Abstract
The human gastrointestinal tract contains distinct microbial communities that differ in composition and function based on their location, as well as age, sex, race/ethnicity, and diet of their host. We describe the bacterial taxa present in different locations of the GI tract, and their specific metabolic features. The distinct features of these specific microbial communities might affect human health and disease. Several bacterial taxa and metabolic modules (biochemical functions) have been associated with human health and the absence of disease. Core features of the healthy microbiome might be defined and targeted to prevent disease and optimize human health.
Keywords: gastrointestinal, HMP, lumen, metagenomics, microbe, mucosa
History and Background
The human microbiome comprises diverse microbiologic ecosystems and is composed of a variety of bacteria, archaea, microeukaryotes (e.g., fungi), and viruses. Although we have appreciated the diversity of our microbial world for decades, scientists were constrained by the inability to culture many bacteria in the laboratory. Paradigms in biomedical science and medicine are changing in a fundamental way as we explore potential contributions of human-associated microbes to health and expand our understanding of disease susceptibility and pathogenesis. In the early 1900s, scientists studied the abilities of microbes to promote longevity (1) and bacteriophages (bacterial viruses) to treat infections (2, 3). During the twentieth century, scientists led military-style assaults on the biosphere with its war on microbes and global campaigns to eradicate infectious diseases. Great progress was made in the discovery and development of novel antimicrobial agents and vaccines to treat and prevent infectious diseases.
In the 21st century, we are attempting to develop a more balanced mindset as we seek to understand the role of the human microbiome in physiology and manipulate it to optimize health and prevent or treat disease. In recent years, we have increased our understanding of microbial communities and their corresponding metagenomes at different human body sites greatly (4). Here, we review the bacterial component of the human gastrointestinal (GI) microbiome in healthy individuals and discuss what we have learned about its role in health (i.e., the condition of being free from disease).
Defining the GI Microbiome in Health
How should biomedical research scientists and gastroenterologists define health and a healthy human microbiome? The microbiome may contribute to human health by increasing relative fitness, resilience, or optimal functioning of the body. The human microbiome has been defined by global ecological parameters such as richness, diversity, and evenness of its microbial communities. Microbial richness and diversity are commonly defined by studies of microbial composition using 16S rRNA gene sequencing or whole-genome sequence-based metagenomic analyses; various indices of richness, diversity, and evenness have been used to compare bacterial populations from stool and intestinal biopsy specimens. Greater richness and diversity of bacterial species in the human intestine may be an indicator of health. Reduced bacterial diversity has been observed in the fecal communities of preterm infants who develop necrotizing enterocolitis, compared with those who do not (5), and reduced GI bacterial diversity during infancy has been associated with increased risk of allergic disease later in life (6). In contrast, greater bacterial richness and diversity were found to correspond with better nutritional status, fewer co-morbidities, and greater overall health in a cohort of elderly individuals (7).
In addition to bacterial species counts and estimates of taxonomic diversity, richness of the human GI microbiome has also been characterized on the basis of microbial functional gene richness, determined by metagenomic DNA sequencing (8). Using this approach, researchers have been able to categorize individuals as either high gene count (HGC) or low gene count (LGC), and these gene counts appear to have implications for health. In a survey of 123 non-obese and 169 obese Danish individuals, LGC and HGC individuals had mean quantities of 380,000 and 640,000 bacterial/phage genes, respectively. HGC individuals are generally considered to have a greater repertoire of microbial metabolic functions, a functionally more robust gut microbiome, and greater overall health, including lower prevalence of obesity and metabolic disorders (9).
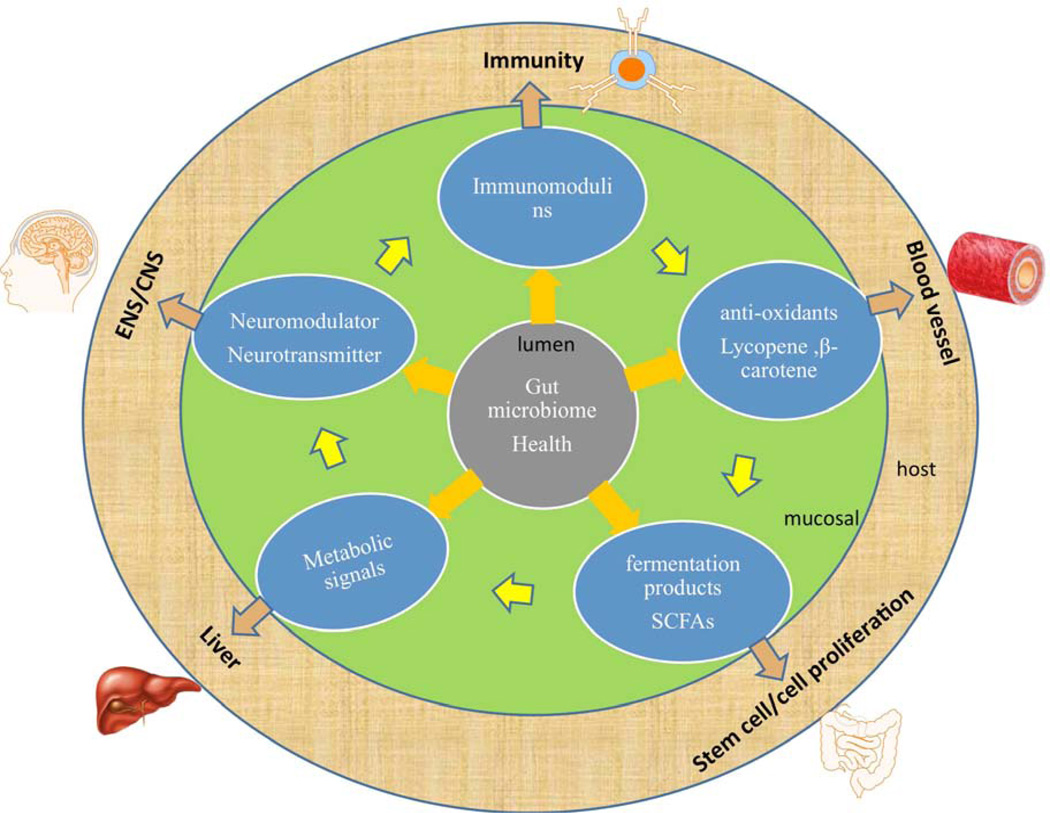
Beyond microbial richness and diversity, a healthy gut microbiome can be defined by the presence of classes of microbes that enhance metabolism, resilience to infection and inflammation, resistance to cancer or autoimmunity, endocrine signaling, and brain function (brain–gut axis). The microbiome may mediate these effects via secretion of factors that modulate intestinal permeability, the mucus layer, epithelial cell function, innate and adaptive immunity, intestinal motility, and neurotransmission (Figures 1, 2). The presence of representative core bacterial genera or species, or core metabolic functions/modules, could help to define a healthy microbiome at a given site in the human body, including the GI tract. Examples of bacterial taxa that have been associated with human health and proper GI function include Bacteroides, Bifidobacterium, Clostridium clusters XIVa and IVa (butyrate producers), Eubacterium, Faecalibacterium, Lactobacillus and Roseburia. Bacterial species that might protect against weight gain and are enriched in HGC individuals include Anaerotruncus colihominis, Butyrovibrio crossotus, Akkermansia spp., and Faecalibacterium spp. (9). Core metabolic modules may include pathways involved in central carbohydrate metabolism and cofactor/vitamin biosynthesis. Metabolic pathways that were consistently present in the intestinal microbiome of healthy adults included spermidine biosynthesis and methionine degradation (4). Among HGC adults, who are relatively healthy, the relative abundance of genes belonging to pathways involved in hydrogen and methane production increased, whereas pathways involved in hydrogen sulphide production decreased (9). These metabolic activities might someday serve as markers of microbiome functionality and be used in routine GI health evaluations.
Figure 1. The Intestinal Microbiome and its Effects in the Intestinal Mucosa.
The intestinal microbiome (top) lies adjacent to the intestinal epithelium. Specific effects are described in text boxes.
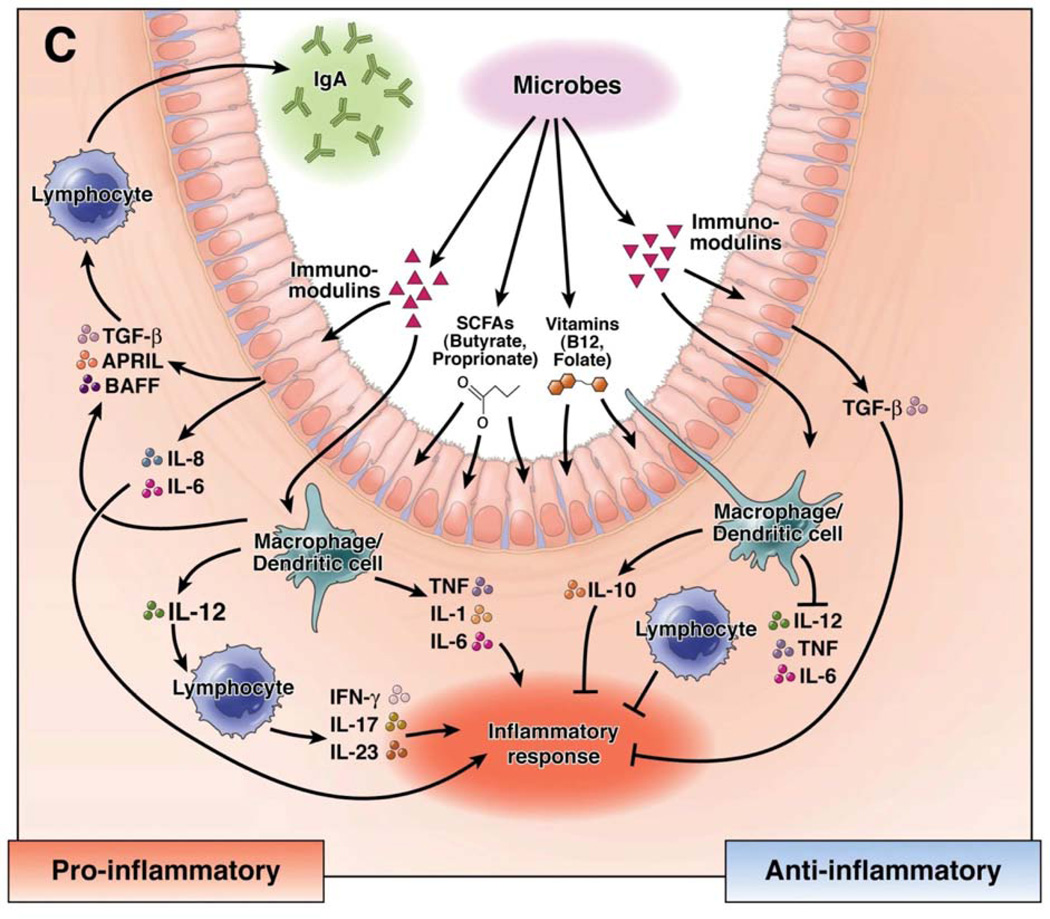
Figure 2. Immunomodulation by the Intestinal Microbiome.
Microbial metabolites regulate the mucosal immune system in the intestine through pro-inflammatory and antiinflammatory mechanisms. Immunostimulation can occur in response to the release of proinflammatory cytokines from epithelial cells, mononuclear cells, and lymphocytes, antiinflammatory responses are driven by the production of TGF-β and IL-10 from epithelial and mononuclear cells.
Reprinted from (69).
Defining the Gut Microbiome - Practical Aspects and Environmental Factors
The human GI microbiome comprises diverse microbial communities that differ based on their location along the length of the GI tract (esophagus, stomach, small intestine, and large intestine or colon). Most human intestinal microbiome studies have relied on stool specimens. Although easily acquired, stool specimens do not provide reliable information about nonpathogenic microbes in the esophageal or gastric microbiome, and, likewise, are considered to be poorly representative of the upper (proximal) GI tract and the mucosa-associated microbiota. The composition of the esophageal and gastric microbiomes is distinct from the microbiome of the colon (4, 10, 11); studies have demonstrated differences in the microbial compositions of biopsy and stool specimens (12). Esophagogastroduodenal endoscopy can be used to collect samples from the proximal gut, but it is a challenge to perform these medical procedures in healthy control populations. Future advances in capsule endoscopy could improve methods for collecting samples from the upper GI tract and small intestine of healthy populations.
In addition to the longitudinal axis, the healthy human microbiome differs axially, from lumen to the mucosal surface. Biopsy or surgical specimens are required to carefully evaluate the composition of mucosa-associated microbiota. Several studies have compared microbial compositional data derived from intestinal biopsy specimens with those from colon content or stool, and reported differences in mucosa-associated microbiota samples collected via un prepped colonoscopy (13, 14). Although these studies are limited by the small numbers of healthy individuals from whom un-prepped colonoscopy and biopsy samples were collected, bacterial composition of the mucosa-associated microbiota in healthy adults seems to differ markedly from that of the luminal microbiome. The phylum Firmicutes was greatly enriched in the mucosa-associated microbiota vs the luminal (largely stool) microbiota (14). One practical consideration is whether individuals have undergone bowel cleansing or colonic lavage prior to sampling. Standard colonic lavage diminishes the diversity of the mucosaassociated microbiota, in addition to the obvious impact on the luminal or less adherent microbiota in the fecal stream (14). As an example, the mucosa-associated genus Mucispirillum consistently vanished following colonic lavage, and this genus is known to colonize the mucus layer of the GI tract in mammals (15).
Practical considerations in any evaluation of the healthy GI microbiome should include age, sex, race/ethnicity, and diet. The Human Microbiome Project documented the importance of considering sex and race/ethnicity in evaluating differences between microbiomes of individuals. In various body sites, several taxonomic clade differences were apparent in microbiota of healthy men vs women (4, 16), although the number of differences attributable to sex in stool was limited. Race and ethnicity were also associated with differences in bacterial composition at multiple body sites. Age appears to be a particularly important consideration with respect to GI microbial community composition. Prime examples of changes in GI microbiome composition with age may be found in the rapid development of the intestinal microbiome early in human life (e.g.,(17)), as well as in comparisons of infant GI communities with those of adults (18).
Diet has also been shown in several studies to shape the composition of the GI microbiome. There are large-scale differences (i.e., detectable at the bacterial phylum level) between fecal microbiomes of children in northern Italy and children in Burkina Faso (19). Healthy children in Burkina Faso consume diets rich in fiber and complex plant carbohydrates; correspondingly, the genera Prevotella and Xylanibacter are detected in greater proportions in African than Italian children. Likewise, the quantities of vegetables, fruit, and meat consumed by elderly individuals correspond with the composition of their fecal microbiomes and their overall health status (7). Changes in diet can produce relatively rapid, reproducible alterations in the composition of the gut microbiome (20), but overarching community features, such as enterotype, remained stable during a 10 day dietary intervention (21).
Studies of mice have suggested that diet helps determine the receptivity of a gut community to modulation and manipulation. When mice are co-housed, their coprophagic habits allow GI microbiota and microbiome-induced phenotypes to be shared with one another. Obese mice fed low-fat diets high in plant polysaccharides readily acquired microbiota characteristic of lean mice when they were co-housed with lean mice (22). Microbial composition was less likely to shift in obese mice fed a high-fat, low-plant-polysaccharide diet and co-housed with lean mice. Likewise, the use of the low-fat, high-plant-polysaccharide diet alone did not result in as great a shift toward a lean microbiome as did the low-fat, high-plantpolysaccharide diet plus co-housing with lean individuals (22). Together, a growing body of evidence, some of which we have highlighted here, indicates that diets of healthy children and adults shape their intestinal microbiome—any specification of a healthy human gut microbiome must be placed in the context of basic dietary patterns. It is also important to note that most published studies of the human microbiome have lacked careful evaluation of dietary history.
Microbiome studies are often performed using specimens collected at a single time point. Temporal stability or variability of the GI microbiome is a primary consideration in healthy individuals. The gut microbiome appears to be stable over time in healthy adults, despite the presence of symptoms such as abdominal pain or bloating (23). However, antibiotics, overseas travel, and temporary (acute) illness can cause variations in the composition of the intestinal microbiome of healthy adults. Most core GI bacterial phylotypes (70% of 288 bacterial taxa) identified by DNA microarray-based hybridization varied less than 10% in terms of relative abundance during a 7 week sampling period (23).
Changes in Bacterial Composition Along the GI Tract
Esophagus, Stomach, and Small Intestine
Although there have been few studies of the microbiota of the proximal esophagus, a variety of bacterial species pass through this tube, which connects the oral cavity and stomach. Insufficient data have been gathered about the stable microbial populations in the proximal esophagus. Distal esophageal microbiome studies (10) have documented a microbial ecosystem that is relatively limited in diversity and dominated by Streptococcus species. In contrast to other body sites, where greater microbial diversity is associated with human health, increased microbial diversity in the distal esophagus has been associated with chronic inflammation and dysplasia (24). Limited diversity and the presence of a few dominant genera, such as Streptococcus, could be important features of the healthy esophagus. Other genera such as Prevotella, Actinomyces, Lactobacillus, and Staphylococcus have been detected in the distal esophagus; these bacterial genera might provide complementary health-associated functions and ecological resilience. Bacterial groups such as streptococci could include strains that extend their habitats from the oral cavity to the esophageal mucosa.
The microbial diversity of the human stomach, like that of the esophagus, is limited. The low pH of the gastric lumen limits the types of microbes that can live there, selecting for acidresistant bacterial populations. The presence or absence of the gastric pathogen Helicobacter pylori strongly affects the composition of the gastric microbiome. When H pylori infection is present, the diversity of the stomach microbiome is severely limited, with H pylori accounting for the vast majority of the taxa detected (25). H pylori can be a commensal species or a pathogen. Martin Blaser was the first to propose that eradication of H pylori by antimicrobial therapy increased susceptibility of individuals to esophagitis and neoplasia (26). Alternatively, the presence of H pylori in the gastric microbiome has been linked with peptic ulcer disease and gastritis; because of this association, antimicrobial therapy to eradicate H pylori has become widely accepted in medical practice. The most prevalent genera in the gastric microbiome, besides H pylori, include several species often found in the oral cavity and esophagus. A total of 10 genera comprise the most predominant genera in the human stomach—these include Prevotella, Streptococcus, Veillonella, and Rothia. Streptococcus is the most dominant genus in distal esophagus and stomach in the absence of H pylori infection (11).
The human small intestine remains a frontier for exploration of healthy human microbiology. It is generally accepted that bacterial communities increase in relative diversity and complexity in the proximal–distal direction, from duodenum through jejunum and ileum. Based on routine microbiology and early molecular studies, the genus Streptococcus appears to be a dominant genus in the duodenum and jejunum (27–29). Small intestinal metagenomic studies in healthy individuals are lacking. Ileostomy studies provide limited information about the healthy small bowel.
Large Intestine (Colon), Distal Gut, and Feces
The Human Microbiome Project, funded by the US National Institutes of Health, included a total of 300 subjects recruited from St. Louis and Houston. Five hundred fifty-four individuals were screened for inclusion in the study. Many criteria were considered in selecting the adults for the final cohort (16). The mean age of these individuals was 26 years, and the mean body mass index was 24 kg m−2. Specimens were collected from a total of 18 body sites, but the human GI tract was represented by a single stool specimen, collected by each individual him or herself.
In the end, analyses of data from 242 healthy participants (18–40 years old) in the Human Microbiome Project were presented in 2 landmark reports, published in 2012 (4, 30). Microbial composition was determined by 16S rRNA gene sequencing and metagenomic profiles were generated by whole-genome shotgun sequencing. Based on the taxonomic profiles of the whole-genome shotgun sequencing, the healthy GI microbiome contains between 70–100 known bacterial species (4, 31). However, 16S rRNA gene-based approaches suggest that the numbers of bacteria in the healthy human gut likely exceed 1000 species (30, 32), particularly when rare, low-abundance, and uncultivated or unclassified bacterial taxa are considered. Microarray-based studies with signals generated by known bacterial taxa reported an average of 470 phylotypes per healthy adult subject; this number represents a likely underestimate of total bacterial taxa per individual (23).
In terms of bacterial composition, the healthy human GI microbiome is dominated by the phyla Firmicutes and Bacteroidetes, followed distantly by Actinobacteria and Verrucomicrobia. The phylum Proteobacteria accounts for only a small proportion of GI bacteria in healthy individuals, but Proteobacteria often account for higher proportions in patients with GI diseases (reviewed in (33)). Firmicutes and Bacteroidetes are the predominant phyla in the large intestine. Although some evidence suggests that the ratio of Firmicutes:Bacteroidetes can be used to determine predisposition to obesity and metabolic syndromes (34), it varies significantly among healthy individuals. As such, the significance of the ratios of Firmicutes:Bacteroidetes remains controversial and its implications for human health are unclear. Less than 0.1% of the human GI microbiota comprises primary, overt pathogens such as Campylobacter jejuni, Salmonella enterica, and Vibrio cholerae. Opportunistic pathogens are also present; 56 of 327 pathogens from the PATRIC database were detected in healthy adults. Species such as Bacteroides thetaiotamicron and the potential pathogen Bacteroides fragilis were detected in 46% and 16% of stool specimens of health adults, accounting for 0.1% or more of the microbiota, respectively (4). Escherichia coli accounted for 0.1% or more of the microbiota in only 15% of stool specimens, although 61% of stool specimens had levels of E coli above the lower limit of detection. Some bacterial species such as Prevotella copri were detected in stool specimens but rarely in samples from other body sites. Because of this, they can be considered signature GI species. The relative abundances of signature commensal taxa (e.g., Bacteroides, Prevotella, Ruminococcus) and low proportions of canonical pathogens and Proteobacteria may serve as key indicators of GI microbiome wellness should be considered in analyses of GI microbiomes with respect to human health.
If the healthy gut microbiome is defined based on total number of bacterial genes, the Human Microbiome Project catalogued 5,140,472 non-redundant bacterial genes in healthy individuals. However, the sum total from the more-recent Human Microbiome Project and MetaHIT studies estimated that there are more than 10 million non-redundant genes in the human microbiome. To extend the relevance of microbial genes in human metagenomes, the MetaHIT European cohort was stratified into HGC and LGC groups. Several features of the HGC microbiome have been reported to indicate digestive health (9). These include a methanogenic/acetogenic ecosystem, increased proportions of butyrate-producing bacteria, increased ratio of Akkermansia (Verrucomicrobia):Ruminococcus torque/gnavus, increased potential for hydrogen production, reduced potential for production of hydrogen sulphide, and reduced numbers of Campylobacter/Shigella.
Three basic enterotypes have been described in healthy adults. Enterotype 1 contains a high proportion of genera Bacteroides, enterotype 2 a high proportion of Prevotella, and enterotype 3 a high proportion of Ruminococcus (35). These enterotypes were also characterized based on proportions of specific metabolic modules. For example, genes belonging to the biotin and riboflavin biosynthesis pathways were detected with greater frequency in enterotype 1, whereas genes involved in the thiamine and folate biosynthesis pathways were detected with greater frequencies in enterotype 2. The concept of enterotypes in healthy humans has been challenged by alternative concepts, such as that of enterogradients (36). Enterogradients could be a useful model to explain the relative distributions of different classes of gut bacteria in different individuals.
The GI Microbiome in the Context of Development and Aging
From our initial colonization through old age, our GI microbiome has important roles in digestion, acquisition of nutrients, and immune regulation (37–40). The microbial communities that inhabit the human GI tract are shaped by multiple environmental influences, including lifestyle, life events, and importantly, the aging process. As studies of the microbiome have expanded in focus beyond healthy adults, we have begun to gain insight into the communities of infants, children, and the elderly. This growing body of research indicates that the gut microbiome changes with development and maturation of its host. However, neither is a discretely linear process; distinct phases of development occur in both the host and its GI microbiome.
It was long believed that humans were sterile before birth, but recent studies have shown that the gut microbiome can be seeded in utero. Bacteria have been cultured successfully from the first meconium of full-term, healthy neonates (41), and 16S rRNA gene surveys of meconium depict a relatively simple community, dominated by genera such as Escherichia-Shigella, Enterococcus, Leuconostoc, Lactococcus, and/or Streptococcus (42). There is additional evidence that mode of birth (vaginal delivery vs Caesarean section) may affect the composition of the early microbiome. Infants who are delivered vaginally have initial microbiomes that contain large numbers of lactobacilli and resemble those of the mothers’ vaginal tract, whereas those delivered by Caesarean section have initial microbiomes containing Staphylococcocus, Corynebacterium, and Propionibibacterium, largely resembling the microbiome the mothers’ skin (43, 44). Notably, these studies included relatively small subject cohorts, so no firm conclusions can be made.
Following birth, the GI microbiome develops rapidly. Bifidobacteria, specialized in the metabolism of milk oligosaccharides, are frequently among the earliest post-birth colonizers. As dietary richness and environmental exposures increase during the first year of life, the richness and complexity of the GI microbiome also increase (17, 18, 44, 45). This process is often characterized as chaotic (46), but cohort and longitudinal studies of the GI microbiome suggest that bacterial community richness increases steadily during the first year of life (17, 18).
Infancy is a distinctive stage in the GI microbiome, based not only on the types of taxa that are present and the rapid rate at which the community develops, but also in terms of its functional repertoire. The infant microbiome appears to be specialized (relative to that of adults) with respect to the acquisition of nutrients, including B complex vitamins and multiple amino acids (18); much of this specialization is rooted in dietary acquisition of nutrients vs de novo synthesis by gut microbes. For example, infant GI communities are enriched with genes that regulate de novo synthesis of folate, whereas the adult microbiome contains greater numbers of genes involved in dietary folate utilization (18). In contrast, the adult microbiome contains greater numbers of genes involved in vitamin B12 (cobalamin) synthesis than that of infants (18).
By the time a child reaches ~3 years of age, his or her GI microbiome is 40%–60% similar to that of a healthy adult (18). This is the same level of similarity as among adults (18, 47), supporting the concept that by the time a child reaches 3 years of age, his or her GI microbiome has reached an adult-like state (17, 46). Evidence to the contrary, however, has been reported for young children (1–4 years old) (48) and adolescents (49). Although the GI microbiomes of children and teens contain many of the same taxa as adults, significant differences in proportions of Bacteroides spp. and Bifidobacterium spp., as well as members of the clostridia, have been reported.
It has been proposed that the adult GI microbiome remains stable from the third through seventh decades of life (50); the composition of the GI microbiome of young adults (25–40 years old) has high degree of similarity to with that of older adults (63–76 years old). More recent studies suggest that even during this relatively stable period of microbiome development, important taxa and their functions could be decreasing. As we age, the proportions of bifidobacteria, Faecalibacterium prausnitzii, and multiple members of the Firmicutes typically decrease, whereas proportions of E coli, other members of the Proteobacteria, and Staphylococcus often increase (50–52). The GI microbiomes of older adults can also be differentiated from those of younger adults based on the reduced potential for vitamin B12 biosynthesis and activities of microbial reductases, as well as increased potential for DNA damage, stress response, and immune system compromise (53). These findings suggest that the GI microbiomes of older adults may represent a pro-inflammatory phenotype.
Aging and inflammation are co-mingled processes, and key hallmarks of aging include declines in gastrointestinal function and host immune response, and the development of chronic, low-grade inflammation. Older age (65–100 years) brings more important transitions in the composition, function, and stability of the GI microbiome (50, 54). The elderly have greater inter-individual variation in their GI microbiomes than younger adults (7). Much of this variation appears to be diet-driven, and it correlates strongly with indicators of relative health, including markers of frailty and inflammation (7).
The Bacterial Metagenome and Functional Connections to Human Health
Carbohydrate Metabolism
Complex carbohydrates and simple sugars provide abundant food sources and substrates for microbial metabolism in the human gut. The influx of carbohydrates from the diet helps to shape the composition and function of the human microbiome in healthy individuals. Comparisons of pediatric populations found large differences in bacterial composition, even at the phylum level, among healthy children who consume diets dominated by plant carbohydrates vs those who consumed a typical Western diet (19). The phylum Bacteroidetes was dominant among children who consumed the plant-rich diets. This phylum contains the members of the genus Bacteroides, many of which are enriched with genes that encode carbohydrate-active enzymes (CAZyme). CAZymes include glycosyltransferases, which assemble carbohydrates, and glycoside hydrolases and polysaccharide lyases, which are involved in carbohydrate catabolism.
The species Bacteroides thetaioatamicron was found in 46% of healthy adults. The genome of this bacterium encodes more than 260 glycoside hydrolases, compared with the 97 glycoside hydrolases encoded by the entire human genome (www.cazy.org/b135.html). Less than 20 human enzymes have been shown to use complex dietary carbohydrates as substrates for hydrolytic degradation. Intestinal bacteria encode a greater number and variety of CAZymes than do the microbes found at other body sites in healthy adults (55). These findings reveal the importance of the intestinal microbiome (vs the human genome) in the metabolism of dietary carbohydrates.
The types of carbohydrates in the human diet can also affect the composition of the bacteria in the human intestinal microbiome. Conversely, the composition of the community can affect its ability to metabolize dietary carbohydrates. Plant starch, which is rich in amylopectin or amylose and a common component of today’s diet, can be metabolized by colonic bacteria such as Bifidobacterium, Bacteroides, and Fusobacterium spp. (56). Individuals who consume diets rich in plant carbohydrates appear to have larger proportions of Prevotella species (21), whereas those who consume animal fat and protein have higher proportions of Bacteroides species. Likewise, relative consumption of vegetables, fruit, and meat has been reported to be an important factor driving the composition of the GI microbiome and health in the elderly (7).
The relative abilities of GI bacteria to metabolize molecules from various food groups may provide opportunities for dietary modification and functional optimization of the microbiome. The original purpose of probiotics was to provide microbial lactase, in dairy microbes, to facilitate digestion of lactose by humans (reviewed in (57)). By-products of carbohydrate metabolism could provide additional opportunities for intestinal microbes to provide health benefits. Carbohydrate oxidation and microbial metabolism in the intestine can lead to production of oxalate; calcium oxalate is the most common form of renal stones. GI bacteria such as Oxalobacter formigenes, Lactobacillus spp., and Bifidobacterium spp. can metabolize oxalate in the intestine and might reduce risk for kidney stones (58, 59). O formigenes is a strict colonic anaerobe that provides one of the few clearly demonstrated health benefits of the intestinal microbiome. Catabolism of resistant starch or indigestible carbohydrates by the microbes eventually yields short chain fatty acids such as butyrate, acetate, and propionate (60). These promote health and provide resistance to infection. Other bacterial populations associated with a healthy intestine include active butyrate-producing microbes (12). Butyrate production can regulate the differentiation of colonic T regulatory cells and influence inflammatory and allergic responses (61). The ability to generate acetate was associated with resistance to damage by Shiga toxin-producing E coli (62).
Protein and Amino Acid Metabolism – Luminal Conversion
GI microbes can catabolize protein to amino acids and participate in the luminal conversion of amino acids to biogenic amines, immunomodulatory compounds, and other signaling molecules (Figure 2). Proteins from the diet may be catabolized by microbial proteinases and peptidases in cooperation with human proteinases. The generation of free amino acids by protein catabolism in the stomach and small intestine provide substrates for luminal conversion of amino acids.
Amino acids can be converted by decarboxylation reactions and catalyzed by GI bacteria to form different signaling molecules. L-histidine can be converted to the biogenic amine, histamine, by histidine decarboxylases (encoded by bacterial hdcA genes) produced by GI bacteria; histamine can then be transported to the intestinal lumen by the histidine:histamine anti-porter. Histamine can suppress production of inflammatory cytokines by signaling through the histamine type 2 receptor, which is present on intestinal epithelial cells (63, 64). Other examples of microbiome-mediated amino acid decarboxylation include the conversion of glutamate to gamma-aminobutyric acid (GABA), tyrosine to tyramine, and phenylalanine to β-phenylethylamine. Bacterial gadB genes encode glutamate decarboxylases, which can generate the neurotransmitter and potential immunomodulin GABA in the intestine (65).
Each of these luminal conversion systems include amino acid transporters that facilitate entry of amino acid from the lumen to the bacterial cells, and promote the extrusion of signaling compounds generated by amino acid decarboxylation.
In addition to the generation of small signaling molecules, bacteria can produce antimicrobial peptides or bacteriocins that confer resilience or resistance to infection by enteric pathogens (66). The production of anti-microbial compounds by commensals in the microbiome might prevent expansion of enteric pathogens or indigenous pathobionts (67). The generation of anti-microbial peptides, immunomodulatory compounds, and neurotransmitters by the microbiome could therefore affect the performance of the mucosal immune system and enteric nervous system, and be used in microbiome-mediated therapies.
Summary and Future Directions
The healthy GI microbiome can be described in terms of global parameters (richness, diversity), compositional features (bacterial phyla, taxa), and functional features (metabolic modules and pathways). Animal models of human disease, including gnotobiotic animals, and advances with organoids (and enteroids) will provide opportunities for scientists to determine the functions and specific health benefits provided different features of the GI microbiome. As more multi-‘omics studies of the human microbiome are completed, scientists may define additional features associated with a healthy microbiome. Microbiomes of healthy individuals vary with sex, race/ethnicity, and age; and personalized microbiome information can be given broader context when compared with databases such as that of the American Gut project (http://americangut.org). Strategies to prevent disease and interventions to improve health might be directed towards a new setpoint for any individual’s intestinal microbiome (68). Dietary modulation, fecal microbiome transplantation, or other efforts to manipulate microbial communities may leverage beneficial features of healthy microbiomes to define treatment goals.
Acknowledgements
This work was supported in part by research support from the National Institutes of Health (NIH; R01 AT004326, UH3 DK083990, U01 CA170930). We also acknowledge the support of the NIH (NIDDK)-funded (DK56338) Texas Medical Center Digestive Diseases Center. J. Versalovic received unrestricted research support from BioGaia AB (Stockholm, Sweden).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Metchnikoff I. The Prolongation of Life Optimistic Studies. Springer Publishing Company; 2004. [Google Scholar]
- 2.Pirisi A. Phage therapy--advantages over antibiotics? Lancet. 2000;356(9239):1418. doi: 10.1016/S0140-6736(05)74059-9. [DOI] [PubMed] [Google Scholar]
- 3.Keen EC. Phage therapy: concept to cure. Front Microbiol. 2012;3:238. doi: 10.3389/fmicb.2012.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huttenhower C, Gevers D, Knight R, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Hoenig JD, Malin KJ, et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3(8):944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisgaard H, Li N, Bonnelykke K, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128(3):646–652. doi: 10.1016/j.jaci.2011.04.060. [DOI] [PubMed] [Google Scholar]
- 7.Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 8.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 10.Pei Z, Bini EJ, Yang L, et al. Bacterial biota in the human distal esophagus. Proc Natl Acad Sci U S A. 2004;101(12):4250–4255. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersson AF, Lindberg M, Jakobsson H, et al. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE. 2008;3(7):e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu S, Wang Y, Lichtenstein L, et al. Regional differences in colonic mucosa-associated microbiota determine the physiological expression of host heat shock proteins. Am J Physiol Gastrointest Liver Physiol. 2010;299(6):G1266–G1275. doi: 10.1152/ajpgi.00357.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrell L, Wang Y, Antonopoulos D, et al. Standard colonic lavage alters the natural state of mucosal-associated microbiota in the human colon. PLoS One. 2012;7(2):e32545. doi: 10.1371/journal.pone.0032545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson BR, O'Rourke JL, Neilan BA, et al. Mucispirillum schaedleri gen. nov, sp. nov, a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol. 2005;55(Pt 3):1199–1204. doi: 10.1099/ijs.0.63472-0. [DOI] [PubMed] [Google Scholar]
- 16.Aagaard K, Petrosino J, Keitel W, et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. 2013;27(3):1012–1022. doi: 10.1096/fj.12-220806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107(33):14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.David LA, Maurice CD, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013 doi: 10.1038/nature12820. (nature12820). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ridaura VK, Faith JJ, Rey FE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jalanka-Tuovinen J, Salonen A, Nikkila J, et al. Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS ONE. 2011;6(7):e23035. doi: 10.1371/journal.pone.0023035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L, Lu X, Nossa CW, et al. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology. 2009;137(2):588–597. doi: 10.1053/j.gastro.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bik EM, Eckburg PB, Gill SR, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006;103(3):732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blaser MJ. Hypothesis: the changing relationships of Helicobacter pylori and humans: implications for health and disease. J Infect Dis. 1999;179(6):1523–1530. doi: 10.1086/314785. [DOI] [PubMed] [Google Scholar]
- 27.Skar V, Skar AG, Bratlie J, et al. Beta-glucuronidase activity in the bile of gallstone patients both with and without duodenal diverticula. Scand J Gastroenterol. 1989;24(2):205–212. doi: 10.3109/00365528909093038. [DOI] [PubMed] [Google Scholar]
- 28.Justesen T, Nielsen OH, Jacobsen IE, et al. The normal cultivable microflora in upper jejunal fluid in healthy adults. Scand J Gastroenterol. 1984;19(2):279–282. [PubMed] [Google Scholar]
- 29.Wilson M. Bacteriology of Humans: an Ecological Perspective. Blackwell Publishing LTD; 2008. [Google Scholar]
- 30.Methe BA, Nelson KE, Pop M, et al. A framework for human microbiome research. Nature. 2012;486(7402):215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type-2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 32.Li K, Bihan M, Yooseph S, et al. Analyses of the microbial diversity across the human microbiome. PLoS One. 7(6):e32118. doi: 10.1371/journal.pone.0032118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pflughoeft KJ, Versalovic J. Human Microbiome in Health and Disease. Annual Review of Pathology: Mechanisms of Disease. 2011;7:99–122. doi: 10.1146/annurev-pathol-011811-132421. [DOI] [PubMed] [Google Scholar]
- 34.Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 35.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Backhed F, Fraser CM, Ringel Y, et al. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12(5):611–622. doi: 10.1016/j.chom.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 37.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 38.Backhed F, Ley RE, Sonnenburg JL, et al. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 39.Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69(5):1046S–1051S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- 40.Hansen CH, Nielsen DS, Kverka M, et al. Patterns of early gut colonization shape future immune responses of the host. PLoS One. 2012;7(3):e34043. doi: 10.1371/journal.pone.0034043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jimenez E, Marin ML, Martin R, et al. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008;159(3):187–193. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 42.Gosalbes MJ, Llop S, Valles Y, et al. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin Exp Allergy. 2013;43(2):198–211. doi: 10.1111/cea.12063. [DOI] [PubMed] [Google Scholar]
- 43.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69(5):1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 45.Enck P, Zimmermann K, Rusch K, et al. The effects of maturation on the colonic microflora in infancy and childhood. Gastroenterol Res Pract. 2009;2009:752401. doi: 10.1155/2009/752401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmer C, Bik EM, DiGiulio DB, et al. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Segata N, Haake SK, Mannon P, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13(6):R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ringel-Kulka T, Cheng J, Ringel Y, et al. Intestinal microbiota in healthy U.S. young children and adults--a high throughput microarray analysis. PLoS One. 2013;8(5):e64315. doi: 10.1371/journal.pone.0064315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agans R, Rigsbee L, Kenche H, et al. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol Ecol. 2011;77(2):404–412. doi: 10.1111/j.1574-6941.2011.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biagi E, Nylund L, Candela M, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5(5):e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Claesson MJ, Cusack S, O'Sullivan O, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mariat D, Firmesse O, Levenez F, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lan Y, Kriete A, Rosen G. Selecting age-related functional characteristics in the human gut microbiome. Microbiome. 2013;1:2. doi: 10.1186/2049-2618-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tiihonen K, Ouwehand AC, Rautonen N. Human intestinal microbiota and healthy ageing. Ageing Res Rev. 2010;9(2):107–116. doi: 10.1016/j.arr.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 55.Cantarel BL, Lombard V, Henrissat B. Complex carbohydrate utilization by the healthy human microbiome. PLoS One. 2012;7(6):e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X, Conway PL, Brown IL, et al. In vitro utilization of amylopectin and highamylose maize (Amylomaize) starch granules by human colonic bacteria. Appl Environ Microbiol. 1999;65(11):4848–4854. doi: 10.1128/aem.65.11.4848-4854.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaukat A, Levitt MD, Taylor BC, et al. Systematic review: effective management strategies for lactose intolerance. Ann Intern Med. 2010;152(12):797–803. doi: 10.7326/0003-4819-152-12-201006150-00241. [DOI] [PubMed] [Google Scholar]
- 58.Sidhu H, Hoppe B, Hesse A, et al. Absence of Oxalobacter formigenes in cystic fibrosis patients: a risk factor for hyperoxaluria. Lancet. 1998;352(9133):1026–1029. doi: 10.1016/S0140-6736(98)03038-4. [DOI] [PubMed] [Google Scholar]
- 59.Magwira CA, Kullin B, Lewandowski S, et al. Diversity of faecal oxalate-degrading bacteria in black and white South African study groups: insights into understanding the rarity of urolithiasis in the black group. J Appl Microbiol. 2012;113(2):418–428. doi: 10.1111/j.1365-2672.2012.05346.x. [DOI] [PubMed] [Google Scholar]
- 60.Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10(4):336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;12721 doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 62.Fukuda S, Toh H, Hase K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469(7331):543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 63.Thomas CM, Hong T, van Pijkeren JP, et al. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS ONE. 2012;7(2):e31951. doi: 10.1371/journal.pone.0031951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sander LE, Lorentz A, Sellge G, et al. Selective expression of histamine receptors H1R, H2R, and H4R, but not H3R, in the human intestinal tract. Gut. 2006;55(4):498–504. doi: 10.1136/gut.2004.061762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Biase D, Pennacchietti E. Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol Microbiol. 2012;86(4):770–786. doi: 10.1111/mmi.12020. [DOI] [PubMed] [Google Scholar]
- 66.O'Shea EF, Cotter PD, Stanton C, et al. Production of bioactive substances by intestinal bacteria as a basis for explaining probiotic mechanisms: bacteriocins and conjugated linoleic acid. Int J Food Microbiol. 2012;152(3):189–205. doi: 10.1016/j.ijfoodmicro.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 67.Kamada N, Seo SU, Chen GY, et al. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 68.Lemon KP, Armitage GC, Relman DA, et al. Microbiota-targeted therapies: an ecological perspective. Sci Transl Med. 2012;4(137):137rv135. doi: 10.1126/scitranslmed.3004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Preidis GA, Versalovic J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology. 2009;136(6):2015–2031. doi: 10.1053/j.gastro.2009.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]