Abstract
The unlimited growth potential of tumors depends on telomere maintenance and typically depends on telomerase, an RNA-dependent DNA polymerase, which reverse transcribes the telomerase RNA template, synthesizing telomere repeats at the ends of chromosomes. Studies in various model organisms genetically deleted for telomerase indicate that several recombination-based mechanisms also contribute to telomere maintenance. Understanding the molecular basis of these mechanisms is critical since some human tumors form without telomerase, yet the sequence is maintained at the telomeres. Recombination-based mechanisms also likely contribute at some frequency to telomere maintenance in tumors expressing telomerase. Preventing telomere maintenance is predicted to impact tumor growth, yet inhibiting telomerase may select for the recombination-based mechanisms. Telomere recombination mechanisms likely involve altered or unregulated pathways of DNA repair. The use of some DNA damaging agents may encourage the use of these unregulated pathways of DNA repair to be utilized and may allow some tumors to generate resistance to these agents depending on which repair pathways are altered in the tumors. This review will discuss the various telomere recombination mechanisms and will provide rationale regarding the possibility that L1 retrotransposition may contribute to telomere maintenance in tumors lacking telomerase.
Keywords: Telomere, DNA repair, Retrotransposons, Tumors
INTRODUCTION
As initially proposed by Herman Muller and Barbara McClintock, the ends of all linear genomes are comprised of a unique and genetically stable structure, termed the telomere [1]. It was later realized that telomeres must require some mechanism (s) to maintain the sequence due to the end-replication problem [2,3]. This is typically achieved by telomerase. However studies using various model organisms, which have been genetically deleted for telomerase, indicate that multiple recombination-based mechanisms can contribute to telomere maintenance. Such mechanisms are likely recombination-based and use repetitive sequences, such as retrotransposons. This review will focus on the current understanding of these recombination-based mechanisms and proposes that retrotransposons may contribute to the mechanisms. Understanding these mechanisms is critical for the eventual development of specific treatments for individuals who develop tumors that lack telomerase. Tumors lacking telomerase can still maintain the telomere sequences and typically include sarcomas and certain types of brain tumors. Tumors lacking telomerase would not be sensitive to telomerase inhibitors, which are currently in development. Insight into how repetitive sequences, such as retrotransposons, impact telomere maintenance in mammalian cells is currently under investigation in our laboratory.
Telomeres & Subtelomeres
Telomeres are nucleoprotein complexes located at the beginning and ends of all linear chromosomes. The microsatellite sequence repeat located at the telomere varies among organisms. In addition in some organisms, such as Drosophila, there is a complete lack of the microsatellite repeat. Instead Drosophila telomeres are comprised of an array of repetitive elements, termed non-LTR retrotransposons. In humans and mice the telomere microsatellite repeat sequence is (5’-TTAGGG)n, however in other organisms, such as yeast, the sequence is (5’-TG1–3). The frequency of the reiteration of the microsatellite repeat contributes to the telomere length. In addition, the telomere sequence is bound by a multiprotein complex termed the shelterin complex. In human cells this consists of the proteins TRF2, TRF1, TIN2, Rap1, TPP1 and Pot1 [4]. The shelterin complex protects the termini from triggering an inappropriate DNA damage response and is thought to contribute to the formation of a potential (telomere) T-loop structure that resembles a Holliday junction intermediate, and is present at the termini of some chromosomes [5]. Since TRF1 bind in a sequence specific manner to the microsatellite repeat, it is thought that the removal of the shelterin complex, due to the loss of the microsatellite repeat (telomere shortening), triggers additional pathways that recruit telomerase to the telomere [6]. In the absence of telomerase, the telomeres become dysfunctional activating a DNA damage response [7].
In addition to the telomeres, chromosomes have additional structural zones known as the subtelomeres. These regions are proximal to the microsatellite telomere repeat and are comprised of a mosaic of different types of repetitive sequences in the genome [8]. In humans, subtelomeres are highly polymorphic due to large segmental duplications, which are likely generated from homologous recombination among repetitive sequences [9–12].
Telomerase
Telomerase is a ribonucleoprotein complex comprised of a reverse transcriptase (in humans, TERT) and an RNA component (in humans, TERC or hTR) that specifically generates the canonical telomere repeat (5’-TTAGGG in humans) by using RNA as a template [13–16]. Telomerase function in vivo also depends on other proteins that bind the RNA, including dyskerin, and various dyskerin binding proteins including NHP2, NOP10 and GAR1 [17–20]. The telomerase ribonucleoprotein complex (RNP) is recruited to short telomeres, and reverse transcribes a portion of the RNA component by adding sequence to the terminal 3’-OH, thereby maintaining the telomere length [6]. The expression of telomerase maintains telomeres in rapidly dividing human primary cells, including most stem cells and germ cells [21,22]. Most other somatic cells lack telomerase activity. Without telomerase, the telomeres progressively shorten, leading to genome instability, senescence, or apoptosis [6]. Thus telomere shortening has been implicated in both cancer due to genome instability, and aging due to cellular senescence and apoptosis. Heterozygous mutations in various genes required for telomerase activity can also contribute to human diseases including: dyskeratosis congenita, aplastic anemia, and pulmonary fibrosis [23–27]. The genetic mechanism associated with telomerase and disease progression is due to haploinsufficiency [28,29]. However, not all reported mutations in telomerase genes cause disease [30]. This could be contributed to various reasons including: genetic anticipation, the sequence change is a variant that was perhaps misclassified as a mutation, or that additional genetic and/or environmental factors contribute to the onset of telomerase-associated diseases.
Human tumors lacking telomerase
The unlimited growth potential of tumors requires telomere maintenance, which is thought to occur by telomerase, since telomerase is detected in most tumor cells [21]. Yet it is peculiar that the telomeres in most human tumor cells are still extremely short compared to normal tissue, and questions whether telomerase solely contributes to telomere maintenance in tumors expressing telomerase [31]. In addition, although telomerase inhibitors have been developed, these inhibitors have yet to impact tumor growth. This is perhaps because mechanisms other than telomerase contribute to telomere maintenance. An estimated 10% of human tumors lack telomerase activity. Despite the lack of telomerase, the telomeres in these tumors are still maintained and are extremely long and heterogeneous in length (ranging from 3kb to 50kb) [32–34]. Tumors lacking telomerase also have the ability to divide indefinitely in culture. Tumors lacking telomerase are typically described as using Alternative Lengthening of Telomeres (ALT) mechanisms or basically recombination-based mechanisms of telomere maintenance. Nearly 50% of sarcomas lack telomerase, including osteosarcomas and soft tissue sarcomas. In addition, certain types of brain tumors often lack telomerase. These include glioblastoma, medulloblastoma, astrocytomas and oligosarcomas [35]. Some epithelial-derived tumors have been reported to lack telomerase, including gastric, ovarian, adrenocortical and breast carcinomas [32,33,36]. Thus most tumors lacking telomerase are derived from certain primary cell types of mesenchymal cell origin [34,36]. Some of our projects are focused on understanding this cell type specificity as it relates to active retrotransposition in primary somatic cells, including neuronal and mesenchymal cells, lacking telomerase.
Telomere maintenance in the absence of telomerase activity was initially detected when primary human foreskin fibroblasts were immortalized with SV40 T antigen [32]. A portion of the immortalized cells were found to lack telomerase, yet had very long heterogeneous telomere lengths [32]. However mouse tumors which lack telomerase can maintain telomeres without extensive lengthening of the bulk telomere lengths [37,38]. Telomere maintenance that occurs without changing the bulk telomere length is analogous to the Type 1 mechanism of recombination used in yeast lacking telomerase [39–41].
Immortalized human cells and tumors lacking telomerase have other general features. Extrachromosomal DNA can be detected in some tumors lacking telomerase and exist as either single or double-stranded circles [42,43]. These circular DNA fragments have the potential to integrate into the telomere using recombination or may serve as a template for rolling circle replication [43]. The recent development of an assay to detect these extrachromosomal DNA fragments, called the C-circle assay, is potentially a useful assay to initially evaluate if a tumor is using telomere recombination for telomere maintenance. Extrachromosomal DNA can sometimes associate with nuclear foci which are essentially aggregates of the protein PML, along with the recombination proteins, extrachromosomal telomeric DNA, and various telomere-binding proteins [32,33,42,44–47]. These foci have been called ALT associated PML bodies or APBs, and are only detected in human tumors and immortalized cells lacking telomerase [44]. The appearance of APBs correlates with the onset of SV40 T-antigen mediated immortalization of primary cells, but are detected in a only small portion of the total cell population [44]. Whether APBs or extrachromosomal DNA are directly involved in telomere recombination mechanisms is uncertain. However telomere maintenance can occur without APBs or extrachromosomal DNA [48,49]. Again these findings illustrate the existence of multiple telomere recombination mechanisms. Tumors lacking telomerase have also been associated with microsatellite instability at certain non-telomeric genomic loci [50] and are increased for telomere sister chromatid exchanges [45,50]. Tumors expressing telomerase may also utilize telomerase-independent mechanisms for telomere maintenance at reduced frequency. Therefore, the treatment of tumors with telomerase inhibitors could select for these additional telomere maintenance mechanisms. In summary, not all tumors express telomerase, and instead these tumors have the potential to utilize various recombination-based mechanisms for telomere length maintenance. Characteristics associated with tumors lacking telomerase include extensive telomere length heterogeneity, the presence of APBs, extrachromosomal DNA, microsatellite instability and increased T-SCEs. Although not all telomerase negative tumors exhibit the same characteristics, these variable features suggest that multiple non-telomerase mechanisms are contributing to telomere maintenance [34,48,49,51].
Overall, studies that can directly test for telomere recombination in human tumors will be critical. We are currently using assays to detect subtelomere recombination in mouse tumors lacking telomerase and plan to develop such assays to detect subtelomere recombination in human cells. These microarrays along with sequencing for copy number changes in the subtelomere will evaluate which mechanisms are operating in tumors lacking telomerase. More specifically these arrays will determine which mammalian genes contribute to BIR and will be critical to develop treatments for tumors lacking telomerase and for treating tumors that are resistant to telomerase inhibitors.
Telomere recombination
Mechanisms other than telomerase that have been proposed to maintain the telomeres include T-loops, hairpin structures, G-quadruplexes, extrachromosomal DNA, break-induced replication, and retrotransposons [52,53]. Telomere maintenance in the absence of telomerase has been studied in various organisms and has been extensively characterized in yeast lacking telomerase. Telomere recombination in yeast can use two different recombination pathways to maintain the telomeres and are typically described as Type I or Type II survivors [39–41]. The dominant mechanism utilized in yeast lacking telomerase is break-induced replication (BIR). BIR involves DNA synthesis coupled with recombination and will generate DNA intermediates including: Holliday junctions and stalled replication forks [54–56]. Although initially described in replicating bacteriophage, studies in yeast indicate BIR also occurs in eukaryotes by multiple pathways [57–64]. Next we will describe in detail the different recombination mechanisms and the mechanism of BIR.
Telomere recombination based on studies in yeast lacking telomerase
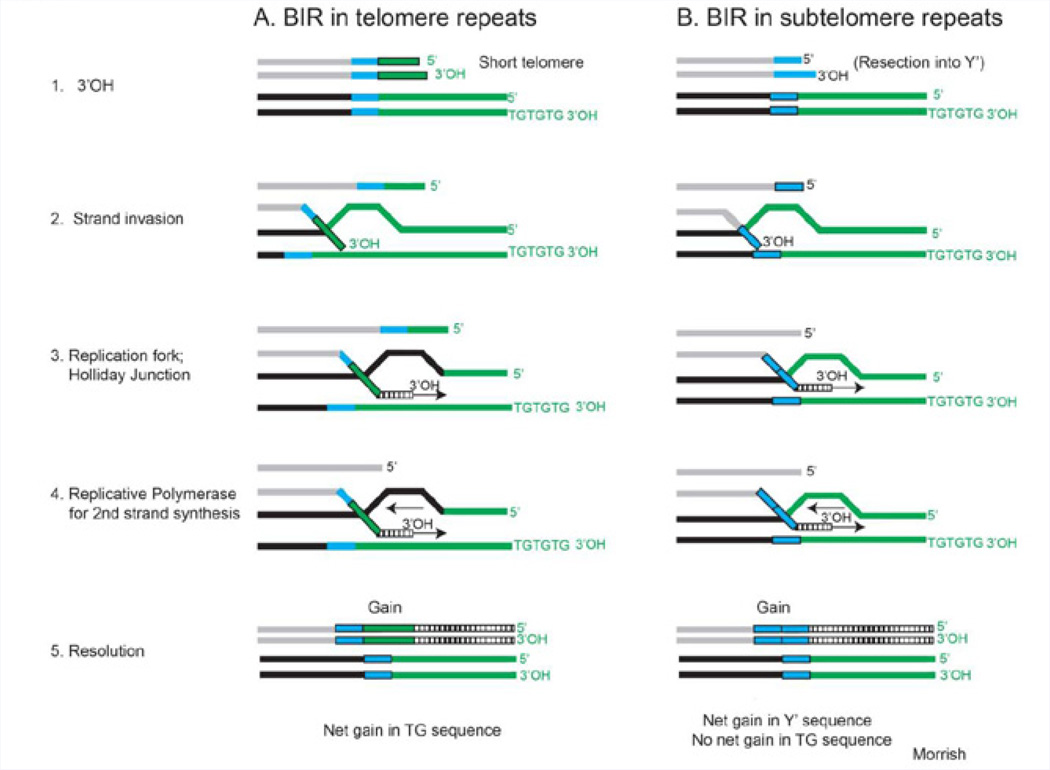
Initial experiments suggesting that recombination can contribute to telomere maintenance, was observed in yeast in which the telomere repeat sequence was transferred to a linearized plasmid DNA [65]. Bona fide evidence that recombination could sufficiently maintain the telomeres was shown when yeast cells were deleted for telomerase. After most cells underwent senescence, survivors were generated that grew without telomerase [39]. Two general types of recombination events were observed 1) amplified Y’ telomere repeats and 2) amplified Y’ subtelomere repeats [40,41] (Figure 1 A&B). Both types depend on recombination as shown by the requirement for Rad52, but occur by different mechanisms since each type had different characteristics and genetic requirements [40,41]. For instance, Type II survivors have long heterogeneous tracts of telomere repeats, require Rad50, and initiate recombination within telomere repeat tracts (Figure 1A). Type I survivors have short telomere repeats, amplified subtelomeric Y’ repetitive elements, require Rad51, and initiate recombination in the subtelomere Y’ sequences (Figure 1B). Interestingly, Type I survivors are more frequent among the initial survivor populations, but these Type I survivors grow slowly and have a growth disadvantage compared to Type II survivors. Studies in both S. cerevisiae and K. lactis, have further shown that BIR mechanisms help maintain the telomeres [54,63,66,67]. Thus, survivor formation in yeast deleted for telomerase occurs by recombination in using different types of BIR mechanisms. From these studies it yeast, it indicates that telomere maintenance can occur by recombination events within the telomere or subtelomere repeats and depends on Rad50 and Rad51 repair proteins, respectively.
Figure 1. Break-induced replication.
Shown are models of break-induced replication based on studies in yeast (54). A. For Type II survivors, the 3’-OH of a short telomere (green) strand invades into another telomere repeat. This sequence can be on the other homolog or on a different chromosome. Strand invasion generates a replication fork and a Holliday junction. Conventional DNA polymerases are used to synthesize both strands, followed by resolution of the Holliday junction. BIR that initiates in the telomere repeat will result in a net gain of telomere sequence. B. For Type I survivors, BIR in subtelomere repeats occurs when telomere shortening proceeds into a Y’ (blue) repeat. BIR that is initiated within the subtelomeres results in a net gain in Y’ sequence.
Break-induced replication (BIR)
Walmsley et al. described a model proposing that telomeres may use a recombination-based replicative mechanism for elongation, similar to BIR [68]. Subsequent studies in yeast indicate BIR is the predominant mechanism to maintain telomeres [54]. BIR is initiated when a 3’-OH of one free end at a damaged chromosome, such as a short telomere, or a stalled replication fork, strands invades into another chromosome, often referred as a “recipient” chromosome (Figure 1). A recipient chromosome can be any other chromosome exhibiting sequence homology with the 3’ end of the damaged chromosome. We propose that the first strand synthesis of a retrotransposon intermediate could also initiate BIR. For instance, during reverse transcription, the first synthesized strand could invade at a recipient chromosome that has a similar type of retrotransposon sequence. Strand invasion of a 3’-OH into another chromosome generates a replication fork and forms a Holliday junction. This replication intermediate allows for conventional polymerases to generate a new sequence, using the recipient chromosome as a template. The new synthesis of this first strand proceeds by copying to the terminal end of the recipient chromosome, followed by second strand synthesis and resolution of the Holliday junction. Consistent with the involvement of conventional DNA polymerases, BIR depends on various DNA replication factors [69]. Furthermore, the non-essential Polδ subunit, Pol32, is required for both BIR and telomere maintenance in yeast [67].
Repair by break-induced replication is error prone, and can generate loss of heterozygosity (LOH) events, non-reciprocal translocations, and copy number changes (duplications or deletions). BIR between two homologs with different alleles, such as a wildtype or mutant p53 allele, could lead to LOH of p53, a common early event during tumorigenesis. Thus LOH generates two copies of the same allele, and is a frequent hallmark in some tumor types. If BIR occurs between different chromosomes this leads to non-reciprocal translocations, duplications, or segmental duplications [70–72]. Consistent with a role of BIR in maintaining telomeres in mammalian tumors lacking telomerase, mouse tumors lacking telomerase have elevated frequencies of copy number changes, non-reciprocal translocations, and loss of heterozygosity compared to tumors expressing telomerase [73–76]. These findings suggest that BIR mechanisms are more frequent in tumors lacking telomerase.
In summary, BIR mechanisms are evolutionarily conserved, involve both replication and recombination, and generate mutational errors characteristic of tumors lacking telomerase. Studies in yeast lacking telomerase find that BIR is the predominant pathway for telomere maintenance and can occur by two different pathways. BIR can initiate within the subtelomere or telomere repeats. If BIR initiates in the subtelomere, the individual chromosome will have increased both the amount of subtelomere and telomere sequences. If BIR initiates within the telomere repeats the individual chromosome will have only increased the amount of telomere repeats. An increase in BIR events during continuous cell divisions could increase the bulk telomere length. Thus telomere maintenance by BIR can lengthen the telomere at an individual chromosome. A further understanding of the different BIR pathways in mammalian cells will be important to elucidate telomere maintenance in the absence of telomerase.
Recombination in human tumors lacking telomerase
Various mechanisms have been proposed for telomere maintenance in tumors lacking telomerase. It seems genetically possible that depending on the tumor and the genetic mutations that have accumulated in different repair genes, that different recombination pathways are possible. Dunham et al., provided direct experimental evidence that mechanisms, similar to BIR, are likely occurring in human tumors lacking telomerase [77]. In this study a selectable marker was targeted within a telomere array of a single chromosome. Clonal cells containing the single targeted end were serially passaged. In these passaged cells it was found that the marker was copied to other chromosomes indicating that an interchromosomal, recombination-based mechanism had occurred consistent with BIR. In this study, the tag was located in the telomere repeat, not within the subtelomere repeats. Therefore, any chromosomes that could initiate BIR, either from a short telomere or from a subtelomere, could strand invade into the tagged telomere, and would copy the marker to an interchromosomal location. Others propose that recombination by T-SCEs are contributing to the mechanism of telomere maintenance, however T-SCEs do not explain how the sequences at the marked chromosome was amplified to a different chromosome, since T-SCEs only occur between sister chromatids. However, it is important to realize a mechanism like T-SCEs cannot result in a net increase in telomere sequence, since there is no DNA synthesis during sister chromatid exchange. Since sister chromatid exchanges are detected by an assay called CO-FISH, it is possible that the Holliday junction intermediates that form during BIR are actually contributing to the presumed increase in T-SCEs that are detected by CO-FISH. There is the possibility that recombination occurs within the T-loop, which involves strand invasion of the terminal 3’-OH into internal regions of the very same chromosome (i. e. intrachromosomal recombination) [78]. However it is important to recognize that T-loop recombination will not amplify sequence to different chromosome ends. Therefore, recombination-based mechanisms that are similar to BIR are likely responsible for the copy number increase and the transfer of sequences to other chromosomes in human tumors lacking telomerase.
Most experiments on the mechanism of BIR are based on studies using yeast. Little is known regarding how the mechanism of BIR occurs in mammalian cells. In addition, homologous recombination is more tightly regulated; with arguably non-homologous ending joining (NHEJ) the more widely used mechanism for DNA repair in mammalian cells. However specific genomic changes associated with BIR in telomerase deficient tumors, indicate that BIR mechanisms occur in telomerase negative tumors including loss of heterozygosity (LOH), non-reciprocal translocations, segmental duplications, and copy number changes. During BIR, repetitive sequences are used as substrates for recombination. Given the significant amount of repetitive sequences in the human genome, we are investigating whether certain types of repetitive sequences contribute to telomere maintenance in cells deficient for telomerase. Specifically we are examining if LINE-1 retrotransposition into DNA breaks contributes to telomere maintenance. We are also testing whether genes involved in telomere recombination impact LINE-1 retrotransposition into the telomere by using a previously published PCR/Southern based assay. This will allow us to test for LINE-1 integration events, using a cell-culture based assay for retrotransposition, coupled with a lentiviral shRNA approach to various candidate genes [79].
Other proposed telomere recombination mechanisms in human tumors lacking telomerase include recombination at T-loops, which are potential secondary structures present at some telomeres in vivo [5]. As described earlier, telomere sister chromatid exchanges (T-SCEs) have been proposed to maintain the telomeres [45,78]. Extrachromosomal DNA can also maintain telomeres in the yeast strain Kluyveromyces lactis when cells are deleted for telomerase either by rolling circle replication or by direct integration into the telomere; a similar mechanism likely occurs in human tumors since many have detectable extrachromosomal DNA [42,80,81]. Lastly telomere studies in Drosophila, find that the ends of chromosomes are completely devoid of a microsatellite repeat. Instead Drosophila telomeres are exclusively comprised of non-LTR retrotransposons [82]. The telomeres of other organisms also harbor non-LTR retrotransposons within the telomere [83]. Furthermore a number of eukaryotes harbor actively mobile retrotransposons that lack the endonuclease and reside at the telomeres [84,85]. Given that we also have found that mammalian cells with dysfunctional telomeres will result in retrotransposition into telomere sequences, we suspect that in some cases retrotransposons may be contributing to telomere maintenance in tumors lacking telomerase [79,86]. Specifically we will be testing if the mechanism of LINE-1 integration by target-primed reverse transcription (TPRT), involves the repair mechanism of BIR.
Retrotransposons
The bulk of repetitive sequences in the human genome are derived from transposable elements [87,88]. Transposons can be classified into three general groups and include DNA-based transposons, and two different types of RNA-based transposons. The RNA-based transposons include both LTR-retrotransposons (ie. Human Endogenous Retroviruses, HERVs) and non-LTR retrotransposons (ie. LINEs). Briefly, LTR-retrotransposons are structurally similar to a retrovirus and encode for a reverse transcriptase in addition to other proteins. LTR retrotransposons are inactive in the human genome but are active in many non-mammalian genomes [89]. Active in the human genome are non-LTR retrotransposons including LINEs, SINEs, and SVA elements [90–94]. SINEs and SVAs do not encode for a reverse transcriptase, and instead depend on the L1 reverse transcriptase for the mobility of the RNA [95,96]. These various types of mobile genetic elements have the potential to impact telomere maintenance by active reverse transcription. In addition, these sequences along with other repetitive sequences likely provide sequence substrates for homologous recombination.
The most distal sequences located at the telomeres in human chromosomes are tandem arrays of perfect hexameric 5’-TTAGGG, microsatellite repeats. This hexameric repeat degenerates (ie. 5’-TTTGGG, 5’-TTAGGC) into the proximal areas of the subtelomere. Thus, human subtelomeres are a patchwork of unique and repetitive sequences [11]. Given that mammalian genomes have a plethora of repetitive sequences, these repetitive sequences are potentially used for homologous recombination in the subtelomere. However, it is uncertain if certain types of repetitive sequences are more prone to recombination. In particular we plan to focus on repetitive sequences that are known to mobilize in the genome, including LINEs and SINEs. These repetitive elements are likely candidates since the mechanism of mobility, by target primed reverse transcription (TPRT), has many parallels and evolutionary relationships with telomerase [97–99]. In addition retrotransposition can occur into dysfunctional telomeres [79]. To determine if retrotransposons impact telomere maintenance will require a biological approach using conventional assays for telomere biology along with assays to study LINE-1 retrotranspositon. To examine telomere recombination, assays include pq-ratios, CO-FISH, C-circles, Q-FISH, and the selection or detection of subtelomere recombination events (Figure 2). To detect subtelomere recombination events we are using a microarray approach to examine this process using a mouse B-cell lymphoma model lacking telomerase. We also anticipate the need for sequencing to determine if the breakpoints occur within certain types of repetitive sequences. These assays will need to be coupled with computational and statistical approaches to decipher how certain repair and recombination genes impact the frequency of retrotransposition within the telomeres and subtelomere sequences. In addition we are in the early stages of utilizing additional model systems that exclusively utilize retrotransposons for telomere maintenance, such as Drosophila, or organisms that have bona fide telomere microsatellite repeats and retrotransposons at the telomeres, such as the silkworm Bombyx mori, [82,83]. Overall these studies will help elucidate the genes that contribute to LINE-1 retrotransposition into the telomere and will allow us to determine if the signatures of BIR events in the subtelomeres are altered following LINE-1 integration into the telomeres/subtelomeres.
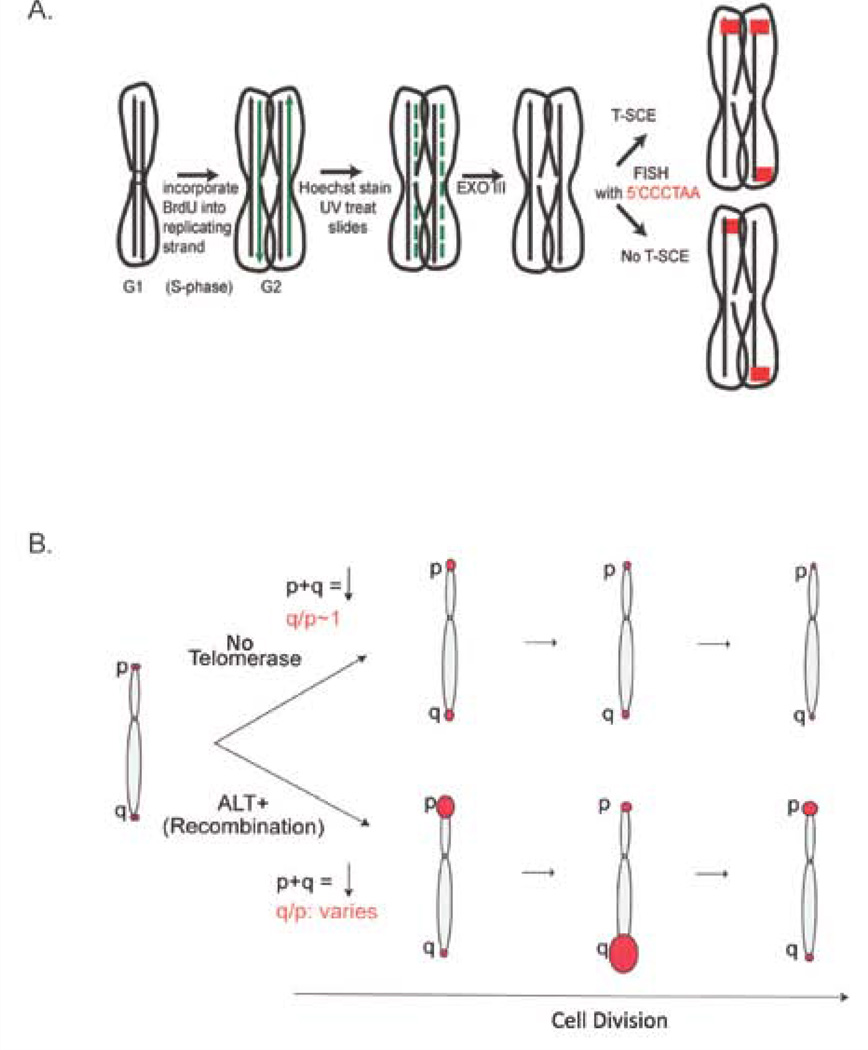
Figure 2. Assays to directly examine telomere recombination.
A. CO-FISH assay: Replicating cells are grown in the presence of the nucleotide analogs BrdU/C, for one complete round of replication. Due to semi-conservative DNA replication, BrdU/C will be incorporated into the newly replicated strand. Metaphase spreads are treated with Hoechst and UV, which nicks the BrdU/C incorporated daughter strand, which is then degraded by exonulcease III (ExoIII). Telomere probes (5’CCCTAA) are then hybridized to the ends. If a sister chromatid exchange occurred then the strand will be protected from degradation and two signals will be detected. B. pq-ratio assay: This assay measures the variation in telomere lengths at the p- and q-arms of a chromosome. The telomeres at both ends of a given chromosome shorten at a similar rate, therefore the telomere ratio for most chromosomes is expected to be q/p~1. If instead recombination is used to maintain the ends, then the amount of lengthening at one end could be different compared to the other end. This results in variable values for the pq-ratio.
Extrachromosomal DNA
Studies using various model organisms suggest extrachromosomal DNA contributes to telomere maintenance by rolling circle replication or by direct integration [80,100]. For instance, K. lactis deleted for telomerase can use extrachromosomal DNA to elongate short telomeres. Once incorporated into the telomere, the sequence is propagated to other ends by BIR mechanisms.
Self-replicating extrachromosomal DNA with telomere repeats and Y’ sequence can also be detected in S. cerevisiae [101,102]. In human tumors extrachromosomal DNA can also be comprised of telomere repeats or different types of repetitive DNA, such as non-LTR retrotransposons [103]. Various kinds of extrachromosomal DNA are detected in human tumors lacking telomerase, including C-circles, G-circles, and T-circles [46,104]. The nomenclature indicates whether the sequence is single-stranded and comprised of sequences: (5’-CCCTAA, C-circles), (5’-TTAGGG, G-circles) or is double-stranded and contains both (5’-CCCTAA/5’-TTAGGG, T-circles). T-circles also likely have nicks at both strands; therefore a polymerase could not use it for a template during rolling circle replication. Instead, T-circles may directly integrate into the ends using BIR mechanisms [46]. C- and G-circles can be amplified by a polymerase indicating that at least one strand lacks a nick; therefore these structures could be primed for rolling circle replication [104]. As with K. lactis, human tumors that incorporate extrachromosomal DNA into the telomeres likely amplify this sequence into other telomeres by BIR.
In addition to the integration and propagation of extrachromosomal DNA into telomeres, the formation of extrachromosomal DNA also likely depends on recombination, perhaps by recombination within a T-loop [5]. In S. cerevisiae recombination between Y’ elements, could also generate extrachromosomal DNA which then autonomously replicates [101]. Interestingly, the formation of some types of extrachromosomal DNA in human cells depends on Xrcc3 (a Rad51 paralog) and also Nbs1 that forms a complex with Rad50 and Mre11 [105,106]. shRNA knockdown of these genes in human tumor cells lacking telomerase reduces the amount of extrachromosomal DNA, and correlates with telomere shortening. However, these shRNAs have no impact on the ability of the cells to grow. This minimal impact on growth suggests that the presence of extrachromosomal DNA may contribute to only some non-telomerase telomere maintenance mechanisms that can be used in the cells [106]. In summary, telomere maintenance in human cells lacking telomerase likely utilizes extrachromosomal DNA, which depends on recombination mechanisms for the generation, integration, and amplification of the telomere sequence. It is uncertain how extrachromosomal DNA is incorporated into the telomeres in human cells, however it has the potential of introducing both telomere and non-telomere repeat sequences into the ends of chromosomes.
Genes contributing to telomere recombination in human tumors lacking telomerase are also involved in BIR mechanisms
A variety of genes have been identified that likely contribute to telomere maintenance in human tumors lacking telomerase. These include Rad50 and the MRN complex, and some Rad51 paralogs [107,108]. These findings are consistent with studies on yeast survivors and these genes play a role in different pathways of BIR [39–41,109,110]. Some genes involved in telomere maintenance in tumors lacking telomerase have orthologs that contribute to BIR in yeast and include SMC5/6 and Mus81-EME1 [111,112]. Additional candidate genes that have putative roles in Holliday junction formation or resolution, and are also likely involved in break-induced replication, include Top3a, FANCD2, and FANCA. Thus understanding the role of various genes in non-telomerase telomere maintenance mechanisms will provide additional insight into telomere maintenance in the absence of telomerase and will help determine what genes contribute to BIR mechanisms in mammalian cells.
Structural Maintenance of Chromosomes: SMC5/6 complex
Studies in human cells lacking telomerase indicate that the structural maintenance of chromosome proteins, SMC5 and SMC6 (SMC5/6), contribute to telomere maintenance in the absence of telomerase [113,114]. The SMC5/6 complex functions in recombination restart of replication forks, and maintenance of repetitive sequence. To function in these roles, the SMC5/6 complex binds the MMS21/NSE2 complex and other components. MMS21/NSE2 has an E3 SUMO (small ubiquitin-like modifier) ligase activity and is involved in sumoylation, a posttranslational modification, and likely sumoylates components of the shelterin complex including RAP1, TRF1, TRF2, and TIN2 in human cells [113]. shRNA knockdown of SMC5/6 causes telomere shortening, senescence, and reduced T-SCEs [113]. However, it is uncertain whether other recombination mechanisms, such as BIR were also affected.
Studies in yeast suggest that the Smc5/6 protein complex plays a role in telomere maintenance and some smc5/6 yeast mutants increase BIR [111,114]. Yeast temperature sensitive smc5/6 mutants, also deleted for telomerase, showed severe growth arrest due to accelerated senescence and delayed survivor formation, yet these mutants had minimal effects on telomere lengths [115]. Thus it was concluded that the role of Smc5/6 in yeast lacking telomerase is recombination-independent. These findings suggest that in yeast Smc5/6, may have different functions compared to human cells. Therefore it will be necessary to sort out what role Smc5/6 plays in telomere maintenance when telomerase is deleted in yeast and human cells. Overall, a potential role of the SMC5/6 sumoylation complex in the maintenance of telomeres in tumors and yeast lacking telomerase further suggests BIR mechanisms contribute to telomere maintenance.
Mus81-Eme1: a structure specific endonuclease
Mus81 may also contribute to telomere maintenance in human tumors deficient for telomerase. Mus81 is a structure specific endonuclease that preferentially cleaves 3’- flaps, Holliday junctions, D-loops, and replication forks [116]. The cleavage activity of Mus81 requires dimerization with Eme1 and forms a multiprotein complex with the mammalian Holliday junction resolvase, BTBD12/SLX4 [117]. Therefore, Mus81 may regulate or inhibit BIR by cleavage of the Holliday junction. Mus81 may also interface with telomere recombination mechanisms by a putative interaction with TRF2. In vitro, Mus81 can bind to TRF2, which reduces the DNA binding and cleavage activity of Mus81. Also, deletion of TRF2 increases the Mus81 endonuclease cleavage activity for D-loops and other cleavage substrates, but does not alter production of extrachromosomal DNA [116]. Thus, TRF2 or the shelterin complex could regulate Mus81, which may in turn regulate BIR at the telomere.
The shRNA knockdown of Mus81 reduces proliferation specifically in human tumors lacking telomerase, and increases the amount of telomeres with signal free ends (SFEs) [116,118]. SFEs are likely ends that are either too short or lack telomere sequence for binding of the fluorescently labeled telomere probes. Surprisingly, Mus81 knockdown did not increase apoptosis, telomere fusions, or impact the telomere length. Knockdown also decreased the frequency of telomere sister chromatid exchanges (T-SCEs) but this result could be an issue with the quantitation in the CO-FISH assay. For instance, Mus81−/− mice have a global increase in sister chromatid exchanges, therefore to correctly score for T-SCEs in the CO-FISH assay, the cells need to be normalized for global sister chromatid exchanges [119]. Mus81 knockdown did not impact the formation of extrachromosomal DNA; therefore it is possible that telomere maintenance mechanisms that involve extrachromosomal DNA could still operate when Mus81 is reduced. Finally, the deletion of Mus81 in yeast increases pol32-dependent BIR events [67,112].
In summary, these studies illustrate that if multiple non-telomerase mechanisms maintain telomeres, the impact on telomere length may not always be detected by disrupting just one pathway. Furthermore, some genes contributing to non-telomerase telomere maintenance mechanisms in human cells also have yeast orthologs, which contribute to BIR.
LTR-retrotransposons can maintain telomeres in yeast survivors
LTR-retrotransposons in the human genome, such as human endogenous retroviruses or, HERVs, can account for an estimated 8% of the human genome; however these elements no longer mobilize in the human genome. In S. cerevisiae, LTR-retrotransposons account for ~3% of the genome, many are still active, and include the various types of Ty elements [89]. Ty elements that transpose encode for an integrase, a reverse transcriptase, and synthesize cDNA in a cytoplasmic particle [120]. Some Ty elements, such as Ty5, normally reside near telomeres in different yeast strains, and preferentially integrate into silenced regions including telomeres [121]. Furthermore, the LTR-retrotransposon Ty1 can contribute to the generation of survivors in yeast deleted for telomerase and are activated by telomere shortening [40,122,123]. Initially, the frequency of survivor formation by Ty1 transposition was found to be rare in telomerase deficient S. cerevisiae grown at 30°C [40]. Using these growth conditions, Ty1 transposition is normally reduced [124]. More recent studies found that a significant portion of survivor formation depended on Ty1 transposition when yeast was grown at 25°C, a temperature favoring transposition. These studies found that Ty1 encoded reverse transcriptase could also reverse transcribe Y’ transcripts, in Ty1 cytoplasmic particles. These Ty1-Y’ chimeric cDNAs were then subsequently integrated into the telomeres [123]. It will be interesting to perform telomere Southern blots on these Ty1-Y’ survivors, to determine if both types of survivors formed, and if the insertions altered the telomere lengths. Integration of the Ty1-Y’ cDNAs at the telomere was also associated with genome instability. It will also be interesting to test if Ty1-Y’ sequence amplifies to other ends by break-induced replication [125] (Figure 3). Overall, these findings indicate that LTR-retrotransposons can maintain telomeres in yeast lacking telomerase. More importantly, it illustrates that other reverse transcriptases can impact telomere maintenance in cells lacking telomerase.
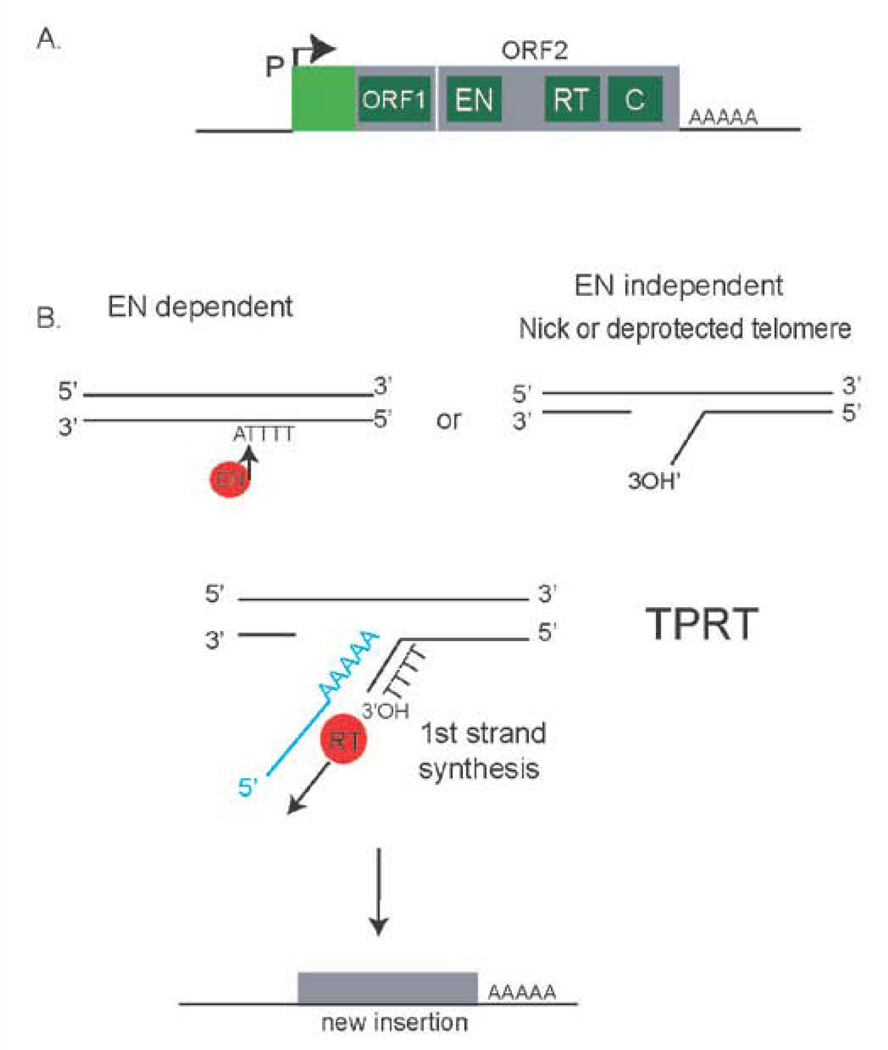
Figure 3. TPRT & ENi-retrotransposition.
A. A full-length retrotransposition competent L1 contains an L1-specific promoter, two intact open reading frames (ORF1 and ORF2), a polyadenylation signal sequence, and is followed by a poly(A)tail. ORF1 encodes for a nucleic acid binding protein and ORF2 encodes for both an endonuclease and a reverse transcriptase, and a C-domain of unknown function. B. Target-primed reverse transcription is a mechanism whereby the L1-encoded endonuclease cleaves the genomic DNA at a consensus cleavage site. The 3’-OH on the nicked DNA is used to prime reverse transcription of the L1 RNA. ENi-retrotransposition can also occur, whereby L1 can insert at DNA lesions or deprotected telomeres. For both conventional TPRT and ENi-retrotransposition, reverse transcription generates an intermediate that could be used during BIR.
Non-LTR retrotransposons mobilize by target primed reverse transcription
Non-LTR retrotransposons are mobile genetic elements, and encode for a reverse transcriptase with homology to telomerase. These elements are evolutionarily conserved, and are present in numerous organisms [16,97,99]. Non-LTR retrotransposons replicate using a mechanism called target-primed reverse transcription, or TPRT [98]. The majority of the human genome is derived from non-LTR retrotransposons (LINEs or SINEs) [87,88]. These elements still mobilize in the human genome, and are major drivers of evolutionary change, genome instability, and human genetic variation [90–92,126,127]. Much of the non-LTR retrotransposon sequences within the human genome cannot mobilize, because the sequence is 5’truncated, rearranged or mutated [128]. Typically, only retrotransposition-competent L1s can mobilize in the genome. Retrotransposition-competent L1s in the human genome have two intact open reading frames (ORFs) (Figure3). ORF1 is a chaperone with nucleic acid binding activity, and ORF2 encodes for an endonuclease, a reverse transcriptase, and a C-domain of unknown function [129–131]. In order to retrotranspose, a full-length L1 RNA is transcribed from an internal promoter [132]. The RNA is then exported to the cytoplasm for translation. The proteins generated from the L1 RNA bind to the L1 RNA in cis to form a ribonucleoprotein complex (RNP). Once in the nucleus, target-primed reverse transcription is primed at a 3’-OH in the genome, which is typically generated by the L1 encoded endonuclease. It is uncertain how integration is completed, however various models can account for the completion of an L1 integration event. Characteristics of conventional L1 integration by TPRT include insertions at a consensus EN cleavage site, 5’truncations of the integrated L1, a 3’ variable length poly (A) tail sequence, and flanking target-site duplications. However, integration can occur by additional mechanisms as exemplified by examination of the human genome sequence compared to other primates, or by the study of L1 integration events in cell culture assays or different mouse models [132–138]. For instance, L1 integration could be coupled with BIR (Figure 4B). In such a model, the first strand of cDNA generated by L1 reverse transcription could strand invade at another genomic location harboring L1 sequence [98]. This may occur in the human genome since in yeast, BIR can initiate by strand invasion into repetitive regions in the yeast genome and copy >130kb to the end of a chromosome [63,139].
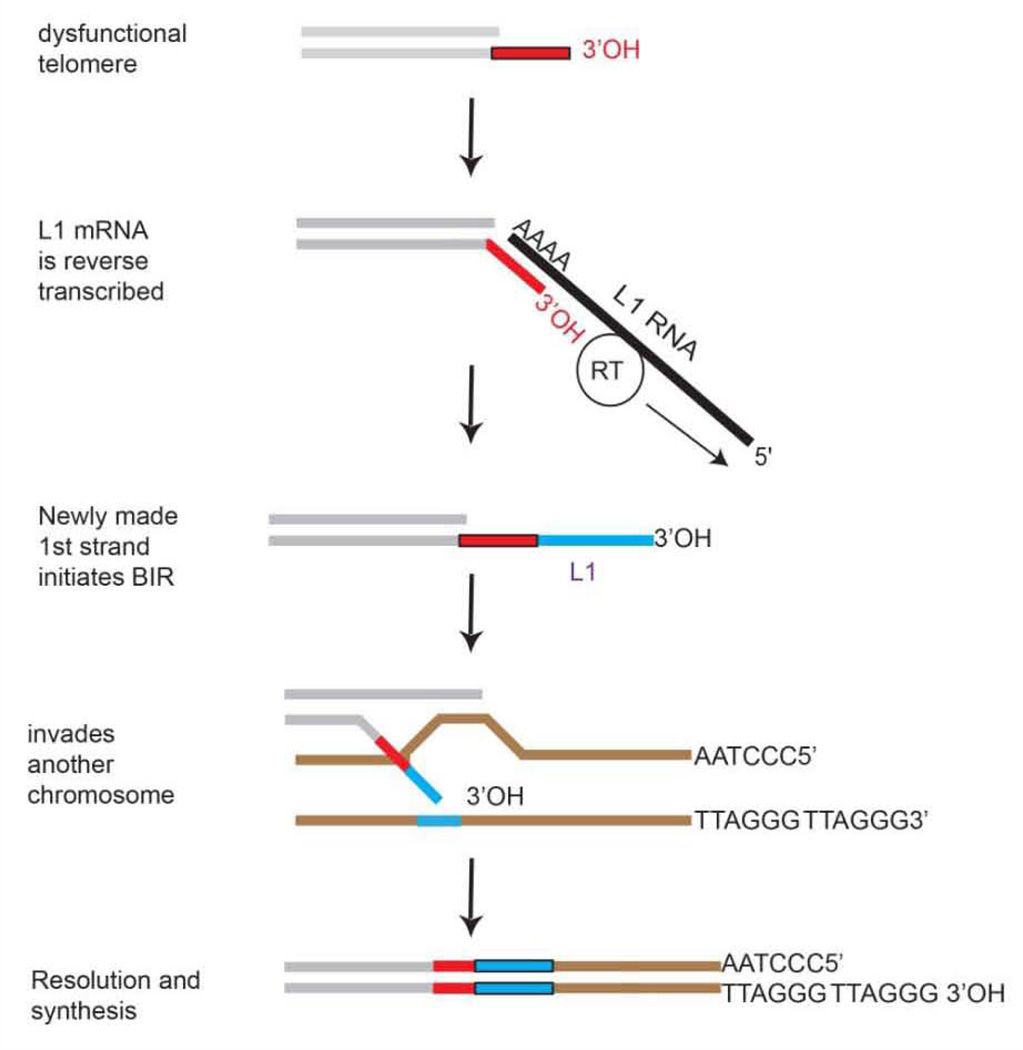
Figure 4. BIR coupled with target-primed reverse transcription (TPRT).
ENi-retrotransposition or TPRT at a dysfunctional telomere will generate by reverse transcription a free-end that could be used to strand invade by BIR. The newly generated cDNA, would strand invade into another chromosome where a non-LTR retrotransposon resides. Conventional polymerases would copy sequences onto the chromosome using the other chromosome as a template. Thus ENi-retrotransposition or conventional TPRT could be coupled with BIR.
Endonuclease-independent (ENi) -retrotransposition can occur at deprotected telomeres and DNA breaks
An additional pathway of integration, endonuclease-independent (ENi) retrotransposition was initially detected in hamster cells defective in non-homologous end-joining, due to the lack of XRCC4 (Figure 4A) [86]. Characterization of ENi integration events indicated that insertions occurred at DNA lesions since integration occurred at atypical endonuclease cleavage sites. The insertions also lacked target-site duplications, and were often both 5’- and 3’-truncated. In addition, the events were associated with insertions or deletions of additional sequences. ENi-retrotransposition was also frequent in DNA-PKcs deficient cells, another component of non-homologous end-joining, and in cells with dysfunctional telomeres, due to the expression of a dominant negative TRF2 [79]. Sequence characterization of these insertions showed some events inserted adjacent to telomere repeats, suggesting that reverse transcription was primed from the telomere 3’-OH. Although initially detected in a cell culture system, integration events with similar characteristics have been observed in some mouse mutants [140]. In addition, analysis of human genomic sequence finds that ENi-retrotransposition events can also occur in vivo and are associated with interchromosomal recombination events [138]. Thus, integration of non-LTR retrotransposons into short or dysfunctional telomeres by ENi-retrotransposition, could contribute to non-telomerase telomere maintenance mechanisms in mammalian tumor cells.
PLE-like elements
Evolutionary studies in other organisms suggest ENi-retrotransposition may be an ancient mechanism of retrotransposition, and provides additional insight into the evolutionary origin of telomerase [85]. Specifically, a certain class of retrotransposons, called Penelope-like elements (PLEs), resides in the genome of bdelloid rotifers, and other eukaryotes [84]. These Penelope-like retrotransposons are embedded within telomere repeats, and have an intact reverse transcriptase but lack an endonuclease domain. PLE transcripts include telomere repeats that could base pair with the terminal 3’-OH at the end of the chromosome, implicating reverse transcription initiates from the terminal 3’-OH. The orientation of the PLEs in the genome also suggests that reverse transcription initiates at the terminal 3’-OH. Lastly, these elements likely function with telomerase since the chromosome ends have a more canonical telomere repeat.
Drosophila telomeres
Telomere maintenance in Drosophila requires non-LTR retrotransposons, and the telomeres are comprised of three different non-LTR retrotransposon sequences, including Het-A, TART, and TAHRE [82,141]. Drosophila telomeres also have capping proteins, which are required to prevent telomere fusions [142–144]. These capping proteins share similarities to a shelterin complex seen in other eukaryotes. In addition, ATM, and the Rad50-MRE11-NBS1 (MRN) complex are required to prevent telomere fusions in Drosophila [145,146]. As with most non-LTR retrotransposons, integration of these retrotransposons into the telomeres occurs by a mechanism termed target-primed reverse transcription or TPRT (Figure 3) [98]. Only TART and TAHRE encode for reverse transcriptase [147,148]. Thus, the mobility of Het-A likely depends on TART or TAHRE reverse transcriptases provided in trans [147]. Recombination likely also contributes at some frequency to the amplification of some of the sequences [149]. The mechanism of transcription of Het-A and TART is unusual. For instance, the mechanism of TPRT orients the retrotransposon with the promoter residing at the very termini of the chromosome. Telomere erosion would destroy the promoter reducing future retrotransposition events. To circumvent this issue, Drosophila non-LTR retrotransposons have evolved to initiate transcription from a promoter residing in the 3’UTR of the adjacent retrotransposon [150]. In summary, telomere specific retrotransposons in Drosophila are a mechanism of telomere maintenance that does not involve telomerase. Some may consider this a Drosophila-specific mechanism; however non-LTR retrotransposons are active in mammalian genomes and under some conditions ENi-retrotransposition can integrate into telomere repeats. These insertions could be capped or could invoke BIR to maintain the ends. Capping of insertions that integrate at the ends would only amplify the retrotransposon sequences, and would not increase the amount of telomere repeats. This would be consistent with Type I survivors seen in yeast, where telomeres are maintained but do not increase the amount of telomere repeats. Alternatively, ENi-retrotransposition at a telomere or even conventional TPRT at subtelomere or internal locations could invoke BIR mechanisms increasing the amount of telomere repeats at an end, eventually increasing the bulk amount of telomere repeats in the population.
In summary, non-LTR retrotransposons and other types of repetitive sequences in the human genome are likely involved in telomere maintenance in the absence of telomerase. These mechanisms of telomere maintenance could couple retrotransposition with BIR. Although these non-telomerase mechanisms have the potential to occur if telomerase is present, the mere efficiency and direct function of telomerase would predominate in maintaining ends. Thus, non-LTR retrotransposons may facilitate telomere maintenance in the absence of telomerase by reverse transcription at DNA breaks and dysfunctional telomeres, or by recombination using BIR-like mechanisms among similar DNA sequences.
CONCLUSIONS
A portion of tumors lack telomerase, add including ~50% of sarcomas, certain types of brain tumors, and some epithelial-derived tumors. Developing treatments for tumors lacking telomerase will depend on understanding non-telomerase mechanisms for telomere maintenance. In addition, some of these mechanisms are likely selected in telomerase positive tumors, when treated with telomerase inhibitors. Tumors lacking telomerase have characteristics associated with BIR mechanisms (i. e., LOH, copy number variations, non-reciprocal translocations), suggesting that BIR mechanisms may contribute to telomere maintenance. It is uncertain whether some BIR mechanisms could be coupled with ENi-retrotransposition or conventional retrotransposition. However, it is curious that LINE-1 retrotransposition is detected in some primary cells types [151,152]. For instance, neuronal progenitor cells readily accommodate L1 retrotransposition, and a portion of tumors lacking telomerase such as glioblastomas, astrocytomas, medulloblastomas, and oligosarcomas represent a large portion of tumors lacking telomerase [35,151,153]. Furthermore, a number of sarcomas lack telomerase, and retrotransposition has also been detected in primary human fibroblasts [152,153]. Given that a significant portion of the human genome is comprised of retrotransposons, it seems possible that BIR mechanisms between these regions of the human genome may facilitate retrotransposition during telomere maintenance. Additional understanding of the genes that contribute to either BIR or retrotransposition will help determine if these mechanisms cooperate to maintain the ends in tumors lacking telomerase.
ACKNOWLEDGEMENTS
I would also like to thank the input from various individuals including Carol Greider, John Moran, Jef Boeke, Sarah Wheelan, John Goodier, Vivek Behera, and Margaret Strong. Various funding has contributed to the development of this project including: the Leukemia and Lymphoma Society, Scholar Award from the American Society of Hematology, the Maryland Stem Research Fund, and the National Cancer Institute.
ABBREVIATIONS
- LINE-1 or L1
Long interspersed nucleotide element-1
- RNP
Ribonucleoprotein
- ALT
Alternative lengthening of telomeres
- SV40 T
Simian Virus 40 large T antigen
- PML
promyelocytic leukemia
- T-SCEs
Telomere-sister chromatid exchanges
- CO-FISH
Chromosome orientation fluorescence in situ hybridization
- APBs
Alternative lengthening of telomeres promyelocytic leukemia bodies
- NHEJ
Non-homologous end-joining
- LOH
loss of heterozygosity
- LTR
long terminal repeat
- HERV
human endogenous retrovirus
- SINEs
short interspersed nucleotide elements
- SVA
SINE-VNTR-Alu
- TPRT
target-primed reverse transcription
- Q-FISH
quantitative fluorescence in situ hybridization
- C-CIRCLES
telomere C-strand circles
- G-CIRCLES
Telomere G-strand circles
- T-CIRCLES
double-stranded circular telomere repeats
- SFEs
signal-free ends
- PLEs
Penelope-like elements
- TAHRE
Telomere-Associated and HeT-A-Related Element
- TRF2
telomeric repeat binding factor 2
- TIN2
(TRF1)-interacting nuclear factor 2
- TPP1
POT1 and TIN2-interacting protein
- Pot1
Protection of telomeres 1
- NBS1
Nijmegen Breakage Syndrome
- MRN
MRE11/Rad50/NBS1 complex
- SMC5/6
Structural maintenance of chromosomes 5/6
- Mus81
Structure-specific endonuclease
- EME1
essential meiotic endonuclease 1
- Top3a
topoisomerase (DNA) III alpha
- FANCD2
Fanconi anemia, complementation group D2
- FANCA
Fanconi anemia, complementation group A
- NSE2
Non-structural maintenance of chromosomes element 2
- E3 SUMO
Small ubiquitin-like modifier 3
- RAP1
Repressor/activator protein 1 homolog
- TRF1
Telomeric repeat-binding factor 1
- TRF2
Telomeric repeat-binding factor 2
- TIN2
(TRF1) -interacting nuclear factor 2
- SLX4
Structure-specific endonuclease subunit
REFERENCES
- 1.Muller HJ. The remaking of chromosomes. Collecting Net. 1938;13:181. [Google Scholar]
- 2.Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 3.Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 4.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 5.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 6.Hemann MT, Strong MA, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 7.d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 8.Riethman H. Human telomere structure and biology. Annu Rev Genomics Hum Genet. 2008;9:1–19. doi: 10.1146/annurev.genom.8.021506.172017. [DOI] [PubMed] [Google Scholar]
- 9.Bailey JA, Liu G, Eichler EE. An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet. 2003;73:823–834. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linardopoulou EV, Williams EM, Fan Y, Friedman C, Young JM, Trask BJ. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambrosini A, Paul S, Hu S, Riethman H. Human subtelomeric duplicon structure and organization. Genome Biol. 2007;8:R151. doi: 10.1186/gb-2007-8-7-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudd MK, Endicott RM, Friedman C, Walker M, Young JM, Osoegawa K, et al. NISC Comparative Sequencing Program. Comparative sequence analysis of primate subtelomeres originating from a chromosome fission event. Genome Res. 2009;19:33–41. doi: 10.1101/gr.083170.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 14.Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 15.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 17.Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell JR, Wood E, Collins K. Telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 19.Wong JM, Collins K. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes Dev. 2006;20:2848–2858. doi: 10.1101/gad.1476206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vulliamy T, Beswick R, Kirwan M, Marrone A, Digweed M, Walne A, et al. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc Natl Acad Sci U S A. 2008;105:8073–8078. doi: 10.1073/pnas.0800042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 22.Chiu CP, Dragowska W, Kim NW, Vaziri H, Yui J, Thomas TE, et al. Differential expression of telomerase activity in hematopoietic progenitors from adult human bone marrow. Stem Cells. 1996;14:239–248. doi: 10.1002/stem.140239. [DOI] [PubMed] [Google Scholar]
- 23.Knight SW, Heiss NS, Vulliamy TJ, Greschner S, Stavrides G, Pai GS, et al. X-linked dyskeratosis congenita is predominantly caused by missense mutations in the DKC1 gene. Am J Hum Genet. 1999;65:50–58. doi: 10.1086/302446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, et al. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi H, Baerlocher GM, Lansdorp PM, Chanock SJ, Nunez O, Sloand E, et al. Mutations of the human telomerase RNA gene (TERC) in aplastic anemia and myelodysplastic syndrome. Blood. 2003;102:916–918. doi: 10.1182/blood-2003-01-0335. [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi H, Calado RT, Ly H, Kajigaya S, Baerlocher GM, Chanock SJ, et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med. 2005;352:1413–1424. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 27.Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 28.Marrone A, Stevens D, Vulliamy T, Dokal I, Mason PJ. Heterozygous telomerase RNA mutations found in dyskeratosis congenita and aplastic anemia reduce telomerase activity via haploinsufficiency. Blood. 2004;104:3936–3942. doi: 10.1182/blood-2004-05-1829. [DOI] [PubMed] [Google Scholar]
- 29.Armanios M, Chen JL, Chang YP, Brodsky RA, Hawkins A, Griffin CA, et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc Natl Acad Sci USA. 2005;102:15960–15964. doi: 10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam HY, Clark MJ, Chen R, Chen R, Natsoulis G, O’Huallachain M, et al. Performance comparison of whole-genome sequencing platforms. Nat Biotechnol. 2011;30:78–82. doi: 10.1038/nbt.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meeker AK, Hicks JL, Platz EA, March GE, Bennett CJ, Delannoy MJ, et al. Telomere shortening is an early somatic DNA alteration in human prostate tumorigenesis. Cancer Res. 2002;62:6405–6409. [PubMed] [Google Scholar]
- 32.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 34.Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 35.Hakin-Smith V, Jellinek DA, Levy D, Carroll T, Teo M, Timperley WR, et al. Alternative lengthening of telomeres and survival in patients with glioblastoma multiforme. Lancet. 2003;361:836–838. doi: 10.1016/s0140-6736(03)12681-5. [DOI] [PubMed] [Google Scholar]
- 36.Henson JD, Hannay JA, McCarthy SW, Royds JA, Yeager TR, Robinson RA, et al. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin Cancer Res. 2005;11:217–225. [PubMed] [Google Scholar]
- 37.Chang S, Khoo CM, Naylor ML, Maser RS, DePinho RA. Telomere-based crisis: functional differences between telomerase activation and ALT in tumor progression. Genes Dev. 2003;17:88–100. doi: 10.1101/gad.1029903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrish TA, Greider CW. Short telomeres initiate telomere recombination in primary and tumor cells. PLoS Genet. 2009;5:e1000357. doi: 10.1371/journal.pgen.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 40.Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Q, Ijpma A, Greider CW. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol Cell Biol. 2001;21:1819–1827. doi: 10.1128/MCB.21.5.1819-1827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AY, Pickett HA, et al. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol. 2009;27:1181–1185. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- 44.Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- 45.Londoño-Vallejo JA, Der-Sarkissian H, Cazes L, Bacchetti S, Reddel RR. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 2004;64:2324–2327. doi: 10.1158/0008-5472.can-03-4035. [DOI] [PubMed] [Google Scholar]
- 46.Pickett HA, Cesare AJ, Johnston RL, Neumann AA, Reddel RR. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 2009;28:799–809. doi: 10.1038/emboj.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cesare AJ, Kaul Z, Cohen SB, Napier CE, Pickett HA, Neumann AA, et al. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat Struct Mol Biol. 2009;16:1244–1251. doi: 10.1038/nsmb.1725. [DOI] [PubMed] [Google Scholar]
- 48.Fasching CL, Bower K, Reddel RR. Telomerase-independent telomere length maintenance in the absence of alternative lengthening of telomeres-associated promyelocytic leukemia bodies. Cancer Res. 2005;65:2722–2729. doi: 10.1158/0008-5472.CAN-04-2881. [DOI] [PubMed] [Google Scholar]
- 49.Jeyapalan JN, Mendez-Bermudez A, Zaffaroni N, Dubrova YE, Royle NJ. Evidence for alternative lengthening of telomeres in liposarcomas in the absence of ALT-associated PML bodies. Int J Cancer. 2008;122:2414–2421. doi: 10.1002/ijc.23412. [DOI] [PubMed] [Google Scholar]
- 50.Jeyapalan JN, Varley H, Foxon JL, Pollock RE, Jeffreys AJ, Henson JD, et al. Activation of the ALT pathway for telomere maintenance can affect other sequences in the human genome. Hum Mol Genet. 2005;14:1785–1794. doi: 10.1093/hmg/ddi185. [DOI] [PubMed] [Google Scholar]
- 51.Johnson JE, Varkonyi RJ, Schwalm J, Cragle R, Klein-Szanto A, Patchefsky A, et al. Multiple mechanisms of telomere maintenance exist in liposarcomas. Clin Cancer Res. 2005;11:5347–5355. doi: 10.1158/1078-0432.CCR-05-0684. [DOI] [PubMed] [Google Scholar]
- 52.Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 53.Zakian VA. Structure and function of telomeres. Annu Rev Genet. 1989;23:579–604. doi: 10.1146/annurev.ge.23.120189.003051. [DOI] [PubMed] [Google Scholar]
- 54.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 55.Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- 56.Karlseder J. Telomeric proteins: clearing the way for the replication fork. Nat Struct Mol Biol. 2006;13:386–387. doi: 10.1038/nsmb0506-386. [DOI] [PubMed] [Google Scholar]
- 57.Meselson M, Weigle JJ. Chromosome brekage accompanying genetic recombination in bacteriophage. Proc Natl Acad Sci U S A. 1961;47:857–868. doi: 10.1073/pnas.47.6.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Formosa T, Alberts BM. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986;47:793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- 59.Malkova A, Ivanov EL, Haber JE. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci U S A. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuzminov A, Stahl FW. Double-strand end repair via the RecBC pathway in Escherichia coli primes DNA replication. Genes Dev. 1999;13:345–356. doi: 10.1101/gad.13.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Motamedi MR, Szigety SK, Rosenberg SM. Double-strand-break repair recombination in Escherichia coli: physical evidence for a DNA replication mechanism in vivo. Genes Dev. 1999;13:2889–2903. doi: 10.1101/gad.13.21.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morrow DM, Connelly C, Hieter P. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malkova A, Signon L, Schaefer CB, Naylor ML, Theis JF, Newlon CS, et al. RAD51-independent break-induced replication to repair a broken chromosome depends on a distant enhancer site. Genes Dev. 2001;15:1055–1060. doi: 10.1101/gad.875901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malkova A, Naylor ML, Yamaguchi M, Ira G, Haber JE. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol Cell Biol. 2005;25:933–944. doi: 10.1128/MCB.25.3.933-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dunn B, Szauter P, Pardue ML, Szostak JW. Transfer of yeast telomeres to linear plasmids by recombination. Cell. 1984;39:191–201. doi: 10.1016/0092-8674(84)90205-8. [DOI] [PubMed] [Google Scholar]
- 66.Hackett JA, Feldser DM, Greider CW. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001;106:275–286. doi: 10.1016/s0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- 67.Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 68.Walmsley RW, Chan CS, Tye BK, Petes TD. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature. 1984;310:157–160. doi: 10.1038/310157a0. [DOI] [PubMed] [Google Scholar]
- 69.Lydeard JR, Lipkin-Moore Z, Sheu YJ, Stillman B, Burgers PM, Haber JE. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 2010;24:1133–1144. doi: 10.1101/gad.1922610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bosco G, Haber JE. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics. 1998;150:1037–1047. doi: 10.1093/genetics/150.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deem A, Barker K, Vanhulle K, Downing B, Vayl A, Malkova A. Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics. 2008;179:1845–1860. doi: 10.1534/genetics.108.087940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 74.Laud PR, Multani AS, Bailey SM, Wu L, Ma J, Kingsley C, et al. Elevated telomere-telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes Dev. 2005;19:2560–2570. doi: 10.1101/gad.1321305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qi L, Strong MA, Karim BO, Huso DL, Greider CW. Telomere fusion to chromosome breaks reduces oncogenic translocations and tumour formation. Nat Cell Biol. 2005;7:706–711. doi: 10.1038/ncb1276. [DOI] [PubMed] [Google Scholar]
- 76.Feldser DM, Greider CW. Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell. 2007;11:461–469. doi: 10.1016/j.ccr.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 78.de Lange T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- 79.Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 80.Natarajan S, McEachern MJ. Recombinational telomere elongation promoted by DNA circles. Mol Cell Biol. 2002;22:4512–4521. doi: 10.1128/MCB.22.13.4512-4521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cesare AJ, Groff-Vindman C, Compton SA, McEachern MJ, Griffith JD. Telomere loops and homologous recombination-dependent telomeric circles in a Kluyveromyces lactis telomere mutant strain. Mol Cell Biol. 2008;28:20–29. doi: 10.1128/MCB.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Biessmann H, Mason JM, Ferry K, d’Hulst M, Valgeirsdottir K, Traverse KL, et al. Addition of telomere-associated HeT DNA sequences “heals” broken chromosome ends in Drosophila. Cell. 1990;61:663–673. doi: 10.1016/0092-8674(90)90478-w. [DOI] [PubMed] [Google Scholar]
- 83.Osanai-Futahashi M, Fujiwara H. Coevolution of telomeric repeats and telomeric repeat-specific non-LTR retrotransposons in insects. Mol Biol Evol. 2011;28:2983–2986. doi: 10.1093/molbev/msr135. [DOI] [PubMed] [Google Scholar]
- 84.Gladyshev EA, Arkhipova IR. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci U S A. 2007;104:9352–9357. doi: 10.1073/pnas.0702741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Curcio MJ, Belfort M. The beginning of the end: links between ancient retroelements and modern telomerases. Proc Natl Acad Sci U S A. 2007;104:9107–9108. doi: 10.1073/pnas.0703224104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 87.Smit AF, Tóth G, Riggs AD, Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- 88.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 89.Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 90.Huang CR, Schneider AM, Lu Y, Niranjan T, Shen P, Robinson MA, et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–1182. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ewing AD, Kazazian HH., Jr Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 2011;21:985–990. doi: 10.1101/gr.114777.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cordaux R, Hedges DJ, Herke SW, Batzer MA. Estimating the retrotransposition rate of human Alu elements. Gene. 2006;373:134–137. doi: 10.1016/j.gene.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 95.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 96.Hancks DC, Kazazian HH., Jr SVA retrotransposons: Evolution and genetic instability. Semin Cancer Biol. 2010;20:234–245. doi: 10.1016/j.semcancer.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eickbush TH. Telomerase and retrotransposons: which came first? Science. 1997;277:911–912. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- 98.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 99.Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 100.Underwood DH, Zinzen RP, McEachern MJ. Template requirements for telomerase translocation in Kluyveromyces lactis. Mol Cell Biol. 2004;24:912–923. doi: 10.1128/MCB.24.2.912-923.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Horowitz H, Haber JE. Identification of autonomously replicating circular subtelomeric Y’ elements in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:2369–2380. doi: 10.1128/mcb.5.9.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Louis EJ, Haber JE. Mitotic recombination among subtelomeric Y’ repeats in Saccharomyces cerevisiae. Genetics. 1990;124:547–559. doi: 10.1093/genetics/124.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schmidt H, Taubert H, Lange H, Kriese K, Schmitt WD, Hoffmann S, et al. Small polydispersed circular DNA contains strains of mobile genetic elements and occurs more frequently in permanent cell lines of malignant tumors than in normal lymphocytes. Oncol Rep. 2009;22:393–400. [PubMed] [Google Scholar]
- 104.Henson JD, Reddel RR. Assaying and investigating Alternative Lengthening of Telomeres activity in human cells and cancers. FEBS Lett. 2010;584:3800–3811. doi: 10.1016/j.febslet.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 105.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 106.Compton SA, Choi JH, Cesare AJ, Ozgür S, Griffith JD. Xrcc3 and Nbs1 are required for the production of extrachromosomal telomeric circles in human alternative lengthening of telomere cells. Cancer Res. 2007;67:1513–1519. doi: 10.1158/0008-5472.CAN-06-3672. [DOI] [PubMed] [Google Scholar]
- 107.Zhong ZH, Jiang WQ, Cesare AJ, Neumann AA, Wadhwa R, Reddel RR. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J Biol Chem. 2007;282:29314–29322. doi: 10.1074/jbc.M701413200. [DOI] [PubMed] [Google Scholar]
- 108.Tarsounas M, Mu�oz P, Claas A, Smiraldo PG, Pittman DL, Blasco MA, et al. Telomere maintenance requires the RAD51D recombination/repair protein. Cell. 2004;117:337–347. doi: 10.1016/s0092-8674(04)00337-x. [DOI] [PubMed] [Google Scholar]
- 109.Krishna S, Wagener BM, Liu HP, Lo YC, Sterk R, Petrini JH, et al. Mre11 and Ku regulation of double-strand break repair by gene conversion and break-induced replication. DNA Repair (Amst) 2007;6:797–808. doi: 10.1016/j.dnarep.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Le S, Moore JK, Haber JE, Greider CW. RAD51 and RAD50 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hwang JY, Smith S, Ceschia A, Torres-Rosell J, Aragon L, Myung K. Smc5–Smc6 complex suppresses gross chromosomal rearrangements mediated by break-induced replications. DNA Repair (Amst) 2008;7:1426–1436. doi: 10.1016/j.dnarep.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ho CK, Mazón G, Lam AF, Symington LS. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol Cell. 2010;40:988–1000. doi: 10.1016/j.molcel.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Potts PR, Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol. 2007;14:581–590. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- 114.Noël JF, Wellinger RJ. Abrupt telomere losses and reduced end-resection can explain accelerated senescence of Smc5/6 mutants lacking telomerase. DNA Repair (Amst) 2011;10:271–282. doi: 10.1016/j.dnarep.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 115.Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, et al. The Smc5–Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9:923–931. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- 116.Zeng S, Xiang T, Pandita TK, Gonzalez-Suarez I, Gonzalo S, Harris CC, et al. Telomere recombination requires the MUS81 endonuclease. Nat Cell Biol. 2009;11:616–623. doi: 10.1038/ncb1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zeng S, Yang Q. The MUS81 endonuclease is essential for telomerase negative cell proliferation. Cell Cycle. 2009;8:2157–2160. doi: 10.4161/cc.8.14.9149. [DOI] [PubMed] [Google Scholar]
- 119.McPherson JP, Lemmers B, Chahwan R, Pamidi A, Migon E, Matysiak-Zablocki E, et al. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science. 2004;304:1822–1826. doi: 10.1126/science.1094557. [DOI] [PubMed] [Google Scholar]
- 120.Boeke JD, Eichinger D, Castrillon D, Fink GR. The Saccharomyces cerevisiae genome contains functional and nonfunctional copies of transposon Ty1. Mol Cell Biol. 1988;8:1432–1442. doi: 10.1128/mcb.8.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zou S, Ke N, Kim JM, Voytas DF. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 1996;10:634–645. doi: 10.1101/gad.10.5.634. [DOI] [PubMed] [Google Scholar]
- 122.Scholes DT, Kenny AE, Gamache ER, Mou Z, Curcio MJ. Activation of a LTR-retrotransposon by telomere erosion. Proc Natl Acad Sci U S A. 2003;100:15736–15741. doi: 10.1073/pnas.2136609100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Maxwell PH, Coombes C, Kenny AE, Lawler JF, Boeke JD, Curcio MJ. Ty1 mobilizes subtelomeric Y’ elements in telomerase-negative Saccharomyces cerevisiae survivors. Mol Cell Biol. 2004;24:9887–9898. doi: 10.1128/MCB.24.22.9887-9898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Paquin CE, Williamson VM. Temperature effects on the rate of ty transposition. Science. 1984;226:53–55. doi: 10.1126/science.226.4670.53. [DOI] [PubMed] [Google Scholar]
- 125.Maxwell PH, Curcio MJ. Incorporation of Y’-Ty1 cDNA destabilizes telomeres in Saccharomyces cerevisiae telomerase-negative mutants. Genetics. 2008;179:2313–2317. doi: 10.1534/genetics.108.089052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 127.Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, et al. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 128.Deininger PL, Moran JV, Batzer MA, Kazazian HH., Jr Mobile elements and mammalian genome evolution. Curr Opin Genet Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 129.Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 130.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 131.Martin SL. Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1. RNA Biol. 2010;7:706–711. doi: 10.4161/rna.7.6.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 133.Ostertag EM, DeBerardinis RJ, Goodier JL, Zhang Y, Yang N, Gerton GL, et al. A mouse model of human L1 retrotransposition. Nat Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 134.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 135.An W, Han JS, Wheelan SJ, Davis ES, Coombes CE, Ye P, et al. Active retrotransposition by a synthetic L1 element in mice. Proc Natl Acad Sci U S A. 2006;103:18662–18667. doi: 10.1073/pnas.0605300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lee J, Han K, Meyer TJ, Kim HS, Batzer MA. Chromosomal inversions between human and chimpanzee lineages caused by retrotransposons. PLoS One. 2008;3:e4047. doi: 10.1371/journal.pone.0004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Myers JS, Vincent BJ, Udall H, Watkins WS, Morrish TA, Kilroy GE, et al. A comprehensive analysis of recently integrated human Ta L1 elements. Am J Hum Genet. 2002;71:312–326. doi: 10.1086/341718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sen SK, Huang CT, Han K, Batzer MA. Endonuclease-independent insertion provides an alternative pathway for L1 retrotransposition in the human genome. Nucleic Acids Res. 2007;35:3741–3751. doi: 10.1093/nar/gkm317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hackett JA, Greider CW. End resection initiates genomic instability in the absence of telomerase. Mol Cell Biol. 2003;23:8450–8461. doi: 10.1128/MCB.23.23.8450-8461.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kojima T, Nakajima K, Mikoshiba K. The disabled 1 gene is disrupted by a replacement with L1 fragment in yotari mice. Brain Res Mol Brain Res. 2000;75:121–127. doi: 10.1016/s0169-328x(99)00313-7. [DOI] [PubMed] [Google Scholar]
- 141.George JA, DeBaryshe PG, Traverse KL, Celniker SE, Pardue ML. Genomic organization of the Drosophila telomere retrotransposable elements. Genome Res. 2006;16:1231–1240. doi: 10.1101/gr.5348806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cenci G, Siriaco G, Raffa GD, Kellum R, Gatti M. The Drosophila HOAP protein is required for telomere capping. Nat Cell Biol. 2003;5:82–84. doi: 10.1038/ncb902. [DOI] [PubMed] [Google Scholar]
- 143.Raffa GD, Raimondo D, Sorino C, Cugusi S, Cenci G, Cacchione S, et al. Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev. 2010;24:1596–1601. doi: 10.1101/gad.574810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Raffa GD, Siriaco G, Cugusi S, Ciapponi L, Cenci G, Wojcik E, et al. The Drosophila modigliani (moi) gene encodes a HOAP-interacting protein required for telomere protection. Proc Natl Acad Sci U S A. 2009;106:2271–2276. doi: 10.1073/pnas.0812702106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ciapponi L, Cenci G, Ducau J, Flores C, Johnson-Schlitz D, Gorski MM, et al. The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr Biol. 2004;14:1360–1366. doi: 10.1016/j.cub.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 146.Gao G, Bi X, Chen J, Srikanta D, Rong YS. Mre11-Rad50-Nbs complex is required to cap telomeres during Drosophila embryogenesis. Proc Natl Acad Sci U S A. 2009;106:10728–10733. doi: 10.1073/pnas.0902707106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Biessmann H, Kasravi B, Bui T, Fujiwara G, Champion LE, Mason JM. Comparison of two active HeT-A retroposons of Drosophila melanogaster. Chromosoma. 1994;103:90–98. doi: 10.1007/BF00352317. [DOI] [PubMed] [Google Scholar]
- 148.Danilevskaya O, Slot F, Pavlova M, Pardue ML. Structure of the Drosophila HeT-A transposon: a retrotransposon-like element forming telomeres. Chromosoma. 1994;103:215–224. doi: 10.1007/BF00368015. [DOI] [PubMed] [Google Scholar]
- 149.Kahn T, Savitsky M, Georgiev P. Attachment of HeT-A sequences to chromosomal termini in Drosophila melanogaster may occur by different mechanisms. Mol Cell Biol. 2000;20:7634–7642. doi: 10.1128/mcb.20.20.7634-7642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Danilevskaya ON, Arkhipova IR, Traverse KL, Pardue ML. Promoting in tandem: the promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell. 1997;88:647–655. doi: 10.1016/s0092-8674(00)81907-8. [DOI] [PubMed] [Google Scholar]
- 151.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kubo S, Seleme MC, Soifer HS, Perez JL, Moran JV, Kazazian HH, Jr, et al. L1 retrotransposition in nondividing and primary human somatic cells. Proc Natl Acad Sci U S A. 2006;103:8036–8041. doi: 10.1073/pnas.0601954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Henson JD, Reddel RR. Assaying and investigating Alternative Lengthening of Telomeres activity in human cells and cancers. FEBS Lett. 2010;584:3800–3811. doi: 10.1016/j.febslet.2010.06.009. [DOI] [PubMed] [Google Scholar]