Abstract
Purpose
We examined in a prospective, randomized, international clinical trial the performance of a previously defined 30-gene predictor (DLDA-30) of pathologic complete response (pCR) to preoperative weekly paclitaxel and fluorouracil, doxorubicin, cyclophosphamide (T/FAC) chemotherapy, and assessed if DLDA-30 also predicts increased sensitivity to FAC-only chemotherapy. We compared the pCR rates after T/FAC versus FAC×6 preoperative chemotherapy. We also performed an exploratory analysis to identify novel candidate genes that differentially predict response in the two treatment arms.
Experimental Design
273 patients were randomly assigned to receive either weekly paclitaxel × 12 followed by FAC × 4 (T/FAC, n=138), or FAC × 6 (n=135) neoadjuvant chemotherapy. All patients underwent a pretreatment FNA biopsy of the tumor for gene expression profiling and treatment response prediction.
Results
The pCR rates were 19% and 9% in the T/FAC and FAC arms, respectively (p<0.05). In the T/FAC arm, the positive predictive value (PPV) of the genomic predictor was 38% (95%CI:21–56%), the negative predictive value (NPV) 88% (CI:77–95%) and the AUC 0.711. In the FAC arm, the PPV was 9% (CI:1–29%) and the AUC 0.584. This suggests that the genomic predictor may have regimen-specificity. Its performance was similar to a clinical variable-based predictor nomogram.
Conclusions
Gene expression profiling for prospective response prediction was feasible in this international trial. The 30-gene predictor can identify patients with greater than average sensitivity to T/FAC chemotherapy. However, it captured molecular equivalents of clinical phenotype. Next generation predictive markers will need to be developed separately for different molecular subsets of breast cancers.
INTRODUCTION
There are several clinical and molecular features that can identify generally more or less chemotherapy sensitive subsets of breast cancers but there are no clinically useful predictive biomarkers to select one chemotherapy regimen over another. Preoperative (neoadjuvant) chemotherapy provides a direct opportunity to assess treatment sensitivity in early stage breast cancers and pathologic complete response (pCR) is a powerful early surrogate of long-term survival (1,2). Recent studies have established that basal-like or triple receptor-negative breast cancers include a greater proportion of highly chemotherapy sensitive tumors, reflected by the significantly higher pCR rates, compared to ER-positive breast cancers (3,4). Among the ER-positive cancers, high Oncotype DX recurrence score (Genomic Health Inc, Redwood City, CA), Luminal-B molecular class, HER-2 overexpression and high histologic or genomic grade are associated with greater chemotherapy sensitivity (5,6,7). However, these molecular and clinicopathologic variables appear to predict general chemotherapy sensitivity, and have limited value in guiding the choice of a specific treatment regimen. Among several other markers, high expression/amplification of Topoisomerase II alpha and low expression of microtubule binding protein Tau have recently been suggested as potential predictors of sensitivity to anthracyclines and taxanes, respectively (8,9). However, neither these nor other proposed markers showed consistent clinically useful predictive value (10,11,12).
We previously developed a genomic predictor of pathologic complete response to preoperative sequential weekly paclitaxel followed by fluorouracil, doxorubicin and cyclophosphamide (T/FAC) chemotherapy from a single-arm retrospective study that included 82 patients in the discovery and 51 in the validation phase (13). The genomic predictor uses information from 30 different probe sets (i.e. genes) and employs diagonal linear discriminant analysis for prediction rules and is therefore referred to as the DLDA-30 predictor. The goal of the current study was to evaluate the predictive performance and assess the regimen specificity of this multi-gene predictor in a prospective, two-arm, randomized, multi-center, international, neoadjuvant clinical trial. Two commonly used, standard chemotherapy regimens including T/FAC and FAC-alone were compared for pCR rates. The predictive performance of the genomic signature was assessed independently in each treatment arm and a marker-treatment-outcome interaction test was performed. We also examined the performance of our previously reported clinical-pathologic variable-based nomogram to predict pCR (14). It remains unknown if the sequential paclitaxel/FAC regimen are superior to 6 courses of FAC or FEC (the control arm of this study) in terms of pathologic response rates or survival. Therefore, we also compared the pCR rates for patients who received T/FAC versus FAC×6 preoperative chemotherapy in this trial.
MATERIALS AND METHODS
Patient eligibility
Patients with clinical stage I–III breast cancer were eligible. Histological diagnosis of invasive cancer and estrogen, progesterone and HER-2 receptor status were determined from a diagnostic core needle or incisional biopsy before therapy. All patients had to agree to a separate, pre-treatment research fine needle aspiration (FNA) of the cancer for gene expression analysis. Patients were accrued at six international sites including The University of Texas M. D. Anderson Cancer Center (MDACC, n=96) and the Lyndon B Johnson General Hospital (LBJ, n=19) in Houston, TX, USA; the Instituto Nacional de Enfermedades Neoplasicas in Lima, Peru (n=79); the Centro Medico Nacional de Occidente in Guadalajara, Mexico (n=19); and the clinical trial group Grupo Español de Investigacion en Cancer de Mama (GEICAM) in Spain (n=60). This study was approved by the institutional review boards of each participating institution and all patients signed an informed consent for voluntary participation. The study was conducted between October 2003 and October 2006.
Treatment
Treatment was not selected based on gene expression results, patients were centrally randomized with blocked randomization at MDACC into one of two treatment arms; Arm A: (T/FAC) weekly paclitaxel 80 mg/m2/week × 12 courses followed by 5-fluorouracil 500 mg/m2, doxorubicin 50 mg/m2, and cyclophosphamide 500 mg/m2 all on day 1 repeated in 21-day cycles × 4 courses. Epirubicin (100 mg/m2) could be substituted for doxorubicin at the discretion of local investigators; Arm B: FAC (or FEC if epirubicin was used) × 6 courses at the same doses and schedule as above. Toxicity information was not collected during this trial because the two treatment arms were considered standard community-based therapy. Any patient developing unacceptable grade 3 or grade 4 toxicity was removed from the study. Patients with clinical or radiological disease progression were considered as having residual disease (RD) in the final analysis. After completion of neoadjuvant chemotherapy, all patients underwent modified radical mastectomy or lumpectomy and sentinel lymph node biopsy or axillary node dissection as determined by the surgeon; pCR was defined as the complete absence of invasive cancer cells in the breast and lymph nodes (15). Pathologic response was centrally reviewed by a breast pathologist (WFS).
Gene expression analysis and response prediction
Two to three FNA passes obtained with 23- or 25-gauge needles were collected into vials containing 1 mL RNA later solution (Ambion, Austin, TX) and stored at 4°C until mailed to MDACC in a cooler pack or dry ice. At MDACC, specimens were stored at −80°C until gene expression profiling on Affymetrix U133A gene chips (Santa Clara, CA). The same array platform, standard operating procedure and normalization method (dCHIP) was used as previously reported (13). The reference chip and normalization procedure are available online1. FNA samples contain on average 80% neoplastic cells and little or no normal breast epithelium or stromal cells (16). Gene expression information generated from FNAs represents the molecular characteristics of the invasive cancer, including the molecular class (3). RNA was extracted from FNA samples using the RNAeasy Kit (Qiagen, Valencia, CA). The amount and quality of RNA were assessed with DU-640 UV Spectrophotometer (Beckman Coulter, Fullerton, CA), and they were considered adequate for further analysis if the optical density260/280 ratio was ≥1.8 and the total RNA yield was ≥1µg. Seventy-five percent of all aspirations yielded at least 1 µg total RNA required for the gene expression profiling. Previously, thirty-one total RNA specimens were split, labeled, and hybridized in duplicates several months apart in the same and in a different laboratory to assess technical reproducibility of gene expression–based predictions, and demonstrated 97% concordance in these replicate experiments (13). Gene expression profiling was performed on Affymetrix U133A gene chips in batches over a 3 year period at MD Anderson Cancer Center. Genomic prediction of response was performed using a standardized computer code (13,17). Expression results from the thirty selected predictor genes were entered into the class prediction algorithm and each case was assigned a response status of “pCR” or “residual disease (RD)” prospectively, before actual pathologic outcome data became available. The complete microarray data of this trial is available at the GEO database accession number GSE20271.
We also evaluated the predictive performance of a previously established multivariate clinical nomogram that is freely available online2 (14). This nomogram combines information from patient age, tumor size, histologic grade and estrogen receptor status to predict probability of pCR after preoperative T/FAC or FAC chemotherapies. None of the current cases were included in the development of the genomic or clinical response predictors.
Predictive performances are presented as sensitivity, specificity, positive and negative predictive values and area under the receiver operating characteristic (ROC) curve (AUC). For the clinical nomogram, we also report calibration (i.e., agreement between observed outcome frequencies and predicted probabilities) and discrimination (i.e., whether the relative ranking of individual predictions is in the correct order).
Statistical design
The primary objective of this study was to establish that patients with DLDA-30-positve tumors are significantly more likely to experience pCR to T/FAC chemotherapy than patients who are predicted to have residual cancer by the genomic predictor. The secondary objectives of the study were to compare the pCR rates between the sequential T/FAC and FAC×6 treatment arms, and to assess interaction between prediction status assigned by the DLDA-30 predictor and pCR to two different neoadjuvant chemotherapies. The primary endpoint of this study was to assess the rate of pCR after completion of preoperative chemotherapy. Sample size was calculated based on the primary objective using computer simulations. Computer simulations were carried out to estimate the power to detect gene expression profile effects and profile-by-treatment interaction effects at different sample sizes. The following assumptions were used based on the original discovery and validation results (13): (i) the prevalence of DLDA-30 marker-positive patients is 30% (since the pCR rate after T/FAC is expected to be approximately 25–30% in unselected patients), (ii) pCR rate to T/FAC treatment in the marker-positive group is ≥ 60% (based on the positive predictive value observed in the previous single arm study) (13), and (iii) pCR rate to FAC treatment in the marker-positive group is between 20–40% (higher than 10–15% seen historically in unselected patients but lower than observed for T/FAC). Repeated fitting of a logistic regression model with 10,000 iterations for each case were performed with pCR as the dependent variable and including terms for treatment, microarray profile group, and treatment-profile interaction. With these assumptions, a study with 210 patient (105 in each arm) would have a ≥95% power to detect a significant marker effect for T/FAC therapy (i.e. significantly higher pCR rates in marker-positive compared to marker-negative cases). However, with this sample size the power varies substantially to detect significant marker-treatment arm interaction effect depending on differences in pCR rates between the treatment arms (Supplementary Table 1). We assumed a 25–30% loss of samples and therefore the maximum sample size was set to 273. In the final analysis we used multivariate logistic regression models with terms for age, treatment, tumor size, grade, ER and HER-2 status and genomic prediction to calculate odds ratios for pCR.
In a hypothesis-generating, exploratory analysis we searched for novel differentially predictive genes by fitting logistic regression models with response (pCR vs. RD) as the outcome and with treatment type (FAC vs. TFAC) and gene expression values of gene i (for each probe set on the U133A chip) as covariates. We included a test for interaction between treatment and gene expression, and calculated the p-value for the gene-treatment interaction term. To adjust for multiple testing, we used a beta-uniform mixture (BUM) model to estimate the false discovery rate (FDR) (18). In order to explore the power of the interaction test in the completed study, we generated 4 random normal distributions representing particular gene expression values - one for each treatment-response group. The values for the means and standard deviations were taken from the observed normalized and log2 transformed microarray data. Each distribution had standard deviation = 0.3 and sample size equal to that from the data (pCR/FAC: n=7, RD/FAC: n=80, pCR/TFAC: n=19, RD/TFAC: n=72), we set the means for pCR/FAC and RD/FAC to 2.5 (i.e. no effect on FAC response by gene expression), we also set the mean for RD/TFAC to 2.5 and varied the mean for pCR/TFAC from 2.5 to 3.2. We fit the logistic regression model described above. Over 50 iterations, we tracked how often the interaction test p-value was <0.05 for a given gene and took this value as the measure of the power of the test. When the pCR/TFAC mean was 2.6 the power was 14%. For 2.7, the power was 30%; for 2.8, the power was 51%; for 2.9, the power was 72%; for 3.0, the power was 86%; for 3.1, the power was 92%; and for 3.2, the power of the interaction test was 98%.
RESULTS
Patient characteristics and response to chemotherapy
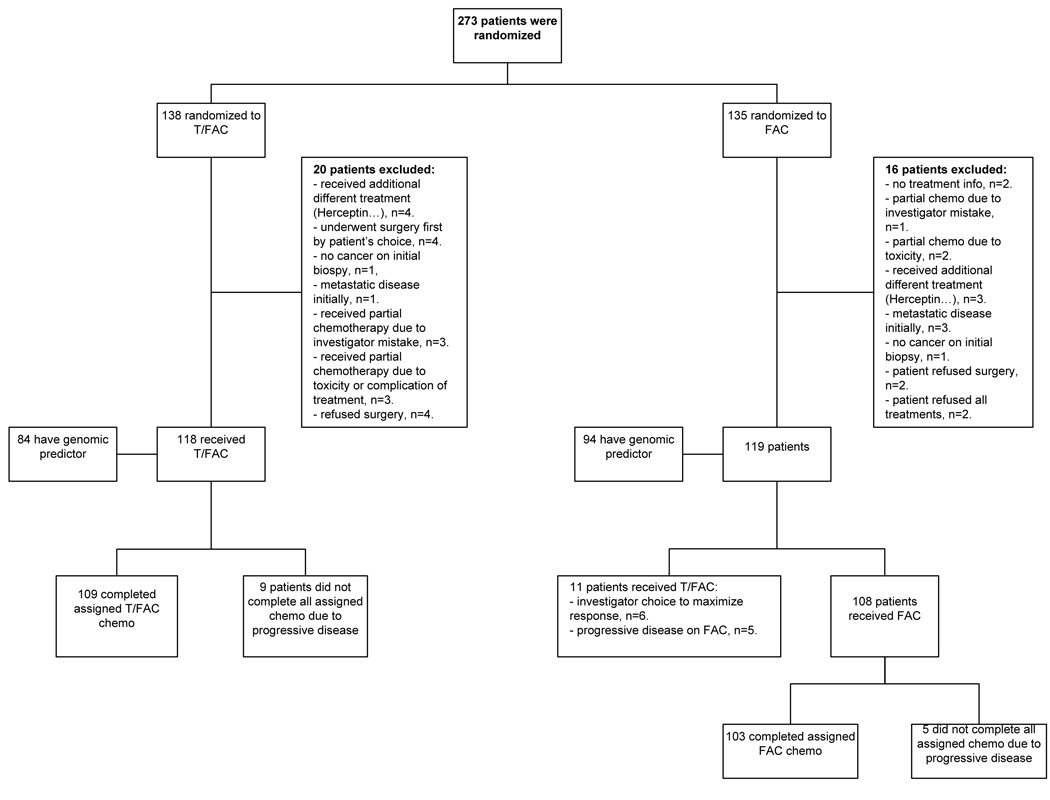
Two hundred and seventy three patients were enrolled, 138 were randomized to T/FAC and 135 to FAC chemotherapy (intent-to-treat population). Twenty (7%) and 16 (6%) patients were excluded from genomic response analysis in each treatment arm respectively due to eligibility violations including non-study treatment regimen, patient withdrawal or lack of pathologic assessment of response. Of the 118 patients who received T/FAC, 9 patients progressed clinically, these were considered as residual disease for response prediction analysis. Of the 119 patients who were assigned to receive FAC chemotherapy, 11 received T/FAC treatment (to maximize response or due to progression on FAC) and these cases were assigned to the T/FAC treatment group for genomic response prediction analysis. The remaining 108 cases, including 5 cases that progressed, comprised the FAC treatment cohort for the final response prediction analysis (Supplementary Table 2). Figure 1 illustrates the flow and assignment of specimens.
Figure 1. Flow chart of patients and samples used in the analysis.
Based on treatment received, the pCR rates were significantly higher 19% (n=24/129) in the T/FAC arm compared to 9% (n=10/113) in the FAC arm (p<0.05). For the calculation of FAC efficacy, the 5 cases that were resistant to FAC and were crossed over to the T/FAC arm were counted as FAC failures, thus n=108+5=113. In the intent-to-treat population, the pCR rates were the same as above, 19.6% (n=27/138) and 9.6% (n=13/135) in the T/FAC and FAC arms (p<0.05), respectively.
Two hundred and four FNA samples (75%) yielded sufficient quality and quantity of RNA to perform gene expression analysis. The main reasons for failure were acellular aspirates and low RNA yield; 5 profiles (2.5%) failed array QC after hybridization. After excluding the patients who had no response information available (Figure 1), 178 cases remained with complete pathologic response and genomic prediction results for final analysis. Of these, 91 received T/FAC and 87 received FAC chemotherapy. Clinical characteristics of these patients are presented in Table 1. Both treatment groups were well balanced for age, race, histological type and grade, tumor size, clinical nodal status, and hormone receptor and HER2 status. The pCR rates were 21% (95%CI:0.13–0.29) in the T/FAC group (n=19) and 8% (95%CI:0.02–0.14) in the FAC group (n=7) (p=0.019).
Table 1. Clincal characteristics of the 178 patients by treatment received for whom an observed pathologic tumor reponse and molecular prediction of tumor response are available.
| Patients received T/FAC | Patients received FAC | |||||
|---|---|---|---|---|---|---|
| Clinical and Pathological characteristics | No of patients | (%) | No of patients | (%) | p-value | |
| No of patients | 91 | 51.1 | 87 | 48.9 | ||
| Pathologic complete response (pCR) | 19 | 20.9 | 7 | 8.0 | 0.02 | |
| Residual disease (RD) | 72 | 79.1 | 80 | 92.0 | 0.02 | |
| Race | 0.42 | |||||
| White | 40 | 44.0 | 41 | 47.1 | ||
| Black | 9 | 9.9 | 4 | 4.6 | ||
| Hispanic | 41 | 45.1 | 42 | 48.3 | ||
| Asian | 1 | 1.1 | 0 | 0.0 | ||
| Mean age, years (range) | 51.5 (26–73) | 50.3 (31–74) | 0.41 | |||
| Menopausal status | 0.63 | |||||
| Premenopausal | 46 | 50.5 | 48 | 55.2 | ||
| Postmenopausal | 44 | 48.4 | 38 | 43.7 | ||
| Unknown | 1 | 1.1 | 1 | 1.1 | ||
| Histology | 0.11 | |||||
| Ductal | 81 | 89 | 83 | 96.7 | ||
| Lobular | 6 | 6.6 | 1 | 1.1 | ||
| Mixed | 4 | 4.4 | 2 | 2.2 | ||
| Clinical T size | 0.89 | |||||
| T0–T1 | 8 | 8.8 | 5 | 5.7 | ||
| T2 | 39 | 42.8 | 37 | 42.6 | ||
| T3 | 19 | 20.9 | 18 | 20.7 | ||
| T4 | 25 | 27.5 | 26 | 29.9 | ||
| unknown | 0 | 0.0 | 1 | 1.1 | ||
| Clinical N stage | 0.76 | |||||
| N0 | 31 | 34.1 | 28 | 32.3 | ||
| N1 | 38 | 41.7 | 33 | 37.9 | ||
| N2–3 | 22 | 24.2 | 25 | 28.7 | ||
| unknown | 0 | 0.0 | 1 | 1.1 | ||
| Grade* | 0.45 | |||||
| 1 | 10 | 11 | 5 | 5.7 | ||
| 2 | 30 | 33 | 31 | 35.6 | ||
| 3 | 36 | 39.5 | 36 | 41.5 | ||
| unknown | 15 | 16.5 | 15 | 17.2 | ||
| ER status** | 0.85 | |||||
| Negative | 42 | 46.2 | 38 | 43.7 | ||
| Positive | 49 | 53.8 | 49 | 55.7 | ||
| PR status** | 0.98 | |||||
| Negative | 49 | 53.8 | 46 | 52.9 | ||
| Positive | 42 | 46.2 | 41 | 47.1 | ||
| HER2 status*** | 0.35 | |||||
| Not overexpressed | 75 | 82.4 | 77 | 88.5 | ||
| Overexpressed | 16 | 17.6 | 10 | 11.5 | ||
| Patients underwent ALND or SLNB | 84 | 92.3 | 81 | 93 | 0.93 | |
| Mean number of LN removed (range) | 15.8 (1–38) | 15.8 (1–43) | 0.97 | |||
| Patients with ALN involvement | 46/84 | 46/81 | 0.9 | |||
| Type of surgery | 0.99 | |||||
| Breast conservation | 11 | 12.1 | 11 | 12.6 | ||
| Mastecotmy | 66 | 72.5 | 65 | 74.8 | ||
| Surgery not done due to PD | 6 | 6.6 | 6 | 6.9 | ||
| Unknown | 8 | 8.8 | 5 | 5.7 | ||
Abbreviations: FAC = 5-Fluorouracil, Doxorubicin and Cyclophosphamide; T/FAC = weekly Paclitaxel and 5-Fluorouracil Doxorubicin and Cyclophosphamide; ER = Estrogen receptor; PR = Progesterone receptor; ALN = axillary lymph node; ALND = ALN Dissection; SLNB = Sentinel lymph node biopsy; PD = Progressive disease;
Histological grade according to the modified Black’s nuclear grade;
Status for ER and PR was determined by immunohistochemistry;
Status for HER2 was determined by immunohiostochemistry or Fluorescence In Situ Hybridization.
Performance of the DLDA-30 genomic predictor and the clinical nomogram to predict pCR
In the T/FAC arm, the positive predictive value (PPV) of the genomic predictor was 38% (95%CI:21–56%), the negative predictive value (NPV) was 88% (CI:77–95%), sensitivity and specificity were 63% (CI:38–84%) and 72% (CI:60–82%), respectively. The observed pCR rate was 38% in the cohort that was predicted to achieve pCR (marker-positive patients), compared to 19% in unselected patients (p=0.032), and 12% in the marker-negative patients (patients predicted to have residual disease) (p=0.006). The AUC was 0.711 (CI:0.570–0.852). In the FAC-only arm, the PPV and NPV were 9% (CI:1–29%), and 92% (CI:83–97%), respectively. The sensitivity and specificity were 29% (CI:4–71%) and 75% (CI:64–84%), and the AUC was 0.584 (CI:0.353–0.815) (Figure 2A and Table 2). The observed pCR rates were identical (9%) in the overall population and in the marker-positive and -negative patient subsets.
Figure 2. Receiver operating characteristic (ROC) and calibration curves measure the performance of the genomic and clinical predictors.
(A) ROC curve for the predictive performance of the DLDA-30 genomic pCR predictor on the 178 cases for whom an observed pathologic tumor response and molecular prediction of tumor response were available. (B) ROC and calibration curves for the predictive performance of the clinical nomogram to predict pCR on the same cases used in the genomic analysis. The corresponding performance metrics of the genomic and clinical predictors are found in Table 2.
Abbreviations: FAC = 5-Fluorouracil, Doxorubicin and Cyclophosphamide; T/FAC = weekly Paclitaxel and 5-Fluorouracil, Doxorubicin and Cyclophosphamide; AUC = Area Under the ROC Curve.
Table 2. Performance metrics of the genomic and clinical predictors.
The corresponding ROC and calibration curves are represented graphically in Figure 2.
| T/FAC (n=91) | FACx6 (n=87) | |||
|---|---|---|---|---|
| Genomic predictor | ROC | AUC | 0.711 (95%CI:0.570–0.852) | 0.584 (95%CI:0.353–0.815) |
| PPV | 38% (95%CI:21–56) | 9% (95%CI:1–29) | ||
| NPV | 88% (95%CI:77–95) | 92% (95%CI:83–97) | ||
| Sensitivity | 63% (95%CI:38–84) | 29% (95%CI:4–71) | ||
| Specificity | 72% (95%CI:60–82) | 75% (95%CI:64–84) | ||
| Clinical predictor | Discrimination (ROC) | AUC | 0.89 (95%CI:0.85–0.93) | 0.82 (95%CI:0.75–0.89) |
| Calibration | p-value | 0.21 | 0.03 | |
| E max | 10.5% | 15.5% | ||
| E average | 4.8% | 9.2% | ||
Abbreviations: FAC = 5-Fluorouracil, Doxorubicin and Cyclophosphamide; T/FAC = weekly Paclitaxel and 5-Fluorouracil, Doxorubicin and Cyclophosphamide; AUC = Area Under the ROC Curve, ROC = receiver operating characteristic, CI = confidence interval; E = difference in predicted probabilities and observed frequencies; E max = maximal error, E average= average error.
We applied the clinical nomogram to the same 178 patients for which microarray-based prediction was available (Figure 2B and Table 2). In the T/FAC arm, the discrimination was high with an AUC of 0.89 (95%CI:0.85–0.93), which was not statistically significantly different from the AUC of the DLDA-30 predictor. The model was also well calibrated with no significant difference between the predicted and the observed probability (p=0.21). When the nomogram was applied to patients who received FAC, the discrimination remained high with an AUC of 0.82 (95%CI: 0.75–0.89), but the calibration was less good, the nomogram significantly (p=0.03) over-predicted pCR. Thus, the clinical predictor was accurate in predicting pCR after T/FAC, and less accurate but still effective in predicting pCR after FAC×6.
Multivariate logistic regression model
In a multivariate analysis using all samples (n= 178) and including treatment arm, age, clinical tumor size, clinical nodal status, grade, ER and HER2 status, and DLDA-30 score as variables, only ER status (p=0.008), tumor size (p=0.018), and treatment arm (p=0.015) were significant independent predictors of pCR. In the T/FAC treatment cohort, only ER status (p=0.022) and tumor size (p=0.046) were independent predictors of pCR. In the FAC treatment cohort, none of the variables was a significant predictor for pCR. The small number of events (seven pCR) and lack of power may have prevented the identification of any significant predictors with confidence within this subset.
New candidate biomarker identification using marker-treatment interaction test
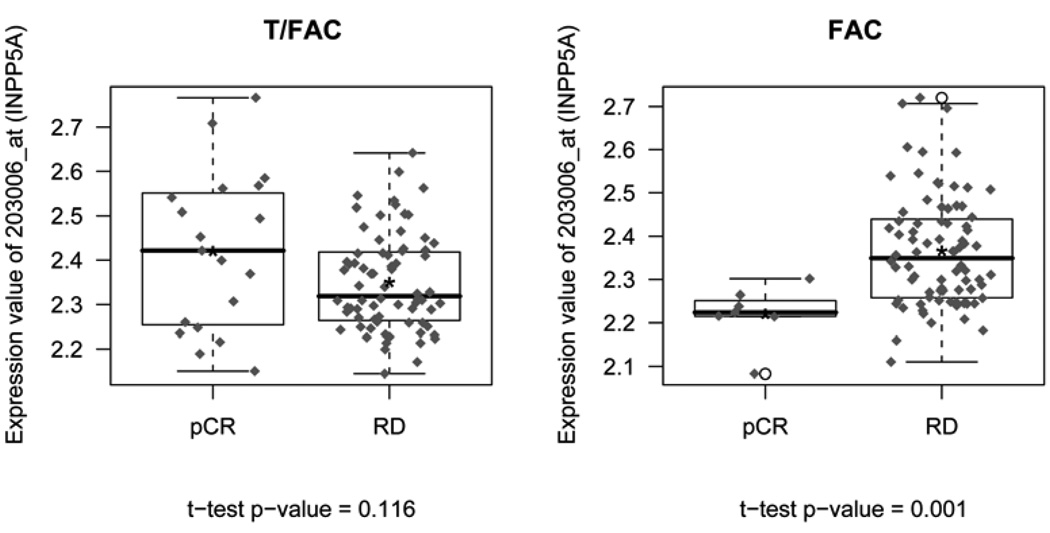
In an exploratory analysis, we examined what genes were differentially predictive of response (pCR vs RD) by treatment arm by testing for gene-treatment interaction. When all probe sets on the U133A microarray were considered individually, logistic regression models identified 206 probe sets with interaction p values ≤0.05 (Supplementary Table 3). The gene with the most significant gene expression-treatment interaction p-value was the inositol polyphosphate-5-phosphatase (INPP5A) (Figure 3). After adjustment for multiple comparisons using the beta uniform mixture model of p-values, no probe sets remained significant at a reasonable false detection rate (FDR). The p-value distribution showed a paucity of low p-values indicating a lack of power for the individual comparisons.
Figure 3. Expression of the inositol polyphosphate-5-phosphatase (INPP5A) gene in the two response cohorts within the two treatment arms.
INPP5A had the most significant gene expression-treatment arm interaction p-value (p= 0.00193, unadjusted for multiple comparisons). “*” indicates the mean value.
Abbreviations: pCR=pathologic complete response, RD=residual cancer, T/FAC= weekly Paclitaxel and 5-Fluorouracil, Doxorubicin and Cyclophosphamide, FAC= 5-Fluorouracil, Doxorubicin and Cyclophosphamide.
Supplementary Table 4 contains the gene-treatment interaction results for the individual 30 probe sets included in the DLDA-30 predictor. The overall DLDA-30 score showed no significant interaction with treatment (p=0.443). Three of the individual probe sets including Na+/K+ transporting ATPase interacting protein 1 (NKAIN1), meteorin (METRN), and delta 2 catenin (CTNND2) showed significant interaction with treatment (p≤0.05), and one probe set (G protein-coupled receptor activity modifying protein 1, RAMP1) had borderline significance (p=0.051). Figure 4 shows the plots of the fitted logistic regression models for the 4 genes and for the overall score. The plots show how the probability of pCR varies by treatment arm as a function of gene expression level or DLDA-30 score values.
Figure 4. Plots of the fitted logistic regression models illustrating the gene-treatment interactions for the 4 most significant genes of the DLDA-30 predictor and the 30-gene prediction score itself.
The plots show how the probability of pCR varies by treatment arm as function of gene expression level or DLDA score values.
Retrospective power calculations for interaction test based on mean gene expression values for the DLDA-30 probe sets and observed response rates using logistic regression model, indicated that the current study had a power between 14%–50% to detect significant interaction effects.
DISCUSSION
We tested a multigene predictor of pCR to preoperative sequential weekly paclitaxel and FAC chemotherapy from fine-needle biopsies of breast cancers in a prospective randomized international trial. pCR is important as a clinical end point because these patients experience excellent cancer-free long-term survival.
The 30-gene predictor was predictive of response to T/FAC chemotherapy. Patients who were predicted to achieve pCR to T/FAC (marker-positive patients) had significantly higher pCR rates (38%, 95%CI:21–56%) than unselected patients (19%, p=0.032), or patients predicted to have residual disease (12%, p=0.006) (marker-negative patients) when treated with this regimen. These results confirm that the multigene predictor can identify patients with greater than average sensitivity to T/FAC chemotherapy; they are consistent with two previous small studies (13,17).
A test that could be used to select one therapy over another needs to have a high positive predictive value and high sensitivity. Our test achieved a PPV of 38%, a sensitivity of 63% and the negative predictive value (NPV) was 88%. These performance measures were less than what we have observed in the earlier validation studies but were still within the 95% confidence intervals of the earlier performance estimates. Therefore, these results are consistent with the previous reports (13,17).
It is increasingly clear that general chemotherapy sensitivity can also be gauged by considering routine clinical variables such as proliferative activity, ER and HER-2 status, molecular subtype and histologic grade. Basal-like (or triple-negative) breast cancers have higher likelihood of pCR after preoperative chemotherapy than other molecular subtypes, and among the ER-positive cancers, high Oncotype DX recurrence score, Luminal-B molecular class, HER-2 overexpression and high genomic grade are each associated with greater chemotherapy sensitivity (3–7). All these assays tend to capture molecular characteristics of similar patients with clinically ER-negative, and/or high grade and high proliferation tumors. When we compared the predictive performance of the DLDA-30 genomic test in T/FAC treated patients with a multivariate clinical prediction model including grade and ER status, the overall predictive performance of the genomic test was similar to the performance of the clinical nomogram. In a multivariate logistic regression analysis that included all patients, only ER-status, tumor size and treatment arm but not the genomic test results were independent predictors of pCR. This indicates that this first generation genomic chemotherapy response predictor mostly captures gene expression information associated with clinical phenotype, particularly ER status and proliferative activity that is reflected in histological grade. This study illustrates an inherent pitfall in developing predictive markers from patient cohorts that include different breast cancer subtypes. Estrogen receptor-positive and -negative cancers have large-scale gene expression differences, and they also have substantially different sensitivities to preoperative chemotherapy and this can lead to confounding of estrogen regulated genes with treatment response genes in studies that consider all breast cancers together. During the development of the DLDA-30 predictor cases with pCR were compared to cases with residual cancer and both ER-positive and -negative cancers were included in the analysis. Inevitably the predictor included many genes that reflected the phenotypic differences between responders (pCR) and non-responders (RD), responders being predominantly triple-negative cancers and highly proliferative ER+ cancers. To illustrate this point we tested the DLDA-30 pCR predictor as a predictor of ER-status: in the current study population it had a PPV of 0.7 and AUC of 0.766 as predictor of ER-negative versus -positive phenotype. Similar phenotype-associated predictive value limits the use of the 70-gene prognostic signature (MammaPrint) or OncotypeDx in ER-negative breast cancers (19). The next generation predictive (and prognostic) markers will need to be developed separately for different molecular subsets of breast cancers in order to increase their clinical utility (20).
A secondary objective of this randomized study was to evaluate the regimen specificity of the DLDA-30 predictor by assessing its performance on the FAC-treated patients. Excellent survival in patients who achieve pCR most likely reflects benefit from chemotherapy since most clinical and molecular characteristics associated with pCR (ie, ER-negative status, high histologic or genomic grade, high Oncotype DX recurrence score, basal-like or Luminal-B molecular class “intrinsic subtype”) predict worse natural history in the absence of chemotherapy (5,6,7). But the above variables predict general chemosensitivity of a tumor, which can be useful clinically. However more useful will be the predictive test that can discriminate between the probability of response to different chemotherapies and thus help guide the rational selection of specific treatment in individual patients. We tested the performance of the genomic predictor in the two treatment arms. The DLDA-30 identified a patient population that is more likely to achieve pCR after T/FAC chemotherapy than the general patient population, but it did not predict pCR in the FAC-only treatment arm. In that arm the PPV and NPV were 9% and 92% respectively and the sensitivity and specificity were 29% and 75% with an AUC of 0.584. This indicates that our genomic predictor may have some-regimen specificity. Alternatively, this may also represent a false-negative finding due to lack of power because of the small number of events in the FAC group (pCR n=7) and small overall sample size. The marker-treatment interaction test was also not significant, but post hoc power calculations showed 14–50% power to detect significant interaction effects for individual probe sets or for the combined DLDA-30 score. Interestingly, although the genomic predictor performed similarly to the clinical pCR predictor nomogram at predicting response to T/FAC, the clinical nomogram retained a comparable predictive accuracy in FAC-treated patients while the genomic test did not. This further supports some degree of regimen specific predictive value for the genomic test that the clinical predictor lacks.
The ideal data to develop treatment specific predictors is a randomized trial that is adequately powered to identify individual genes with significant treatment-marker interaction effect. However, such data sets are rare and prospective power calculations for gene-treatment interaction tests are difficult because the magnitude of effect is unknown and it is likely to be variable for different genes. The current data set provided an opportunity to perform an exploratory analysis. We examined each probe set on the array for potential marker-treatment interaction effect. This analysis was undertaken to assess if we could identify genes that have larger gene-treatment-response interaction effect than the probes sets that were included in the DLDA-30 predictor. We identified numerous, potential treatment-arm specific predictor candidates (Supplementary Table 3). These genes will need to be tested in other independent data sets.
We also show that weekly paclitaxel × 12 followed by FAC × 4 regimen results in significantly higher pCR rate (19% versus 9%, p<0.05) than 6 courses of FAC (or FEC). The pCR rates for T/FAC are consistent with findings from a larger study using the same preoperative therapy (21). These results support the increasing consensus that the addition of a taxane to anthracycline-based therapy improves pCR rates and long-term outcomes in breast cancer.
In summary, prospective gene expression analysis for response prediction was feasible in this randomized, international trial. Seventy-five percent of the FNA specimens mailed to a central laboratory yielded adequate RNA for genomic analysis. A 30-gene molecular test was predictive of pCR to T/FAC and not to FAC chemotherapy, but it did not perform better than a freely available web-based clinical nomogram. The clinical nomogram, however, lacked regimen-specificity and predicted response equally well to both T/FAC and FAC. Like most other currently in use molecular response predictors, that rely on measuring molecular equivalents of clinical phenotype, this first generation genomic predictor derives its predictive value from detecting the large-scale gene expression differences that distinguish ER-negative from ER-positive tumors and high grade from low grade cancers. To improve their clinical utility, second generation genomic predictors will need to be developed separately for the different molecular and phenotypic subsets of breast cancers.
TRANSLATIONAL RELEVANCE
There are several clinical and molecular features that can identify generally more or less chemotherapy sensitive subsets of breast cancers but there are no clinically useful predictive biomarkers that can guide the selection of one chemotherapy regimen over another in individual patients. We tested a 30-gene predictor of pathologic complete response to neoadjuvant sequential weekly paclitaxel and FAC chemotherapy from fine-needle biopsies of breast cancers in a prospective randomized international trial. We also compared the treatment efficacy of T/FAC vs FAC×6 chemotherapies. The results confirm the ability of the genomic predictor to identify patients with higher than average sensitivity to T/FAC chemotherapy but not to FAC treatment. Prospective gene expression biomarker analysis was technically and logistically feasible in this multicenter international trial. Most current molecular response predictors in breast cancer including this one suffer from capturing molecular equivalents of clinical phenotype. To improve their clinical utility, second generation genomic predictors will need to be developed separately for the different molecular and phenotypic subsets of breast cancers.
Supplementary Material
Acknowledgments
Grant support: Grants from the Breast Cancer Research Foundation and the Commonwealth Foundation to LP. There was no direct support from the pharmaceutical industry to conduct this trial.
Footnotes
Presented in part as an abstract at the 2009 32nd Annual AACR San Antonio Breast Cancer Symposium (SABCS), on Dec 9, 2009, in San Antonio, Texas. For this, the first author AT received an AACR Translational Research Scholar Award for Breast Cancer Research, supported by a grant from Susan G. Komen for the Cure.
Disclosure of Potential Conflicts of Interest:
Employment (other than primary affiliation, e.g., consulting): none.
Commercial Research Grant: none.
Other Commercial Research Support: none.
Honoraria from Speakers Bureau: none.
Ownership Interest (including patents): Minor <$10,000, W Fraser Symmans, stock ownership, Nuvera Biosciences.
Consultant/Advisory Board: Lajos Pusztai, Nuvera Biosciences (Uncompensated); W Fraser Symmans, Nuvera Biosciences (Uncompensated).
Other (e.g. expert testimony – please be specific): none.
REFERENCES
- 1.Fisher B, Bryant J, Wolmark N, et al. Effect of preoperative chemotherapy on the outcome of women with operable breast cancer. J Clin Oncol. 1998;16:2672–2685. doi: 10.1200/JCO.1998.16.8.2672. [DOI] [PubMed] [Google Scholar]
- 2.Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26(8):1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 3.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 4.Carey LA, Dees EC, Sawyer L, et al. The Triple Negative Paradox: Primary Tumor Chemosensitivity of Breast Cancer Subtypes. Clin Cancer Res. 2007;13:2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 5.Gianni L, Zambetti M, Clark K, et al. Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. J Clin Oncol. 2005;23:7265–7277. doi: 10.1200/JCO.2005.02.0818. [DOI] [PubMed] [Google Scholar]
- 6.Andre F, Mazouni C, Liedtke C, et al. HER2 expression and efficacy of preoperative paclitaxel/FAC chemotherapy in breast cancer. Breast Cancer Res Treat. 2008;108(2):183–190. doi: 10.1007/s10549-007-9594-8. [DOI] [PubMed] [Google Scholar]
- 7.Liedtke C, Hatzis C, Symmans WF, et al. Genomic grade index is associated with response to chemotherapy in patients with breast cancer. J Clin Oncol. 2009;27(19):3185–3191. doi: 10.1200/JCO.2008.18.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knoop AS, Knudsen H, Balslev E, et al. Retrospective analysis of Topoisomerase IIa amplification and deletions as predictive markers in primary breast cancer patients randomly assigned to cyclophosphamide, methotrexate, and fluorouracil or cyclophosphamide, epirubicin or fluorouracil: Danish Breast Cancer cooperative Group. J Clin Oncol. 2005;23:7483–7490. doi: 10.1200/JCO.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Rouzier R, Rajan R, Hess KR, et al. Microtubule associated protein tau is a predictive marker and modulator of response to paclitaxel-containing preoperative chemotherapy in breast cancer. Proc Natl Acad Sci U S A. 2005;102:8315–8320. doi: 10.1073/pnas.0408974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esteva FJ, Hortobagyi GN. Topoisomerase II alpha amplification and anthracycline-based chemotherapy: the jury is still out. J Clin Oncol. 2009;27:3416–3417. doi: 10.1200/JCO.2009.22.6449. [DOI] [PubMed] [Google Scholar]
- 11.Pusztai L, Jeong JH, Gong Y, et al. Evaluation of microtubule-associated protein-tau expression as a prognostic and predictive marker in NSABP-B 28 randomized clinical trial. J Clin Oncol. 2009;27(26):4287–4292. doi: 10.1200/JCO.2008.21.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pusztai L. Markers predicting clinical benefit in breast cancer from microtubule-targeting agents. Ann Oncol. 2007;18(12):xii15–xii20. doi: 10.1093/annonc/mdm534. [DOI] [PubMed] [Google Scholar]
- 13.Hess KR, Anderson K, Symmans WF, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24(26):4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 14.Rouzier R, Pusztai L, Delaloge S, et al. Nomograms to predict pathologic complete response and metastasis-free survival after preoperative chemotherapy for breast cancer. J Clin Oncol. 2005;23:8331–8339. doi: 10.1200/JCO.2005.01.2898. [DOI] [PubMed] [Google Scholar]
- 15.Mazouni C, Peintinger F, Kau SW, et al. Residual ductal carcinoma in situ in patients with complete eradication of invasive breast cancer after neoadjuvant chemotherapy does not adversely affect patient outcome. J Clin Oncol. 2007;25(19):2650–2655. doi: 10.1200/JCO.2006.08.2271. [DOI] [PubMed] [Google Scholar]
- 16.Symmans WF, Ayers M, Clark EA, et al. Total RNA yield and microarray gene expression profiles from fine needle aspiration and core needle biopsy samples of breast cancer. Cancer. 2003;97:2960–2971. doi: 10.1002/cncr.11435. [DOI] [PubMed] [Google Scholar]
- 17.Peintinger F, Anderson K, Mazouni C, et al. Thirty-gene pharmacogenomic test correlates with residual cancer burden after preoperative chemotherapy breast cancer. Clin Cancer Res. 2007;13(14):4078–4082. doi: 10.1158/1078-0432.CCR-06-2600. [DOI] [PubMed] [Google Scholar]
- 18.Pounds S, Morris SW. Estimating the occurrence of false positive and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics. 2003;19:1236–1242. doi: 10.1093/bioinformatics/btg148. [DOI] [PubMed] [Google Scholar]
- 19.Sotiriou C, Pusztai L. Gene expression signatures in breast cancer. N Engl J Med. 2009;360(8):790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 20.Hatzis C, Symmans WF, Lin F, et al. Genomic predictors of pathologic response to preoperative chemotherapy for triple-negative and ER-positive/HER2-negative breast cancers. Proc Am Soc Clin Oncol. 2008;26(15S):23s. (abstr 571) [Google Scholar]
- 21.Green MC, Buzdar AU, Smith T, et al. Weekly paclitaxel improves pathologic complete remission in operable breast cancer when compared with paclitaxel once every 3 weeks. J Clin Oncol. 2005;23:5983–5992. doi: 10.1200/JCO.2005.06.232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.