Abstract
The present studies were to determine whether the multi-kinase inhibitor sorafenib or its derivative regorafenib interacted with the ERBB1/ERBB2 inhibitor lapatinib to kill CNS tumor cells. In multiple CNS tumor cell types sorafenib and lapatinib interacted in a greater than additive fashion to cause tumor cell death. Tumor cells lacking PTEN, and anoikis or lapatinib resistant cells were as sensitive to the drug combination as cells expressing PTEN or parental cells, respectively. Similar data were obtained using regorafenib. Treatment of brain cancer cells with [sorafenib + lapatinib] enhanced radiation toxicity. The drug combination increased the numbers of LC3-GFP vesicles; this correlated with a reduction in endogenous LC3II, and p62 and LAMP2 degradation. Knock down of Beclin1 or ATG5 significantly suppressed drug combination lethality. Expression of c-FLIP-s, BCL-XL or dominant negative caspase 9 reduced drug combination toxicity; knock down of FADD or CD95 was protective. Expression of both activated AKT and activated MEK1 or activated mTOR was required to strongly suppress drug combination lethality. As both lapatinib and sorafenib are FDA approved agents, our data argue for further determination as to whether lapatinib and sorafenib is a useful glioblastoma therapy.
Keywords: Sorafenib, Lapatinib, Autophagy, Glioma, AKT, ERK1/2, mTOR, PTEN, p70 S6K, Necrosis
Introduction
The ERBB receptor family consists of four members, ERBB1-4. Ligand-bound receptors homo- or hetero-dimerize and through this process trans-phosphorylate themselves on tyrosine residues (Valerie et al, 2007). Many cancer types have elevated ERBB receptor signaling leading to aberrant/constitutive downstream pro-survival signaling e.g. (Pietrantonio et al, 2013). In some tumor cells mutant activated forms of ERBB1 are expressed, notably in lung cancer and in glioblastoma (Gong et al, 2013). Because of the propensity of tumor cells to over-express this receptor family multiple small molecule inhibitors of ERBB1 have been developed including Gefitinib and Tarceva. Lapatinib is a reversible tyrosine kinase inhibitor that suppresses ERBB1 / 2 / 4 activation and is approved for the treatment of ERBB2+ breast cancer in combination with capecitabine (Quatrale et al, 2011).
Sorafenib and regorafenib are multi-kinase inhibitors approved for the treatment of liver and kidney, and colon cancers, respectively (Carr et al, 2013). Sorafenib was originally developed as an inhibitor of RAF-1 in the ERK1/2 pathway. The steady state (7 day) Cmax for sorafenib is ~21 μM in plasma, with > 95% of the drug protein bound based on in vitro human serum binding assays; none the less patient response data would argue that a significant amount of the drug has to be bioavailable in a tumor based on its single agent effects by decreasing both ERK1/2 phosphorylation through inhibition of RAF-1 and reducing MCL-1 protein expression via the induction of endoplasmic reticulum stress in tumor cells that are not specifically oncogene addicted (Elser et al, 2007; Hotte and Hirte, 2002). Furthermore, it is known that certain sorafenib metabolites are much more biologically active than sorafenib itself (Pratz et al, 2010). Our prior in vitro and in vivo data have argued using several sorafenib + “drug” combinations that the PDGFRβ, and also FLT3, is a major target of sorafenib for its interactions with other agents e.g. with histone deacetylase inhibitors and with pemetrexed (Park et al, 2010; Park et al, 2008; Bareford et al, 2011).
A major biological effect of sorafenib is the induction of an endoplasmic reticulum (ER) stress / unfolded protein response (UPR), with reduced expression of proteins that have short half-lives such as MCL-1 and BCL-XL (e.g. Martin et al, 2009; Rahmani et al, 2007). Reduced MCL-1 levels due to sorafenib exposure have been linked in many tumor types to increased levels of apoptosis. Studies by our group have also linked high dose single agent sorafenib exposure to an increase in the levels of autophagic markers including increased numbers of LC3-GFP vesicles and elevated expression of Beclin1 and ATG5; however lower sorafenib concentrations only caused a modest transient alteration in autophagy flux (Park et al, 2010; Park et al, 2008; Bareford et al, 2011). Other studies from our groups have shown that based on the sorafenib dose the induction of endoplasmic reticulum stress may be a “protective” or a “toxic” event in the cellular response to the drug (Rahmani et al, 2005).
In the United States, glioblastoma (GBM) is diagnosed in ~20,000 patients per annum. High-grade tumors such as anaplastic astrocytoma and glioblastoma account for the majority of tumors (Ramirez et al, 2013; Omuro and De Angelis, 2013). Even under optimal circumstances, in which all of the tumor can be surgically removed and the patients are maximally treated with radiation and chemotherapy, the mean survival is only extended from ~3 months to 1 year. There is a major unmet need for new approaches to treat this lethal disease.
The present studies determined whether the ERBB1/2/4 inhibitor lapatinib interacted with the multi-kinase inhibitors sorafenib / regorafenib to kill glioblastoma and other tumor cell types. Our data demonstrate a strong interaction between these drugs in multiple tumor cell types with killing that is due to both death receptor activation and a toxic form of autophagy.
Materials and Methods
Materials
Phospho-/total- antibodies were purchased from Cell Signaling Technologies (Danvers, MA) and Santa Cruz Biotech. (Santa Cruz, CA). All drugs were purchased from Selleckchem (Houston, TX). Commercially available validated short hairpin RNA molecules to knock down RNA / protein levels were from Qiagen (Valencia, CA). Antibody reagents, other kinase inhibitors, caspase inhibitors cell culture reagents, and non-commercial recombinant adenoviruses have been previously described (Bareford et al, 2011; Cruickshanks et al, 2012; Cruickshanks et al, 2013; Booth et al, 2012; Bareford et al, 2012). Previously characterized semi-established GBM5 / GBM6 / GBM12 / GBM14 primary glioblastoma cells were supplied by Dr. C.D. James (University of California, San Francisco) and Dr. J.N. Sarkaria (Mayo Clinic, Rochester MN) and were not further characterized by ourselves. The low passage primary human glioblastoma isolates (patient 1; patient 2; patient 3) were obtained / isolated from discarded tumor tissue after standard of care surgery. Patients had previously given informed under IRB approved consent to the use of tumor tissue. Tumor samples were made anonymous of all patient identifiers by the VCU TDAAC prior to hand-over to the Dent laboratory.
Methods
Cell culture and in vitro exposure of cells to drugs. All fully established cancer lines were cultured at 37 °C (5% (v/v CO2) in vitro using RPMI supplemented with 10% (v/v) fetal calf serum and 10% (v/v) Non-essential amino acids. All primary human glioblastoma cells were cultured at 37 °C (5% (v/v CO2) in vitro using RPMI supplemented with 2% (v/v) fetal calf serum and 10% (v/v) Non-essential amino acids. at 37 °C (5% (v/v CO2) in vitro using RPMI supplemented with 5% (v/v) fetal calf serum and 10% (v/v) Non-essential amino acids. For short-term cell killing assays and immunoblotting, cells were plated at a density of 3 × 103 per cm2 and 24h after plating were treated with various drugs, as indicated. In vitro small molecule inhibitor treatments were from a 100 mM stock solution of each drug and the maximal concentration of Vehicle (DMSO) in media was 0.02% (v/v). Cells were not cultured in reduced serum media during any study.
Cell treatments, SDS-PAGE and Western blot analysis
Cells were treated with various drug concentrations, as indicated in the Figure legends. SDS PAGE and immunoblotting was performed as described (Bareford et al, 2011; Cruickshanks et al, 2012; Cruickshanks et al, 2013; Booth et al, 2012; Bareford et al, 2012).
Recombinant adenoviral vectors; infection in vitro
We generated and purchased previously noted recombinant adenoviruses as per refs. Cells were infected with these adenoviruses at an approximate m.o.i. as indicated in the Figure / Legend (usually 50 m.o.i.). Cells were incubated for 24 h to ensure adequate expression of transduced gene products prior to drug exposures.
Detection of cell death by Trypan Blue, Hoechst, and live/dead assays
For trypan blue and Hoechst assays floating cells were isolated along with attached cells that were harvested by trypsinization with Trypsin/EDTA for ~10 min at 37 °C. For live/dead assays in 96 well plates, plates were gently spun to sediment detached dead cells onto the plate. Cells were then incubated with di-ethidium bromide to detect cells with disrupted plasma membranes and cells visualized using a Hermes Wiscan microscope with imaging software to permit cell counting and determination of the percentage dead cells.
Assessment of autophagy
Cells were transfected with a plasmid to express a green fluorescent protein (GFP) tagged form of LC3 (ATG8). For analysis of cells transfected with the GFP-LC3 construct, the GFP-LC3 - positive vesicularized cells were examined under the X40 objective of a Zeiss Axiovert fluorescent microscope.
Plasmid transfection
Plasmids
Cells were plated as described above and 24h after plating, transfected. Plasmids (0.5 μg) expressing a specific mRNA or appropriate vector control plasmid DNA was diluted in 50 μl serum-free and antibiotic-free medium (1 portion for each sample). Concurrently, 2 μ l Lipofectamine 2000 (Invitrogen), was diluted into 50 μl of serum-free and antibiotic-free medium. Diluted DNA was added to the diluted Lipofectamine 2000 for each sample and incubated at room temperature for 30 min. This mixture was added to each well / dish of cells containing 200 μl serum-free and antibiotic-free medium for a total volume of 300 μl and the cells were incubated for 4h at 37°C. An equal volume of 2X medium was then added to each well. Cells were incubated for 48h, then treated with drugs. To assess transfection efficiency of plasmids we used a plasmid to express GFP and defined the percentage of cells being infected as the percentage of GFP+ cells. For all cell lines the infection efficiency was > 70%.
siRNA
Cells were plated in 60 mm dishes from a fresh culture growing in log phase as described above, and 24h after plating transfected. Prior to transfection, the medium was aspirated and 1 ml serum-free medium was added to each plate. For transfection, 10 nM of the annealed siRNA, the positive sense control doubled stranded siRNA targeting GAPDH or the negative control (a “scrambled” sequence with no significant homology to any known gene sequences from mouse, rat or human cell lines) were used (predominantly Qiagen, Valencia, CA; occasional alternate siRNA molecules were purchased from Ambion, Inc., Austin, Texas). Ten nM siRNA (scrambled or experimental) was diluted in serum-free media. Four μl Hiperfect (Qiagen) was added to this mixture and the solution was mixed by pipetting up and down several times. This solution was incubated at room temp for 10 min, then added drop-wise to each dish. The medium in each dish was swirled gently to mix, then incubated at 37 °C for 2h. One ml of 10% (v/v) serum-containing medium was added to each plate, and cells were incubated at 37 °C for 24-48h before re-plating (50 × 103 cells each) onto 12-well plates. Cells were allowed to attach overnight, then treated with drugs (0-48h). Trypan blue exclusion assays and SDS PAGE / immunoblotting analyses were then performed at the indicated time points.
Data analysis
Comparison of the effects between various in vitro drug treatments was performed after analysis of variance using the Student’s t test. Differences with a p value of <0.05 were considered statistically significant. Experiments shown are the means of multiple individual points from multiple studies (± S.E.M.). Median dose-effect isobologram colony-formation analyses to determine synergism of drug interaction were performed according to the methods of Chou and Talalay using the CalcuSyn program for Windows (Biosoft, Cambridge, UK). Cells were treated with agents at an escalating fixed concentration drug dose. A combination index of <1.00 indicates synergy of interaction between the two drugs; a combination index of ~1.00 indicates an additive interaction; a combination index (CI) value of >1.00 indicates antagonism of action between the agents.
Results
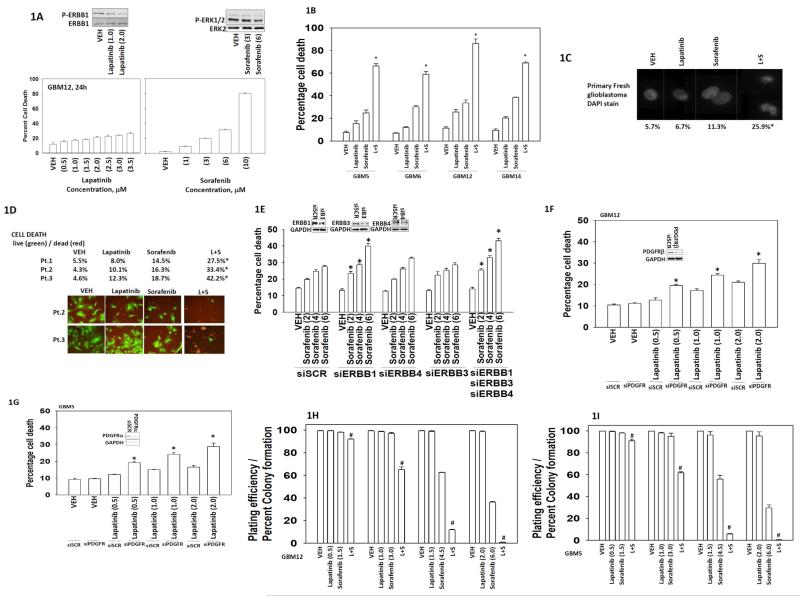
Initial studies examined the dose-response of glioblastoma cells to increasing concentrations of sorafenib or lapatinib; demonstrating that low microM doses of the drugs have modest effects on tumor cell viability (Figure 1A). Of note in our hands sorafenib weakly inhibited ERK1/2 phosphorylation at concentrations at or below 3 μM. Lapatinib and sorafenib interacted in a greater than additive fashion to kill semi-established primary human glioblastoma cells (Figure 1B). Killing was also observed in glioblastoma cells very recently isolated from the tumors of patients (patients 1-3) (Figures 1C and 1D). Of note, based on use of the live/dead assay as well as based on the nuclear morphology of cells treated with the drug combination, our killing process was most likely “necro-apoptotic” rather than purely apoptotic (Figure 1D). Lapatinib is an inhibitor of ERBB1, ERBB2 and ERBB4, and knock down of ERBB1, and to a greater extent ERBB1/3/4, enhanced sorafenib lethality, at doses as low as 2 μM, in GBM12 cells (Figure 1E).
Figure 1. Sorafenib and lapatinib interact to kill glioblastoma cells.
(A) GBM12 cells were treated with sorafenib (1.0-10.0 μM) or lapatinib (0.5-3.5 μM) as indicated. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM). Upper blots: lapatinib reduces ERBB1 phosphorylation; sorafenib modestly reduces ERK1/2 phosphorylation. (B) GBM5 / GBM6 / GBM12 / GBM14 cells were treated with vehicle (DMSO), sorafenib (6.0 μM) and/or lapatinib (2.0 μM) as indicated. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM) *p < 0.05 greater than vehicle control. (C) Freshly isolated glioblastoma cells from the operating room were treated with vehicle (DMSO), sorafenib (3.0 μM) and/or lapatinib (2.0 μM) as indicated. Cells were isolated 24h after exposure and spun onto glass slides and DAPI stained. Representative images are shown from each treatment condition. (D) Low passage primary human glioblastoma cells (patient 1; patient 2; patient 3) were treated with vehicle (DMSO), sorafenib (3.0 μM) and/or lapatinib (2.0 μM) as indicated. Cells were isolated 24h after exposure and viability determined by live = green / dead = red assay using a Hermes WiScan platform (n = 3, +/− SEM) *p < 0.05 greater than vehicle control. (E) GBM12 cells were transfected to knock down the expression of ERBB1, ERBB3 and/or ERBB4. Thirty six h after transfection cells were treated with vehicle (DMSO), or sorafenib (2.0-6.0 μM) as indicated. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM) *p < 0.05 greater than corresponding siSCR control. (F) GBM12 cells were transfected to knock down expression of PDGFRβ. Thirty six h after transfection cells were treated with vehicle (DMSO), or lapatinib (0.5-2.0 μM) as indicated. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM) *p < 0.05 greater than corresponding siSCR control. (G) GBM5 cells were transfected to knock down expression of PDGFRα. Thirty six h after transfection cells were treated with vehicle (DMSO), or lapatinib (0.5-2.0 μM) as indicated. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM) *p < 0.05 greater than corresponding siSCR control. (H) and (I) GBM12 or GBM5 cells, as indicated in the Figure, were plated (250-1,500 single cells / well) in six well plates. Cells were permitted to attach and after 12h treated with drugs. Cells were treated with vehicle (DMSO), sorafenib (1.5-6.0 μM) and/or lapatinib (0.5-2.0 μM) as indicated for 24h. Media was removed and replaced with drug free media and cells permitted to grow and form colonies for the next 14 days (n = 3 in sextuplicate, +/− SEM) #p < 0.05 less than vehicle control.
In prior studies we have shown that key survival regulatory proteins whose signaling functions are blocked by sorafenib at low concentrations are the platelet derived growth factor receptor (PDGFR) and FLT3. In the present studies we focused on manipulating the expression / function of the PDGFR. Knock down of the PDGFRβ enhanced lapatinib lethality in GBM12 cells at lapatinib concentrations as low as 0.5 μM (Figure 1F). Knock down of the PDGFRα enhanced lapatinib lethality in GBM5 cells (Figure 1G). In colony formation assays lapatinib and sorafenib interacted in a synergistic fashion to kill glioblastoma cells, at sorafenib concentrations as low as 1.5 μM and lapatinib concentrations as low as 0.5 μM, with combination index values of less than 0.70 at sorafenib concentrations below the IC50 killing for sorafenib as a single agent (Figures 1H and 1I).
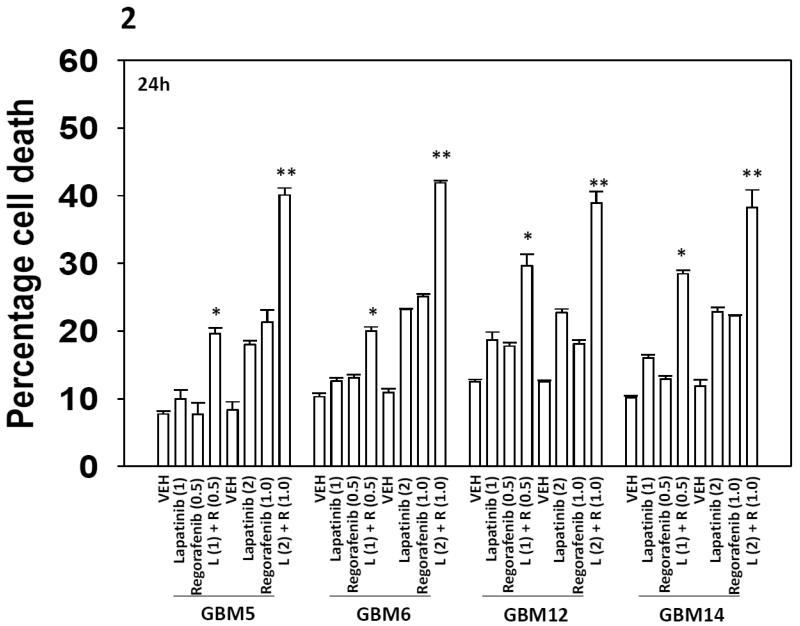
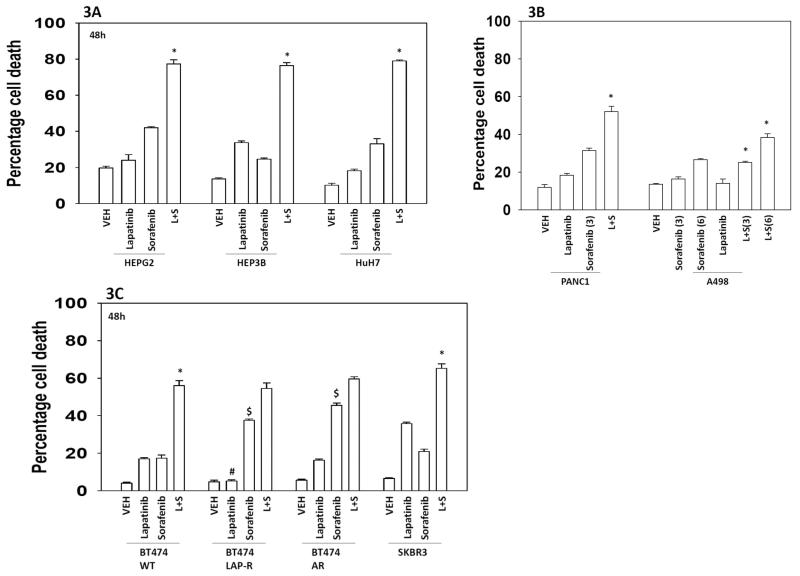
Regorafenib is a derivative of sorafenib with greater solubility and potency in vitro and in vivo that the parent compound sorafenib. Regorafenib is approved for the treatment of colon cancer. Regorafenib interacted with lapatinib in a dose-dependent fashion to kill multiple primary human glioblastoma isolates (Figure 2). Lapatinib and sorafenib also interacted to kill multiple other tumor cell types (hepatoma; renal; pancreatic), including breast cancer cells made anoikis resistant or resistant to lapatinib as a single agent (Figures 3A-3C). Radiotherapy is used in the treatment of glioblastoma, and [lapatinib + sorafenib] treatment radiosensitized brain cancer cells (Figure 4).
Figure 2. Regorafenib and lapatinib interact to kill GBM cells.
(A) GBM5 / GBM6 / GBM12 / GBM14 cells were treated with vehicle (DMSO), regorafenib (0.5-1.0 μM) and/or lapatinib (1.0-2.0 μM) as indicated. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM) *p < 0.05 greater than vehicle control; ** p < 0.05 greater than value in [L(1) + R (1)] cells.
Figure 3. Sorafenib and lapatinib interact to kill multiple tumor types.
(A) HEPG2 / HEP3B / HuH7 hepatoma cells were treated with vehicle (DMSO), sorafenib (6.0 μM) and/or lapatinib (2.0 μM) as indicated. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM) *p < 0.05 greater than vehicle control. (B) PANC1 pancreatic and A498 renal carcinoma cells were treated with vehicle (DMSO), sorafenib (3.0, 6.0 μM as indicated) and/or lapatinib (2.0 μM) as indicated. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM) *p < 0.05 greater than vehicle control. (C) Breast cancer cells: SKBR3; parental wild type BT474; lapatinib-resistant BT474; anoikis resistant BT474; were treated with vehicle (DMSO), sorafenib (6.0 μM) and/or lapatinib (2.0 μM) as indicated. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM) *p < 0.05 greater than vehicle control; #p < 0.05 less than corresponding value in wild type BT474 cells; $ p < 0.05 greater than corresponding value in wild type BT474 cells.
Figure 4. Treatment with [lapatinib + sorafenib] radiosensitizes GBM cells.
GBM5 and GBM12 cells were plated as single cells in sextuplicate. Twelve h after plating cells were treated with vehicle (DMSO), sorafenib (2 μM; 4 μM), lapatinib (1 μM; 2 μM) or the drugs in combination as indicated. Thirty min after drug treatment cells were irradiated (2 Gy) or mock exposed (time out of incubator ~20 min). The media was replaced after 12h with drug free media and colonies permitted to form over the following 10 days. Colonies were fixed, stained and counted (n = 3 +/− SEM). # p < 0.05 less than corresponding value in unirradiated cells.
In prior studies we have shown that in a dose dependent fashion sorafenib can modulate the levels of autophagosome formation. In the present studies we found that sorafenib and lapatinib interacted to increase early autophagosome formation and for over a prolonged period of time (Figure 5A). Lapatinib caused increased LC3I and LC3II, and p62 expression and reduced LAMP2 expression (smeared double band) indicative of stalled autophagic flux and clearance of acidic endosomes (Figure 5B). Combined treatment of lapatinib exposed cells with sorafenib resulted in reduced LC3I/LC3II and p62 expression, arguing that sorafenib has stimulated autophagic flux. Sorafenib activated PKR-like endoplasmic reticulum kinase (PERK) and elevated eIF2α phosphorylation whilst it inhibited expression of MCL-1, an effect that was prolonged by co-exposure to lapatinib. Combined exposure to lapatinib and sorafenib reduced the phosphorylation / activity of ERK1/2, AKT, p70 S6K and mTOR. Based on these observations we defined the roles of endoplasmic reticulum stress, autophagic and apoptotic pathways in the lethal interaction between sorafenib and lapatinib. Expression of dominant negative PERK suppressed the induction of LC3-GFP vesicles / autophagosomes and protected cells from [lapatinib + sorafenib] toxicity which correlated with sustained MCL-1 expression after drug exposure (Figure 5C). Knock down of Beclin1 or ATG5 suppressed the lethal effect of [sorafenib + lapatinib] treatment (Figures 5D and 5E).
Figure 5. Treatment with [lapatinib + sorafenib] kills in part through toxic autophagy.
(A) GBM12 cells were transfected with a plasmid to express LC3-GFP. Twenty four h after transfection cells were treated with vehicle (DMSO), sorafenib (6 μM), lapatinib (2 μM) or the drugs combined. The numbers of intense staining GFP+ punctae were counted 6h, 12h and 24h after drug treatment. (n = 3 +/− SEM). * p < 0.05 greater than value in SOR treatment alone cells. (B) GBM12 cells were treated with vehicle (DMSO), sorafenib (6 μM), lapatinib (2 μM) or the drugs combined. Cells were isolated 6h and 12h after drug treatment and lysates subjected to SDS PAGE followed by immunoblotting to determine the expression / phosphorylation of the indicated proteins (representative blots are shown, n = 3). (C) Left Graph: GBM12 cells were transfected with empty vector (CMV) or a plasmid to express dominant negative PERK, and in parallel transfected with a plasmid to express LC3-GFP. Twenty four h after transfection cells were treated with vehicle (DMSO), sorafenib (6 μM), lapatinib (2 μM) or the drugs combined. The numbers of intense staining GFP+ punctae were counted 12h after drug treatment. (n = 3 +/− SEM). # p < 0.05 less than corresponding value in CMV transfected cells. Right Graph: GBM12 cells were transfected with empty vector (CMV) or a plasmid to express dominant negative PERK. Twenty four h after transfection cells were treated with vehicle (DMSO), sorafenib (6 μM), lapatinib (2 μM) or the drugs combined. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3 +/− SEM). # p < 0.05 less than corresponding value in CMV transfected cells. (D) and (E) GBM5 and GBM12 cells were transfected with a scrambled siRNA (siSCR) or siRNA molecules to knock down expression of Beclin1 (siBeclin1) or ATG5 (siATG5). Thirty six h after transfection cells were treated with vehicle (DMSO), sorafenib (6 μM), lapatinib (2 μM) or the drugs combined. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM) #p < 0.05 less than corresponding value in siSCR control.
Although nuclear morphology argued that killing was not “purely” apoptotic, in Figure 5B we noted that the drug combination increased cleavage of caspase 3, suggestive of some input from apoptotic pathways. Hence we next explored whether cell death signaling by the drug combination utilized the extrinsic (caspase 8) or the intrinsic (caspase 9) pathways. Expression of dominant negative caspase 9, the mitochondrial protective protein BCL-XL or the caspase 8 inhibitor c-FLIP-s protected cells against the toxic effects of [sorafenib + lapatinib] (Figures 6A and 6B). Combined drug treatment increased NOXA expression and over-expression of BCL-XL protected cells; thus we next determined the relative roles of BAX, BAK, NOXA and PUMA in the killing response. Knock down of BAX+BAK but not of NOXA+PUMA significantly reduced killing by sorafenib and lapatinib treatment (Figure 6C). As expression of the caspase 8 inhibitor c-FLIP-s was protective, we determined whether death receptors upstream of caspase 8 were playing a role in the killing process. Knock down of CD95 death receptor expression or knock down of the linker protein FADD expression reduced [lapatinib + sorafenib] toxicity (Figure 6D). Quenching of reactive oxygen species or of de novo ceramide synthesis prevented CD95 activation (Figure 6E).
Figure 6. Sorafenib and lapatinib treatment kills through the extrinsic and intrinsic apoptosis pathways.
(A) and (B) GBM5 and GBM12 cells were infected with a control empty vector adenovirus (CMV, 50 m.o.i.) or viruses to express dominant negative caspase 9, BCL-XL or c-FLIP-s. Twenty four h after infection cells were treated with vehicle (DMSO), sorafenib (6 μM), lapatinib (2 μM) or the drugs combined. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM) #p < 0.05 less than corresponding value in CMV control; ## p < 0.05 less than corresponding value in dominant negative caspase 9 expressing cells. (C) GBM12 cells were transfected with a control scrambled siRNA (siSCR) or siRNA molecules to knock down the expression of BAX, BAK, NOXA, PUMA. Thirty six h after transfection cells were treated with vehicle (DMSO), sorafenib (6 μM), lapatinib (2 μM) or the drugs combined. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM) #p < 0.05 less than corresponding value in siSCR control. (D) GBM12 cells were transfected with a scrambled siRNA (siSCR) or siRNA molecules to knock down expression of CD95 (siCD95) or FADD (siFADD). Thirty six h after transfection cells were treated with vehicle (DMSO), sorafenib (6 μM) and lapatinib (2 μM) combined. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM) #p < 0.05 less than corresponding value in siSCR control. (E) GBM12 cells in 96 well plates were pre-treated with vehicle, the ceramide synthase inhibitor Fumonisin B1 (25 μM) or the ROS quenching agent N-acetyl cysteine (10 mM). Cells were then treated with vehicle (DMSO), sorafenib (6 μM) and lapatinib (2 μM) combined. Six h after treatment cells were fixed in situ without permeabilization. The levels of cell surface CD95 were assessed after immunohistochemical detection of CD95 using a Hermes Wiscan instrument and associated WiSoft densitometry software (n = 3, +/− SEM) #p < 0.05 less than corresponding value in vehicle control. (F) GBM12 cells were infected with a control empty vector adenovirus (CMV, 50 m.o.i.) or viruses to express activated AKT (caAKT) and/or activated MEK1 (caMEK1). Twenty four h after infection cells were treated with vehicle (DMSO), sorafenib (6 μM), lapatinib (2 μM) or the drugs combined. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM) #p < 0.05 less than corresponding value in CMV control; ## p < 0.05 less than corresponding value in caAKT or caMEK1 expressing cells. (G) GBM12 cells were transfected with a control empty vector plasmid (CMV) or plasmids to express activated p70 S6K (ca-p70) and/or activated mTOR (ca-mTOR). Twenty four h after infection cells were treated with vehicle (DMSO), sorafenib (6 μM), lapatinib (2 μM) or the drugs combined. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM) #p < 0.05 less than corresponding value in CMV control; ## p < 0.05 less than corresponding value in ca-p70 expressing cells. (H) GBM6 and GBM12 cells were pre-treated with vehicle (DMSO) or with the JNK inhibitory peptide (10 μM). Cells were then treated with vehicle (DMSO), sorafenib (6 μM), lapatinib (2 μM) or the drugs combined. Cells were isolated 24h after exposure and viability determined by trypan blue exclusion (n = 3, +/− SEM) #p < 0.05 less than corresponding value in vehicle control. (I) Upper: GBM12 cells were pre-treated with vehicle (DMSO) or with the JNK inhibitory peptide (10 μM). Cells were then treated with vehicle (DMSO), sorafenib (6 μM), lapatinib (2 μM) or the drugs combined. Cells were isolated 12h after exposure and lysates generated in CHAPS buffer followed by immunoprecipitation and SDS PAGE to determine activated BAX levels. Lower: GBM12 cells were transfected with a scrambled siRNA (siSCR) or an siRNA molecule to knock down expression of CD95 (siCD95). Thirty six h after transfection cells were treated with vehicle (DMSO), sorafenib (6 μM), lapatinib (2 μM) or the drugs combined. Cells were isolated 12h after treatment and the levels of P-JNK determined.
As the drug combination inhibited both AKT and ERK1/2 phosphorylation we introduced activated forms of AKT and MEK1 into cells and determined the response to [lapatinib + sorafenib] treatment. Activation of either the AKT or the MEK1 pathway resulted in a modest protection from drug combination toxicity (Figure 6F). Combined activation of the AKT and MEK1 pathways resulted in enhanced cytoprotection. As the drug combination also inhibited both p70 S6K and mTOR phosphorylation we introduced activated forms of p70 S6K and mTOR into cells and determined the response to [lapatinib + sorafenib] treatment. Activation of the p70 S6K pathway resulted in a modest protection from drug combination toxicity (Figure 6G). Activation of mTOR protected cells to a significantly greater extent than p70 S6K; expression of activated mTOR abolished the drug-induced increase in autophagosomes (data not shown). Inhibition of the JNK pathway protected cells from drug combination toxicity (Figure 6H). Inhibition of JNK signaling blocked drug-induced activation of BAX; knock down of CD95 reduced JNK activation (Figure 6I).
Discussion
The present studies were initiated to determine whether the ERBB1/2/4 kinase inhibitor lapatinib interacted with the multi-kinase inhibitors sorafenib/regorafenib to kill glioblastoma and other solid tumor cell types in vitro. Collectively our data demonstrated that lapatinib and sorafenib/regorafenib interact in a greater than additive fashion to kill a genetically diverse set of primary human glioblastoma isolates; isolates with altered PDGFR, ERBB1 and PTEN function, as well as other tumor cell types.
Despite the fact that ERBB family receptor function is commonly dysregulated in glioblastoma cells, ERBB receptor inhibitors have not yet been approved as a standard of care treatment for this malignancy. There are several possibilities why a lack of effect has been observed in patients: (1) approved receptor inhibitors are at sub-therapeutic concentrations in glioblastoma tumors due to blood-brain barrier effects. This possibility was most recently noted by Reardon et al (Reardon et al, 2013); (2) Glioblastoma cells can rapidly switch their signaling addiction from reliance on ERBB family receptors to other receptor tyrosine kinases to survive. For example, Akhavan et al recently showed that de-repression of PDGFRβ, predominantly due to ERK1/2 activity, promotes resistance to ERBB1 inhibitors in glioblastoma (Akhavan et al, 2013). Since sorafenib is a potent PDGFRα/β inhibitor; has the ability to down-regulate MCL-1 expression and can suppress signaling through the ERK1/2 pathway, a logical hypothesis would be that sorafenib could enhance lapatinib toxicity; (3) as best described in lung cancer, ERBB receptors can undergo secondary mutations in the presence of ERBB inhibitors rendering them inhibitor resistant (Roengvoraphoj et al, 2013). In addition to receptor effects, Fenton et al suggested that acquired resistance to ERBB1 inhibitors in glioblastoma is mediated by phosphorylation of PTEN at tyrosine 240 which leads to loss of PTEN function, through loss of membrane interaction, and consequent activation of PI3K/AKT pathway (Fenton et al, 2012); (4) by the sustained up-regulation of pro-survival proteins such as BCL-XL and MCL-1; these proteins are independently targets of sorafenib and lapatinib.
With regard to expression of pro-survival proteins, it has been demonstrated that down-regulation of MCL-1 by sorafenib is independent of caspase activation and MEK1/2 / ERK1/2 pathway, and suggested that sorafenib exerts this effect in a PERK dependent manner (Rahmani et al, 2005; Rahmani et al, 2007). In these studies it was proposed that activation of PERK followed by subsequent phosphorylation of eIF2α inhibited protein translation leading to a rapid decline of MCL-1 due to its short half-life. In the present studies we noted that the drug-induced increase in phosphorylation of eIF2α was accompanied by a decrease in MCL-1 levels in GBM12 cells. Over-expression of the parallel mitochondrial protective protein BCL-XL protected cells from [lapatinib + sorafenib] lethality.
As the levels of MCL-1 were declining, which correlated with caspase 3 cleavage, we defined in detail the apoptotic pathways that were activated in response to sorafenib and lapatinib treatment. Expression of dominant negative caspase 9, over-expression of BCL-XL or over-expression of the caspase 8 inhibitor c-FLIP-s reduced the toxicity of the drug combination. Elevated levels of NOXA, a toxic BCL-2 family member was observed, as was activation of BAX, and knock down of (BAX+BAK), but surprisingly not (NOXA+PUMA), was protective. As inhibition of caspase 8 was protective we further investigated whether inhibition of death receptor signaling played any role in drug combination lethality. Knock down of the death receptor CD95 or of the docking protein FADD reduced cell killing. Thus lapatinib and sorafenib promote activation of both the extrinsic and intrinsic apoptosis pathways. Of note, however, was that data from fresh and primary glioblastoma cells indicated that the nuclear morphology of dead cells was not wholly apoptotic; appearing more necro-apoptotic.
Thus in addition to apoptosis, the induction of autophagy was also investigated through use of LC3-GFP vesicle formation, western blots analyses and knock down of ATG5 and Beclin 1. The LC3-GFP vesicle formation assay demonstrated a significant increase in the formation of autophagosomes as early as 6 hours after treatment with sorafenib and with the combination of sorafenib and lapatinib. Western blot analysis also showed an increase in endogenous total LC3 (isoforms I and II) and Beclin 1 expression in response to the drugs, both individually and in combination. Surprisingly, accumulation of p62 was observed within the first 6 hours of lapatinib (single agent) exposure showing a stall in lysosomal degradation of ubiquitin-tagged proteins. This phenomenon was not observed in cells exposed to sorafenib and the combination of both agents. In order to determine the role of autophagy in drug-mediated cell death, ATG5 and Beclin 1 were knocked down individually and inhibition of autophagy reduced drug combination-induced cell death.
Both lapatinib and sorafenib are protein kinase inhibitors and it would be a logical hypothesis that multiple intracellular pathways would exhibit some form of reduced activity following drug combination treatment. Drug combination exposure rapidly reduced the phosphorylation, at activating sites, of ERK1/2, AKT, p70 S6K and mTOR. All of these kinases through a variety of mechanisms can act to maintain tumor cell viability and growth. Expression of activated forms of AKT, MEK1, p70 S6K and mTOR individually reduced lapatinib and sorafenib toxicity; activation of mTOR exhibited the greatest protective effect. JNK signaling played a toxic role, facilitating BAX activation in a CD95-dependent manner. As mTOR acts as a gate-keeper kinase for the formation of autophagic vesicles, these findings are consistent with the regulation of autophagy as a key player in the mechanism of cell killing by lapatinib and sorafenib.
The clinical relevance of many in vitro studies using sorafenib has recently been the subject of controversy, with an editorial article in Clinical Cancer Research highlighting the general poor understanding of sorafenib pharmacology with regard to pre-clinical in vitro testing (Smith et al, 2013). And, many correlative papers suggest that whilst the C max for sorafenib is ~20 μM, the free concentrations of sorafenib are 1% or less of this total in human plasma (Villarroel et al, 2012). In studies by Pratz et al, using tumor cells cultured in human plasma the IC50 of sorafenib for inhibiting the RTK FLT3 was ~300 nM and of inhibiting ERK1/2 phosphorylation ~800 nM. These studies also discovered that a metabolized form of the drug, nitro-sorafenib, was ~15 times more potent at inhibiting FLT3 and ERK1/2 phosphorylation than the parent compound. It is also not known at present how prolonged exposure to sorafenib alters the tissue / plasma distribution of the drug which could be higher than the levels in plasma. We found that in colony formation assays at sorafenib concentrations as low as 1.5 μM a synergy was observed of cell killing in two GBM isolates. The IC50 for sorafenib –induced killing as a single agent for both isolates was between 4.5 μM and 6.0 μM. Thus the drug combinatorial effects are observed at far below the IC50 of sorafenib. And, as this was only a 24h exposure to the drug and in patients one would be treating with sorafenib and lapatinib QD for weeks at a time we would argue our data has physiologic / clinical relevance. Furthermore, we have previously shown in liver cancer cells that key sorafenib targets for its interaction with vorinostat are the wild type PDGFR and FLT3 receptors (Park et al, 2010). We presented data showing that knock down of the PDGFR significantly enhanced lapatinib toxicity in two isolates. It should also be noted that in Figure 1A a concentration of 3 μM sorafenib in our hands did not significantly alter ERK1/2 phosphorylation again implying that it was the effect of sorafenib on class III receptor tyrosine kinases that played the major role in mediating sorafenib combinatorial effects.
In conclusion, we have demonstrated that sorafenib in combination with lapatinib kills multiple primary human glioblastoma tumor isolates in a greater than additive manner in a process that involves induction of endoplasmic reticulum stress, autophagy, and intrinsic and extrinsic apoptotic pathways. Sorafenib induces cell death mainly through activation of CD95 and recruitment of FADD whereas lapatinib triggers mitochondria-mediated cell death. Inhibition of autophagy reduced combination-mediated cell death suggesting killing can proceed either through cytotoxic autophagy or activation of caspase-dependent pathways.
Acknowledgements
Support for the present study was funded from PHS grants from the National Institutes of Health [R01-CA141704, R01-CA150214, R01-DK52825]; the Department of Defense [W81XWH-10-1-0009]. PD is the holder of the Universal Inc. Professorship in Signal Transduction Research. The authors have no conflicts of interest to report. The concept of combining lapatinib and sorafenib as an anti-cancer therapy was the original idea of Dr. Hossein Hamed.
Support for the present study was funded from PHS grants from the National Institutes of Health [R01-CA141704, R01-CA150214, R01-DK52825]; the Department of Defense [W81XWH-10-1-0009]. PD is the Universal Inc. Professorship in Signal Transduction Research.
Abbreviations
- ERK
extracellular regulated kinase
- MEK
mitogen activated extracellular regulated kinase
- PI3K
phosphatidyl inositol 3 kinase
- MAPK
mitogen activated protein kinase
- ca
constitutively active
- dn
dominant negative
- CMV
empty vector plasmid or virus
- si
small interfering
- SCR
scrambled
- Ad
adenovirus
- VEH
vehicle
- SOR
sorafenib
- REGO
regorafenib
- LAP
lapatinib
Footnotes
The authors have no conflicts of interest.
References
- Akhavan D, Pourzia AL, Nourian AA, Williams KJ, Nathanson D, Babic I, Villa GR, Tanaka K, Nael A, Yang H, Dang J, Vinters HV, Yong WH, Flagg M, Tamanoi F, Sasayama T, James CD, Kornblum HI, Cloughesy TF, Cavenee WK, Bensinger SJ, Mischel PS. De-repression of PDGFRβ transcription promotes acquired resistance to EGFR tyrosine kinase inhibitors in glioblastoma patients. Cancer Discov. 2013;3:534–47. doi: 10.1158/2159-8290.CD-12-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bareford MD, Park MA, Yacoub A, Hamed HA, Tang Y, Cruickshanks N, Eulitt P, Hubbard N, Tye G, Burow ME, Fisher PB, Moran RG, Nephew KP, Grant S, Dent P. Sorafenib enhances pemetrexed cytotoxicity through an autophagy-dependent mechanism in cancer cells. Cancer Res. 2011;71:4955–67. doi: 10.1158/0008-5472.CAN-11-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bareford MD, Hamed HA, Allegood J, Cruickshanks N, Poklepovic A, Park MA, Ogretmen B, Spiegel S, Grant S, Dent P. Sorafenib and pemetrexed toxicity in cancer cells is mediated via SRC-ERK signaling. Cancer Biol Ther. 2012;13:793–803. doi: 10.4161/cbt.20562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L, Cruickshanks N, Ridder T, Chen CS, Grant S, Dent P. OSU-03012 interacts with lapatinib to kill brain cancer cells. Cancer Biol Ther. 2012;13:1501–11. doi: 10.4161/cbt.22275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr BI, D’Alessandro R, Refolo MG, Iacovazzi PA, Lippolis C, Messa C, Cavallini A, Correale M, Di Carlo A. Effects of low concentrations of regorafenib and sorafenib on human HCC cell AFP, migration, invasion, and growth in vitro. J Cell Physiol. 2013;228:1344–50. doi: 10.1002/jcp.24291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshanks N, Tang Y, Booth L, Hamed H, Grant S, Dent P. Lapatinib and obatoclax kill breast cancer cells through reactive oxygen species-dependent endoplasmic reticulum stress. Mol Pharmacol. 2012;82:1217–29. doi: 10.1124/mol.112.081539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshanks N, Hamed HA, Booth L, Tavallai S, Syed J, Sajithlal GB, Grant S, Poklepovic A, Dent P. Histone deacetylase inhibitors restore toxic BH3 domain protein expression in anoikis-resistant mammary and brain cancer stem cells, thereby enhancing the response to anti-ERBB1/ERBB2 therapy. Cancer Biol Ther. 2013;14:982–96. doi: 10.4161/cbt.26234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elser C, Siu LL, Winquist E, Agulnik M, Pond GR, Chin SF, Francis P, Cheiken R, Elting J, McNabola A, Wilkie D, Petrenciuc O, Chen EX. Phase II trial of sorafenib in patients with recurrent or metastatic squamous cell carcinoma of the head and neck or nasopharyngeal carcinoma. J Clin Oncol. 2007;25:3766–73. doi: 10.1200/JCO.2006.10.2871. [DOI] [PubMed] [Google Scholar]
- Fenton TR, Nathanson D, Ponte de Albuquerque C, Kuga D, Iwanami A, Dang J, Yang H, Tanaka K, Oba-Shinjo SM, Uno M, Inda MM, Wykosky J, Bachoo RM, James CD, DePinho RA, Vandenberg SR, Zhou H, Marie SK, Mischel PS, Cavenee WK, Furnari FB. Resistance to EGF receptor inhibitors in glioblastoma mediated by phosphorylation of the PTEN tumor suppressor at tyrosine 240. Proc Natl Acad Sci U S A. 2012;109:14164–9. doi: 10.1073/pnas.1211962109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H, Kovar JL, Cheung L, Rosenthal EL, Olive DM. A comparative study of affibody, panitumumab, and EGF for near-infrared fluorescence imaging of EGFR- and EGFRvIII-expressing tumors. Cancer Biol Ther. 2013;15(2) doi: 10.4161/cbt.26719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotte SJ, Hirte HW. BAY 43-9006: early clinical data in patients with advanced solid malignancies. Curr Pharm Des. 2002;8:2249–53. doi: 10.2174/1381612023393053. [DOI] [PubMed] [Google Scholar]
- Martin AP, Park MA, Mitchell C, Walker T, Rahmani M, Thorburn A, Häussinger D, Reinehr R, Grant S, Dent P. BCL-2 family inhibitors enhance histone deacetylase inhibitor and sorafenib lethality via autophagy and overcome blockade of the extrinsic pathway to facilitate killing. Mol Pharmacol. 2009;76:327–41. doi: 10.1124/mol.109.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–50. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- Park MA, Reinehr R, Häussinger D, Voelkel-Johnson C, Ogretmen B, Yacoub A, Grant S, Dent P. Sorafenib activates CD95 and promotes autophagy and cell death via Src family kinases in gastrointestinal tumor cells. Mol Cancer Ther. 2010;9:2220–31. doi: 10.1158/1535-7163.MCT-10-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MA, Zhang G, Martin AP, Hamed H, Mitchell C, Hylemon PB, Graf M, Rahmani M, Ryan K, Liu X, Spiegel S, Norris J, Fisher PB, Grant S, Dent P. Vorinostat and sorafenib increase ER stress, autophagy and apoptosis via ceramide-dependent CD95 and PERK activation. Cancer Biol Ther. 2008;7:1648–62. doi: 10.4161/cbt.7.10.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrantonio F, Perrone F, Biondani P, Maggi C, Lampis A, Bertan C, Venturini F, Tondulli L, Ferrari D, Ricci V, Villa F, Barone G, Bianco N, Ghidini A, Bossi I, Fanetti G, Di Bartolomeo M, de Braud F. Single agent panitumumab in KRAS wild-type metastatic colorectal cancer patients following cetuximab-based regimens: Clinical outcome and biomarkers of efficacy. Cancer Biol Ther. 2013 Sep 4;14(12) doi: 10.4161/cbt.26343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratz KW, Cho E, Levis MJ, Karp JE, Gore SD, McDevitt M, Stine A, Zhao M, Baker SD, Carducci MA, Wright JJ, et al. A pharmacodynamic study of sorafenib in patients with relapsed and refractory acute leukemias. Leukemia. 2010;24:1437–1444. doi: 10.1038/leu.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quatrale AE, Porcelli L, Silvestris N, Colucci G, Angelo A, Azzariti A. EGFR tyrosine kinases inhibitors in cancer treatment: in vitro and in vivo evidence. Front Biosci. 2011;16:1962–72. doi: 10.2741/3833. [DOI] [PubMed] [Google Scholar]
- Rahmani M, Davis EM, Crabtree TR, Habibi JR, Nguyen TK, Dent P, Grant S. The kinase inhibitor sorafenib induces cell death through a process involving induction of endoplasmic reticulum stress. Mol Cell Biol. 2007;27:5499–513. doi: 10.1128/MCB.01080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani M, Davis EM, Bauer C, Dent P, Grant S. Apoptosis induced by the kinase inhibitor BAY 43-9006 in human leukemia cells involves down-regulation of Mcl-1 through inhibition of translation. J Biol Chem. 2005;280:35217–27. doi: 10.1074/jbc.M506551200. [DOI] [PubMed] [Google Scholar]
- Ramirez YP, Weatherbee JL, Wheelhouse RT, Ross AH. Glioblastoma Multiforme Therapy and Mechanisms of Resistance. Pharmaceuticals (Basel) 2013;6:1475–1506. doi: 10.3390/ph6121475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon DA, Groves MD, Wen PY, Nabors L, Mikkelsen T, Rosenfeld S, Raizer J, Barriuso J, McLendon RE, Suttle AB, Ma B, Curtis CM, Dar MM, de Bono J. A phase I/II trial of pazopanib in combination with lapatinib in adult patients with relapsed malignant glioma. Clin Cancer Res. 2013;19:900–8. doi: 10.1158/1078-0432.CCR-12-1707. [DOI] [PubMed] [Google Scholar]
- Roengvoraphoj M, Tsongalis GJ, Dragnev KH, Rigas JR. Epidermal growth factor receptor tyrosine kinase inhibitors as initial therapy for non-small cell lung cancer: focus on epidermal growth factor receptor mutation testing and mutation-positive patients. Cancer Treat Rev. 2013;39:839–50. doi: 10.1016/j.ctrv.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Smith MA, Houghton P. A proposal regarding reporting of in vitro testing results. Clin Cancer Res. 2013;19:2828–33. doi: 10.1158/1078-0432.CCR-13-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerie K, Yacoub A, Hagan MP, Curiel DT, Fisher PB, Grant S, Dent P. Radiation-induced cell signaling: inside-out and outside-in. 2007;6:789–801. doi: 10.1158/1535-7163.MCT-06-0596. [DOI] [PubMed] [Google Scholar]
- Villarroel MC, Pratz KW, Xu L, Wright JJ, Smith BD, Rudek MA. Plasma protein binding of sorafenib, a multi kinase inhibitor: in vitro and in cancer patients. Invest New Drugs. 2012;30:2096–102. doi: 10.1007/s10637-011-9767-5. [DOI] [PMC free article] [PubMed] [Google Scholar]