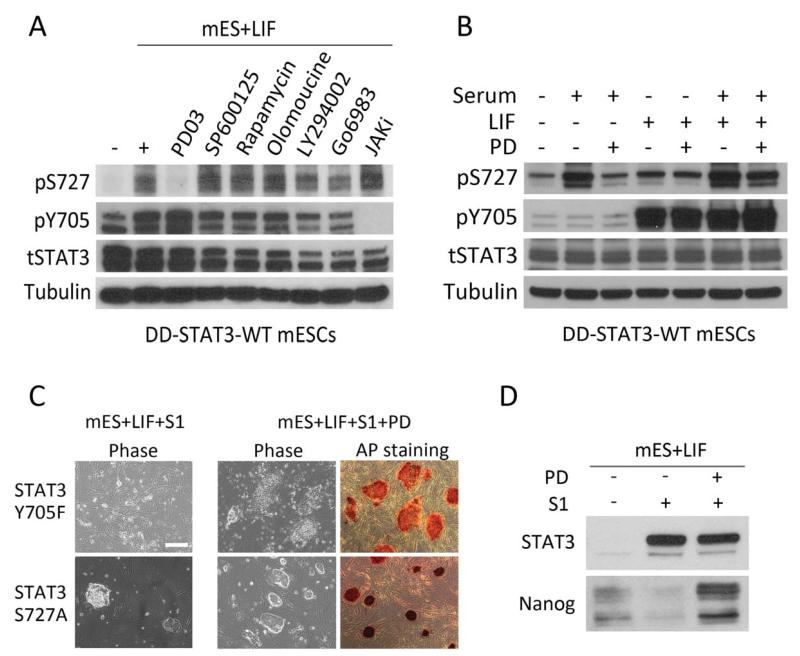
Many growth factors and cytokines, including LIF, induce phosphorylation of S727 through different serine-threonine kinases. To identify the kinase(s) responsible for S727 phosphorylation in mESCs, we treated STAT3−/− + DD-STAT3-WT mESCs with different kinase inhibitors and monitored the changes in pS727. STAT3−/− + DD-STAT3-WT mESCs cultured in serum+LIF for 1h contained a dramatically elevated level of pS727 (Fig. 5A). Of the various kinase inhibitors tested, only the Erk1/2 inhibitor PD0325901 effected a reduction in pS727. Noticeably, JAK inhibitor 1, which completely prevented Y705 phosphorylation, had no effect on the level of pS727, suggesting that phosphorylation of Y705 and S727 is regulated via independent mechanisms. Similar results were obtained in the wild-type R1 mESC line (Supporting Information Fig. S3A). LIF stimulation activates a number of different signaling molecules, including STAT3, PI3K-Akt, and Ras/Erk. To elucidate whether LIF contributes to the elevated level of STAT3 pS727, we stimulated cells with either serum or LIF and tested the effect of PD0325901 addition on STAT3 phosphorylation. The addition of PD0325901 effectively inhibited serum-induced phosphorylation of S727, while Y705 phosphorylation was unaffected. In comparison, LIF stimulation led to a significant increase in the level of pY705, but not pS727 and treatment with PD0325901 had no effect (Fig. 5B; Supporting Information S3B).
Figure 5.
PD0325901 inhibition of serum-induced phosphorylation of S727 prevents mouse embryonic stem cell (mESC) differentiation. (A): Phosphorylation of S727 in mESCs was specifically inhibited by PD0325901. After overnight starvation with basal medium+S1, STAT3−/− + destabilizing domain (DD)-STAT3-WT mESCs were pre-treated with various inhibitors for 2 hours and then stimulated with mESC+LIF for 1h. (B): PD0325901 blocked serum-, but not leukemia inhibitory factor (LIF)-, induced phosphorylation of S727. Serum and LIF stimulation was applied to STAT3−/− + DD-STAT3-WT mESCs separately before immunoblot analysis. (C): Alkaline phosphatase staining and morphology of STAT3−/− + DD-STAT3 mESCs in mESC+LIF+S1 with or without addition of PD0325901. Scale bar = 50 μm (D). PD0325901 can augment Nanog expression in STAT3−/− + DD-STAT3-S727A mESCs. Abbreviations: DD, destabilizing domain; LIF, leukemia inhibitory factor; mESC, mouse embryonic stem cell.
Activation of Erk1/2 signaling leads to mESC differentiation and its suppression can promote self-renewal [34, 35]. In our system, STAT3−/− and STAT3−/− + DD-STAT3-Y705F mESCs were able to grow in the mESC+LIF+S1+PD0325901 condition. Cultured on feeder layers, they formed flattened, AP-positive colonies before eventually differentiating after 3–5 passages (Fig. 5C). This suggested that PD0325901 could reduce dependence on STAT3 pY705 in the maintenance of pluripotency by blocking S727 phosphorylation. On the other hand, administration of PD0325901 to STAT3−/− + STAT3-S727A mESCs cultured in the LIF+S1 condition resulted in improved cell survival and consequently more AP-positive colonies (Fig. 5C). PD0325901 also augmented Nanog expression in STAT3−/− + STAT3-S727A mESCs (Fig. 5D). Therefore, PD0325901 might promote mESC self-renewal by blocking STAT3 pS727-induced differentiation and strengthening core factor-sustained pluripotency.
Discussion
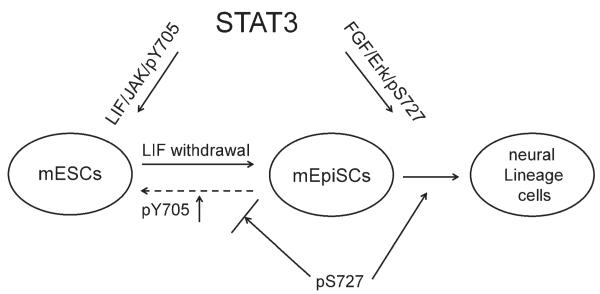
The LIF/STAT3 pathway has been shown to play an important role in the pluripotency of mESCs cultured in serum-supplemented medium. In this study, we demonstrated, for the first time, the involvement of STAT3 pS727 in regulating the homeostasis of mESCs. Using STAT3−/− mESCs maintained in N2B27 + 2i, we were able to manipulate the STAT3 signaling pathway and functionally test the phenotypic outcomes by transferring cells to LIF-dependent culture condition. The resulting data suggest the presence of a dynamic equilibrium of STAT3 Y705 and S727 phosphorylation in controlling the stem cell identities (Fig. 6).
Figure 6.
Proposed model for the role of differential STAT3 phosphorylation in regulating stem cell fates. In mouse embryonic stem cells (mESCs), activation of leukemia inhibitory factor (LIF)/JAK signaling induces phosphorylation of STAT3 Y705 to maintain self-renewal. Upon withdrawal of LIF from the culture environment, mESCs differentiate into mouse epiblast stem cells (mEpiSCs) concomitant with a switch from LIF/JAK-mediated phosphorylation of Y705 to FGF/Erk-mediated phosphorylation of S727. In the mEpiSC stage, STAT3 pS727 promotes neural commitment and secures this differentiation-primed state by inhibiting JAK/pY705-induced reprogramming. Abbreviations: FGF, fibroblast growth factor; LIF, leukemia inhibitory factor; mESC, mouse embryonic stem cell; mEpiSC, mouse epiblast stem cell.
Foshay and Gallicano recently reported that CCE mESCs overexpressing STAT3β, a STAT3 isoform lacking the S727 residue, produced about 40% fewer neural progenitor cells than wild-type CCE cells did [36]. In our experiments, we observed a greater reduction in neural differentiation potential in the absence of S727 (Fig. 3), likely due to lack of endogenous STAT3 in the STAT3−/− mESCs. Considering that STAT3β was previously described as a dominant-negative regulator in STAT3 transactivation [37], the decrease in neural differentiation was thus interpreted as evidence of the necessity of STAT3 activity in this process [36], while leaving any specific contribution from pS727 unexamined. Our results demonstrate that pS727 actually plays a key role in mESC neural differentiation.
In the absence of other self-renewal inputs, mouse ESCs exit pluripotency after LIF stimulation ceases, and can be directed to a neuroectodermal fate after progressing to the mEpiSC stage. This transition is often considered to be the result of a switch in signaling dependence from LIF to FGF. Consistent with the finding that FGF-induced Erk1/2 activation is required for neural specification and lineage commitment of pluripotent mESCs [38, 39], our results showed that Erk1/2 is the primary kinase controlling STAT3 S727 phosphorylation in mESCs, thus further demonstrating the association between S727 phosphorylation and mESC neural differentiation. It has also been suggested that neural induction of ESCs represents a cell-intrinsic fate commitment in a differentiation-permissive culture condition of minimized exogenous signals [40]. This model would seem to account for the observed efficiency with which STAT3−/− mESCs underwent differentiation into neural progenitor cells in our experiments. Our results indicated that neither STAT3 pY705 nor STAT3 itself was required for the induction of differentiation. However, in ESCs with abundant STAT3 expression, the STAT3 pool needs to be switched from LIF-induced pY705-dominant status to FGF/Erk-induced pS727-dominant status to initiate the differentiation programs, a change that is typically achieved by the removal of supplemental LIF from the culture environment. These findings suggest an underlying mechanism in which LIF signaling and FGF signaling converge on a single molecule, STAT3, to regulate the self-renewal and fate commitment of mESCs.
Previous studies showed that mEpiSCs engineered to induce highly increased, sustained and exclusive activation of JAK/STAT3/pY705 signaling could be reprogrammed back to a state of naïve pluripotency under N2B27 condition. LIF treatment, on the other hand, led only to modest activation of JAK/STAT3, along with MAPK and PI3K pathways, and failed to reprogram mEpiSCs [32, 33]. This was verified by our observation that STAT3−/− + STAT3-WT mEpiSCs still differentiated in the N2B271LIF condition. However, loss of STAT3 S727 phosphorylation endowed LIF the capacity to reprogram mEpiSCs to naïve pluripotency, indicating the opposing functions of STAT3 Y705 and S727 in this process. Therefore, we propose that loss of STAT3 S727 phosphorylation relieves the requirement for hyperactivation of JAK/STAT3/pY705 to overcome the EpiSC reprogramming block.
Mouse ESCs grown in the mESC+LIF condition exhibit greater morphological and transcriptional heterogeneity than those grown in the chemically defined N2B27 + 2i condition. Given that LIF stimulation antagonizes differentiation cues induced by FGF/Erk signaling in the serum condition, phosphorylation of STAT3, which is dynamically regulated by both pathways, might contribute to the population heterogeneity. Moreover, several studies have reported that the expression level of Nanog, in Oct4-positive mESCs, regulates the equilibrium between naïve pluripotency and lineage-primed states [41-43]. STAT3−/− + STAT3-S727A mESCs showed lower Nanog expression than STAT3−/− + STAT3-WT mESCs, despite being Oct4-positive and morphologically indistinguishable from those cells. Thus, it is possible that STAT3 pS727 enables the maximal transcription of pluripotency factors while also preserves susceptibility to differentiation triggers.
Conclusion
Our results showed the presence of a shifting equilibrium of STAT3 phosphorylation that controls mESC fates (Fig. 6). In an environment conducive to self-renewal (serum medium supplemented with LIF), a high level of STAT3 pY705 is essential to counteract differentiation cues from FGF/Erk signaling; pS727 is dispensable, serving only to promote proliferation and optimal pluripotency. On the other hand, in differentiative culture, a high level of STAT3 pS727 is critical for the initiation of mESC differentiation and neural commitment, and inhibits JAK/STAT3-pY705-induced reprogramming to naïve pluripotency.
Supplementary Material
Acknowledgments
We thank Hoon Kim, Ping Li, and Chih-I Tai for technical assistance and Charles Ashton for critical reading of the manuscript. This work was supported by the California Institute for Regenerative Medicine (CIRM) New Faculty Award II (RN2-00938-1).
Footnotes
Author Contributions
G.H.: conception and design, collection and assembly of data, manuscript writing, H.Y.: conception and design, collection of data, S.Y. and C.T.: collection of data, Q.-L.Y.: conception and design, financial support, manuscript writing.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Bromberg J, Darnell JE., Jr. The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 2.Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 3.Qin HR, Kim HJ, Kim JY, et al. Activation of signal transducer and activator of transcription 3 through a phosphomimetic serine 727 promotes prostate tumorigenesis independent of tyrosine 705 phosphorylation. Cancer Res. 2008;68:7736–7741. doi: 10.1158/0008-5472.CAN-08-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazan-Halevy I, Harris D, Liu Z, et al. STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells. Blood. 2010;115:2852–2863. doi: 10.1182/blood-2009-10-230060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aggarwal BB, Kunnumakkara AB, Harikumar KB, et al. Signal transducer and activator of transcription-3, inflammation, and cancer: How intimate is the relationship? Ann N Y Acad Sci. 2009;1171:59–76. doi: 10.1111/j.1749-6632.2009.04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi X, Zhang H, Paddon H, et al. Phosphorylation of STAT3 serine-727 by cyclin-dependent kinase 1 is critical for nocodazole-induced mitotic arrest. Biochemistry. 2006;45:5857–5867. doi: 10.1021/bi052490j. [DOI] [PubMed] [Google Scholar]
- 7.Burdon T, Smith A, Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 2002;12:432–438. doi: 10.1016/s0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- 8.Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251:199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 9.de la Iglesia N, Konopka G, Puram SV, et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22:449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman RS, Lourenco P, Tonner E, et al. The role of Stat3 in apoptosis and mammary gland involution. Conditional deletion of Stat3. Adv Exp Med Biol. 2000;480:129–138. doi: 10.1007/0-306-46832-8_16. [DOI] [PubMed] [Google Scholar]
- 11.Chapman RS, Lourenco PC, Tonner E, et al. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev. 1999;13:2604–2616. doi: 10.1101/gad.13.19.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Farrell AM, Liu Y, Moore KW, et al. IL-10 inhibits macrophage activation and proliferation by distinct signaling mechanisms: Evidence for Stat3-dependent and -independent pathways. EMBO J. 1998;17:1006–1018. doi: 10.1093/emboj/17.4.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutchins AP, Diez D, Takahashi Y, et al. Distinct transcriptional regulatory modules underlie STAT3’s cell type-independent and cell type-specific functions. Nucl Acid Res. 2013;41:2155–2170. doi: 10.1093/nar/gks1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niwa H, Burdon T, Chambers I, et al. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raz R, Lee CK, Cannizzaro LA, et al. Essential role of STAT3 for embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 1999;96:2846–2851. doi: 10.1073/pnas.96.6.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeda K, Noguchi K, Shi W, et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci U S A. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong C, Huang G, Ashton C, et al. Generating gene knockout rats by homologous recombination in embryonic stem cells. Nat Protoc. 2011;6:827–844. doi: 10.1038/nprot.2011.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tompers DM, Labosky PA. Electroporation of murine embryonic stem cells: A step-by-step guide. Stem Cells. 2004;22:243–249. doi: 10.1634/stemcells.22-3-243. [DOI] [PubMed] [Google Scholar]
- 20.Banaszynski LA, Chen LC, Maynard-Smith LA, et al. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyakoshi M, Yamamoto M, Tanaka H, et al. Serine 727 phosphorylation of STAT3: An early change in mouse hepatocarcinogenesis induced by neonatal treatment with diethylnitrosamine. Mol Carcinog. 2012 doi: 10.1002/mc.21949. [DOI] [PubMed] [Google Scholar]
- 22.Sakaguchi M, Oka M, Iwasaki T, et al. Role and regulation of STAT3 phosphorylation at Ser727 in melanocytes and melanoma cells. J Invest Dermatol. 2012;132:1877–1885. doi: 10.1038/jid.2012.45. [DOI] [PubMed] [Google Scholar]
- 23.Gartsbein M, Alt A, Hashimoto K, et al. The role of protein kinase C delta activation and STAT3 Ser727 phosphorylation in insulin-induced keratinocyte proliferation. J Cell Sci. 2006;119:470–481. doi: 10.1242/jcs.02744. [DOI] [PubMed] [Google Scholar]
- 24.Cartwright P, McLean C, Sheppard A, et al. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 25.Kidder BL, Yang J, Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. Plos One. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Woo AJ, Chu J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 28.Qin S, Zhang CL. Role of Kruppel-like factor 4 in neurogenesis and radial neuronal migration in the developing cerebral cortex. Mol Cell Biol. 2012;32:4297–4305. doi: 10.1128/MCB.00838-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao F, Hata R, Zhu P, et al. Conditional deletion of Stat3 promotes neurogenesis and inhibits astrogliogenesis in neural stem cells. Biochem Biophys Res Commun. 2010;394:843–847. doi: 10.1016/j.bbrc.2010.03.092. [DOI] [PubMed] [Google Scholar]
- 30.Tsai SY, Yang LY, Wu CH, et al. Injury-induced Janus kinase/protein kinase C-dependent phosphorylation of growth-associated protein 43 and signal transducer and activator of transcription 3 for neurite growth in dorsal root ganglion. J Neurosci Res. 2007;85:321–331. doi: 10.1002/jnr.21119. [DOI] [PubMed] [Google Scholar]
- 31.Ying QL, Smith AG. Defined conditions for neural commitment and differentiation. Methods Enzymol. 2003;365:327–341. doi: 10.1016/s0076-6879(03)65023-8. [DOI] [PubMed] [Google Scholar]
- 32.Yang J, van Oosten AL, Theunissen TW, et al. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell. 2010;7:319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Oosten AL, Costa Y, Smith A, et al. JAK/STAT3 signalling is sufficient and dominant over antagonistic cues for the establishment of naive pluripotency. Nat Commun. 2012;3:817. doi: 10.1038/ncomms1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamazaki T, Kehoe SM, Nakano T, et al. The Grb2/Mek pathway represses Nanog in murine embryonic stem cells. Mol Cell Biol. 2006;26:7539–7549. doi: 10.1128/MCB.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burdon T, Stracey C, Chambers I, et al. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev Biol. 1999;210:30–43. doi: 10.1006/dbio.1999.9265. [DOI] [PubMed] [Google Scholar]
- 36.Foshay KM, Gallicano GI. Regulation of Sox2 by STAT3 initiates commitment to the neural precursor cell fate. Stem Cells Dev. 2008;17:269–278. doi: 10.1089/scd.2007.0098. [DOI] [PubMed] [Google Scholar]
- 37.Caldenhoven E, van Dijk TB, Solari R, et al. STAT3beta, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. J Biol Chem. 1996;271:13221–13227. doi: 10.1074/jbc.271.22.13221. [DOI] [PubMed] [Google Scholar]
- 38.Kunath T, Saba-El-Leil MK, Almousailleakh M, et al. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- 39.Stavridis MP, Lunn JS, Collins BJ, et al. A discrete period of FGF-induced Erk1/2 signalling is required for vertebrate neural specification. Development. 2007;134:2889–2894. doi: 10.1242/dev.02858. [DOI] [PubMed] [Google Scholar]
- 40.Kamiya D, Banno S, Sasai N, et al. Intrinsic transition of embryonic stem-cell differentiation into neural progenitors. Nature. 2011;470:503–509. doi: 10.1038/nature09726. [DOI] [PubMed] [Google Scholar]
- 41.Chambers I, Silva J, Colby D, et al. Nanog safeguards pluripotency and mediates germ-line development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 42.Galvin-Burgess KE, Travis ED, Pierson KE, et al. TGF-beta-Superfamily Signaling Regulates Embryonic Stem Cell Heterogeneity: Self-Renewal as a Dynamic and Regulated Equilibrium. Stem Cells. 2013;31:48–58. doi: 10.1002/stem.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh AM, Hamazaki T, Hankowski KE, et al. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.