Abstract
Cutaneous leishmaniasis has various phenotypic aspects consisting of polymorphic amastigotes with different genetic ranges. Samples were collected from suspected patients of Khuzestan province. Prepared smears were stained, scaled, and measured using ocular micrometer. The Cyt b, ITS-rDNA, and microsatellite genes of Leishmania were amplified and Leishmania species were identified by molecular analyses. Of 150 examined suspected patients, 102 were identified to Leishmania species (90 L. major, nine L. tropica, and three unidentified). The amastigotes of 90 L. major had regular and different irregular shapes within three clinical lesions with no and/or low genetic diversity. Three haplotypes of Cyt b of L. major were found but no variation was observed using ITS-rDNA gene. Interesting findings were that all nine L. tropica had regular amastigote shapes with more genetic variations, also a patient which had coinfection of L. major, L. tropica, and Crithidia. At least two L. major and L. tropica were identified in suspected patients of the regions. Different irregular amastigotes' shapes of L. major can be explained by various reservoir hosts and vectors. In contrast, more molecular variations in L. tropica could be justified by genetic characters. Unidentified Leishmania could be mixed pathogens or nonpathogens with mammals' Leishmania or Crithidia.
1. Introduction
Cutaneous leishmaniasis (CL) is caused by parasites belonging to the genus Leishmania parasitic and referred to neglected tropical diseases [1, 2]. CL is distributed in more than half of the Iranian provinces (20 out of 32) with different prevalence (1.8%–37.9%), also, the number of reported CL cases/year and estimated annual CL incidence are 26,630 and 69,000 to 113,300, respectively [3, 4].
In order of epidemiological importance, three well known principle ethioparasitological agents, Leishmania (Leishmania) major; Yakimoff and Schokhor, 1914, L. (L.) tropica; Wright, 1903, and less frequently L. (L.) infantum; Nicolle, 1908, unequivocally caused CL in Iran [5, 6]. However, recently nonpathogenic L. (L.) turanica was reported from human with ZCL (zoonotic cutaneous leishmaniasis) in Iran but authors did not judge that as a causative agent of CL [7].
Different clinical aspects are caused by CL and are observed in several parts of the world wide but many influencing factors of CL have not absolutely understood [8, 9]. The spectrum of genetic diversity in the genus Leishmania is described as one of the most controversial issues [10–16]. The genetic heterogeneity/homogeneity of protozoan parasite may bring diverse phenotypic ranges including clinical manifestations or even diversity of morphometric characters in Trypanosomatidae [12, 17–19].
Currently, Leishmania species are unambiguously circulating in Khuzestan province, where it is a neglected focus of leishmaniasis in southwestern Iran, coborder with Iraq. The different Leishmania parasites' aspects have not been precisely elucidated yet such as ultrataxonomy (strain/haplotype), phenotypic features, and their phylogenetic inferences in Khuzestan province [20–23]. The complexities of these Leishmania criteria are associated with the intricacy of leishmaniasis epizoology, migration of people to Iraq and vice versa, recent reactivation of old foci, lack of using accurate diagnostic markers, developing geographical distribution with high temperature (50°C) and humidity (95%), long distance, remote distance from the equipped laboratory, and unsafe sampling in Iraq border [21].
To assess genetic diversity analysis, a number of nuclear and extranuclear DNA markers were employed, including mini-exon, 18S-rRNA, gp63 gene locus, microsatellites, HSP-70, ribosomal internal transcribed spacer regions (ITS-rDNA), minicircles of kinetoplast DNA (kDNA), and cytochrome b (Cyt b) (kDNA maxicircle) [24–32].
Cyt b is the central redox catalytic subunit of quinol which belongs to the mitochondrial genome and it is valuable in understanding phylogenetic study among individuals [33–35].
The ITS-rDNA marker is utilized to infer evolutionary relationships in genus Leishmania because of having conserved regions and low intracellular polymorphism [30, 36].
Microsatellites are short tandemly repeated DNA sequence motifs which are distributed plentifully in Leishmania species. Microsatellites have a high specificity in identification of Leishmania infections owing to their allelic characters [37].
Molecular identification of Leishmania species has been conducted by targeting different DNA markers in Iran [29, 38–40]. But in ignored ZCL foci of Khuzestan province, few extensive studies were done on epidemiological and molecular aspects on morphological features of leishmaniases [20–23]. In this research, epidemiological, molecular, and morphological methods were combined with more details on morphological characters of Leishmania amastigote shapes, clinical appearances of lesions, and the type of ulcers using nuclear (ITS-rDNA) and mitochondrial (Cyt b) genes. The finding of prevalence of CL, molecular variations, and different morphological shapes of Leishmania parasite can height our knowledge about behavioral aspects of L. major and L. tropica in southwestern Iran.
2. Materials and Methods
2.1. Sampling and Identifying of Leishmania Parasites
In southwest of Iran, within the ZCL foci, three geographical locations, in west (Dashte azadegan, Sosangerd and Hoveyze), central (Ahvaz), and east (Ramhormoz and Ramshir) along with their neighboring villages were sampled (31°7′N to 32°5′N and 47°41′E to 48°30′E) (Figure 1). Some surveyed locations are situated in the border of Iran and Iraq within about 10,406 km2 with 12–210 meters above sea level (A.S.L.). The environmental and climate conditions of the studied regions are different from other CL foci in Iran with temperature range: 20–60°C, humidity: 50–85%, and monthly average rainfall: 17–25 mm (Figure 1, Table 1).
Figure 1.
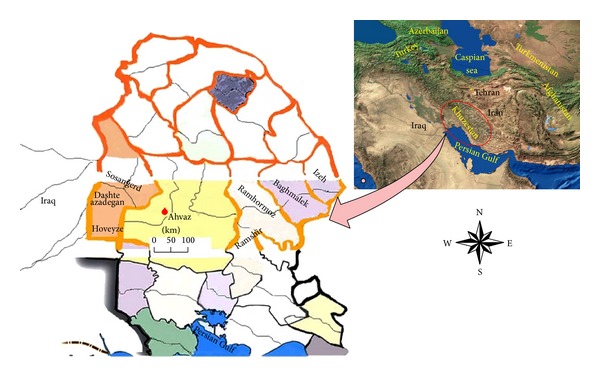
Sampled regions from suspected patients in foci of cutaneous leishmanisis along with their neighboring villages in three geographical locations of Khuzestan province, Iran (western, Dashte azadegan, Sosangerd, and Hoveyze; central, Ahvaz and eastern, Ramhormoz and Ramshir).
Table 1.
Characteristics of confirmed patients to cutaneous leishmaniasis based on collection study site, personal characters, microscopic observation, and molecular methods.
| Province regions (positions) |
Collection site | Characteristics | Microscopic observation | Molecular methods | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cities and villages |
Sex | Age groups (years) |
Lesion duration (month) |
Lesion number | Lesion types |
Leishmania amastigotes' positive grading numbers: 1+ to 6+ |
Leishmania amplification by targeted genes | RFLP with BsuRI & SspI, sequencing and phylogenetic analyses | ||||||||||||||||||||||||||
| Female | Male | <1 | 1–3 | 3–5 | 5–10 | 10–15 | 15–25 | >25 | <1 | 2-3 | >3 | Single | Double | Multiple | Wet | Dry | Mixed | 1+ | 2+ | 3+ | 4+ | 5+ | 6+ | Total |
ITS-rDNA/ & Cyt b & ITS-microsatellite +ve |
L.
major +ve |
L.
tropica +ve |
Unidentified; Leishmania sp |
||||||
| −ve | +ve | ITS-rDNA | Cyt b |
∗Mic | −ve | |||||||||||||||||||||||||||||
| Khuzestan | ||||||||||||||||||||||||||||||||||
| Ahvaz (central) |
Kotabdullah | 4 | ∗∗1 | 0 | 1 | 1 | 1 | 2 | 0 | 0 | 2 | 2 | 1 | 3 | 1 | 1 | 4 | 1 | 0 | 0 | 1 | 0 | 2 | 1 | 1 | 0 | 5 | 3 | 1 | 1 | 0 | 2 | 2 | 1 |
| Damane kochak | 4 | 6 | 0 | 1 | 2 | 1 | 1 | 4 | 1 | 7 | 1 | 2 | 6 | 2 | 2 | 7 | 2 | 1 | 1 | 1 | 1 | 4 | 2 | 1 | 3 | 10 | 8 | 0 | 2 | 3 | 5 | 0 | 0 | |
| Ashari bozorg | 3 | 7 | 0 | 1 | 1 | 3 | 0 | 3 | 2 | 3 | 4 | 3 | 2 | 5 | 3 | 7 | 2 | 1 | 1 | 1 | 0 | 2 | 2 | 4 | 1 | 10 | 7 | 1 | 0 | 1 | 6 | 0 | 0 | |
| Bahonar | 2 | 2 | 0 | 0 | 0 | 0 | 2 | 1 | 1 | 3 | 1 | 0 | 0 | 0 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 4 | 3 | 1 | 0 | 1 | 3 | 1 | 0 | |
| Sheyban | 2 | 3 | 0 | 0 | 0 | 2 | 0 | 2 | 1 | 3 | 2 | 0 | 2 | 2 | 1 | 1 | 4 | 0 | 2 | 1 | 1 | 0 | 1 | 0 | 0 | 5 | 4 | 1 | 0 | 0 | 5 | 0 | 0 | |
| Koye abozar | 0 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 1 | 1 | 0 | 0 | 2 | 0 | 0 | |
| Kian abad | 0 | 5 | 0 | 0 | 0 | 2 | 0 | 3 | 0 | 2 | 2 | 1 | 5 | 0 | 0 | 3 | 2 | 0 | 0 | 1 | 1 | 1 | 2 | 0 | 0 | 5 | 3 | 2 | 0 | 0 | 2 | 0 | 0 | |
| Shahrake bargh | 2 | 6 | 0 | 1 | 1 | 2 | 0 | 2 | 2 | 6 | 0 | 2 | 4 | 3 | 1 | 7 | 0 | 1 | 1 | 2 | 1 | 1 | 0 | 3 | 1 | 8 | 3 | 2 | 2 | 1 | 3 | 1 | 0 | |
| Padad | 2 | 3 | 0 | 0 | 1 | 2 | 0 | 0 | 2 | 4 | 1 | 0 | 4 | 1 | 0 | 2 | 2 | 1 | 0 | 0 | 4 | 1 | 0 | 0 | 0 | 5 | 4 | 1 | 0 | 0 | 3 | 0 | 0 | |
| Sadeghiyeh | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 2 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | |
| Malashiye | 0 | 3 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 2 | 1 | 0 | 2 | 1 | 0 | 1 | 2 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 3 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | |
| Amaniyeh | 1 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 1 | 2 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 3 | 3 | 1 | 1 | 0 | 2 | 0 | 0 | |
| 400 dastghah | 1 | 3 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 3 | 0 | 1 | 1 | 3 | 0 | 1 | 3 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 4 | 2 | 2 | 0 | 0 | 3 | 0 | 0 | |
| Koye bonovat | 1 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 2 | 1 | 0 | 2 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 3 | 2 | 1 | 0 | 0 | 2 | 1 | 0 | |
| Dashteazadegan (western) |
Hajiyeh | 1 | 2 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 2 | 1 | 1 | 2 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 3 | 2 | 1 | 0 | 0 | 2 | 0 | 1 |
| Soydani | 0 | 4 | 0 | 1 | 0 | 1 | 2 | 0 | 0 | 2 | 1 | 1 | 1 | 1 | 2 | 3 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 4 | 3 | 2 | 0 | 1 | 3 | 1 | 0 | |
| Bostan centre | 3 | 3 | 0 | 0 | 0 | 0 | 2 | 2 | 2 | 2 | 3 | 1 | 0 | 3 | 3 | 2 | 3 | 1 | 1 | 2 | 1 | 1 | 1 | 0 | 0 | 6 | 5 | 1 | 4 | 0 | 3 | 0 | 0 | |
| Shakeriye | 2 | 3 | 0 | 0 | 0 | 1 | 2 | 2 | 0 | 2 | 2 | 1 | 2 | 3 | 0 | 3 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 2 | 0 | 5 | 3 | 2 | 0 | 0 | 4 | 0 | 0 | |
| Jalaliye | 1 | 2 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 2 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 3 | 4 | 1 | 1 | 1 | 2 | 1 | 0 | |
| Albo afri | 1 | 3 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 1 | 3 | 3 | 0 | 1 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 4 | 2 | 2 | 0 | 0 | 4 | 0 | 0 | |
| Malekiyeh | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 1 | 1 | 2 | 0 | 0 | |
| Koye abodi | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 2 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | |
| Hoveyze | 3 | 7 | 1 | 0 | 1 | 1 | 0 | 3 | 4 | 5 | 2 | 3 | 1 | 4 | 5 | 5 | 5 | 0 | 1 | 2 | 2 | 2 | 1 | 2 | 0 | 10 | 3 | 2 | 1 | 0 | 5 | 1 | 1 | |
| Farhangian | 1 | 6 | 0 | 1 | 1 | 1 | 1 | 3 | 0 | 2 | 5 | 0 | 1 | 5 | 1 | 5 | 1 | 1 | 0 | 4 | 1 | 2 | 0 | 0 | 1 | 7 | 3 | 1 | 1 | 1 | 5 | 1 | 0 | |
| Ramhormoz (eastern) |
Darre khosh | 3 | 6 | 1 | 0 | 0 | 0 | 2 | 5 | 1 | 8 | 1 | 0 | 3 | 2 | 4 | 7 | 2 | 0 | 0 | 1 | 0 | 5 | 1 | 2 | 0 | 9 | 5 | 1 | 2 | 0 | 7 | 0 | 0 |
| Shardin | 1 | 4 | 1 | 1 | 0 | 0 | 0 | 1 | 2 | 0 | 2 | 3 | 2 | 2 | 1 | 3 | 1 | 1 | 0 | 1 | 0 | 1 | 3 | 0 | 0 | 5 | 4 | 1 | 1 | 0 | 3 | 0 | 0 | |
| Shamilan | 3 | 3 | 1 | 2 | 0 | 0 | 1 | 2 | 0 | 3 | 2 | 1 | 1 | 2 | 3 | 5 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | 2 | 1 | 1 | 1 | 5 | 0 | 0 | |
| Total | 43 | 94 | 9 | 12 | 13 | 21 | 19 | 37 | 26 | 66 | 47 | 24 | 51 | 47 | 39 | 86 | 38 | 11 | 7 | 20 | 22 | 45 | 21 | 22 | 13 | 137 | 90 | 30 | 19 | 11 | 90 | 9 | 3 | |
| 137 | 137 | 137 | 137 | 137 | 137 | 150 | 139 | 102 | ||||||||||||||||||||||||||
* = microsatellite, ∗∗1 = One patient had coinfections with L. major, L. tropica, and Crithidia.
The lesions of suspected patients to CL were sampled in 2012-2013. The smears were obtained and prepared in two methods of passive and active for Leishmania parasites. The personal information was recorded for each suspected patient (sex, age, lesion duration, type and number of lesion, patients' travelling to endemic regions of ZCL, and drug consumption) (Table 1) [7].
The samples were smeared on two microscopic slides, dried, and stained by Giemsa and Leishmania infections were identified under light microscope with high magnification (×1000). The positive slides of each patient were graded to order leishman bodies' density from +1 to +6 [41]. Various morphometric shapes and the sizes of amastigotes of Leishmania species were accurately assessed (30 min. per slide) by ocular micrometer (×100 objective = 1 μm per unit space) and Dino Capture 2.0.
Serous of some samples were subcutaneously inoculated into the base tail of the BALB/c and then was examined weekly with the intent of inspecting the appearance of lesion at the position of injection for 6 months. Some serous from active lesion of suspected patients were cultured in Novy-MacNeal-Nicolle (NNN) medium and incubated in 22°C for 6 weeks. The samples were subcultured weekly and checked regularly to monitor the growth and the presence of promastigotes. All experiments on the humans and animals were performed according to the guidelines of the Ethical Board of Pasteur Institute of Iran.
2.2. Extraction of Total Genomic DNA
DNA of parasites was directly extracted from graded slides based on modified Phenol-chloroform protocol [42]. All Giemsa-stained slides were washed with ethanol and covered with 300 μL lysis buffer (50 mM NaCl, 50 mM Tris, 10 mM EDTA, pH 7.4, 1% v/v Triton x-100). Giemsa-stained slides should be free from blood in order to prevent misdiagnosis of visualized amastigotes from platelets as well as adverse effects of protoporphyrin on amplified DNA by PCR. The extraction of the genomic DNA of each Giemsa-stained slide was followed by the protocol of Bordbar and Parvizi [29]. DNA was resuspended in 50 μL 1X TE and stored at −20°C. The DNA concentration of each extraction was measured using a NanoDrop (Thermo Scientific Inc., Wilmington, DE). DNA extraction of Leishmania parasites was carried out in a molecular biology laboratory where amplified and cloned DNAs had never been performed [30].
2.3. PCR Amplification and RLFP for ITS-rDNA, Cyt b, and microsatellite Genes of Leishmania Parasites from Suspected Patients
The standard PCR was employed to detect Leishmania parasites in suspected patients by targeting three genes, ITS-rDNA about 480 bp, Cyt b about 880 bp, and microsatellite about 180 bp. PCR products were subjected to electrophoresis in 1.5% agarose gel and were observed under ultraviolet light after staining for 15 min with (0.5 g/mL) ethidium bromide.
For digesting the PCR amplicons in RFLP, the suitable enzymes were selected. The sequences of ITS-rDNA and Cyt b genes of Old World Leishmania were retrieved (ITS-rDNA: L. major: EF413078, L. tropica: KC540906, and L. infantum: EU330402; Cyt b: L. major: AB095961, L. tropica: EF579904, and L. infantum: EF579895). To standardize the number and the size of DNA fragments for each Leishmania species, the retrieved sequences were designed by in-silico analysis software CLC DNA Workbench 5.2 (CLC bio A/S, Aarhus, Denmark) (Figure 2). Endonuclease reaction of ITS-rDNA and Cyt b genes was performed in a volume of 30 μL containing 2 μL of BsuRI (HaeIII) with cut site GG↓CC and 2 μL of Ssp1 with cut site AAT↓ATT, 10 μL of PCR products, 2 μL of 10x buffer, and 16 μL of distilled water for 4 h (delayed digestion) at 37°C for ITS-rDNA gene and 5 min. (rapid digestion) at 37°C for Cyt b gene. Standard strains of WHO for L. major (HOM/SU/73/5ASKH), L. infantum (MHOM/TN/80/IPT1), and L. tropica (MHOM/PS/2008/344Jn SF53) were used as positive controls. The negative controls were applied, one of which had no PCR product and the other one had no restriction enzyme. After digesting PCR products, the fragments were analyzed using electrophoresis on agarose gel 3% containing ethidium bromide and ladder of 50 bp (Fermentas) (Figure 3).
Figure 2.
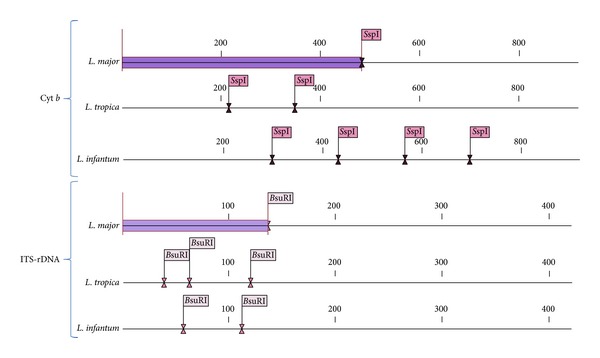
In-silico prediction of SspI and BsuR1 restriction enzymes for Cyt b and ITS-rDNA genes in the Leishmania species analyzed in this study.
Figure 3.
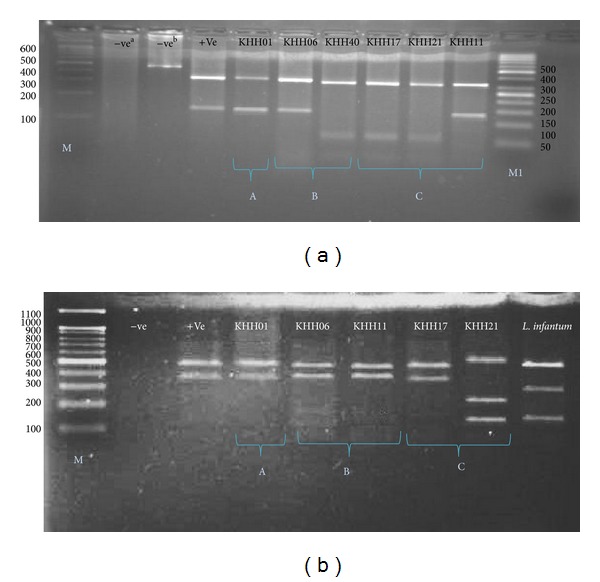
PCR-RFLP observations in Khuzestan isolates based on ITS-rDNA (a) and Cyt b (b) genes. A; Lane 1 (−vea: negative control containing BsuR1 without PCR product), lane 2 (−veb: negative control containing PCR product without BsuR1), lane 3 (+ve: positive control for L. major), lanes 4, 5; KHH 01 and 06 (L. major): isolated from Eastern and central Khuzestan, respectively. Lanes 6–8; KHH 40, KHH 17, and KHH 21 (L. tropica): isolated from central and western Khuzestan, lane 9; KHH 11; L. major isolated from western Khuzestan. M: 100 bp size marker. M1: 50 bp size marker. −ve: negative and +ve: positive. B; lane 1 (−ve: negative control containing BsuR1 without PCR product), lane 2 (+ve: positive control for L. major), lanes 3–6; KHH 01 and 06 KHH 11 and 17 (L. major): isolated from eastern and central and western Khuzestan, respectively. Lane 7; KHH 21 (L. tropica): isolated from western Khuzestan, lane 8; Leishmania infantum with reference strain; MHOM/TN/80/IPT1. A; isolated samples from eastern Khuzestan, B; isolated samples from central Khuzestan and C; isolated samples from western Khuzestan.
2.4. Evaluation of PCR Band Sensitivities in Three Implemented Genes: ITS-rDNA, Microsatellite, and Cyt b
In order to analyze and compare the sensitivity of PCR bands for three genes of ITS-rDNA, microsatellite, and Cyt b, the serial dilutions of target DNAs were established in order of 1 : 1, 1 : 2, 1 : 4, 1 : 8, and 1 : 16 for each mentioned gene, afterward, the PCR band intensities of each gene were distinctly calculated in different dilutions based on their chromatogram length using Gel Analyzer software version 3.0.
2.5. DNA Sequencing and Phylogeny Analyzing ITS-rDNA and Cyt b Genes
To reconfirm the results of RFLP, some amplified PCR products were directly sequenced by targeting ITS-rDNA and Cyt b genes in both directions using the ITS1F, ITS2R4 and LCBF1, LCBR2 primers by ABIPRISMTM 3130 Genetic Analyzer automated sequencer (Applied Biosystem, USA). Individual sequences were aligned, justified, and edited in consensus positions compared to GenBank sequences of all regional species in case of homology and congruency of new haplotypes with the use of Sequencher v.4.1.4 Software for PC (Gene Codes Corporation).
Maximum likelihood (ML) tree was constructed via MEGA v5.05 for showing the phylogenetic position of common and new haplotypes of the ITS-rDNA and Cyt b sequences based on the Kimura 2-parameter model of nucleotide substitution search by stepwise addition of 100 random replicates and bootstrap values with 1000 replicates [43].
3. Results
3.1. Morphometric Characterization of Leishmania Species and Their Epidemiological Features in Suspected Patients
In total, 150 suspected patients were examined microscopically for Leishmania infections among 81,000 population in three different study sites with acute and/or chronic lesions' signs. 137 out of 150 samples (91.3%) were observed microscopically positive with Leishmania parasites and cultured in NNN medium (Table 1).
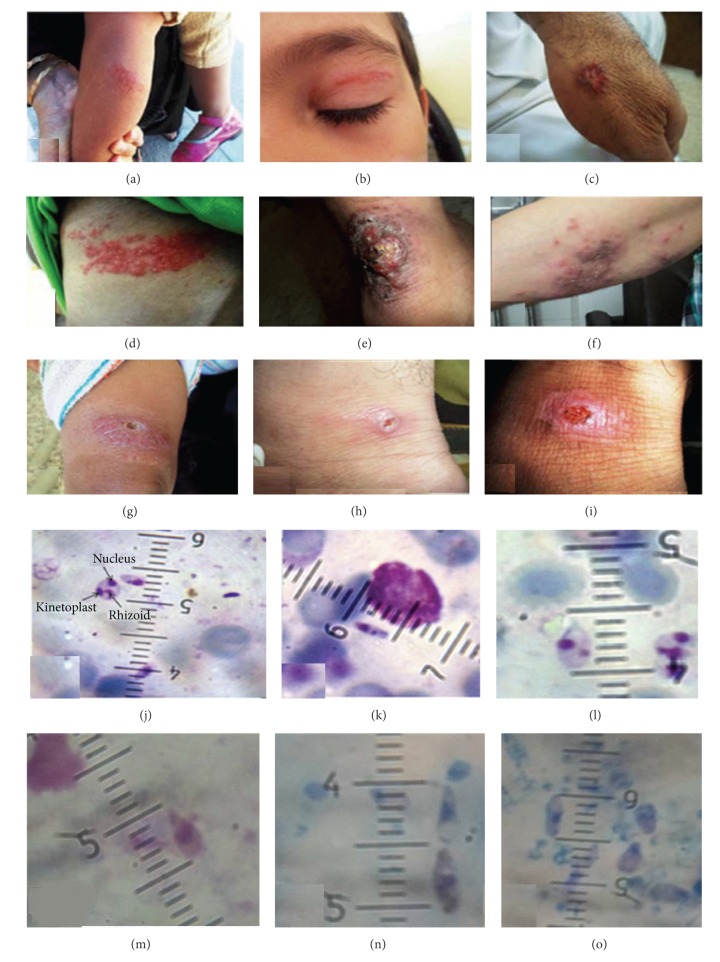
Leishmania infections were found more in 15–25 years old (37/137: 27%) than other groups and in males (94/137: 68.6%). The grades of positive smears were arranged from +1 to +6 (WHO 1991). More infected patients with L. major had grade +4 (45/137: 32.8%). The average size of L. major amastigotes was in range 3–5 ± 0.31 μm in length and 2.15 ± 0.21 μm in width (Table 2). L. major amastigotes were observed with obvious vacuole and polymorphic shapes (regular, oval, and round; 74/90: 82.2%) (Table 2). The irregular polymorphic shapes of L. major amastigotes were classified into Cigarette, Spindle, and Pear; 16/90: 17.7% and visualized in three lesion types of wet, dry, and mixed. Interestingly, the sizes of regular and irregular amastigote shapes were approximately 3-4 and 4-5 μm, respectively (Figures 4(k)–4(o)). It is noteworthy that polymorphic amastigote shapes were observed only in one lesion form. The wet lesions were grouped in classic (volcanic sign: 56/70: 80%) and nonclassic (herpetiform, erythematous papulonodules, hyperkeratotic, eczematoid, zosteriform, and psoriasiform patterns: 17/70: 24.3%) (Figures 4(d)–4(i)). In contrast, L. tropica amastigotes were sized approximately 2-3 ± 0.23 μm in length and 2.2 ± 0.11 μm in width with amastigotes' shapes of round and/or oval within dry lesions (100%) (Figures 4(a), 4(b), and 4(j)). The grade of positive smears of L. tropica showed that all observed smears were +6.
Table 2.
Personal information, lesion characteristics, shapes, and sizes of amastigotes in L. major and L. tropica isolated from patients in three locations of study sites.
| Criteria | Lesion type in L. tropica | Lesion types in L. major |
Morphometric characteristics of L. tropica amastigotes |
Morphometric characteristics of L. major amastigotes | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size of amastigotes (µm) |
Shapes of amastigotes |
Sizes of amastigotes (µm) |
Shapes of amastigotes | |||||||||||||
| Regular | Regular | Irregular | ||||||||||||||
| Age group (years) | Dry | Wet | Dry | Mixed | 2-3 | Round | Oval | 3-4 | >4 | Round | Oval | Pear | Cigarette | Spindle | ||
| Age ranges |
Total | ∗C | ∗NC | |||||||||||||
| <1 | 4 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 1 | 2 | 0 | 1 | 0 |
| 1–3 | 9 | 1 | 5 | 2 | 1 | 0 | 1 | 1 | 0 | 6 | 2 | 2 | 3 | 2 | 1 | 0 |
| 3–5 | 11 | 1 | 5 | 1 | 2 | 1 | 1 | 1 | 0 | 7 | 3 | 1 | 9 | 0 | 0 | 0 |
| 5–10 | 12 | 2 | 5 | 2 | 3 | 1 | 2 | 1 | 1 | 7 | 3 | 2 | 6 | 1 | 1 | 0 |
| 10–15 | 18 | 2 | 10 | 3 | 2 | 1 | 2 | 1 | 1 | 12 | 4 | 3 | 11 | 1 | 1 | 0 |
| 15–25 | 27 | 2 | 17 | 3 | 3 | 2 | 2 | 1 | 1 | 20 | 5 | 2 | 20 | 1 | 1 | 1 |
| >25 | 18 | 1 | 12 | 4 | 1 | 0 | 1 | 0 | 1 | 15 | 2 | 1 | 11 | 1 | 2 | 2 |
|
| ||||||||||||||||
| Total (%) | 99 | 9 | 56 | 17 | 12 | 5 | 9 | 5 (55.5) | 4 (44.4) | 71 (78.9) | 19 (21.1) | 12 (13.3) | 62 (68.9) | 6 (6.7) | 7 (7.7) | 3 (3.3) |
| 70 (77.7) | 17 (18.9) | 74 (85.5) | 16 (17.7) | |||||||||||||
| 90 | 9 | 90 | 90 | |||||||||||||
*C: Classic and ∗NC: Nonclassic.
Figure 4.
Clinical manifestations of cutaneous leishmaniasis cases in Khuzestan. (a) Dry lesion: isolated from Ahvaz, (b) Dry lesion: isolated from Sosangerd, (c) Mixed lesion: isolated from kotabdollah, Ahvaz, (d) Nonclassical wet-type: Erythematous papulonodules lesion isolated from Ahvaz, (e) Nonclassical wet-type: hyperkeratotic lesion isolated from Ahvaz, (f) Nonclassical wet-type: herpetiform lesion isolated from Hoveyze, ((g)–(i)) classical wet-type form: volcanic lesion isolated from Sosangerd, Ahvaz and Ramhormoz, respectively. (j) The amastigote of Leishmania tropica with size 2-3 μm. ((k)–(o)) Polymorphic amastigotes of Leishmania major with size 3–5 μm. (k) Cigarette form with size 3-4 μm, ((l)–(o)) pear shapes with size 5 μm.
Details of frequency of Leishmania lesions in suspected patients, lesion duration after adult sandflies' biting, lesion types, and lesions' numbers are shown in Table 1.
3.2. The Pixel Intensity of Leishmania by Targeting Three Genes of Cyt b, Microsatellite, and ITS-rDNA
The pixel intensity of applied genes was examined. The PCR products that were tested in gel electrophoresis rely on their chromatogram length. The sensitivity of PCR bands showed that the Cyt b gene has more pixel intensity (140) than microsatellite (125) and ITS-rDNA (110) genes in a minimum dilution, 1 : 16 (Figure 5(a)).
Figure 5.
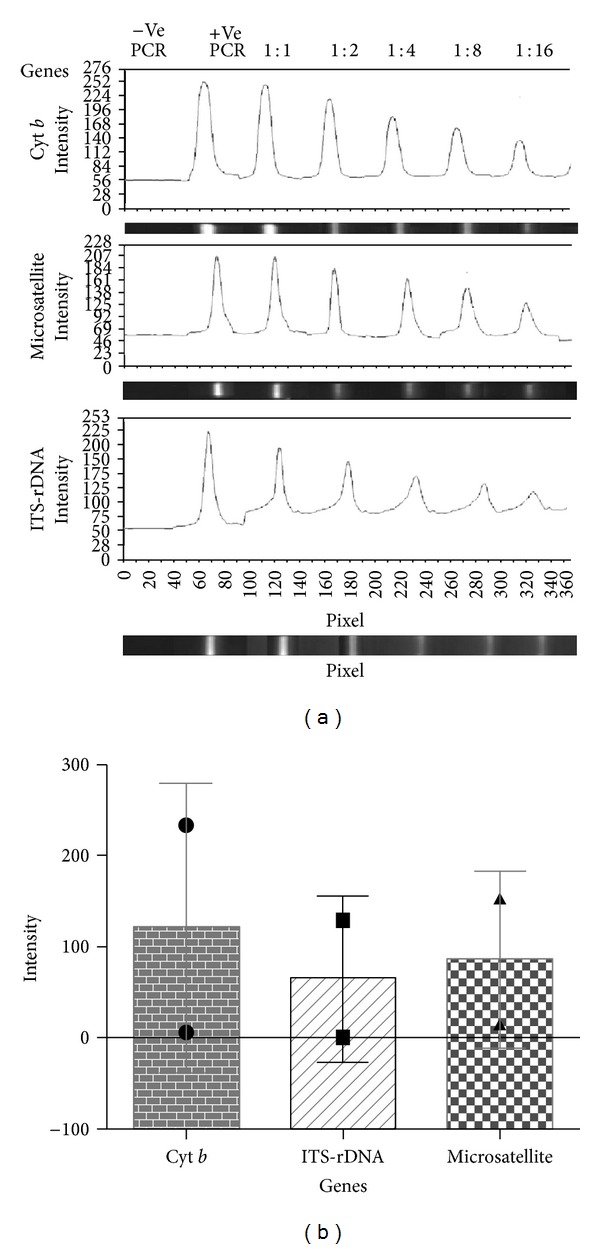
(a) Evaluating of three amplified PCR bands sensitivity based on provided serial dilution of target DNAs in ITS-rDNA, microsatellite, and Cyt b genes. Purified DNA (10 ng) from L. major was used as positive PCR control (+ve PCR). A negative PCR control (−ve PCR) without DNA was included in each reaction. Pixel intensity of PCR bands was calculated using Gel Analyser software (2010a) in different genes. (b) To assess the average of different dilutions of PCR bands based on three applied genes in this study, a graph presents the pixel intensity of Cyt b; 110, ITS-rDNA; 82 and ITS2-microsatellite; 90 (from left to right). Bar graph indicates the mean ± SEM.
The average of different PCR dilutions was statistically analyzed and the mean of PCR bands demonstrated that Cyt b gene has more intensity (110) than microsatellite (90) and ITS-rDNA (82) (Figure 5(b)).
3.3. Identification of Leishmania Species by Targeting ITS-rDNA and Cyt b Genes Using RFLP
In this investigation, L. major and L. tropica were identified and typed molecularly in suspected patients in neglected foci of Khuzestan province. 139 out of 150 suspected patients were found infected with Leishmania parasites using three different genes of ITS-rDNA (90/139: 64.7%), Cyt b (30/139: 21.5%), and microsatellite (19/139: 13.6%) (Table 1).
37 out of 139 (26.6%) PCR products did not have enough DNA for digesting by BsuRI (HaeIII) for ITS-rDNA and Ssp1 for Cyt b (maxicircle) enzymes and/or sequencing of these genes. 102 positive samples were digested and some directly sequenced to identify Leishmania species. Of 139 suspected patients, 102 cases had 90 L. major (64.7%), nine had L. tropica (6.47%), and three were unidentified (2.15%).
The digestion of BsuRI (HaeIII) of ITS-rDNA and Ssp1 of Cyt b enzymes for detection of different Leishmania species was performed by CLC DNA Workbench 5.2 software which are shown in Figure 2.
3.4. Sequencing and Phylogenetic Analyses of Leishmania Species by Targeting ITS-rDNA and Cyt b Genes
Only 25 out of 90 L. major and all nine L. tropica were sequenced from both Cyt b and ITS-rDNA genes. All sequences of Leishmania parasites in this study were aligned, edited, analyzed, and compared with some Leishmania sequences of Cyt b and ITS-rDNA genes with those which had been previously submitted to the GenBank. L. tropica had more molecular variation than L. major (25 L. major sequences versus nine L. tropica). Cyt b had a little more variation in comparison with ITS-rDNA gene for both L. major and L. tropica.
L. major had three haplotypes for Cyt b gene. One common haplotype was reported in 23 L. major sequences which were similar with haplotype KHUZ57 (GenBank Accession no. AB095961). Two other haplotypes (haplotypes KHUZ17 and KHUZ19) were novel and unique (KM393221 and KM393217). For ITS-rDNA gene, all 25 L. (L.) major sequences were similar (GenBank Accession nos. EF413075 and AJ300481) and no variation was found.
Of our nine L. tropica sequences, three haplotypes were identified distinctively including a common and two new haplotypes for Cyt b gene. The common haplotype included six L. tropica sequences which were similar with haplotype KHUZ22 (GenBank Accession no. EF579904). Two new haplotypes were seen in three remained sequences. Haplotype KHUZ02 had two similar sequences (KM393218) and the other one (haplotype KHUZ05) was unique (KM393219). For ITS-rDNA gene of L. tropica, two haplotypes were found. One common haplotype KHUZ11 had eight similar sequences (GenBank Access no. FN677343) and one new haplotype KHUZ07 which was unique (KM393220). Three Leishmania parasites were unidentified because of not enough DNA for direct sequencing or unreadable sequences and/or mixed infections (Table 1).
A coinfection was found from a man (age about 24), resident of Kotabdullah village (Ahvaz region) who had L. major in left hand, L. tropica in face, and a Crithidia in the lesion of right hand. This could be very interesting to notice when different parasites are existed in one patient; therefore, each lesion should be identified to species and examined separately.
Phylogenetic analyses were conducted using our new and common haplotypes of both genes and Leishmania species with those which were submitted in GenBank using MEGA v5.05. Trypanosoma brucei was considered as an out group branch (GenBank Accession nos. M94286 and JN673390) (Figures 6(a) and 6(b)).
Figure 6.
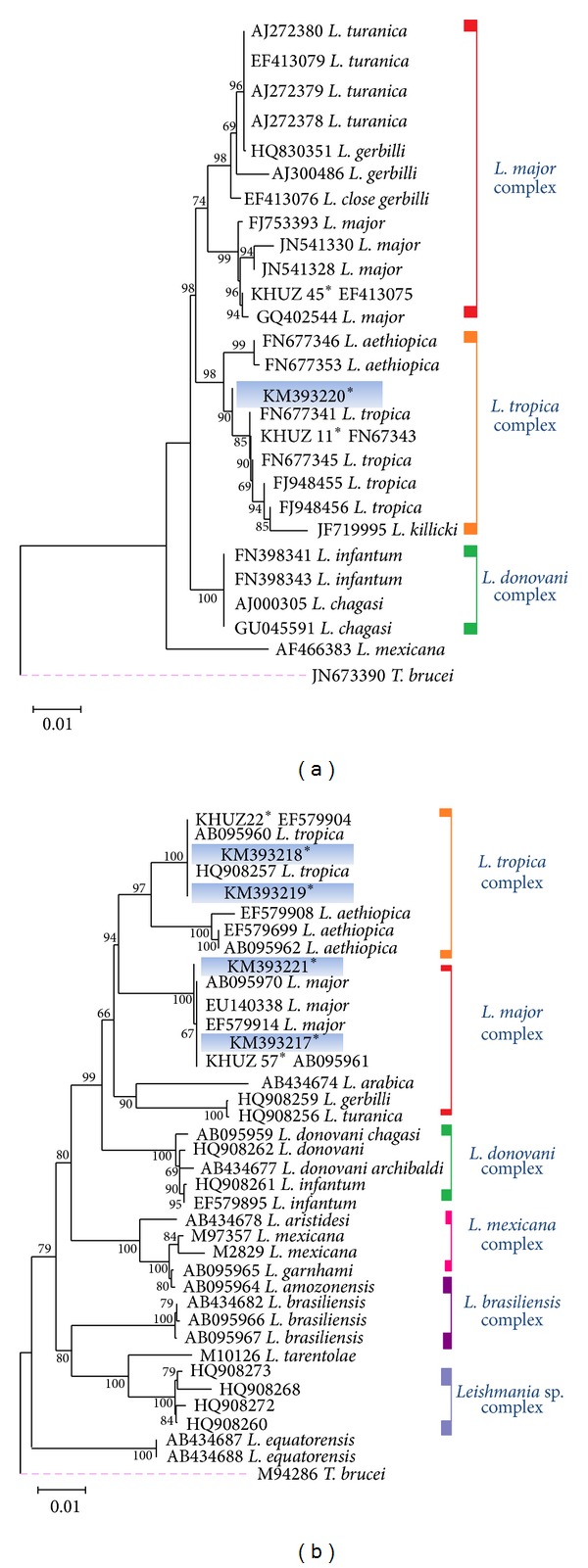
The Phylogeny of Leishmania species according to the maximum-likelihood (ML), tree was conducted based on the multiple sequence alignment (haplotypes) of ITS-rDNA (a) and Cyt b (b) genes by MEGA5.05. Only bootstrap values of higher than 70% are indicated on each branch. Distance represents the number of base substitutions per site. Trypanosoma brucei (M94286; Cyt b and JN673390; ITS-rDNA) is the outgroup branch. ∗ = Analyzed sequences in this study are shown in (a) and (b) for L. major and L. tropica complexes.
4. Discussion
In this investigation, two Leishmania species (L. major and L. tropica) were unequivocally identified in suspected patients of the three different districts including Sosangerd, Dashte Azadegan, and Hoveyze in western bordering with Iraq, Ahvaz as capital city of Khuzestan province in central and also Ramhormoz and Ramshir in eastern Khuzestan province where all these three different regions are not considered widely as ZCL foci (Figure 1) [4]. In north of Khuzestan province, Shush is well known as an endemic focus of ZCL [44]. In our study, prevalence of ZCL in ignored locations of disease was unexpectedly high (17.1 per 10,000) to be compared with previous report, 8 per 10,000 [4]. Also, L. major and L. tropica were found in Sosangerd, Dashte Azadegan, Hoveyze, and Ahvaz, while L. major was only found in Ramhormoz and Ramshir. This maybe because of sandflies' species or reservoir hosts distribution in different regions. Phlebotomus papatasi and P. (Paraphlebotomus) sergenti were collected and identified in these two first locations (Sosangerd, Dashte Azadegan, and Ahvaz). Those are the vectors which circulate in these areas but only P. papatasi was found in the third region (Ramhormoz and Ramshir) whilst the rodent Tatera indica and humans/dogs are reservoir hosts of L. major and L. tropica, respectively in the region [45, 46].
In this study, three different genes were employed to detect Leishmania species. The advantages of RFLP for Cyt b gene were sharp PCR bands with high sensitivity in the shortest possible time (5 min., rapid digestion) compared with ITS-rDNA and microsatellite (Figures 3(a), 3(b), 5(a), and 5(b)). These could depend on high multicopy sequences of Cyt b per cell (comprised of 20–50 maxicircles, 20–35 kb in size) and presence of an unusually long noncoding region (5–20 kb), including five nonedited MURF1, 12S, COI, ND1, and 9S genes [32].
Regular and different irregular shapes of amastigotes of L. major were found in various clinical patterns, this can be stated by various reservoir hosts and vectors compared to L. tropica in the region. Only regular shape of amastigotes was identified in L. tropica, in contrast, more molecular variation was observed in L. tropica sequences. This could be justified by the genetic characters of this parasite, because GC content of L. tropica is lower than L. major [10, 47, 48].
Unidentified Leishmania species could be inducted by mixed pathogens or non-pathogens with L. major, L. tropica, and/or Crithidia [49–51]. Unidentified and mixed infections could be resolved by cloning of genes which was not the purpose of our approved research project [52].
A coinfection of L. major, L. tropica, and Crithidia in different sites on body of the man in our research from Khuzestan province can approve the existence of these pathogens and nonpathogens in suspected patents, but the surprising matter is that how nonpathogen of Crithidia made acute lesion in right hand of patient. More remarkable finding of co-infection were wet lesion for both L. tropica and Crithidia which expecting only for L. major ulcer.
L. major was isolated and clearly identified in three clinical forms (wet, dry, and mixed ulcers), while we expected to observe only in wet lesions. These unexpected observations of three different ulcers should be described by the ability of L. major isolates to have tropism to clinical patterns isolated in southwestern Iran [18, 53].
No correlation was found between the phenotypic character (amastigote morphometrics) and genotypic features in L. major (P > 0.05) using ITS-rDNA (nucleus) and Cyt b (mitochondrial) and microsatellite in the most neglected regions of the middle Khuzestan, Iran. Because different amastigotes' shapes of L. major had the same sequences and no molecular variation was detected. The amastigote shape of L. tropica was observed only in regular shape but they had more molecular variations based on mentioned genes. High GC content of L. major (52.5%) is more stable than low GC content of L. tropica and it should be considered because of triple hydrogen bonds of the GC pair and mainly due to stacking interaction [10, 47, 48, 54].
More variation and heterogeneity of L. tropica in our study also could explain the same as ITS-1 ribosomal DNA sequence of Echinococcus which showed there are two turnover mechanisms, namely, unequal crossing over/transposition and slippage [55].
These findings attenuate explicitly relationship between genotypic and phenotypic characters of CL in L. major and L. tropica, which potentially indicates that genetic variation aspects are not always able to influence the formation of morphometric features or even different clinical appearances [8, 9, 56, 57].
A comprehensive analysis on limits of morphological diversity in any protozoan parasite showed that morphogenetic constraints and extrinsic selection pressures are associated with the full diversity of trypanosomatid morphology including juxtaform and liberform superclasses [19].
Several factors can separately affect the clinical features of L. major lesions. These are including immunosuppressive diseases, migration of nonindigenous populations, host factors, the number of the inoculated parasites, using oral steroids, the site of inoculation, and even wound contamination with inorganic ingredients [8, 9]. Considering challenge on phenotypic and genotypic patterns of L. tropica in suspected patients in Khuzestan was that L. tropica had no polymorphic amastigotes but have more molecular variations with many differentiations in nucleotides of each sequences (haplotype) [10, 17].
Some studies revealed molecular diversity in mini-exon gene of CL agents (L. major and L. tropica) in Khuzestan isolates but the heterogeneity was not coordinated with the size of lesions [22, 23].
Most publications showed that causative agent of cutaneous leishmaniasis is L. major; however Maraghi et al. (2007) identified both L. major (90%) and L. tropica (10%) in this regions [12, 44, 58].
Hajjaran et al. (2013) using only ITS-rDNA gene showed L. tropica with 3.6% polymorphisms and L. major with 7.3% genetic variations in Iran [59], whilst we found more variations in L. tropica to be compared with L. major using Cyt b mitochondrial and ITS-rDNA nucleus genes which are able to draw a real conception of genetic diversity among Leishmania species.
Many studies were carrying out regarding identifications of Leishmania parasites in human, reservoirs hosts, and vectors in Iran and the world which some of them correlated the molecular variations and heterogeneity of Leishmania species with clinical signs [59–62].
5. Conclusions
In this study, L. major and L. tropica were firmly identified in suspected patients with a coinfection of L. major, L. tropica, and Crithidia and some unidentified Leishmania which could be mixed of these three or with the expectation of the existence of L. turanica which recently was reported from human in north of Iran [7]. It is crucial that simultaneous screening of both valuable mitochondrial (Cyt b) and nucleus (ITS-rDNA) genes with morphometric, molecular, and phylogenetic analyses can provide an appropriate approach and reliable methods to identify Leishmania haplotypes, strains, and species.
Acknowledgments
This study was supported by the Pasteur Institute of Iran, Grant no. 605 and was awarded to Dr. Parviz Parvizi. The collections of human samples were made possible by the assistance of the Centre of Health Services Dashte azadegan, Ahvaz and Ramhormoz. The authors thank Mehdi Baghban for helping with the field work and Elnaz AlaeeNovin for helping in Molecular Systematics Laboratory. This research was also supported by PhD studentships to Mr. Adel Spotin, who is based at the Pasteur Institute of Iran, Tehran, and registered at the University of Shahid Beheshti Medical Sciences, Tehran, Iran.
Ethical Approval
The ethical approval was received from the Research Council of Pasteur Institute of Iran, Tehran, Iran.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contribution
Adel Spotin and Soheila Rouhani contributed equally to this work.
References
- 1.Desjeux P. Leishmaniasis: current situation and new perspectives. Comparative Immunology, Microbiology and Infectious Diseases. 2004;27(5):305–318. doi: 10.1016/j.cimid.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 2.WHO Technical Report Series. Geneva, Switzerland: WHO Expert Committee on the Control of Leishmaniases; 2010. Control of the Leishmaniases. [Google Scholar]
- 3.Alvar J, Vélez ID, Bern C, et al. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE. 2012;7(5) doi: 10.1371/journal.pone.0035671.e35671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shirzadi M. Instruction of Cutaneous Leishmaniasis Care (Salak) in Iran. 1st edition. Tehran, Iran: Ministry of health, Treatment and Medical Education, Deputy of Health, Administration Center of Contagious Diseases, Office of Transmissible Disease between Humans and Animal; 2012. [Google Scholar]
- 5.Mohebali M. Visceral leishmaniasis in Iran: review of the epidemiological and clinical features. Iranian Journal of Parasitology. 2013;8(3):348–358. [PMC free article] [PubMed] [Google Scholar]
- 6.Ready PD. Biology of phlebotomine sand flies as vectors of disease agents. Annual Review of Entomology. 2013;58:227–250. doi: 10.1146/annurev-ento-120811-153557. [DOI] [PubMed] [Google Scholar]
- 7.Bordbar A, Parvizi P. High infection frequency, low diversity of Leishmania major and first detection of Leishmania turanica in human in northern Iran. Acta Tropica. 2014;133:69–72. doi: 10.1016/j.actatropica.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Convit J, Ulrich M, Pérez M, et al. Atypical cutaneous leishmaniasis in Central America: possible interaction between infectious and environmental elements. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2005;99(1):13–17. doi: 10.1016/j.trstmh.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Manzur A, Butt UA. Atypical cutaneous leishmaniasis resembling eczema on the foot. Dermatology Online Journal. 2006;12(3) article 18 [PubMed] [Google Scholar]
- 10.Schönian G, Schnur L, El Fari M, et al. Genetic heterogeneity in the species Leishmania tropica revealed by different PCR-based methods. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2001;95(2):217–224. doi: 10.1016/s0035-9203(01)90173-7. [DOI] [PubMed] [Google Scholar]
- 11.Elfari M, Schnur LF, Strelkova MV, et al. Genetic and biological diversity among populations of Leishmania major from Central Asia, the Middle East and Africa. Microbes and Infection. 2005;7(1):93–103. doi: 10.1016/j.micinf.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Mahnaz T, Katrin K, Amer A-J, et al. Leishmania major: genetic heterogeneity of Iranian isolates by single-strand conformation polymorphism and sequence analysis of ribosomal DNA internal transcribed spacer. Acta Tropica. 2006;98(1):52–58. doi: 10.1016/j.actatropica.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Parvizia P, Alaeenovin E, Kazerooni PA, Ready PD. Low diversity of Leishmania parasites in sandflies and the absence of the great gerbil in foci of zoonotic cutaneous leishmaniasis in Fars province, southern Iran. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2013;107(6):356–362. doi: 10.1093/trstmh/trt027. [DOI] [PubMed] [Google Scholar]
- 14.Parvizi P, Alaeenovin E, Mohammadi S, Baghban N. Occurrence of low density of Leishmania infantum in sandflies from a new focus of visceral leishmaniasis in Northwest of Iran. Journal of Vector Borne Diseases. 2013;50(2):127–132. [PubMed] [Google Scholar]
- 15.Rouhani S, Mirzaei A, Spotin A, Parvizi P. Novel identification of Leishmania major in Hemiechinus auritus and molecular detection of this parasite in Meriones libycus from an important foci of zoonotic cutaneous leishmaniasis in Iran. Journal of Infection and Public Health. 2014;7(3):210–217. doi: 10.1016/j.jiph.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Sharbatkhori M, Spotin A, Taherkhani H, Roshanghalb M, Parvizi P. Molecular variation in Leishmania parasites from sandflies species of a zoonotic cutaneous leishmaniasis in northeast of Iran. Journal of Vector Borne Diseases. 2014;51(1):16–21. [PubMed] [Google Scholar]
- 17.Schönian G, Akuffo H, Lewin S, et al. Genetic variability within the species Leishmania aethiopica does not correlate with clinical variations of cutaneous leishmaniasis. Molecular and Biochemical Parasitology. 2000;106(2):239–248. doi: 10.1016/s0166-6851(99)00216-9. [DOI] [PubMed] [Google Scholar]
- 18.Oryan A, Shirian S, Tabandeh M-R, Hatam G-R, Randau G, Daneshbod Y. Genetic diversity of Leishmania major strains isolated from different clinical forms of cutaneous leishmaniasis in southern Iran based on minicircle kDNA. Infection, Genetics and Evolution. 2013;19:226–231. doi: 10.1016/j.meegid.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 19.Wheeler RJ, Gluenz E, Gull K. The limits on trypanosomatid morphological diversity. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0079581.e79581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khademvatan S, Neisi N, Maraghi S, Saki J. Diagnosis and identification of Leishmania spp. from giemsa-stained slides, by real-time PCR and melting curve analysis in south-west of Iran. Annals of Tropical Medicine and Parasitology. 2011;105(8):559–565. doi: 10.1179/2047773211Y.0000000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maraghi S, Mardanshah O, Rafiei A, Samarbafzadeh A, Vazirianzadeh B. Identification of cutaneous leishmaniasis agents in four geographical regions of khuzestan province using nested PCR. Jundishapur Journal of Microbiology. 2013;6(4) [Google Scholar]
- 22.Saki J, Khademvatan S. A molecular study on cutaneous leishmaniasis lesions in Khuzestan province (South West of Iran) Jundishapur Journal of Microbiology. 2011;4(4):283–288. [Google Scholar]
- 23.Saki J, Meamar AR, Oormazdi H, et al. Mini-exon genotyping of Leishmania species in Khuzestan Province, southwest Iran. Iranian Journal of Parasitology. 2010;5(1):25–34. [PMC free article] [PubMed] [Google Scholar]
- 24.Cupolillo E, Brahim LR, Toaldo CB, et al. Genetic polymorphism and molecular epidemiology of Leishmania (Viannia) braziliensis from different hosts and geographic areas in Brazil. Journal of Clinical Microbiology. 2003;41(7):3126–3132. doi: 10.1128/JCM.41.7.3126-3132.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Eys GJJM, Schoone GJ, Kroon NCM, Ebeling SB. Sequence analysis of small subunit ribosomal RNa genes and its use for detection and identification of Leishmania parasites . Molecular and Biochemical Parasitology. 1992;51(1):133–142. doi: 10.1016/0166-6851(92)90208-2. [DOI] [PubMed] [Google Scholar]
- 26.Victoir K, Bañuls AL, Arevalo J, et al. The gp63 gene locus, a target for genetic characterization of Leishmania belonging to subgenus Viannia. Parasitology. 1998;117(1):1–13. [PubMed] [Google Scholar]
- 27.Bulle B, Millon L, Bart J-M, et al. Practical approach for typing strains of Leishmania infantum by microsatellite analysis. Journal of Clinical Microbiology. 2002;40(9):3391–3397. doi: 10.1128/JCM.40.9.3391-3397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montalvo AM, Fraga J, Monzote L, et al. Heat-shock protein 70 PCR-RFLP: a universal simple tool for Leishmania species discrimination in the New and Old World. Parasitology. 2010;137(8):1159–1168. doi: 10.1017/S0031182010000089. [DOI] [PubMed] [Google Scholar]
- 29.Bordbar A, Parvizi P. High density of Leishmania major and rarity of other mammals' Leishmania in zoonotic cutaneous leishmaniasis foci, Iran. Tropical Medicine and International Health. 2014;19(3):355–363. doi: 10.1111/tmi.12258. [DOI] [PubMed] [Google Scholar]
- 30.Parvizi P, Ready PD. Nested PCRs and sequencing of nuclear ITS-rDNA fragments detect three Leishmania species of gerbils in sandflies from Iranian foci of zoonotic cutaneous leishmaniasis. Tropical Medicine and International Health. 2008;13(9):1159–1171. doi: 10.1111/j.1365-3156.2008.02121.x. [DOI] [PubMed] [Google Scholar]
- 31.Yurchenko V, Kolesnikov AA, Lukeš J. Phylogenetic analysis of Trypanosomatina (Protozoa: Kinetoplastida) based on minicircle conserved regions. Folia Parasitologica. 2000;47(1):1–5. doi: 10.14411/fp.2000.001. [DOI] [PubMed] [Google Scholar]
- 32.Yatawara L, Le TH, Wickramasinghe S, Agatsuma T. Maxicircle (mitochondrial) genome sequence (partial) of Leishmania major. Gene content, arrangement and composition compared with Leishmania tarentolae . Gene. 2008;424(1-2):80–86. doi: 10.1016/j.gene.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 33.Howell N, Gilbert K. Mutational analysis of the mouse mitochondrial cytochrome b gene. Journal of Molecular Biology. 1988;203(3):607–617. doi: 10.1016/0022-2836(88)90195-7. [DOI] [PubMed] [Google Scholar]
- 34.Degli Esposti M, de Vries S, Crimi M, Ghelli A, Patarnello T, Meyer A. Mitochondrial cytochrome b: evolution and structure of the protein. Biochimica et Biophysica Acta. 1993;1143(3):243–271. doi: 10.1016/0005-2728(93)90197-n. [DOI] [PubMed] [Google Scholar]
- 35.Luyo-Acero GE, Uezato H, Oshiro M, et al. Sequence variation of the Cytochrome b gene of various human infecting members of the genus Leishmania and their phylogeny. Parasitology. 2004;128(5):483–491. doi: 10.1017/s0031182004004792. [DOI] [PubMed] [Google Scholar]
- 36.Hillis DM, Moritz C, Porter CA, Baker RJ. Evidence for biased gene conversion in concerted evolution of ribosomal DNA. Science. 1991;251(4991):308–310. doi: 10.1126/science.1987647. [DOI] [PubMed] [Google Scholar]
- 37.Jamjoom MB, Ashford RW, Bates PA, Kemp SJ, Noyes HA. Towards a standard battery of microsatellite markers for the analysis of the Leishmania donovani complex. Annals of Tropical Medicine and Parasitology. 2002;96(3):265–270. doi: 10.1179/000349802125000790. [DOI] [PubMed] [Google Scholar]
- 38.Mirzaei A, Rouhani S, Taherkhani H, et al. Isolation and detection of Leishmania species among naturally infected Rhombomis opimus, a reservoir host of zoonotic cutaneous leishmaniasis in Turkemen Sahara, North East of Iran. Experimental Parasitology. 2011;129(4):375–380. doi: 10.1016/j.exppara.2011.08.020. [DOI] [PubMed] [Google Scholar]
- 39.Mahnaz T, Al-Jawabreh A, Kuhls K, Schönian G. Multilocus microsatellite typing shows three different genetic clusters of Leishmania major in Iran. Microbes and Infection. 2011;13(11):937–942. doi: 10.1016/j.micinf.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 40.Sharifi F, Sharifi I, Zarean M, et al. Spatial distribution and molecular identification of Leishmania species from endemic foci of South-Eastern Iran. Iranian Journal of Parasitology. 2012;7(1):45–52. [PMC free article] [PubMed] [Google Scholar]
- 41.WHO. Basic Laboratory Methods in Medical Parasitology. 1st edition. Geneva, Switzerland: World Health Organization (WHO); 1991. [Google Scholar]
- 42.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maraghi S, Samarbaf Zadeh A, Sarlak AA, Ghasemian MB, Vazirianzadeh B. Identification of cutaneous leishmaniases agents by nested polymerase chain reaction (Nested-PCR) in shush city, khuzestan province, Iran. Iranian Journal of Parasitology. 2007;2(3):13–15. [Google Scholar]
- 45.Javadian E, Mesghali A. Studies on cutaneous leishmaniasis in Khuzestan, Iran. Part I. The leptomonad infection of sandflies. Bulletin de la Societe de Pathologie Exotique et de ses Filiales. 1974;67(5):513–516. [PubMed] [Google Scholar]
- 46.Killick-Kendrick R. Phlebotomine vectors of the leishmaniases: a review. Medical and Veterinary Entomology. 1990;4(1):1–24. doi: 10.1111/j.1365-2915.1990.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 47.Ivens AC, Peacock CS, Worthey EA, et al. The genome of the kinetoplastid parasite, Leishmania major . Science. 2005;309(5733):436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bañuls A-L, Hide M, Prugnolle F. Leishmania and the leishmaniases : a parasite genetic update and advances in taxonomy, epidemiology and pathogenicity in humans. Advances in Parasitology. 2007;64:1–109. doi: 10.1016/S0065-308X(06)64001-3. [DOI] [PubMed] [Google Scholar]
- 49.Strelkova MV, Eliseev LN, Ponirovsky EN, et al. Mixed leishmanial infections in Rhombomys opimus: a key to the persistence of Leishmania major from one transmission season to the next. Annals of Tropical Medicine and Parasitology. 2001;95(8):811–819. doi: 10.1080/00034980120111154. [DOI] [PubMed] [Google Scholar]
- 50.Shirian S, Oryan A, Hatam GR, Daneshbod Y. Mixed mucosal leishmaniasis infection caused by Leishmania tropica and leishmania major . Journal of Clinical Microbiology. 2012;50(11):3805–3808. doi: 10.1128/JCM.01469-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ravel C, Cortes S, Pratlong F, Morio F, Dedet J-P, Campino L. First report of genetic hybrids between two very divergent Leishmania species: Leishmania infantum and Leishmania major . International Journal for Parasitology. 2006;36(13):1383–1388. doi: 10.1016/j.ijpara.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 52.Babiker AM, Ravagnan S, Fusaro A, et al. Concomitant infection with leishmania donovani and L. major in single ulcers of cutaneous leishmaniasis patients from sudan. Journal of Tropical Medicine. 2014;2014:8 pages. doi: 10.1155/2014/170859.170859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McMahon-Pratt D, Alexander J. Does the Leishmania major paradigm of pathogenesis and protection hold for New World cutaneous leishmaniases or the visceral disease? Immunological Reviews. 2004;201:206–224. doi: 10.1111/j.0105-2896.2004.00190.x. [DOI] [PubMed] [Google Scholar]
- 54.Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Research. 2006;34(3):564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Herwerden L, Gasser RB, Blair D. ITS-1 ribosomal DNA sequence variants are maintained in different species and strains of Echinococcus . International Journal for Parasitology. 2000;30(2):157–169. doi: 10.1016/s0020-7519(00)00002-3. [DOI] [PubMed] [Google Scholar]
- 56.Motta ACF, Arruda D, Souza CS, Foss NT. Disseminated mucocutaneous leishmaniasis resulting from chronic use of corticosteroid. International Journal of Dermatology. 2003;42(9):703–706. doi: 10.1046/j.1365-4362.2003.01730.x. [DOI] [PubMed] [Google Scholar]
- 57.Puig L, Pradinaud R. Leishmania and HIV co-infection: dermatological manifestations. Annals of Tropical Medicine and Parasitology. 2003;97:S107–S114. doi: 10.1179/000349803225002589. [DOI] [PubMed] [Google Scholar]
- 58.Motazedian H, Noamanpoor B, Ardehali S. Characterization of Leishmania parasites isolated from provinces of the Islamic Republic of Iran. Eastern Mediterranean Health Journal. 2002;8(2-3):338–344. [PubMed] [Google Scholar]
- 59.Hajjaran H, Mohebali M, Mamishi S, et al. Molecular identification and polymorphism determination of cutaneous and visceral leishmaniasis agents isolated from human and animal Hosts in Iran. BioMed Research International. 2013;2013:7 pages. doi: 10.1155/2013/789326.789326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baghaei M. Intraspecific variation in Leishmania major isolated from different forms of zoonotic cutaneous leishmaniasis. Iranian Journal of Medical Sciences. 2005;30(2):51–54. [Google Scholar]
- 61.Dabirzadeh M, Sadeghi HM, Baghaie M, Hejazi H. Genetic polymorphism of Leishmania major in two hyper endemic regions of Iran revealed by PPIP-PCR and ITS- RFLP. Archives of Iranian Medicine. 2012;15(3):151–156. [PubMed] [Google Scholar]
- 62.Ajaoud M, Es-Sette N, Hamdi S, El-Idrissi AL, Riyad M, Lemrani M. Detection and molecular typing of Leishmania tropica from Phlebotomus sergenti and lesions of cutaneous leishmaniasis in an emerging focus of Morocco. Parasites and Vectors. 2013;6(1, article 217) doi: 10.1186/1756-3305-6-217. [DOI] [PMC free article] [PubMed] [Google Scholar]