Abstract
The heart is the first organ required to function during embryonic development and is absolutely necessary for embryo survival. Cardiac activity is dependent on both the sinoatrial node (SAN), which is the pacemaker of heart's electrical activity, and the cardiac conduction system which transduces the electrical signal though the heart tissue, leading to heart muscle contractions. Defects in the development of cardiac electrical function may lead to severe heart disorders. The Erbb2 (Epidermal Growth Factor Receptor 2) gene encodes a member of the EGF receptor family of receptor tyrosine kinases. The Erbb2 receptor lacks ligand-binding activity but forms heterodimers with other EGF receptors, stabilising their ligand binding and enhancing kinase-mediated activation of downstream signalling pathways. Erbb2 is absolutely necessary in normal embryonic development and homozygous mouse knock-out Erbb2 embryos die at embryonic day (E)10.5 due to severe cardiac defects. We have isolated a mouse line, l11Jus8, from a random chemical mutagenesis screen, which carries a hypomorphic missense mutation in the Erbb2 gene. Homozygous mutant embryos exhibit embryonic lethality by E12.5-13. The l11Jus8 mutants display cardiac haemorrhage and a failure of atrial function due to defects in atrial electrical signal propagation, leading to an atrial-specific conduction block, which does not affect ventricular conduction. The l11Jus8 mutant phenotype is distinct from those reported for Erbb2 knockout mouse mutants. Thus, the l11Jus8 mouse reveals a novel function of Erbb2 during atrial conduction system development, which when disrupted causes death at mid-gestation.
Introduction
The heart is needed to pump blood through the blood vessels by repeated, rhythmic contractions, carrying oxygen and nutrients to various parts of the body. In addition to contracting muscle cells, the adult heart requires the correct function of two systems: the sinoatrial node (SAN), which is the impulse-generating (pacemaker) tissue located in the right atrium of the heart, and the cardiac conduction system which consists of the atrioventricular node (AVN), atrioventricular bundle (AVB), bundle branches (BB) and Purkinje fibers (subendocardial branches). The cardiac conduction system transduces the electrical signals from SAN to heart muscle cells. This system consists of specialised cardiomyocytes that are able to conduct cardiac action potentials more quickly and efficiently than any other cells in the heart, allowing synchronised contractions of the atria and ventricles, and is therefore essential for maintaining a consistent heart rhythm. Developmental defects in the formation of the conduction system may lead to heart diseases such as heart block, long Q-T syndrome, atrial and ventricular fibrillation and tachycardia [1], [2].
The heart is the first functional organ to be formed during embryonic development (22 days after conception in humans, 8–8.5 days in mice) and is absolutely necessary for embryo survival and correct development. Cardiac contractions start immediately after the linear heart tube forms [3]; the sinus venosus myocardium located next to the primitive atrium acts as a primary pacemaker which later develops into the SAN [4], [5]. In contrast to the pacemaker, the cardiac conduction system develops progressively. The atrioventricular canal (AVC) at E9.5 gives rise to AVN. The AVB is formed from the crest of the developing ventricular septum; Purkinje fibers develop in the trabecular myocardium from E11.5 until birth, with further postnatal maturation (reviewed in [4], [5]. Until the mature conduction system is formed, cardiac action potentials are transduced through the myocardium directly from one cardiomyocyte to another via intercellular gap junctions [6], [7]. In vertebrates, gap junctions are composed primarily of homo- or hetero-hexamers of connexins [8]. The phosphorylation of connexins plays an important role in regulating the different properties of gap junctions, such as the trafficking, assembly/disassembly, degradation and gating of gap junction channels [6], [7]. During mouse embryonic development, five major connexins are expressed in the heart, including Cx45 (the first connexin expressed in developing mouse heart) [9], [10], Cx43 (the main connexin in chamber myocardium) [11], [12] and Cx40 (found in atrial and ventricular chamber myocardium) [13], [14]. Connexin abnormalities contribute to congenital heart defects, illustrating the importance of electrical signal propagation during cardiac development [15].
The Erbb2 (Mus musculus v-erb-b2 erythroblastic leukemia viral oncogene homolog 2) gene encodes a member of the epidermal growth factor receptor (EGFR) family of tyrosine kinases. Due to its extracellular protein structure, the Erbb2 receptor itself does not bind ligands [16]. Instead, it binds other members of EGF receptor family (Egfr, Erbb3 and Erbb4) to form heterodimers, stabilising their binding to ligands and enhancing kinase-mediated activation of downstream signalling pathways (such as MAP and phosphatidylinositol-3 kinase) [17]–[19]. Erbb2 is able to form heterodimers with any of the other three EGF receptors and is believed to be the preferred dimerisation partner [20]. In humans, ERBB2/HER2 is commonly amplified in breast cancer, leading to receptor over expression and driving the activation of downstream pathways that stimulate malignant cell proliferation [21]. ERBB2/HER2 amplification is a marker of aggressive disease, although the development of the trastuzumab monoclonal antibody targeting the ERBB2/HER2 receptor has revolutionised breast cancer treatment regimes, leading to improved survival rates [21].
In addition to its pathological role in breast cancer, Erbb2 is essential for normal embryonic development. Homozygous knock-out mice lacking Erbb2 are lethal at mid-gestation (embryonic day 10.5) and die due to severe cardiac defects, including an enlarged heart with thin ventricular myocardium, absent trabeculae and decreased atrioventricular cushions, leading to poor circulation and irregular heartbeat [22], [23]. Conditional Erbb2 mutation in ventricular cardiomyocytes leads to a severe dilated cardiomyopathy associated with the occurrence of sudden death, partially due to an impaired cardiac conduction system [24]. Absence of Erbb2 also results in neural defects such as degeneration of motor neurons and absence of Schwann cells (Table 1) [22], [25], [26]. It has been shown that Erbb2 participates in ventricular conduction system development and maturation via its involvement in cardiac trabeculation [23], [27]. However, the role of Erbb2 in the atrial conduction system development and atrial action potential propagation is unclear.
Table 1. Comparison Erbb2 null [22], [25] and l11Jus8 phenotypes.
| Erbb2 null | l11Jus8 -/- | |
| lethality | E10.5- E11.0 | E12.5-13 |
| general | degeneration of motor nerves, absence of Schwann cells, impairment of junctional folds at the neuromuscular synapse | no gross morphological internal organ defects |
| cardiac | absent myocardial trabeculae, thin ventricular wall, enlarged heart, poor circulation, irregular heartbeat, lack of cardiac ventricular myocyte differentiation, decreased endocardial cushion size | no ventricular, AV cushion or OFT defects, thickened, folded atrial myocardium which becomes stretched when atria stop contracting, atria inflated with blood, haemorrhage, heart rate normal but abruptly ceases without progressive decline |
From an N-ethyl-N-nitrosourea (ENU) mutagenesis screen [28] we have isolated a mouse line, l11Jus8, carrying a hypomorphic missense mutant allele of Erbb2. These mice are homozygous lethal by embryonic day (E)12.5-13 and display morphological defects in the developing atria; similar defects have not been reported in Erbb2 targeted deletion mouse mutants (Table 1). We show that l11Jus8 homozygous mutants exhibit compromised atrial function due to defects in atrial electrical signal propagation, leading to an atrial-specific conduction block that does not affect ventricular conduction. As relatively few studies have identified specific defects in atrial development, research on the l11Jus8 mouse model extends our knowledge of the molecular requirements for atrial function. The l11Jus8 mouse line reveals a novel role for Erbb2 during atrial conduction system development, demonstrating the importance of atrial function for embryonic survival.
Materials and Methods
Mice
The l11Jus8 mouse strain was generated by random ENU mutagenesis [29]. Mice were maintained in the Biological Service Facility of the University of Manchester, UK, with local ethical approval and according to UK Home Office requirements (Home Office project 40/2912). Mice used for electrical readings were maintained in the Animal Facility of the Institute of Anatomy, Charles University, Prague, Czech Republic and experiments were carried out in accordance with Czech law governing animal care and experimentation. Mice were out-crossed a minimum of 6 times to 129S5/SvEvBrd animals, although the use of the Chr11 balancer ensured that the candidate region on chromosome 11 remained heterozygous for C57BL/6 and 129S5/SvEvBrd DNA in viable animals. The K14-agouti transgene integrated on the balancer chromosome [29] allowed the identification of animals without molecular genotyping. Embryo genotyping to distinguish between the 129S5/SvEvBrd and C57BL/6 chromosomes was performed by PCR analysis of genomic DNA using STS markers D11Mit4, D11Mit35, D11Mit327 and D11Mit99. STS marker sequences were obtained from the Ensembl database.
Next-Generation Sequencing
The Next-Generation Sequencing was performed in the Manchester Centre for Genomic Medicine (Manchester Academic Health Science Centre). Genomic DNA was isolated from a single E12.5 homozygous l11Jus8 embryo (C57BL/6J background) using ISOLATE II Genomic DNA Kit (Bioline). 1 µg of DNA was used to create the genomic library using the TruSeq DNA Sample Prep Kit. Whole genome sequencing was performed on one lane of a HiSeq 2500 sequencer (Illumina, San Diego, CA, USA), following the manufacturer's protocols. Reads were mapped to the GRCm38/mm10 mouse genome using BWA mapping software [30] and analysed using online Galaxy sequencing analysis tool (main.g2.bx.psu.edu; [31]–[33]). Positions between 69–104 Mb on mouse chromosome 11 were selected and MAQ model with default parameters was applied to identify positions with nucleotides different from the reference. Ensembl Variant Effect Predictor [34] was used to annotate SNPs and eliminate known SNPs. Novel non-coding SNPs were inspected manually to identify those in RNA genes (Table S1). Sequence variants were confirmed in exome sequence data from a single heterozygous l11Jus8 embryo. Exome sequencing was performed at the University of Liverpool Centre for Genomic Research. Sanger sequencing was performed on an additional 3 homozygous mutant l11Jus8 embryos using BigDye v3.1 reagents (Applied Biosystems) in the University of Manchester Sequencing Facility according to manufacturer's instructions to verify base changes in coding regions and non-coding RNA genes. Only those variants confirmed by Sanger sequencing in an additional 3 mutant embryos were considered as candidate mutations. Primers for Sanger sequencing: Med31-Forward AGTAGTCCAGCGGTTGGTTG; Med31-Reverse TCATTTTCCATTCCCACACA; Nf1-Forward CCAGCAACTCAATAATGAGCC; Nf1-Reverse TCAAGTTGTGCACCATGCTAG; Erbb2-Forward TCCCTCTGTTCCCTTGTCTG; Erbb2-Reverse AACCCCCAAAGCACATACCT.
Expression analysis
To test for the expression of Erbb2 genes in heart tissues, atria, ventricles and erythrocytes were isolated from E12.5 embryos, collected and stored at −20°C in RNAlater solution (Ambion). RNA was isolated from tissue samples (2 hearts were pooled together prior to isolation) using Isolate RNA mini kit (Bioline; genomic DNA degradation step is included in the protocol) and the cDNA was prepared with Tetro cDNA Synthesis kit (Bioline). RT-PCR was performed with following primers: Erbb2-Forward ATGGACAGCACCTTCTACCG; Erbb2-Reverse GGAGCAACGTAGCCATCAGT; Prpf8-Forward TCCTGGACTTATTGGTGAAGTG; Prpf8-Reverse GACACGCCAGCCTTTATCAT. Primers were designed in different exons to allow the transcript to be distinguished from genomic product. Expected products sizes for Erbb2: 392 bp for cDNA, 607 bp for genomic DNA; for Prpf8: 464 bp for cDNA, 749 bp for genomic DNA. The Erbb2 fragment was gel purified and sequenced to confirm it was specific for Erbb2.
Protein Structural Modelling
The three dimensional structure of residues 713-1020 of mouse Erbb2 were predicted using homology modelling. The sequences of mouse Erbb2 and human ERBB2 were aligned using ClustalW [35], and structure predicted by Modeller [36] using the known human ERBB2 structure (PDB id 3CRD) [37] as a template. Twenty models were built, and the one with best score was used for further analysis.
In order to predict the likely effects of the Met-802-Arg point mutation, hydrogen atoms were added using Reduce software [38]. The mutation was modelled using KiNG software [39]. All low energy side chain conformation (rotamers) [40] were modelled, and their interactions with the rest of the protein structure assessed using the all-atom contact method [41]. Substantial van der Waals overlaps were found with all rotamers, indicating that the mutant protein cannot fold to the same native structure without additional structural accommodation. The rotamer with the smallest volume of van der Waals overlaps is shown in Figure 1.
Figure 1. Analysis of DNA, protein, and morphological defects in the l11Jus8 mutant mouse.
(A) Three coding mutations found in the 35 Mb candidate intervals on mouse chromosome 11 are located in the Med31, Nf1 and Erbb2 genes. Only recombinant embryos with the C57BL/6 homozygous DNA fragment inherited from the mutagenised parent which contained the Erbb2 mutation were lethal at weaning, whereas embryos with homozygous Med31 and Nf1 and heterozygous Erbb2 mutations were viable. Straight and dotted lines depict homo- and heterozygous DNA, respectively. (B) Confirmation of the T98,433,986G base change in the Erbb2 gene in l11Jus8 mutants by Sanger sequencing. (C) Phenotype of wild type, homozygous l11Jus8 and Erbb2M802R mutant embryos and hearts. Scale bars: 2 mm for embryos, 500 µm for hearts. (D) Erbb2 protein structure: FLD - Furin-Like Domain, STKD - Serine/Threonine-Tyrosine Kinase Domain. Partial sequence of the STK domain (60 amino acids around the mutation point) is shown for mouse (M), human (H), chick (C) and zebrafish (Z) Erbb2 protein. Asterisks indicate conserved amino acids, colon and period indicate conservation between groups of strongly and weakly similar properties, respectively. Arrowhead demarcates the location of the amino acid change. (E) Predicted changes in the protein structure of the Erbb2M802R conserved kinase catalytic domain based on the reported crystal structure of the Erbb2 protein. (F) Top panel: expression of the Erbb2 mRNA in atria (A) and ventricles (V) of hetero- and homozygous l11Jus8, and embryonic erythrocyte (blood) samples. Prpf8 mRNA expression is used as a positive control for cDNA isolation in blood sample. Bottom panel: Erbb2 expression in the yolk sack (YS) of homozygous l11Jus8 embryo, “-RT” control, genomic DNA (gen) control. A no template negative control (neg) is shown in both panels. (G) Erbb2 immunohistochemistry in heterozygote and l11Jus8 atria (Atr) and ventricles (Ven). Embryonic erythrocytes within the cardiac chambers are indicated with an arrowhead. Scale bar = 200 µm.
Histology
Tissues were fixed in Bouin's fixative in PBS and dehydrated through an ethanol gradient. Samples were embedded in paraffin (Paraplast, Sigma), sectioned at 7 µm thickness and stained with Harris' hematoxylin and 1% eosin Y.
Immunostaining
Paraffin-embedded tissue sections were de-waxed and rehydrated prior to immunohistochemistry. Antibodies detecting α-Smooth Muscle Actin (1∶450, Sigma-Aldrich), Laminin α1 (1∶1000, gift from Prof Ray Boot-Handford, University of Manchester, UK), Fibronectin (1∶50, Santa Cruz Biotechnology, Heidelberg, Germany), Phospho-histone H3 (1∶1000, Millipore, Watford, UK), Erbb2 (1∶100, gift from Prof Keith Brennan, University of Manchester UK), and Activated Caspase-3 (1∶500, R & D Systems, Abingdon, UK) were incubated for a minimum of 1 hour at room temperature. Samples were then washed in PBS, incubated with secondary antibody (biotinylated horse anti-mouse 1∶250, biotinylated goat anti-rabbit 1∶1000, biotinylated horse anti-goat 1∶200), and protein localisation visualised using DAB Peroxidase Substrate Kit (SK-4100, Vector Labs). Immunostaining detecting Connexins 40 and 43 was done using Cx40 (1∶100, Santa Cruz) and Cx43 (1∶100, Sigma) antibodies on PFA fixed, paraffin-embedded sections. Antigen retrieval was by boiling in sodium citrate buffer (10 mM pH 6.0) for 20 minutes, samples were then blocked with 3% goat serum for 1 hour at room temperature before incubating with the primary antibody O/N at 4°C (Cx43 1∶100 in PBS and Cx40 1∶100 in 10% goat serum/PBS). Samples were washed the next day in PBS, and incubated with secondary antibody (Cx43 - goat anti-rabbit-FITC 1∶250, Sigma 2 hours at room temperature; Cx40 - anti-goat biotinylated, 1∶250, Vector Labs for 2 hours at room temperature, followed by a 30 minute incubation with Streptavidin-Cy3, 1∶500, GE Healthcare).
For visualisation of cell nuclei, sections were mounted with Vectashield mounting medium containing DAPI (Vector Labs) immediately followed by imaging.
For detection of necrotic cells, freshly dissected hearts were placed in 1 µg/ml Propidium Iodide solution in PBS and incubated for 30 minutes at room temperature, washed for 5 minutes in fresh PBS and imaged using fluorescent microscopy.
In situ hybridization
Whole mount in situ hybridization was carried out as previously described [42]. The Myl4 probe was generated by cloning a PCR product containing the Myl4 cDNA into the PCRII dual promoter plasmid (Invitrogen). PCR primer sequences were Myl4 exon2: GCAACCGACAGTGTCCATATA; Myl4 3′UTR: CATGTGAGTCCAATACTCCGT.
Optical mapping
Embryos were decapitated; hearts and the adjacent posterior thoracic wall were isolated and stained with voltage-sensitive dye 4-{-[2(di-n-butylamino)-6-napthyl]vinyl} pyridiniumin (di-4-ANEPPS, Invitrogen) in Tyrodes solution for 5 minutes at room temperature [43]. E12.5 embryos with visible thoracic haemorrhage and/or yolk sac vascular defects (6/21) were excluded from analysis. Optical mapping was performed at 37°C in oxygenated Tyrodes with added blebbistatin (1∶1000 of 14 mM stock in DMSO) to stop muscle contractions and heart movements. Recordings were done with Ultima L high-speed camera (SciMedia Ltd., Japan) at 0.25, 0.5 and 1 kHz and the analysis of recordings resulting in the generation of spatio-temporal epicardial activation maps was performed using the BV_Analyzer software. The protocol and processing algorithms for generation of activation maps were described in detail previously [44]. At least three recordings were made for each sample. Activation maps were compared together with the rhythm strip of atrial and ventricular rate between genotypes essentially as described [45]. Parameters analysed presence, direction and extent of atrial and ventricular activation and propagation. Gaps in atrial signal propagation were detected by varying threshold of signal/noise ratio during electrical map reconstruction (Figure S1, S2). The presence of the gaps was judged by two parameters: first, a map was considered to contain “gaps” if there was an area without clear signal found at the threshold level when significant noise was still observed in the rest of the image (Figure S1). Secondly, gaps were confirmed by the absence of the signal in one area of the atrium and presence in the other one (Figure S2B–C and F–G).
Results
l11Jus8 mouse line: mutation isolation and analysis
The l11Jus8 mutant mouse line was created by random ENU mutagenesis and isolated from a balancer chromosome screen for mutants with embryonic lethality [28], [29]. The l11Jus8 line was selected for further study because it displayed embryonic lethality at E12.5 arising from suspected cardiac defects, including vascular and cardiac haemorrhage. Due to the use of a balancer chromosome in the mutagenesis screen, the candidate interval for the l11Jus8 mutation was known to be within a 35 Mb region of mouse chromosome 11 between the Trp53 and Wnt3 loci [29]. Since homozygous mutant l11Jus8 embryos do not survive past day E13.5 of embryonic development, we hypothesised that an important protein function would be disrupted in these mutants. In order to find the causative mutation, we isolated genomic DNA from homozygous l11Jus8 embryos, performed genomic Next Generation Sequencing (NGS), and used bioinformatic analysis to identify single nucleotide variations within the 35 Mb candidate interval. Using the GRCm38 mouse genome assembly, three coding changes on mouse chromosome 11 were found: T72,213,810C in the Med31 gene resulting in Tyr-57-Cys missense change, T79,447,501A located within the Nf1 gene resulting in a Met-1113-Lys missense mutation and T98,433,986G within the Erbb2 gene causing a Met-802-Arg missense change (Figure 1A, Table S1). These changes were confirmed by Sanger sequencing on three additional homozygous l11Jus8 mutant embryos. Additionally, annotated novel and non-coding RNAs (microRNAs, lncRNAs, anti-sense RNAs) were also checked for sequence variations specific to the l11Jus8 mutant. Predicted base changes within RNA genes were not confirmed by further Sanger sequencing on additional mutant embryos (Table S1).
To determine which sequence variant best represented the l11Jus8 causative mutation, we bred animals carrying recombinant chromosomes to l11Jus8 heterozygotes, and analysed the viability at weaning of offspring carrying homozygous C57BL/6 DNA (inherited from the mutagenised parent) in smaller intervals within the candidate region. Based on the analysis of genomic DNA from recombinant animals, only the Erbb2 mutation is a valid genetic candidate for causing the l11Jus8 phenotype (Figure 1A), because lethality was only seen in recombinants homozygous for the Erbb2 mutation. We have confirmed that no animals homozygous for the Erbb2 mutation survived to weaning age, showing a significant deviation from expected Mendelian frequency (Chi-squared test; Table S2). Sanger sequencing demonstrated that the Erbb2 mutation was present in homozygous l11Jus8 mutants (Figure 1B). Dissections at E12.5 confirmed that embryos homozygous for the mutation in the Erbb2 gene exhibit the l11Jus8 mutant phenotype (Figure 1C) while heterozygous animals were viable, fertile, and showed no defects during development. Animals double homozygous for both the Med31 and Nf1 missense mutations were viable at weaning age (3 pups in 1 litter). Additionally, a non-coding mutation in Zzef1 previously identified in this mouse line [46] was excluded as a candidate based on recombination mapping.
The mutation in the Erbb2 gene is located in the conserved Serine-Threonine/Tyrosine protein kinase catalytic domain (Figure 1D). Utilising the reported crystal structure of the human ERBB2 protein (PDB id 3RCD [37]), we predict from an analysis of side-chain rotamer conformations [40] that the M802R amino acid change will create van der Waals clashes, impeding folding and likely causing a loss of kinase catalytic function (Figure 1E). It is not known how the M802R affects Erbb2 protein function. We did not detect any differences in the phosphorylation of Erk, one Erbb2 target, in l11Jus8 mutants (Figure S3), so at present cannot determine the biochemical nature of the M802R mutation.
To confirm that Erbb2 is expressed in cardiac tissues and to determine if the missense mutation affects gene expression, we performed RT-PCR with cDNA prepared from RNA isolated from E12.5 atria and ventricles of l11Jus8 heterozygote and homozygote hearts. Since the heart lumen is filled with erythrocytes that are difficult to separate completely from the surrounding cardiac tissue, as a control we also prepared cDNA from embryonic erythrocytes collected during dissections. Clear bands indicating the expression of Erbb2 were found in atrial and ventricular samples of both genotypes but not in the erythrocyte sample (Figure 1F top panel). The expression of the Prpf8 gene (encoding a core component of the spliceosome) [47] was used as a positive control to confirm the presence of cDNA in erythrocyte sample. We also identified Erbb2 expression in the developing E12.5 wild type yolk sac (Figure 1F bottom panel). To determine if Erbb2 protein distribution was altered in l11Jus8 mutants, we examined the localisation of Erbb2 in E12.5 heterozygote and homozygous mutant hearts. No alterations in the expression pattern in the ventricles or atria were detected (Figure 1G). Notably, Erbb2 was expressed throughout the cardiac tissue, but not within blood cells, supporting our RT-PCR data. The presence of Erbb2 protein in l11Jus8 mutants supports our hypothesis that the M802R mutation does not create a null allele.
Morphological characterisation of the l11Jus8 mutant phenotype
Heterozygous l11Jus8 embryos were viable and had no overt developmental defects (Figure S4A, D, B, E). Homozygous mutants were lethal with embryonic death observed between days E12.5-13 of development; mutants exhibited severe haemorrhages in the thoracic cavity and head (Figure 1C). Mutant yolk sacks (YS) display reduced YS vessel volume becoming progressively more severe from E11.5 – E12.5, resulting in visible absence of blood in YS vessels (Figure S5). Notably, yolk sack defects were only visible in the embryos with haemorrhages in the thoracic cavity. Approximately 29% of mutant embryos possessed yolk sac defects at E12.5, indicating that this aspect of the phenotype is not fully penetrant, while lethality by E13.5 was fully penetrant. Histological examination of l11Jus8 mutant embryos at E11.5 revealed subtle defects in atrial wall structure; no other cardiac abnormalities were detected at this stage, and hearts were beating visibly during dissections (data not shown). Because mice homozygous for a targeted deletion of Erbb2 display a reduction in endocardial cushion size [22], we analysed atrioventricular cushion development in l11Jus8 mutant embryos at E12.5 (Figure 2A–B) but found no differences as compared to control littermates. Similarly, no defects were observed in outflow tract (OFT) vessel septation (Figure 2C–D), despite the reported role for Erbb2 in this process [48]. Erbb2 knockout embryos also display a lack of ventricular trabeculation leading to death by E11.0 [22], however l11Jus8 mutant embryos survive past this time point, and have clear evidence of trabeculation at E12.5 that is indistinguishable from control littermates (Figure 2E–F).
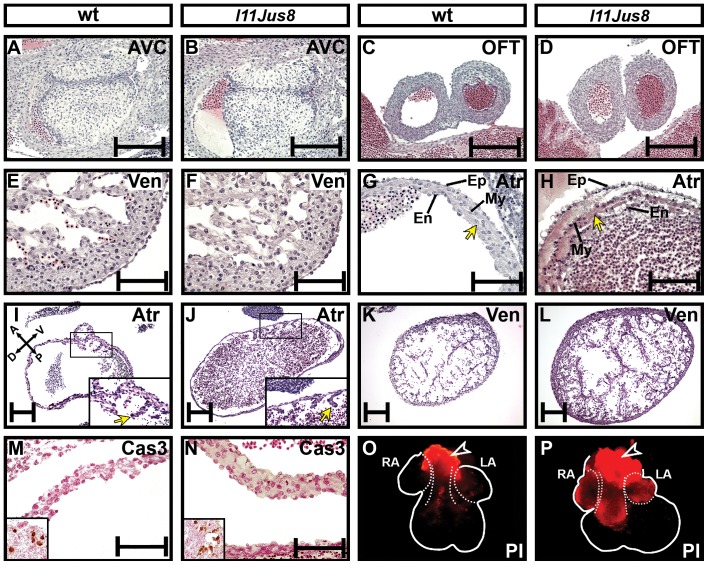
Figure 2. Effect of the homozygous Erbb2M802R mutation on heart morphology in l11Jus8 hearts.
(A–L) Representative H and E stained coronal sections of E12.5 hearts. (A–B) Atrioventricular cushions, (C–D) dorsal outflow tract, (E–F) ventricles, (G–H) atria. Arrows in (G–H) point to the atrial wall. AVC, atrioventricular cushion; OFT, outflow tract; Ven, ventricle; Atr, atrium; Ep, epicardium; En, endocardium; My, myocardium. (I–J) Longitudial atrial and (K–L) ventricular sections; homozygous l11Jus8 heart had distended atria. Embryos in I–L are littermates. Magnified areas in (I–J) show the developing pectinate muscles (arrows). (M–N) Activated Caspase 3 (Cas3) staining of atrial wall at E11.5. Inset shows positive activated Caspase3 staining from trigeminal ganglion on same embryo section. (O–P) Propidium Iodide (PI) labelling of the necrotic cells in E12.5 hearts. RA, right atrium; LA, left atrium. Arrowheads point to the unspecific PI staining resulting from cells of the OFT damaged during dissection. Scale bars: 200 µm in A–H and M–N, 50 µm in I–L.
Although ventricular myocardium, outflow tract and atrioventricular cushion morphology was normal, 5/15 homozygous l11Jus8 E11.5 embryos exhibited collapsing of atria such that the chamber lumen appeared decreased in size and the atrial myocardium appeared thickened and irregularly folded (data not shown). Highly eosinophilic stained patches containing fragmented irregularly shaped cell nuclei were present, and the epi- and endocardium were detached from the myocardium; by E12.5 these changes were more pronounced (Figure 2G–H arrows). In mutant hearts with distended atria (Figure 1C), the atrial wall was noticeably thinner in comparison to wild type (Figure 2I–J) while ventricles remained unchanged (Figure 2K–L). The developing pectinate muscles, found specifically in the atria, were present in both heterozygous and homozygous mutant atria (magnified areas in Figure 2I–J). Aside from cardiac abnormalities, no defects in the development of internal organs or embryonic vasculature were found in l11Jus8 mutants (Figure S4C, F). Notably, the atrial phenotype observed in homozygous l11Jus8 mutants cannot be examined in Erbb2 null embryos (phenotype summarised in Table 1) [22], [25] due to the early gestation lethality of Erbb2 null mutants.
Molecular characterisation of the l11Jus8 mutants
To further characterise the l11Jus8 mutant phenotype, we analysed the expression of Myl4, a marker of cardiomyocyte differentiation, in wild type, l11Jus8 heterozygous and homozygous E10.5 embryos (Figure S6A–C). We also performed immunohistochemistry on atrial cross-sections from wild type, l11Jus8 heterozygous and homozygous E12.5 hearts for Smooth Muscle α-Actin (smooth muscle cell marker), Laminin α 1 (basement membrane component), Fibronectin (cardiac ECM component) and E11.5 hearts for phospho-histone H3 (marker of cell proliferation). Surprisingly, expression of all markers was similar between all three genotypes suggesting the presence of correct cell types and cellular functions in l11Jus8 homozygous mutant hearts (Figure S6D–O).
DAPI staining of cell nuclei in E12.5 atrial sections (Figure S6P–R), revealed signs of fragmented DNA in the eosinophilic patches in l11Jus8 mutants (shown in Figure 2H), which may indicate extensive cell death in these areas. We tested this hypothesis and found that activated Caspase3 expression was identical between wild type and l11Jus8 mutants (Figure 2M–N), suggesting that there was no increased apoptosis in l11Jus8 mutants. However, staining with propidium iodide revealed necrotic cells in E12.5 l11Jus8 mutant atria (3/6 mutant hearts) but no necrosis in heterozygous atria (0/10 heterozygous hearts; Figure 2O–P).
Cardiac function in l11Jus8 mutant embryos
Since the l11Jus8 hearts do not show significant defects at the morphological or molecular level but the embryos display mid-gestation lethality consistent with cardiac defects, we next investigated heart function in the mutant embryos. The heart rate of E10.5, 11.5 and 12.5 embryos was assessed visually during dissections and on ultrasound movies of embryos in utero. No differences were observed between wild type, heterozygous and viable homozygous l11Jus8 embryos (data not shown). However, only 8/10 E11.5 and 4/12 E12.5 homozygous l11Jus8 embryos were viable with a heartbeat present at the time of dissection (Table S3). Moreover, during dissections l11Jus8 mutant hearts with normal macroscopic atrial morphology always contracted normally but hearts with distended atria (confirmed via genotyping as homozygous l11Jus8 mutants) only had ventricles contracting; atria in these hearts either did not contract at all or in rare cases, twitched slightly but lacked proper blood movement through the heart.
Optical mapping of action potential propagation
To investigate cardiac function of the l11Jus8 mutants in detail, we performed optical mapping of the electrical activity in wild type and mutant hearts using a voltage-sensitive dye di-4-ANEPPS to visualise the propagation of the electrical signal through the tissue [43]. In E11.5 hearts of all three genotypes the electrical signal was detected in both atria and ventricles (Table 2; Figure 3A–B; signal in heterozygous hearts not shown). Occasionally, partial atrioventricular block (AV block, defined as signal present in atria but not observed in ventricles) was detected (AVb in Table 2). AV block is often found in optical mapping experiments and results from experimental manipulations [43]. Notably, the l11Jus8 mutant hearts did not display distended atria at E11.5 (Table 2). In E12.5 samples, 9/21 (43%) homozygous l11Jus8 hearts had distended atria and did not contract; mapping these hearts did not result in clear reading of the electrical signal and they were excluded from further analysis (labelled “distended” in Table 2). Only l11Jus8 mutant E12.5 hearts without atrial distension were used in subsequent analysis of electrical activity. At this stage wild type hearts displayed consistent electrical activity in both the atria and ventricles (Figure 3C), or the presence of AV block (Figure 3D white arrowhead). Overall, AV block was found in 58%, 46% and 50% of wild type, heterozygous and homozygous E12.5 l11Jus8 hearts, respectively (AVb in Table 2). Surprisingly, we also found partial (Figure 3E white arrowhead) or full atrial block (Figure 3F white arrowheads): while the electrical signal was present in ventricular myocardium (Figure 3E–F black arrowheads), the corresponding atrial signal was not detected. Atrial block was observed in 2/50 (4%) heterozygous and 6/12 (50%) homozygous l11Jus8 hearts (Ab in Table 2) but not in any of the 29 wild type hearts analysed. An atrial block phenotype has not been described before in wild type mouse or chick hearts, or Cx40 mutant mouse hearts (55 chick hearts ages 3–10 days; 19 wild type mouse hearts, 12 Cx40+/− and 18 Cx40−/− hearts; [45], [49]). Notably, the percentage of absent atrial peaks (as compared to the presence of ventricular peaks) was 13% and 36% in two heterozygous embryos. However, in l11Jus8 homozygous hearts a range of 3.3–100% of atrial peaks were missing (one sample had full atrial block with no atrial signals detected while regular ventricular signal was observed). Statistical analysis shows that the proportion of embryos with atrial block is significantly different between all genotypes in pair-wise comparisons (heterozygous and homozygous l11Jus8 samples Chi square, p<0.0001). The observation that atrial block was present in homozygous l11Jus8 mutant hearts lacking yolk sac vessel defects, cardiac distension, or haemorrhage suggests that atrial electrical alterations precede or occur independently of morphological abnormalities of the atria, yolk sack or necrosis in the mutant hearts. Thus impaired atrial electrical function is the most likely reason for embryonic lethality in l11Jus8 mutants.
Table 2. Numbers of distended hearts, hearts with atrioventricular block (AVb), atrial block (Ab), and atrial signal conduction gaps in optical mapping experiments.
| E11.5 | E12.5 | |||||
| Wild type | het | l11Jus8 | Wild type | het | l11Jus8 | |
| embryos (total) | 6 | 6 | 6 | 29 | 50 | 21 |
| distended heart | 0 | 0 | 0 | 0 | 0 | 9 |
| AVb | 2 | 1 | 1 | 17 | 23 | 6 (of 12) |
| Ab | 0 | 0 | 0 | 0 | 2 | 6 (of 12) |
| A gaps | 0 | 0 | 3 | 0 | 7 | 10 (of 12) |
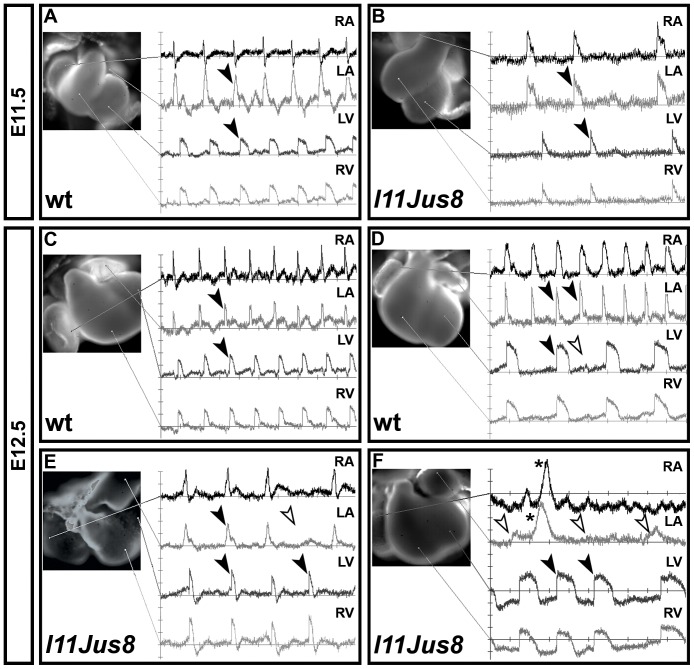
Figure 3. Optical mapping of the cardiac electrical activity obtained with di-4-ANEPPS voltage sensitive dye.
(A–B) and (C–F) show reading of E11.5 and E12.5 hearts, respectively. The location from which the electrical reading was taken is shown with lines onto the cardiac dissection image. Black arrowheads point to matched atrial and ventricular signals. (D) White arrowhead indicates absent signal in left ventricle corresponding to a present signal in left atria (black arrowhead), indicative of AV block. (E–F) White arrowheads indicate absent atrial signals corresponding to present ventricular signals (A block), a phenotype only present in mutant samples. Asterisks on RA and LA traces in (F) indicate signal described in Figure 5C . RA, right atrium; LA, left atrium; LV, left ventricle; RV, right ventricle.
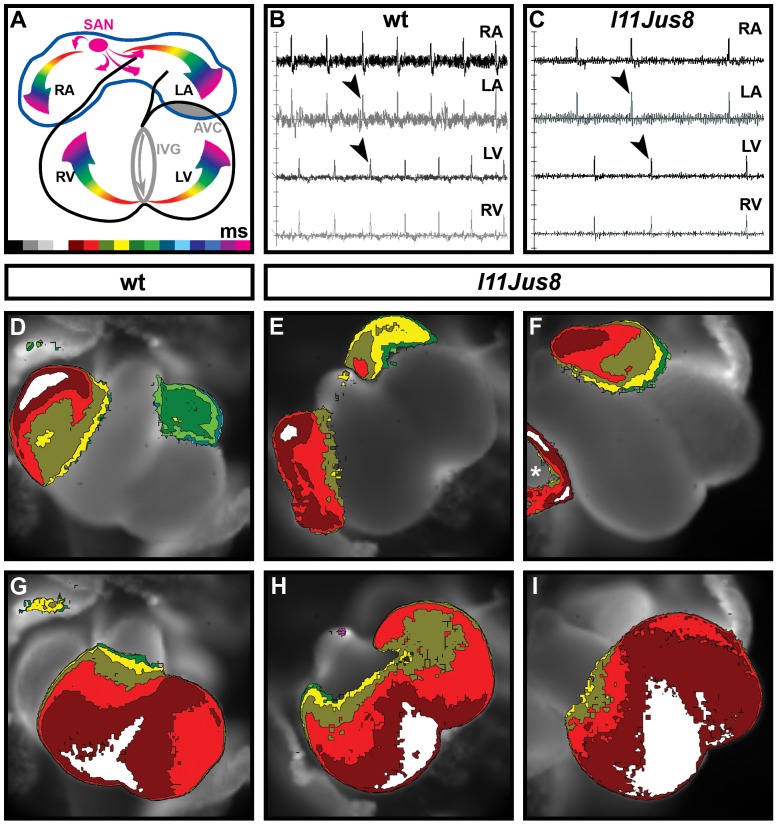
We next investigated the spatiotemporal propagation of the electrical signal through the atrial and ventricular tissue using maps reconstructed from the optical recordings [43]. Electrical signals travel through the developing heart in a characteristic pattern (Figure 4A [45], [50], and alterations in the signal propagation pattern could indicate cardiac dysfunction. Signal propagation maps were reconstructed using the derivatives (Figure 4B–C) calculated from the graphs of the electrical activity shown previously (Figure 3A–B). At E11.5 in hearts of all genotypes without atrial block, the electrical signal originated in the roof of the right atrium, spread within 3–5 ms through the right atrium, and travelled with a slight delay (approximately 2 ms) across the left atrium (Figure 4D–F). After 50–130 ms delay due to passing through the slow-conducting AV canal tissue (AVC), the signal appeared in the interventriclular groove (IVG) from where it moved laterally along ventricles (Figure 4G–I). In E11.5 wild type, heterozygous and 3/6 homozygous l11Jus8 hearts complete atrial activation maps without gaps were reconstructed (Figure 4D–E). However, in 3/6 homozygous l11Jus8 E11.5 hearts, gaps indicating regions of tissue lacking electrical conduction (further defined in Materials and Methods and Figures S1 and S2) were found (asterisk in Figure 4F), implying aberrant signal propagation in mutants. Ventricular signal propagation was complete without gaps in all samples (Figure 4G–I).
Figure 4. Mapping of electrical signal propagation in E11.5 hearts.
(A) Pattern of wild type electrical signal propagation in the E11.5-12.5 heart. Different colours encode the time scale of the signal conduction; each colour represents a one-millisecond interval with black as the initial time point and fuchsia as the final time point. (B–C) Derivatives of the signal recorded in wt and homozygous mutant l11Jus8 E11.5 hearts. Black arrowheads point to the peaks used for map generation. (D–F) Optical maps showing spatio-temporal propagation of the electrical signal through the atria. Asterisk on (F) points to the gap in normal electrical activity in homozygous mutant l11Jus8 atria. (G–I) Optical maps showing electrical signal propagation through the ventricles. Maps are similar for all genotypes. SAN, sinoatrial node; RA, right atrium; LA, left atrium; LV, left ventricle; RV, right ventricle; AVC, atrioventricular canal; IVG, interventricular groove.
Derivatives of the electrical signal were also used to reconstruct signal propagation maps at E12.5 (Figure 5A–C, corresponding to electrical signal shown in Figure 3C, E, F). In E12.5 hearts, no atrial gaps could be detected in wild type samples (Figure 5D). However, regions of atrial tissue lacking electrical signal were observed in 7/50 (14%) heterozygous (data not shown) and 10/12 (83%) homozygous hearts (Figure 5E–F). The proportion of samples demonstrating atrial conduction gaps is statistically significant when compared between wild type and homozygous l11Jus8 samples, as well as between heterozygous and homozygous l11Jus8 samples (p<0.0001). All 6 homozygous l11Jus8 hearts with partial atrial block exhibited gaps in the maps reconstructed from the detected atrial signals, with the majority of atrial tissue completely lacking electrical activity (Figure 5F). Moreover, all maps reconstructed from the sequence of signals in the same sample showed gaps in conduction (Figure S7). In samples of all genotypes, ventricular propagation of the signal was complete over the entire tissue without gaps in conduction (Figure 5G–I).
Figure 5. Mapping of electrical signal propagation in E12.5 hearts.
(A–C) Derivatives of the signal recorded in wt and homozygous l11Jus8 E12.5 hearts. Black arrowheads point to the present peaks used for map generation, white arrowheads point to the missing peaks. Asterisk on (C) depicts the missing derivative for signal present on Figure 3F . The derivatives used in each map image are indicated on the electrical traces. (D–F) Optical maps showing spatio-temporal propagation of the electrical signal through the atria. Asterisks indicate gaps in normal electrical activity. White arrow shows the direction of the signal propagation in wild type and homozygous l11Jus8 atria. (G–I) Optical maps showing electrical signal propagation through the ventricles. Maps are similar for all genotypes. Each colour (D–I) represents a one-millisecond interval with black as the initial time point and fuchsia as the final time point. RA, right atrium; LA, left atrium; LV, left ventricle; RV, right ventricle.
During embryonic development, prior to the formation of the mature cardiac conduction system, cardiac action potentials are transduced between cardiomyocytes via intercellular gap junctions [6], [7]. Connexin protein localisation serves as an indicator of cellular gap junctions, and alterations in connexin protein distribution could result in aberrant gap junctions leading to errors in electrical signal propagation in the embryonic heart [15]. Due to the electrical signal propagation defects detected in homozygous l11Jus8 hearts, we therefore examined the distribution of Connexin 40 and Connexin 43 in wild type and l11Jus8 mutant hearts. However, in both the atria and ventricles of wild type and l11Jus8 mutant hearts no differences in the distribution of either protein could be identified (Figure S8). It has been reported that heterozygosity for either Cx40 or Cx43 reduces atrial conduction [51]. Because we detect both Cx40 and Cx43 in mutant hearts, we hypothesise that a loss of Cx40 or Cx43 is not the underlying mechanism for the atrial conduction defects of the l11Jus8 mutant.
Overall, the optical mapping data above demonstrate that atria of l11Jus8 mutant hearts begin to lose the ability to conduct electrical signal at E11.5, leading to a block in atrial conduction by E12.5-13. The loss of gap junctions due to abnormalities in connexin protein distribution is unlikely to underlie these conduction defects. We propose that the embryonic lethality detected in l11Jus8 mutants arises due to atrial conduction failure and subsequent atrial necrosis, as no other significant cardiovascular abnormalities have been found in mutant embryos.
Discussion
We present data to demonstrate that a missense mutation in Erbb2 results in a specific defect in atrial electrical conduction during embryonic development. The M802R mutation affects a residue within the protein kinase domain of Erbb2 that is conserved across multiple species, including human. Our analysis of human sequence variations from the 1000 genomes project [52] reveals that although this particular mutation is not found in the dataset, variants at positions nearby are likely to be deleterious. The 1000 genomes dataset contains 68 missense variants within the kinase catalytic domain of the ERBB2 protein (ENSP00000269571), of these 45 are predicted by SIFT [53] to be deleterious, and 46 are predicted by PolyPhen [54] to be probably damaging. Additionally, our structural modelling of the M802R mutation indicates that protein stability may be altered due to side chain conformation clashes.
Although the extracellular domain structure of Erbb2 precludes ligand binding [16], the Erbb2 protein functions through heterodimerisation with other members of the EGFR family to achieve ligand-dependent activation of tyrosine kinase activity [16], [55], [56]. There are a series of five autophosphorylation sites within the Erbb2 cytoplasmic tail, one of which has been demonstrated to negatively regulate Erbb2 signalling [57]. Knock-in mice have been generated carrying mutations within these autophosphorylation sites. Notably, homozygous knock-in animals for Y1028F, Y1144F, and Y1227F Erbb2 missense mutations were viable, with an apparently normal phenotype. Animals carrying a hypomorphic Erbb2 allele expressing 10% of wild-type protein levels in trans to a null allele demonstrated perinatal lethality due to impaired diaphragm innervation causing respiratory failure [58]. The Y1028F allele when placed in trans to a null allele was found to rescue the perinatal lethality by reducing Erbb2 protein turnover [58]. Cardiac embryonic development can be assumed to be sufficient in the Erbb2 hypomorphic mutants due to their survival into the perinatal period. Therefore, the atrial electrical alterations and death detected in the l11Jus8 homozygous mutant embryos may indicate that the M802R mutation within the kinase domain of Erbb2 severely compromises Erbb2 function in a manner distinct from the autophosphorylation site missense mutations. In comparison to the less severe phenotypes generated from autophosphorylation site mutants, our data indicate that kinase activity is of greater importance than autophosphorylation for Erbb2 function. Many of the Erbb2 null mutants were analysed on a mixed 129 and C57BL/6 background [22], [25], [26]. The l11Jus8 mice were also maintained on a mixed 129 and C57BL/6 background, although there are likely to be differences to the prior knockouts in the percentages and areas of the genome that are comprised from each strain. Thus, we cannot formally exclude genetic background differences as a confounding factor in comparing the M802R allele to other Erbb2 mutant alleles.
Because the Erbb2 receptor lacks ligand-binding activity, its signalling function must be mediated through interactions with other family member proteins. Therefore, the phenotype we detect in the l11Jus8 mutant mouse might arise due to impaired signal transduction specific to one interacting partner. An examination of the null phenotypes of Egfr/Erbb1, Erbb3 and Erbb4 mutant mice indicates that the cardiac defects present are dissimilar to the l11Jus8 mutant [56]. For example, Erbb4 null mutants display cardiac trabeculation defects and lethality at E10.5 [59] that is similar to the Erbb2 null phenotype. Erbb3 null mutants display a failure to form endocardial cushions, linked to lethality at E13.5 [25]. Egfr/Erbb1 null mutants display early embryonic lethality, but this is not primarily linked to cardiac dysfunction [60], [61]. Thus, the null phenotypes of other EGFR family proteins are not identical to the l11Jus8 phenotype, although genetic background may also contribute to phenotype differences. A more detailed investigation of the function and downstream signalling activities of the other EGFR family members in l11Jus8 mutants may determine if moderation of the function of a specific family member or downstream signalling effector contributes to the l11Jus8 phenotype.
It is unclear why the electrical conduction defect seen in the l11Jus8 mutants is restricted to the atria, as we have shown that Erbb2 is expressed in both the atria and ventricles of the developing heart. Prior studies on a zebrafish Erbb2 mutant demonstrated an aberrant pattern of electrical activity which indicated the ventricular conduction system failed to develop [27]. Additionally, the EGFR ligand Neuregulin 1 (Nrg1) has been shown to promote cardiomyocyte differentiation into cardiac conduction system cells [62]. Hearts cultured with exogenous Nrg1 demonstrate an altered pattern of ventricular conduction activity, however atrial electrical mapping was not presented in the study [62]. Nrg1 treatment of adult rat ventricular cardiomyocytes in primary culture increases focal adhesion formation, resulting in synchronous beating [63], demonstrating another link between Erbb2 and ventricular conduction. It is noted that Nrg1 has higher expression in the ventricles and Nrg2 expression levels are greater in the atria [62]. However, a critical role for Nrg2 in atrial conduction system development can be ruled out from studies on the Nrg2 null mouse, which is viable and presents no cardiac conduction system defects [64].
Embryonic cardiac defects, including atrial conduction failure, are not found in mice with cardiomyocyte-specific deletions of Erbb2 [24], [65]. At present our experiments cannot determine whether a non-cardiomyocyte tissue source requires Erbb2 function for cardiac conduction system development, or whether the M802R missense mutation interferes with a key factor for cardiac conduction system function in a manner that is not replicated in null models. In addition to the embryonic requirement for Erbb2 for cardiac function, it has been proposed that Erbb2 functions in a cardioprotective capacity in adult cardiomyocytes [66]. Notably, the use of the anti-cancer drug trastuzumab, an ERBB2 specific-inhibitor, causes cardiotoxicity and congestive heart failure in some breast cancer patients [55]. Similarly, the deletion of Erbb2 in adult cardiomyocytes causes early cardiac dysfunction and severe dilated cardiomyopathy in the absence of readily demonstrable cardiomyocyte apoptosis [24], [65]. In humans and mice dilated cardiomyopathy contributes to impairment of the conduction system, causing arrhythmias and sudden death [24]. Deletion of Erbb2 in both atria and ventricles results in an earlier onset of cardiac pathology than the deletion of Erbb2 solely in ventricular cardiomyocytes [65]. A requirement for Erbb2 function in the atria may persist into the adult given the increased severity of cardiomyopathy arising from deletion of Erbb2 in cardiomyocytes of all chambers.
The mechanism of the atrial block resulting from the Erbb2 mutation remains unclear. We speculate that the possible mechanism involves abnormalities in action potential transduction through the atrial myocardium rather than the generation of the signal by the SAN, because normal ventricular signals are observed at a regular pace independently of the presence or absence of atrial signals. The shape and duration of the action potentials present in the l11Jus8 mutants do not appear to be altered in comparison to the signals in the control hearts. Given that Erbb2 is a transmembrane protein, it could be involved (directly or indirectly) in the restoration of the membrane potential after action potential transduction. Thus, ablation of Erbb2 activity in l11Jus8 mutants may lead to the slowdown and eventually complete block of the membrane potential restoration and abortion of signal transduction. The hearts used in electrical mapping did not have any apparent atrial distension or necrosis, and were beating at the time of analysis. However, it remains possible that a subtle alteration in atrial cell function occurs prior to the detection of necrosis in mutant embryos, and this alteration subsequently contributes to the cardiac conduction system defects.
The embryonic lethality observed in the l11Jus8 mutant also suggests that ERBB2 mutations may contribute to miscarriages, and the identification in the 1000 genomes dataset of multiple variations within the ERBB2 kinase domain predicted to be detrimental confirms that potentially damaging alleles are present in the human population. Additionally, our research on the l11Jus8 mutant may inform future investigations into chemotherapy-induced cardiotoxicity associated with inhibition of ERBB2 signalling, as new anti-cancer therapeutics are being designed to target the ERBB2 kinase domain [67].
The use of a random mutagenesis approach, which prompted the characterisation of a missense mutation within the kinase domain of Erbb2, led to the discovery of a requirement for Erbb2 in atrial electrical conduction. This role for Erbb2 had not been identified from prior studies on null or phosphorylation site mutants, demonstrating the advantages of using a random mutagenesis approach to annotate gene function. Given the dearth of research investigations into atrial development, our work demonstrates the importance of atrial function for embryonic survival, and highlights the need for additional studies on embryonic atrial function to link Erbb2 kinase domain defects to cardiac conduction system development.
Supporting Information
Series of atrial conduction maps with different thresholds. Wild type (wt): Background noise, clearly visible at threshold value 0, 1 and 2, disappears at threshold 3, and first gap in conduction map is visible at threshold 4 (denoted by asterisk). Heterozygote (het no gaps): background noise disappears at threshold 2 and first gaps are visible at threshold 3 (asterisk). Heterozygote (het gaps) and l11Jus8 mutant (hz gaps): gaps (asterisk) are detected at any threshold value including 0, when no noise is cut off. Maps reconstructed from E12.5 embryonic hearts.
(TIF)
Electrical signals in E12.5 hearts with complete, gapped and absent maps. (A, E, I) Whole heart view. Square denotes the area shown in (B, D, F, H, J, L). (B, F, J) Magnified atria are shown. Black, red and blue asterisks denote the points where the electrical readings were taken. (C, G, K) Electrical readings at different areas of the atria shown in (B, F, J). Colour of the reading corresponds to the colour of the asterisk. White arrowhead denotes the area where ventricular signal was present (not shown) but no atrial signal was detected. (D, H, L) Conduction maps reconstructed from the readings on (C, G, K), respectively. Note the presence of clear signal in all atrium in (B-C) and complete conduction map on (D) while for atrium in (F), strong signal is present in the posterior end (blue), weak signal in the central area (red) and virtually no signal in the anterior area (black). Corresponding conduction map shows significant gap in anterior and central area of the atrium (H). No map (L) could be reconstructed from the reading where atrial signal was absent, white arrowhead in (K).
(TIF)
Western Blot analysis of p-ERK in l11Jus8 hetero- and homozygous animals. Total protein extracts from 6 E13.5 atria and ventricle (pooled separately) for each genotype were probed with Sigma activated mouse α MAP Kinase (M8159) at 1:2000 overnight at 4°, followed by a secondary incubation with Goat α Mouse-HRP (Thermo Scientific 32430) at 1∶5000 for 1 hour at room temperature. Bands were normalized against the loading control α MYH10 11-B (Covance PRB-445P).
(TIF)
Comparison of the wt, l11Jus8 hetero- and homozygous phenotypes at E12.5. Images of internal organs within the thoracic cavity. Structures are labelled in wt images. Lu, lungs; Li, liver; Du, duodenum; Ki, kidney bud; Pa, pancreatic primordium; St, stomach. Scale bars = 1 mm.
(TIF)
Characterisation of yolk sac vessels in wt, l11Jus8 hetero- and homozygous embryos. Embryos within the yolk sac (A-C) show a lack of blood within the vessels in l11Jus8 mutants. PECAM staining of yolk sac vessels (D-L) shows a progressive narrowing or regression of vessels specifically in l11Jus8 mutant yolks sacs from E10.5 to E12.5. Genotypes and developmental stages are labelled on the figure. Scale bars: 2 mm in (A–C), 1 mm in ((D–L).
(TIF)
Characterisation of the atrial wall of the wt, l11Jus8 hetero- and homozygous hearts. (A–C) In situ hybridisation for cardiomyocyte marker Myl4. (D–F) α-SMA immunostaining. (G–I) Laminin immunostaining. (J–L) Fibronectin immunostaining. (M–O) Phospho-histone H3 immunostaining. Arrowheads point to the positively-stained cells. (P–R) DAPI labelling of the nuclei. En, endocardium; Ep, epicardium; My, myocardium. Arrowheads in (R) denote the areas containing cells with fragmented nuclei. Scale bars: 1 mm in (A–C), 100 µm in (D–L, P–R), 40 µm in (M–O). Developmental stage of embryos: (A–C) E10.5, (D–L) E12.5, (M–R) E11.5.
(TIF)
Series of conduction maps reconstructed for the E12.5 heart with atrial block. No map could be reconstructed for the missing atrial signal (A4) while corresponding ventricular signal was present and complete ventricular map was reconstructed (V4). Notably, all atrial map reconstructed from present atrial signals (A1–A3, A5) contain significant gaps while corresponding ventricular maps are complete (V1–V3, V5).
(TIF)
Expression of Connexins 43 and 40 in E12.5 hetero- and homozygous l11Jus8 hearts. (A–B) and (C–D) Sagittal sections of E12.5 hetero- and homozygous l11Jus8 hearts stained with Cx43 and Cx40 antibodies, respectively. Atr, atrium; Ven, ventricle. Arrow heads point to the autofluorescent erythrocytes.
(TIF)
List of sequenced positions on chr11:69–103 Mb in l11Jus8 mouse.
(DOCX)
Analysis of lethality in recombinant animals.
(DOCX)
Viability of the l11Jus8 embryos.
(DOCX)
Acknowledgments
We thank Monica Justice of the Baylor College of Medicine Mouse Mutagenesis Center for Developmental Defects for providing l11Jus8 mutants. We thank James O'Sullivan of the Manchester Centre for Genomic Medicine for Next-Generation Sequencing, Prof Mark Boyett and Dr Halina Dobrzynski (University of Manchester) for providing Connexin 40 and Connexin 43 antibodies, Prof Keith Brennan (University of Manchester) for providing the Erbb2 and phosphor-Erk antibodies, and Prof Ray Boot-Handford (University of Manchester), for providing the Laminin α1 antibody. We thank Prof Neil Hall and Dr Christiane Hertz-Fowler of the University of Liverpool Centre for Genomic Research for performing exome sequencing.
Funding Statement
This work was funded by British Heart Foundation (www.bhf.org/uk) Project Grant PG/08/007/24400, Wellcome Trust (http://www.wellcome.ac.uk/funding/) grant 097820/Z/11/Z to KEH, BBSRC (http://www.bbsrc.ac.uk/home/home.aspx) project grant BB/I020489/1 to SCL, and Grant Agency of the Czech Republic grants P302/11/1308 and 13-12312S to DS, who is additionally supported by Ministry of Education PRVOUK P35/LF1/5 and Academy of Sciences RVO: 67985823. KW was supported by a BBSRC Industrial CASE studentship to Syngenta UK. Exome sequencing was supported by an MRC (http://www.mrc.ac.uk/index.htm) pump-priming grant from the Liverpool Centre for Genomic Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dobrzynski H, Boyett MR, Anderson RH (2007) New insights into pacemaker activity: promoting understanding of sick sinus syndrome. Circulation 115: 1921–1932. [DOI] [PubMed] [Google Scholar]
- 2. Wolf CM, Berul CI (2006) Inherited conduction system abnormalities—one group of diseases, many genes. J Cardiovasc Electrophysiol 17: 446–455. [DOI] [PubMed] [Google Scholar]
- 3. Harvey RP (2002) Patterning the vertebrate heart. Nat Rev Genet 3: 544–556. [DOI] [PubMed] [Google Scholar]
- 4. Christoffels VM, Moorman AF (2009) Development of the cardiac conduction system: why are some regions of the heart more arrhythmogenic than others? Circ Arrhythm Electrophysiol 2: 195–207. [DOI] [PubMed] [Google Scholar]
- 5. Christoffels VM, Smits GJ, Kispert A, Moorman AF (2010) Development of the pacemaker tissues of the heart. Circ Res 106: 240–254. [DOI] [PubMed] [Google Scholar]
- 6. Lampe PD, Lau AF (2000) Regulation of gap junctions by phosphorylation of connexins. Arch Biochem Biophys 384: 205–215. [DOI] [PubMed] [Google Scholar]
- 7. Lampe PD, Lau AF (2004) The effects of connexin phosphorylation on gap junctional communication. Int J Biochem Cell Biol 36: 1171–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lodish H, Berk A, Matsudaira P, Kaiser CA, Krieger M, et al. (2004) Molecular Cell Biology. New York: W.H. Freeman and Company. [Google Scholar]
- 9. Alcolea S, Theveniau-Ruissy M, Jarry-Guichard T, Marics I, Tzouanacou E, et al. (1999) Downregulation of connexin 45 gene products during mouse heart development. Circ Res 84: 1365–1379. [DOI] [PubMed] [Google Scholar]
- 10. Kumai M, Nishii K, Nakamura K, Takeda N, Suzuki M, et al. (2000) Loss of connexin45 causes a cushion defect in early cardiogenesis. Development 127: 3501–3512. [DOI] [PubMed] [Google Scholar]
- 11. Severs NJ, Coppen SR, Dupont E, Yeh HI, Ko YS, et al. (2004) Gap junction alterations in human cardiac disease. Cardiovasc Res 62: 368–377. [DOI] [PubMed] [Google Scholar]
- 12. Vaidya D, Tamaddon HS, Lo CW, Taffet SM, Delmar M, et al. (2001) Null mutation of connexin43 causes slow propagation of ventricular activation in the late stages of mouse embryonic development. Circ Res 88: 1196–1202. [DOI] [PubMed] [Google Scholar]
- 13. Miquerol L, Moreno-Rascon N, Beyer S, Dupays L, Meilhac SM, et al. (2010) Biphasic development of the mammalian ventricular conduction system. Circ Res 107: 153–161. [DOI] [PubMed] [Google Scholar]
- 14. van Kempen MJ, ten Velde I, Wessels A, Oosthoek PW, Gros D, et al. (1995) Differential connexin distribution accommodates cardiac function in different species. Microsc Res Tech 31: 420–436. [DOI] [PubMed] [Google Scholar]
- 15. Salameh A, Blanke K, Daehnert I (2013) Role of connexins in human congenital heart disease: the chicken and egg problem. Front Pharmacol 4: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holbro T, Hynes NE (2004) ErbB receptors: directing key signaling networks throughout life. Annu Rev Pharmacol Toxicol 44: 195–217. [DOI] [PubMed] [Google Scholar]
- 17. Carraway KL 3rd, Cantley LC (1994) A neu acquaintance for erbB3 and erbB4: a role for receptor heterodimerization in growth signaling. Cell 78: 5–8. [DOI] [PubMed] [Google Scholar]
- 18. Carraway KL 3rd, Sliwkowski MX, Akita R, Platko JV, Guy PM, et al. (1994) The erbB3 gene product is a receptor for heregulin. J Biol Chem 269: 14303–14306. [PubMed] [Google Scholar]
- 19. Tzahar E, Levkowitz G, Karunagaran D, Yi L, Peles E, et al. (1994) ErbB-3 and ErbB-4 function as the respective low and high affinity receptors of all Neu differentiation factor/heregulin isoforms. J Biol Chem 269: 25226–25233. [PubMed] [Google Scholar]
- 20. Olayioye MA (2001) Update on HER-2 as a target for cancer therapy: intracellular signaling pathways of ErbB2/HER-2 and family members. Breast Cancer Res 3: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mohd Sharial MS, Crown J, Hennessy BT (2012) Overcoming resistance and restoring sensitivity to HER2-targeted therapies in breast cancer. Ann Oncol 23: 3007–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee KF, Simon H, Chen H, Bates B, Hung MC, et al. (1995) Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature 378: 394–398. [DOI] [PubMed] [Google Scholar]
- 23. Negro A, Brar BK, Lee KF (2004) Essential roles of Her2/erbB2 in cardiac development and function. Recent Prog Horm Res 59: 1–12. [DOI] [PubMed] [Google Scholar]
- 24. Ozcelik C, Erdmann B, Pilz B, Wettschureck N, Britsch S, et al. (2002) Conditional mutation of the ErbB2 (HER2) receptor in cardiomyocytes leads to dilated cardiomyopathy. Proc Natl Acad Sci U S A 99: 8880–8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Erickson SL, O'Shea KS, Ghaboosi N, Loverro L, Frantz G, et al. (1997) ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2-and heregulin-deficient mice. Development 124: 4999–5011. [DOI] [PubMed] [Google Scholar]
- 26. Garratt AN, Voiculescu O, Topilko P, Charnay P, Birchmeier C (2000) A dual role of erbB2 in myelination and in expansion of the schwann cell precursor pool. J Cell Biol 148: 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu J, Bressan M, Hassel D, Huisken J, Staudt D, et al. (2010) A dual role for ErbB2 signaling in cardiac trabeculation. Development 137: 3867–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kile BT, Hentges KE, Clark AT, Nakamura H, Salinger AP, et al. (2003) Functional genetic analysis of mouse chromosome 11. Nature 425: 81–86. [DOI] [PubMed] [Google Scholar]
- 29. Hentges KE, Nakamura H, Furuta Y, Yu Y, Thompson DM, et al. (2006) Novel lethal mouse mutants produced in balancer chromosome screens. Gene Expr Patterns 6: 653–665. [DOI] [PubMed] [Google Scholar]
- 30. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, et al. (2010) Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol Chapter 19: Unit 19 10 11–21. [DOI] [PMC free article] [PubMed]
- 32. Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, et al. (2005) Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15: 1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goecks J, Nekrutenko A, Taylor J, Galaxy T (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, et al. (2010) Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26: 2069–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2: Unit 2 3. [DOI] [PubMed]
- 36. Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234: 779–815. [DOI] [PubMed] [Google Scholar]
- 37. Ishikawa T, Seto M, Banno H, Kawakita Y, Oorui M, et al. (2011) Design and synthesis of novel human epidermal growth factor receptor 2 (HER2)/epidermal growth factor receptor (EGFR) dual inhibitors bearing a pyrrolo[3,2-d]pyrimidine scaffold. J Med Chem 54: 8030–8050. [DOI] [PubMed] [Google Scholar]
- 38. Word JM, Lovell SC, Richardson JS, Richardson DC (1999) Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J Mol Biol 285: 1735–1747. [DOI] [PubMed] [Google Scholar]
- 39. Chen VB, Davis IW, Richardson DC (2009) KING (Kinemage, Next Generation): a versatile interactive molecular and scientific visualization program. Protein Sci 18: 2403–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lovell SC, Word JM, Richardson JS, Richardson DC (2000) The penultimate rotamer library. Proteins 40: 389–408. [PubMed] [Google Scholar]
- 41. Word JM, Lovell SC, LaBean TH, Taylor HC, Zalis ME, et al. (1999) Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogen atoms. J Mol Biol 285: 1711–1733. [DOI] [PubMed] [Google Scholar]
- 42. Henrique D, Adam J, Myat A, Chitnis A, Lewis J, et al. (1995) Expression of a Delta homologue in prospective neurons in the chick. Nature 375: 787–790. [DOI] [PubMed] [Google Scholar]
- 43. Sankova B, Benes J Jr, Krejci E, Dupays L, Theveniau-Ruissy M, et al. (2012) The effect of connexin40 deficiency on ventricular conduction system function during development. Cardiovasc Res 95: 469–479. [DOI] [PubMed] [Google Scholar]
- 44. Reckova M, Rosengarten C, deAlmeida A, Stanley CP, Wessels A, et al. (2003) Hemodynamics is a key epigenetic factor in development of the cardiac conduction system. Circ Res 93: 77–85. [DOI] [PubMed] [Google Scholar]
- 45. Sedmera D, Wessels A, Trusk TC, Thompson RP, Hewett KW, et al. (2006) Changes in activation sequence of embryonic chick atria correlate with developing myocardial architecture. Am J Physiol Heart Circ Physiol 291: H1646–1652. [DOI] [PubMed] [Google Scholar]
- 46. Boles MK, Wilkinson BM, Wilming LG, Liu B, Probst FJ, et al. (2009) Discovery of candidate disease genes in ENU-induced mouse mutants by large-scale sequencing, including a splice-site mutation in nucleoredoxin. PLoS Genet 5: e1000759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Grainger RJ, Beggs JD (2005) Prp8 protein: at the heart of the spliceosome. RNA 11: 533–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Camenisch TD, Schroeder JA, Bradley J, Klewer SE, McDonald JA (2002) Heart-valve mesenchyme formation is dependent on hyaluronan-augmented activation of ErbB2-ErbB3 receptors. Nat Med 8: 850–855. [DOI] [PubMed] [Google Scholar]
- 49.Benes JJ, Ammirabile G, Sankova B, Campione M, Krejci E, et al. (2014) The role of connexin40 in developing atrial conduction. FEBS Letters In press. [DOI] [PubMed]
- 50. Sedmera D, Reckova M, DeAlmeida A, Coppen SR, Kubalak SW, et al. (2003) Spatiotemporal pattern of commitment to slowed proliferation in the embryonic mouse heart indicates progressive differentiation of the cardiac conduction system. Anat Rec A Discov Mol Cell Evol Biol 274: 773–777. [DOI] [PubMed] [Google Scholar]
- 51. Kirchhoff S, Kim JS, Hagendorff A, Thonnissen E, Kruger O, et al. (2000) Abnormal cardiac conduction and morphogenesis in connexin40 and connexin43 double-deficient mice. Circ Res 87: 399–405. [DOI] [PubMed] [Google Scholar]
- 52. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 54. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pentassuglia L, Sawyer DB (2009) The role of Neuregulin-1beta/ErbB signaling in the heart. Exp Cell Res 315: 627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fuller SJ, Sivarajah K, Sugden PH (2008) ErbB receptors, their ligands, and the consequences of their activation and inhibition in the myocardium. J Mol Cell Cardiol 44: 831–854. [DOI] [PubMed] [Google Scholar]
- 57. Dankort DL, Wang Z, Blackmore V, Moran MF, Muller WJ (1997) Distinct tyrosine autophosphorylation sites negatively and positively modulate neu-mediated transformation. Mol Cell Biol 17: 5410–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chan R, Hardy WR, Dankort D, Laing MA, Muller WJ (2004) Modulation of Erbb2 signaling during development: a threshold level of Erbb2 signaling is required for development. Development 131: 5551–5560. [DOI] [PubMed] [Google Scholar]
- 59. Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, et al. (1995) Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 378: 390–394. [DOI] [PubMed] [Google Scholar]
- 60. Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, et al. (1995) Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 269: 230–234. [DOI] [PubMed] [Google Scholar]
- 61. Sibilia M, Wagner EF (1995) Strain-dependent epithelial defects in mice lacking the EGF receptor. Science 269: 234–238. [DOI] [PubMed] [Google Scholar]
- 62. Rentschler S, Zander J, Meyers K, France D, Levine R, et al. (2002) Neuregulin-1 promotes formation of the murine cardiac conduction system. Proc Natl Acad Sci U S A 99: 10464–10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kuramochi Y, Guo X, Sawyer DB (2006) Neuregulin activates erbB2-dependent src/FAK signaling and cytoskeletal remodeling in isolated adult rat cardiac myocytes. J Mol Cell Cardiol 41: 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Britto JM, Lukehurst S, Weller R, Fraser C, Qiu Y, et al. (2004) Generation and characterization of neuregulin-2-deficient mice. Mol Cell Biol 24: 8221–8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Crone SA, Zhao YY, Fan L, Gu Y, Minamisawa S, et al. (2002) ErbB2 is essential in the prevention of dilated cardiomyopathy. Nat Med 8: 459–465. [DOI] [PubMed] [Google Scholar]
- 66. Doggen K, Ray L, Mathieu M, Mc Entee K, Lemmens K, et al. (2009) Ventricular ErbB2/ErbB4 activation and downstream signaling in pacing-induced heart failure. J Mol Cell Cardiol 46: 33–38. [DOI] [PubMed] [Google Scholar]
- 67. Nielsen DL, Kumler I, Palshof JA, Andersson M (2013) Efficacy of HER2-targeted therapy in metastatic breast cancer. Monoclonal antibodies and tyrosine kinase inhibitors. Breast 22: 1–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Series of atrial conduction maps with different thresholds. Wild type (wt): Background noise, clearly visible at threshold value 0, 1 and 2, disappears at threshold 3, and first gap in conduction map is visible at threshold 4 (denoted by asterisk). Heterozygote (het no gaps): background noise disappears at threshold 2 and first gaps are visible at threshold 3 (asterisk). Heterozygote (het gaps) and l11Jus8 mutant (hz gaps): gaps (asterisk) are detected at any threshold value including 0, when no noise is cut off. Maps reconstructed from E12.5 embryonic hearts.
(TIF)
Electrical signals in E12.5 hearts with complete, gapped and absent maps. (A, E, I) Whole heart view. Square denotes the area shown in (B, D, F, H, J, L). (B, F, J) Magnified atria are shown. Black, red and blue asterisks denote the points where the electrical readings were taken. (C, G, K) Electrical readings at different areas of the atria shown in (B, F, J). Colour of the reading corresponds to the colour of the asterisk. White arrowhead denotes the area where ventricular signal was present (not shown) but no atrial signal was detected. (D, H, L) Conduction maps reconstructed from the readings on (C, G, K), respectively. Note the presence of clear signal in all atrium in (B-C) and complete conduction map on (D) while for atrium in (F), strong signal is present in the posterior end (blue), weak signal in the central area (red) and virtually no signal in the anterior area (black). Corresponding conduction map shows significant gap in anterior and central area of the atrium (H). No map (L) could be reconstructed from the reading where atrial signal was absent, white arrowhead in (K).
(TIF)
Western Blot analysis of p-ERK in l11Jus8 hetero- and homozygous animals. Total protein extracts from 6 E13.5 atria and ventricle (pooled separately) for each genotype were probed with Sigma activated mouse α MAP Kinase (M8159) at 1:2000 overnight at 4°, followed by a secondary incubation with Goat α Mouse-HRP (Thermo Scientific 32430) at 1∶5000 for 1 hour at room temperature. Bands were normalized against the loading control α MYH10 11-B (Covance PRB-445P).
(TIF)
Comparison of the wt, l11Jus8 hetero- and homozygous phenotypes at E12.5. Images of internal organs within the thoracic cavity. Structures are labelled in wt images. Lu, lungs; Li, liver; Du, duodenum; Ki, kidney bud; Pa, pancreatic primordium; St, stomach. Scale bars = 1 mm.
(TIF)
Characterisation of yolk sac vessels in wt, l11Jus8 hetero- and homozygous embryos. Embryos within the yolk sac (A-C) show a lack of blood within the vessels in l11Jus8 mutants. PECAM staining of yolk sac vessels (D-L) shows a progressive narrowing or regression of vessels specifically in l11Jus8 mutant yolks sacs from E10.5 to E12.5. Genotypes and developmental stages are labelled on the figure. Scale bars: 2 mm in (A–C), 1 mm in ((D–L).
(TIF)
Characterisation of the atrial wall of the wt, l11Jus8 hetero- and homozygous hearts. (A–C) In situ hybridisation for cardiomyocyte marker Myl4. (D–F) α-SMA immunostaining. (G–I) Laminin immunostaining. (J–L) Fibronectin immunostaining. (M–O) Phospho-histone H3 immunostaining. Arrowheads point to the positively-stained cells. (P–R) DAPI labelling of the nuclei. En, endocardium; Ep, epicardium; My, myocardium. Arrowheads in (R) denote the areas containing cells with fragmented nuclei. Scale bars: 1 mm in (A–C), 100 µm in (D–L, P–R), 40 µm in (M–O). Developmental stage of embryos: (A–C) E10.5, (D–L) E12.5, (M–R) E11.5.
(TIF)
Series of conduction maps reconstructed for the E12.5 heart with atrial block. No map could be reconstructed for the missing atrial signal (A4) while corresponding ventricular signal was present and complete ventricular map was reconstructed (V4). Notably, all atrial map reconstructed from present atrial signals (A1–A3, A5) contain significant gaps while corresponding ventricular maps are complete (V1–V3, V5).
(TIF)
Expression of Connexins 43 and 40 in E12.5 hetero- and homozygous l11Jus8 hearts. (A–B) and (C–D) Sagittal sections of E12.5 hetero- and homozygous l11Jus8 hearts stained with Cx43 and Cx40 antibodies, respectively. Atr, atrium; Ven, ventricle. Arrow heads point to the autofluorescent erythrocytes.
(TIF)
List of sequenced positions on chr11:69–103 Mb in l11Jus8 mouse.
(DOCX)
Analysis of lethality in recombinant animals.
(DOCX)
Viability of the l11Jus8 embryos.
(DOCX)