Abstract
Background
Death-associated protein kinase1 (DAPK1) is an important tumor suppressor gene. DNA methylation can inactivate genes, which has often been observed in the carcinogenesis of cervical cancer. During the past several decades, many studies have explored the association between DAPK1 promoter methylation and cervical cancer. However, many studies were limited by the small samples size and the findings were inconsistent among them. Thus, we conducted a meta-analysis to assess the association between DAPK1 promoter methylation and cervical cancer.
Methods
We systematically searched eligible studies in the PubMed, Web of Science, EMBASE and CNKI databases. Using meta-regression, subgroup analysis and sensitivity analysis, we explored the potential sources of heterogeneity. The odds ratio (OR) and 95% confidence interval (95% CI) were calculated by Meta-Analysis in R.
Results
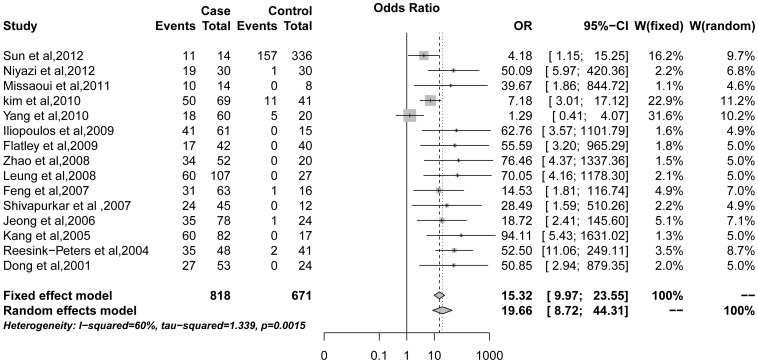
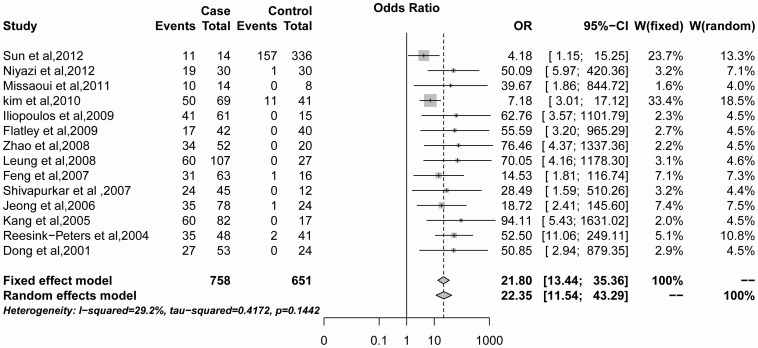
A total of 15 studies from 2001 to 2012, comprising 818 tumor tissues samples and 671 normal tissues samples, were analyzed in this meta-analysis. The frequencies of DAPK1 promoter methylation ranged from 30.0% to 78.6% (median, 59.3%) in cervical cancer tissue and 0.0% to 46.7% (median, 7.8%) in normal cervical tissue. The pooled OR was 19.66 (95%CI = 8.72–44.31) with the random effects model, and heterogeneity was found through the sensitivity analysis. The I2 = 60% (P = 0.002) decreased to I2 = 29.2% (P = 0.144) when one heterogeneous study was excluded, and the pooled OR increased to 21.80 (95%CI = 13.44–35.36) with the fixed effects model.
Conclusion
The results suggested a strong association between DAPK1 promoter methylation and cervical cancer. This study also indicated that DAPK1 promoter methylation may be a biomarker during cervical carcinogenesis that might serve as an early indication of cervical cancer.
Introduction
Cervical cancer is the third most common cancer, after breast and colorectal cancer, among women worldwide, with 529,500 estimated new cases and 275,000 deaths in 2008 according to Ferlay et al. [1]. The development of invasive cervical cancer is a gradual process that occurs over a long period, from cervical intraepithelial neoplastic (CIN) lesions to cervical cancer. Thus, it is critically important to detect precancerous lesions to prevent the development of cervical cancer. Although infection with the human papillomavirus (HPV) is an accepted major risk factor for cervical cancer [2], only a small proportion of HPV infected patients develop invasive cervical cancer [3]. Other risk factors may also contribute to the genesis of this cancer type.
Hypermethylation of the promoter regions of tumor suppressor genes can cause gene inactivation, which is important in the pathogenesis of cancers, and usually occurs in the early stages of cancer development in various types of cancer, including cervical cancer [4], [5]. DNA rmethylation is an early event in carcinogenesis, and is often related to a transcriptional block and the loss of a relevant protein [6]. Because DAPK1 is an important tumor suppressor gene that has been studied extensively, we performed a meta-analysis to assess the association between DAPK1 promoter methylation and cervical cancer.
Materials and Methods
Study search and selection criteria
We systematically reviewed the studies of DAPK1 promoter methylation in cervical cancer, and attempted to find the eligible studies within PubMed, EMBASE, Web of Science and CNKI, using various combinations of Medical Subject Headings (MeSH) and non-MeSH terms. The keywords were “cervical cancer”, “DAPK1”, and “methylation”, while the search strategy was performed in PubMed with “uterine cervical neoplasms” (MeSH), “DAPK1”and“methylation”. The study was conducted till November 1, 2013 without any language limitation.
The studies for inclusion in this meta-analysis had to meet the following standards: (i) the studies assessed the association of DAPK1 methylation and cervical cancer, (ii) the studies provided detailed information about the frequency of DAPK1 methylation for both the cancer group and the normal control group, (iii) methods for the detection of DAPK1 methylation were limited to the methylation-specific polymerase chain reaction (MSP) and real-time quantitative polymerase chain reaction (QMSP).
Studies were excluded based on the following criteria: (i) the studies did not have a normal group (control group), (ii) the raw data could not be isolated from the studies in which the cancer group (case group) also contained individuals with various types of precancerous lesions such as Atypical Squamous Cells of Undetermined Significance (ASCUS), Low-grade Squamous Intraepithelial Lesions (LSIL), and High-grade Squamous Intraepithelial Lesions(HSIL), (iii) a case-control study did not feature the frequency of DAPK1 methylation.
Data Extraction
Two authors independently conducted the extraction of data from the selected studies. The extracted information contained the following: first author's name, publication year, the patients' ethnicities, the methods used in the measurement of DAPK1 methylation, the tissue source of the control group, the mean age of the case group, and the number of participants in the case and control groups. All the information was verified by three reviewers.
To assess the quality of the studies, The Newcastle–Ottawa scale (NOS) (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp) was implemented for quality assessment of observational studies. The NOS is a quality assessment tool which is often used for nonrandomized studies, specifically case-control and cohort studies, included in systematic reviews. It has also been widely used in systematic reviews of nonrandomized studies by The Cochrane Collaboration.
There is a maximum of nine ‘stars’ for each item: four stars to the selection of the study groups, two stars to the comparability of the groups, and three stars to the ascertainment of the outcome of interest. The evaluation was performed independently by two reviewers. Studies with quality scores greater than or equal to 6 were included.
Statistical analysis
The ORs and 95% CIs were calculated to assess the association between DAPK1 promoter methylation and cervical cancer risk. The x 2-based Cochran Q statistic test and I 2 statistics were used to test the heterogeneity among the included studies [7]. Significant heterogeneity was confirmed if P<0.05; I 2>50% was also considered to demonstrate significant heterogeneity [8]. Then, a random effects model (the DerSimonian-Laird estimator) was used to calculate the pooled ORs; otherwise, a fixed-effects model (the Mantel-Haenszel method) was applied [9]. A meta-regression (restricted maximum-likelihood estimator method) was employed to explore the source of the heterogeneity. Furthermore, a subgroup analysis was performed to evaluate the source of the heterogeneity, and t2 was used to determine how much heterogeneity could be explained by subgroup differences. A sensitivity analysis was used to find relatively poor-quality studies by the omission of a single study at a time and to see whether a particular omission could affect the overall OR value. The funnel plots [10] and Egger's test were used to evaluate publication bias. The fail-safe number was also an indicator to assess publication bias. An asymmetric plot suggested a possible publication bias and the P value of Egger's test less than 0.05 was considered to be representative of a statistically significant publication bias [10].
All statistical analyses were calculated with the Meta package (version 2.5–1) in R (version 3.0.1; http://www.r-project.org/).
Results
Search Results and Study Characteristics
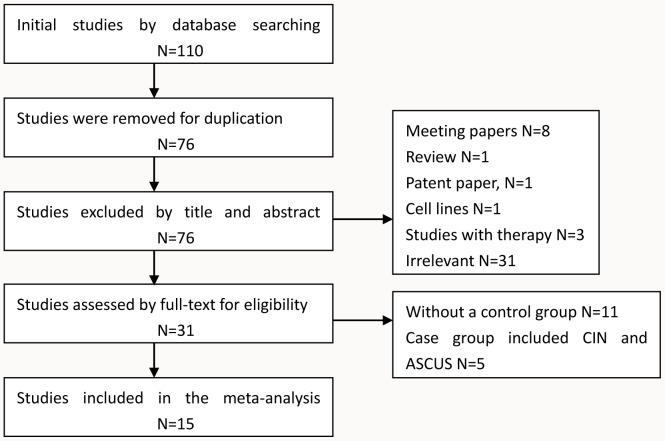
A total of 15 studies that incorporated 1489 patients were included in this meta-analysis (Fig 1). In all, 110 studies were initially found after a search of the above databases, but 34 studies were excluded because of duplication. By screening the titles and abstracts of the remaining76 studies, a further 45 studies were excluded (8 meeting papers, 1 review, 1 patent paper, 1 cell lines, 3 studies with therapy, and 31 irrelevant articles). Eleven studies without a control group and 5 studies that included precancerous tissues such as ASCUS, LSIL, and HSIL in the case group, which meant that the raw data of the cancer patients could not be isolated, were excluded during the process of full-text review. Finally, 15 studies were included in this meta-analysis [3], [11]–[24]. The number of cases ranged from 22 to 350 among the studies with participants from, Asia (8 studies), North Africa (1 study), Europe and North America (7 studies). For the 15 studies, 5 studies used real-time quantitative polymerase chain reaction (QMSP) and the other 10 studies used methylation-specific polymerase chain reaction (MSP) to detect DAPK1 methylation in the case group and in the control group (Table 1).
Figure 1. Flow chart of study selection for the meta-analysis.
Table 1. Information of the studies included in the meta-analysis.
| Tumor | Control | |||||||
| Author | Year | Country | Ethnicity | Method | Age (y) | M+/N | M+/N | Source of Controls |
| Sun et al. [21] | 2012 | China | Asian | MSP | 39.3(18–65) | 11/14 | 157/336 | NT |
| Niyazi et al. [19] | 2012 | China | Asian | MSP | 41.4(27–62) | 19/30 | 1/30 | BCT |
| Missaoui et al. [18] | 2011 | Tunisia | African | MSP | NA | 10/14 | 0/8 | BCT |
| Kim et al. [16] | 2010 | Korea | Asian | MSP | NA | 50/69 | 11/41 | BCT |
| Yang et al. [22] | 2010 | NE | Caucasian | QMSP | 47(38–57) | 18/60 | 5/20 | BCT |
| Iliopoulos et al. [13] | 2009 | Greece | Caucasian | QMSP | 41(18–62) | 41/61 | 0/15 | NT |
| Flatley et al. [3] | 2009 | UK | Caucasian | MSP | 34.3(20–84) | 17/42 | 0/40 | NT |
| Zhao et al. [23] | 2008 | China | Asian | MSP | 42(25–71) | 34/52 | 0/20 | BCT |
| Leung et al. [17] | 2008 | China | Asian | MSP | 52.5(23–85) | 60/107 | 0/27 | AT |
| Feng et al. [12] | 2007 | USA | Caucasian | QMSP | 46.8 | 31/63 | 1/16 | NT |
| Shivapurkar et al. [20] | 2007 | USA | Caucasian | QMSP | NA | 24/45 | 0/12 | BCT |
| Jeong et al. [14] | 2006 | Korea | Asian | MSP | 48.9(24–79) | 35/78 | 1/24 | BCT |
| Kang et al. [15] | 2005 | Korea | Asian | MSP | NA | 60/82 | 0/17 | BCT |
| Reesink-Peters et al. [24] | 2004 | NE | Caucasian | QMSP | NA | 35/48 | 2/41 | NT |
| Dong et al. [11] | 2001 | Korea | Asian | MSP | NA | 27/53 | 0/24 | BCT |
Note: NE: Netherlands; UK: United Kingdom; MSP: methylation-specific polymerase chain reaction; QMSP: real-time quantitative methylation-specific polymerase chain reaction; NA: not applicable; M+: the number of methylations; N: number of total; NT: normal cervical tissues from healthy people; BCT: normal cervical tissues from patients who had benign gynecological diseases such as uterine myoma, adenomyoma, and uterine prolapse; AT: normal cervical tissues adjacent to the tumor.
Quality assessment
The result of NOS demonstrated that the lowest score was 6 and highest score was 9 with a median score of 7.2. Most studies used healthy volunteers from the hospital as controls except those of Feng et al. [12] and Sun et al. [21]. The study by Leung et al. [17] was the only one where the control tissues were derived from adjacent normal tissues (Table 2).
Table 2. Quality assessment of included studies in the meta-analysis.
| Newcastle-Ottawa Scale | ||||||||||
| Author | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | Total |
| Sun et al. [21] | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 9 |
| Niyazi et al. [19] | Yes | Yes | NO | Yes | Yes | NO | Yes | Yes | Yes | 7 |
| Missaoui et al. [18] | Yes | Yes | NO | Yes | Yes | NO | Yes | Yes | Yes | 7 |
| Yang et al. [22] | Yes | Yes | NO | Yes | Yes | NO | Yes | Yes | Yes | 7 |
| kim et al. [16] | Yes | Yes | NO | Yes | Yes | Yes | Yes | Yes | Yes | 8 |
| Iliopoulos et al. [13] | Yes | Yes | NO | Yes | Yes | NO | Yes | Yes | Yes | 7 |
| Flatley et al. [3] | Yes | Yes | NO | Yes | Yes | NO | Yes | Yes | Yes | 7 |
| Zhao et al. [23] | Yes | Yes | NO | Yes | Yes | NO | Yes | Yes | Yes | 7 |
| Leung et al. [17] | Yes | Yes | NO | NO | Yes | NO | Yes | Yes | Yes | 6 |
| Feng et al. [12] | Yes | Yes | Yes | Yes | Yes | NO | Yes | Yes | Yes | 8 |
| Shivapurkar et al. [20] | Yes | Yes | NO | Yes | Yes | NO | Yes | Yes | Yes | 7 |
| Jeong et al. [14] | Yes | Yes | NO | Yes | Yes | NO | Yes | Yes | Yes | 7 |
| Kang et al. [15] | Yes | Yes | NO | Yes | Yes | NO | Yes | Yes | Yes | 7 |
| Reesink-Peters et al. [24] | Yes | Yes | NO | Yes | Yes | NO | Yes | Yes | Yes | 7 |
| Dong et al. [11] | Yes | Yes | NO | Yes | Yes | NO | Yes | Yes | Yes | 7 |
1, indicates case definition and appropriate diagnosis; 2, consecutive patients or cases have a good representation; 3, community controls; 4, controls with no history of study disease; 5, according to the most important factor (patients in the control group were not diagnosed with cervical cancer or any other cancers) to select and analyze the controls; 6, according to the second most important factor (the age of the control group should not have significant heterogeneity compared with the case group) to select and analyze the controls; 7, ascertainment of exposure by blinded interview or record; 8, same method of ascertainment used for cases and controls; and 9, non-response rate the same for cases and controls.
Meta-regression and subgroup analyses
The x2-based Cochran Q statistic test and I 2 statistics found significant heterogeneity among the 15 studies (I2 = 60.0%, P = 0.002). A strong association was observed between DAPK1 promoter methylation and cervical cancer with a pooled OR of = 19.66 (95%CI = 8.72–44.31) based on the random effects model (Fig. 2). For this result, we tried to find the possible source of the heterogeneity. Based on previous studies and our present knowledge, we first used a multiple regression model with five variables based on publication year, ethnicity, method, source of controls, and case sample size. Case groups whose sample size was less than 60, were classified as group A, while the other groups were classified as group B. Through the regression model, we did not find a significant heterogeneity for the five variables listed above (Table 3). We then conducted a subgroup analysis to further assess the source of the heterogeneity.
Figure 2. Pooled OR value for 15 selected studies.
Table 3. Meta-regression analysis on 15 selected studies (Table 1).
| Sources of heterogeneity | Coefficient | 95%CI | P | |
| Lower | Upper | |||
| Publication year | 1.2831 | −0.3213 | 2.8875 | 0.1170 |
| Ethnicity | 1.2830 | −2.5690 | 5.1351 | 0.5139 |
| Method | −1.9797 | −5.9054 | 1.9461 | 0.3230 |
| Source of controls | 0.3772 | −1.3829 | 2.1373 | 0.6745 |
| Case sample size | −0.7374 | −2.1433 | 0.7964 | 0.3460 |
We performed a subgroup analysis according to ethnicity, method, and the case sample size. The OR was 18.22 in Caucasians (95%CI = 3.35–99.03; random effects model) and 17.88 (95%CI = 10.29–31.07; fixed effects model) in Non-Caucasians, the I2 value were obtained separately and were determined to be 75.9% and 42.6% compared with the whole study group (I2 = 60%). With this method, the ORs of the studies that used MSP was 19.10 (95%CI = 11.11–32.84; fixed effects model) and 15.30 (95%CI = 2.34–99.66; random effects model). Similarly, the OR in group A was 25.80 (95%CI = 12.56–53.02; fixed effects model) while the OR in group B was 13.55 (95%CI = 3.93–46.73; random effects model) (Table 4).
Table 4. Subgroup meta-analysis of the association between DAPK1 promoter methylation and cervical cancer.
| Group | Tumor | Control | M-H pooled OR | D+L pooled OR | Heterogeneity | ||||
| M+ | N | M+ | N | OR(95%CI) | OR(96%CI) | I2(%) | P | τ2 | |
| Total | 472 | 818 | 178 | 671 | 15.32(9.97–23.55) | 19.66(8.72–44.31) | 60.0 | 0.002 | 1.34 |
| Ethnicity | |||||||||
| Non-Caucasians | 306 | 499 | 170 | 527 | 17.88(10.29–31.07) | 19.82(8.12–48.35) | 42.6 | 0.083 | 0.70 |
| Caucasians | 166 | 319 | 8 | 144 | 12.27(6.25–24.10) | 18.22(3.35–99.03) | 75.9 | <0.001 | 3.18 |
| Method | |||||||||
| MSP | 323 | 541 | 170 | 567 | 19.10(11.11–32.84) | 21.07(8.98–49.43) | 40.3 | 0.089 | 0.67 |
| QMSP | 149 | 277 | 8 | 104 | 10.46(5.21–21.01) | 15.30(2.34–99.66) | 78.8 | <0.001 | 3.41 |
| Sample size | |||||||||
| A | 177 | 298 | 160 | 513 | 25.80(12.56–53.02) | 27.25(10.67–69.58) | 29.3 | 0.195 | 0.51 |
| B | 295 | 520 | 18 | 158 | 10.70(6.22–18.42) | 13.55(3.93–46.73) | 69.3 | 0.003 | 1.71 |
Notes: Non-Caucasians included Asians and Africans; A: The case sample size was less than 60; B: The case sample size was larger or equal to 60; M-H: the fixed effects mode; D+L: the random effects model.
Sensitivity analysis and subgroup analyses
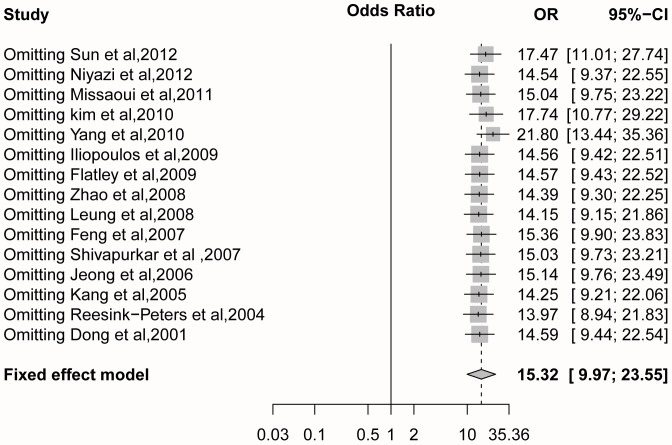
The result of the sensitivity analysis showed that the OR value ranged from 13.97 (95%CI = 8.94–21.83) to 21.80 (95%CI = 13.44–35.36) with a pooled OR of = 15.32 (95%CI = 9.97–23.66) with the fixed effects model (Fig. 3). After the omission of the heterogeneous study (Yang et al., 2010), the pooled OR changed dramatically compared to when other studies were removed. Additionally, the initial heterogeneity (I2 = 60.0%, P = 0.002) decreased to I2 = 29.2% (P = 0.144) with a pooled OR of = 21.80 (95%CI = 13.44–35.36; fixed effects model) when the heterogeneous study was removed (Yang et al., 2010) (Fig. 4). When, we made a further analysis based on ethnicity, method, and the case sample size, the results showed that the heterogeneity in Caucasians, QMSP method and larger sample size disappeared when the data from the heterogeneous study was removed (Yang et al., 2010) (Table 5).
Figure 3. Sensitivity analysis of 15 studies with the fixed effects model.
Figure 4. Pooled OR value of 14 studies omitting one heterogeneous study (Yang et al., 2010).
Table 5. Subgroup meta-analysis of the association between DAPK1 promoter methylation and cervical cancer omitting one heterogeneous study (Yang et al.).
| Group | Tumor | Control | M-H pooled OR | D+L pooled OR | Heterogeneity | ||||
| M+ | N | M+ | N | OR(95%CI) | OR(96%CI) | I2(%) | P | τ2 | |
| Total | 454 | 758 | 173 | 651 | 21.80(13.44–35.36) | 22.35(11.54–43.29) | 29.2 | 0.144 | 0.42 |
| Ethnicity | |||||||||
| Non-Caucasians | 306 | 499 | 170 | 527 | 17.88(10.29–31.07) | 19.82(8.12–48.35) | 42.6 | 0.083 | 0.70 |
| Caucasians | 148 | 259 | 3 | 124 | 37.07(13.37–102.76) | 37.44(13.82–101.40) | 0.0 | 0.877 | 0.00 |
| Method | |||||||||
| MSP | 323 | 541 | 170 | 567 | 19.10(11.11–32.84) | 21.07(8.98–49.43) | 40.3 | 0.089 | 0.67 |
| QMSP | 131 | 217 | 3 | 104 | 34.29(11.53–101.97) | 35.44(12.24–102.63) | 0.0 | 0.771 | 0.00 |
| Sample size | |||||||||
| A | 177 | 298 | 160 | 513 | 25.80(12.56–53.02) | 27.25(10.67–69.58) | 29.3 | 0.195 | 0.51 |
| B | 277 | 460 | 13 | 138 | 18.57(9.63–35.81) | 18.96(6.98–51.53) | 32.7 | 0.191 | 0.49 |
Notes: Non-Caucasians included Asians and Africans; A: The case sample size was less than 60; B: The case sample size was larger or equal to 60; M-H: the fixed effects mode; D+L: the random effects model.
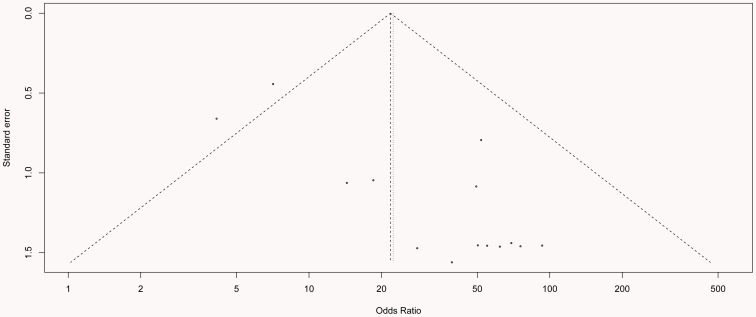
Publication bias
Funnel plots and Egger's test were performed to assess the publication bias of the literature. The shape of the funnel plot in Figure 5 shows a possible asymmetry, but Egger's test resulted in P = 0.551, which indicates that publication bias was very low; no significant bias was found among the included studies. The fail-safe number (Z = 61.12, Nfs0.05 = 1374.98, Nfs0.01 = 674.13) also indicated that the degree of publication bias was very small.
Figure 5. Egger's funnel plot for assessment of publication bias for the remaining 14 studies in the meta-analysis (each study is represented by a point).
Discussion
Death-associated protein kinase1 (DAPK1) could mediate cell death via IFN-gamma and could lead to tumor pathogenesis and metastasis when inactivated [25]. Recently, many studies have shown that DNA methylation alterations are involved in cancer initiation and progression, and could be used to predict the diagnosis and prognosis of human diseases and malignancies [26], [27]. The loss of DAPK1 expression, mainly by hypermethylation of its promoter region, enhances the metastatic potential of cancer cells and has been proven to occur in a variety of cancers, including cancer of the uterine cervix [14], [24]. Although HPV infection is one of the most important risk factors, the majority of patients with HPV infection do not develop cervical cancer. HPV infection alone is insufficient for malignant transformation of cervical cells, which suggest potential roles of other genetic and epigenetic events in cervical carcinogenesis [11].
the result of the pooled OR was 19.66 (95%CI = 8.72–44.31) with the random effects model (Fig 1), which showed that DAPK1 promoter methylation is associated with cervical cancer and therefore, that it might play an important role in the pathogenesis of cervical cancer. This result was consistent with the findings of previous studies [11], [20]. However, significant heterogeneity was observed in those 15 studies, and the reason for heterogeneity could not be explained at the beginning. To explore the possible source of the heterogeneity, we implemented a meta-regression and subgroup analysis. The results showed some heterogeneity in Caucasians, the QMSP method and in a larger sample size through the subgroup analysis (Table 4); then, we conducted a sensitivity analysis to find the source of the heterogeneity. The study by Yang et al. (2010) seems to be the heterogeneous study that affected the meta-analysis, as I2 = 60% (P = 0.002) was reduced to I2 = 29.2% (P = 0.144) when this study was omitted (Fig 4). In addition, further statistical analysis confirmed the heterogeneity of the study of Yang et al. (2010), and no significant heterogeneity of the remaining 14 studies (Table 5).
When the heterogeneous study was omitted, the pooled OR value was increased from 15.32 to 21.80 (the fixed effects model), which suggested a stronger association between DAPK1 promoter methylation and cervical cancer. The heterogeneity presented in QMSP was decreased from I2 = 78.8% to I2 = 0.0%, which indicated that the method of QMSP is better than that of MSP. This conclusion was consistent with the study by Eads et al. [28]. The heterogeneity in Caucasians also decreased from I2 = 75.9% to I2 = 0.0%, which might have been caused by two major reasons. First, the detection of DAPK1 promoter methylation in Caucasians by the QMSP method excluded the study by Flatley et al.. The other reason may be that the heterogeneity in Caucasians is relatively small. Publication bias was evaluated through Funnel plots and Egger's test, and the Egger's test showed P = 0.551, which indicated that there was no significant publication bias. The fail-safe number further confirmed that the trend for publication bias was very small.
This meta-analysis had some limitations. The first limitation was that some studies did not provide detailed information regarding the age of individuals in the case groups and control groups. The second limitation in this meta-analysis was that some studies did not reveal the stage of the cervical cancers or the subtype, which might also be sources of the heterogeneity. Considering the small number of articles that described the stage and type of cervical cancer, the power was too small to make a subgroup for them. Other confounding variables such as method, ethnicity, sample size, and the source of control may also exist. Publication bias was the third limitation. Some unpublished and negative studies may contribute to some bias though no significant publication bias was detected according to Egger's test.
In conclusion, a strong association was observed between DAPK1 promoter methylation and cervical cancer, and therefore, DAPK1 promoter methylation may be valuable as a biomarker. Considering that the quality and quantity of the reviewed articles were limited, larger and well-designed studies should be employed in the future for further confirmation of the association between DAPK1 promoter methylation and cervical cancer.
Supporting Information
PRISMA Checklist.
(DOC)
Acknowledgments
We would like to thank Shen Wei, Wang Man, Xiaofang Du and Deng Song for their useful assistance.
Funding Statement
This work was supported by the National Natural Science Foundation of China (No. 81071663). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, et al. (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127: 2893–2917. [DOI] [PubMed] [Google Scholar]
- 2. Schiffman MH, Castle P (2003) Epidemiologic studies of a necessary causal risk factor: human papillomavirus infection and cervical neoplasia. J Natl Cancer Inst 95: E2. [DOI] [PubMed] [Google Scholar]
- 3. Flatley JE, McNeir K, Balasubramani L, Tidy J, Stuart EL, et al. (2009) Folate status and aberrant DNA methylation are associated with HPV infection and cervical pathogenesis. Cancer Epidemiol Biomarkers Prev 18: 2782–2789. [DOI] [PubMed] [Google Scholar]
- 4. Hesson LB, Cooper WN, Latif F (2007) The role of RASSF1A methylation in cancer. Dis Markers 23: 73–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barton CA, Hacker NF, Clark SJ, O'Brien PM (2008) DNA methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecol Oncol 109: 129–139. [DOI] [PubMed] [Google Scholar]
- 6. Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, et al. (2001) Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet 10: 687–692. [DOI] [PubMed] [Google Scholar]
- 7. Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- 8. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stuck AE, Rubenstein LZ, Wieland D (1998) Bias in meta-analysis detected by a simple, graphical test. Asymmetry detected in funnel plot was probably due to true heterogeneity. BMJ 316: 469; author reply 470–461. [PMC free article] [PubMed] [Google Scholar]
- 10. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dong SM, Kim HS, Rha SH, Sidransky D (2001) Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clin Cancer Res 7: 1982–1986. [PubMed] [Google Scholar]
- 12. Feng Q, Hawes SE, Stern JE, Dem A, Sow PS, et al. (2007) Promoter hypermethylation of tumor suppressor genes in urine from patients with cervical neoplasia. Cancer Epidemiol Biomarkers Prev 16: 1178–1184. [DOI] [PubMed] [Google Scholar]
- 13. Iliopoulos D, Oikonomou P, Messinis I, Tsezou A (2009) Correlation of promoter hypermethylation in hTERT, DAPK and MGMT genes with cervical oncogenesis progression. Oncol Rep 22: 199–204. [DOI] [PubMed] [Google Scholar]
- 14. Jeong DH, Youm MY, Kim YN, Lee KB, Sung MS, et al. (2006) Promoter methylation of p16, DAPK, CDH1, and TIMP-3 genes in cervical cancer: correlation with clinicopathologic characteristics. Int J Gynecol Cancer 16: 1234–1240. [DOI] [PubMed] [Google Scholar]
- 15. Kang S, Kim JW, Kang GH, Park NH, Song YS, et al. (2005) Polymorphism in folate- and methionine-metabolizing enzyme and aberrant CpG island hypermethylation in uterine cervical cancer. Gynecol Oncol 96: 173–180. [DOI] [PubMed] [Google Scholar]
- 16. Kim JH, Choi YD, Lee JS, Lee JH, Nam JH, et al. (2010) Assessment of DNA methylation for the detection of cervical neoplasia in liquid-based cytology specimens. Gynecol Oncol 116: 99–104. [DOI] [PubMed] [Google Scholar]
- 17. Leung RC, Liu SS, Chan KY, Tam KF, Chan KL, et al. (2008) Promoter methylation of death-associated protein kinase and its role in irradiation response in cervical cancer. Oncol Rep 19: 1339–1345. [PubMed] [Google Scholar]
- 18. Missaoui N, Hmissa S, Trabelsi A, Traore C, Mokni M, et al. (2011) Promoter hypermethylation of CDH13, DAPK1 and TWIST1 genes in precancerous and cancerous lesions of the uterine cervix. Pathol Res Pract 207: 37–42. [DOI] [PubMed] [Google Scholar]
- 19. Niyazi M, Liu XW, Zhu KC (2012) [Death-associated protein kinase promoter (DAPK) hypermethylation in uterine cervical cancer and intraepithelial neoplasia in Uyghur nationality women]. Zhonghua Zhong Liu Za Zhi 34: 31–34. [PubMed] [Google Scholar]
- 20. Shivapurkar N, Sherman ME, Stastny V, Echebiri C, Rader JS, et al. (2007) Evaluation of candidate methylation markers to detect cervical neoplasia. Gynecol Oncol 107: 549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun LL, Cao DY, Yang JX, Li H, Zhou XR, et al. (2012) Population-based case-control study on DAPK1, RAR-beta2 and MGMT methylation in liquid-based cytology. Arch Gynecol Obstet 285: 1433–1439. [DOI] [PubMed] [Google Scholar]
- 22. Yang N, Nijhuis ER, Volders HH, Eijsink JJ, Lendvai A, et al. (2010) Gene promoter methylation patterns throughout the process of cervical carcinogenesis. Cell Oncol 32: 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao XL, Meng ZY, Qiao YH, Zhang HL (2008) [Promoter methylation of DAPK gene in cervical carcinoma]. Ai Zheng 27: 919–923. [PubMed] [Google Scholar]
- 24. Reesink-Peters N, Wisman GB, Jeronimo C, Tokumaru CY, Cohen Y, et al. (2004) Detecting cervical cancer by quantitative promoter hypermethylation assay on cervical scrapings: a feasibility study. Mol Cancer Res 2: 289–295. [PubMed] [Google Scholar]
- 25. Inbal B, Cohen O, Polak-Charcon S, Kopolovic J, Vadai E, et al. (1997) DAP kinase links the control of apoptosis to metastasis. Nature 390: 180–184. [DOI] [PubMed] [Google Scholar]
- 26. Hoque MO, Feng Q, Toure P, Dem A, Critchlow CW, et al. (2006) Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. J Clin Oncol 24: 4262–4269. [DOI] [PubMed] [Google Scholar]
- 27. Hiraga J, Kinoshita T, Ohno T, Mori N, Ohashi H, et al. (2006) Promoter hypermethylation of the DNA-repair gene O6-methylguanine-DNA methyltransferase and p53 mutation in diffuse large B-cell lymphoma. Int J Hematol 84: 248–255. [DOI] [PubMed] [Google Scholar]
- 28. Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, et al. (2000) MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res 28: E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA Checklist.
(DOC)