Abstract
We report herein the selective hydroxylation of 10-undecenoic acid with a light-activated hybrid P450 BM3 enzyme. Under previously developed photocatalytic reaction conditions, only a monohydroxylated product is detected by gas chromatography. Hydroxylation occurs exclusively at the allylic position as confirmed from a synthesized authentic standard. Investigation into the stereochemistry of the reaction indicates that the R enantiomer is obtained in 85% ee. The (R)-9-hydroxy-10-undecenoic acid obtained enzymatically is a valuable synthon en route to various natural products further expanding the light-activated P450 BM3 biocatalysis and highlighting the advantages over traditional methods.
Keywords: Light-driven P450 BM3 biocatalyst, Stereoselective hydroxylation, Allylic oxidation, (R)-9-hydroxy-10-undecenoic acid, Natural product synthesis
1. Introduction
Biocatalytic processes are gaining increasing importance in organic synthesis from their unique selectivity advantages over traditional methods.1 Over the last two decades, biocatalysis has emerged as an important technology for meeting the demand for green and sustainable synthesis of pharmaceuticals and other fine chemicals. Among the various biocatalytic reactions comprising hydrolytic, reductive and oxidative reactions, the selective oxidations of unactivated C-H bonds catalyzed by cytochrome P450 enzymes are particularly attractive.
The cytochrome P450 superfamily of heme-thiolate enzymes is known to perform a myriad of C-H bond functionalizations, often with high regio- and stereoselectivity, using molecular dioxygen and two reducing equivalents.2 The consensus mechanism involves two successive one-electron transfer steps from the electron providing reductase to the heme center and activation of molecular dioxygen to form several well-characterized intermediates.3 The high-valent Fe(IV)-oxo porphyrin radical species, Compound I, achieves the desired C-H bond functionalization via a rebound mechanism.4
Among the large and diverse family of P450 enzymes, the fatty acid hydroxylase CYP102A1 or P450 BM3 from Bacillus megaterium with its fused reductase has been shown to be an important design platform for biocatalysis due to the plasticity of its active site and its ability to accommodate a wide range of substrates upon protein engineering.5 In addition, P450 reactions have recently been extended beyond monooxygenation toward olefin cyclopropanation6 and amination.7
The synthetic potential of P450 enzymes has long been recognized8 and recently exploited in the synthesis of several drugs such as the antimalarial artemisinin,9 important metabolites10 and other fine chemicals.11 However their industrial and biotechnological applications is still limited mainly due to the dependence on the electron providing reductase and the need of stoichiometric amounts of the expensive NAD(P)H cofactor. To overcome these limitations, various approaches have emerged. Great advances have been made in the use of whole cell biocatalysis, for example in the large-scale production of nootkatone from valencene by P450 enzymes12 or in the oxidation of alpha-pinene.13 Other approaches have employed NAD(P)H regeneration systems,14 artificial fusion P450 proteins,15 direct chemical16 or electrochemical reduction17, and light-activated systems18 to drive P450 reactions.
We have recently reported a light-driven P450 BM3 biocatalyst (WT/L407C-Ru1) displaying high photocatalytic activity and initial reaction rate in the hydroxylation of lauric acid.19 This hybrid P450 BM3 enzyme exploits the photophysical properties of a strategically positioned photosensitizer, i.e. [Ru(OMebpy)2PhenA]2+ (Ru1) with OMebpy = 4,4’-methoxy-2,2’-bipyridine and phenA = 5-acetamido-1,10-phenanthroline. A highly reductive Ru(I) species is generated, under flash quench reductive conditions, which provides the necessary two electrons in successive one-electron transfer steps to the heme domain in order to activate molecular dioxygen at the heme center and sustain catalysis upon light irradiation.
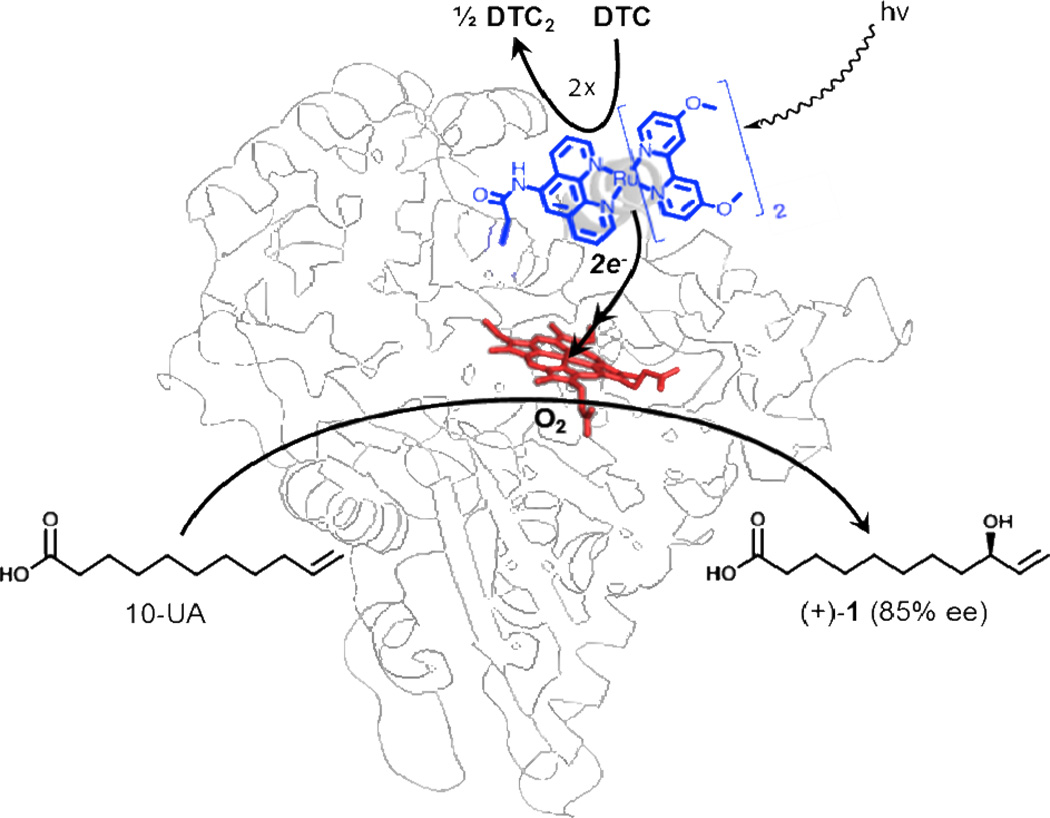
Our current efforts to expand the scope of reactivity and substrate recognition of the light-driven enzymes led to the investigation of 10-undecenoic acid (10-UA) as a substrate. With its terminal double bond, this substrate presents the opportunity to probe epoxidation and hydroxylation reactions. Under the current photochemical conditions, the hydroxylation of 10-UA resulted in only one monohydroxylated product (Figure 1). In addition, the determination of its absolute configuration renders the oxidized product a valuable synthon for the synthesis of various natural products.
Figure 1.
Schematic representation of the light-driven hybrid WT/L407C-Ru1 P450 BM3 heme domain biocatalyst. Upon light activation, the Ru(II) photosensitizer is quenched by sodium diethyldithiocarbamate (DTC) to provide the necessary electrons to the heme domain, which performs the selective hydroxylation of 10-undecenoic acid (10-UA).
2. Materials and Methods
All reagents and solvents used in this work were of analytical grade and purchased from Thermo Fisher Scientific and Sigma-Aldrich. 1H and 13C NMR were recorded on a Varian 400 MHz NMR spectrometer. The fatty acid analysis was performed, after silylation of the acid and alcohol groups, on an Agilent 6890N gas chromatography (GC) system equipped with a flame ionization detector and an HP-5MS capillary column. The stereochemistry of the fatty acid Mosher esters was determined on a Chiralpak AD column (0.46 × 25 cm) connected to a Dionex Summit HPLC system.
2.1. Hybrid enzyme generation
The purification of the P450 BM3 heme domain mutant (WT/L407C) expressed in E.coli and the generation of the hybrid enzyme have been reported previously.19,20 Upon gel purification of the WT/L407C heme domain, fractions with a Rz ratio (A420/A280) greater than 1.5 were pooled. The hybrid enzyme WT/L407C-Ru1 is assembled by reacting the iodoacetamide derivative of the [Ru(OMebpy)2PhenA]2+ (Ru1) complex (with OMebpy = 4,4’-methoxy-2,2’-bipyridine and phenA = 5-acetamido-1,10-phenanthroline) with an engineered non-native cysteine residue (L407C) of P450 BM3 heme domain mutant. The covalent attachment was confirmed by mass spectrometry, UV-vis and fluorescence spectroscopic techniques, as well as by chymotrypsin digestion of the hybrid enzymes.19,20
2.2. Synthesis of the racemic 9-hydroxy-10-undecenoate ((±)-1) and its methyl ester ((±)-2)
The proposed compounds were prepared by either of two routes: 1) allylic oxidation of 10-UA using stoichiometric amounts of SeO2 and tert-butyl peroxide21 followed by methylation of the acid moiety using trimethylsilyldiazomethane 22 or 2) methylation of 10-UA followed by allylic oxidation of the ester.23 Both routes led to valuable intermediates, i.e. racemic mixture of 9-hydroxy-10-undecenoic acid ((±)-1) as well as its methyl ester ((±)-2), for the characterization of the enzymatic product. The 1H and 13C NMR spectra for all the synthesized compounds match those previously reported.
2.3. Light-driven enzymatic substrate oxidation
An aerated solution (10 mL) containing 2 µM of the hybrid enzymes with 500 µM of 10-undecenoic acid (10-UA) and 100 mM sodium diethyldithiocarbamate (DTC) was exposed to visible light irradiation from an Orion 1000 W Xenon arc lamp with IR- and UV-cutoff filters. The solution was maintained at 20 °C using a water bath. After 90 min of light exposure, the reaction mixture was quenched by addition of HCl and the product was extracted with dichloromethane. A small aliquot (500 µL) was evaporated and the residue was silylated using N,O-Bis(trimethylsilyl)trifluoroacetamide (BSTFA)/Pyridine for GC/MS analysis.20 The remainder of the solution was subjected to methylation using trimethylsilyldiazomethane. The methylated product was separated from the starting material by flash chromatography (Elution solvent: hexane/ethyl acetate 10:1, Rf=0.4). Authentic samples prepared in Section 2.2 were used as reference.
2.4. Determination of the stereoselectivity of the enzymatic reaction
Both the racemic methanoate ester ((±)-2) from Section 2.2. and the enzymatic hydroxylated methanoate ester from Section 2.3. were transformed to their Mosher ester following reported procedure24 using (S)-(+)-α-Methoxy-α-trifluoromethylphenylacetyl chloride (MTPA-Cl). Briefly, dry pyridine (2 µL) and MTPA-Cl (2 µL) were added to a CDCl3 solution of the hydroxylated methanoate ester. The progress of the reaction was followed by TLC (hexane/ethyl acetate 10:1). At the end of the reaction, the products were purified by flash chromatography (hexane/ethyl acetate 10:1). The resulting Mosher esters were submitted to chiral analysis using the Chiralpak AD column connected to a Dionex Summit HPLC system (Elution solvent: 92% hexane, 8% isopropylalcohol).
3. Results
3.1. Regioselective oxidation of 10-undecenoic acid by light-driven WT/L407C-Ru1 enzyme
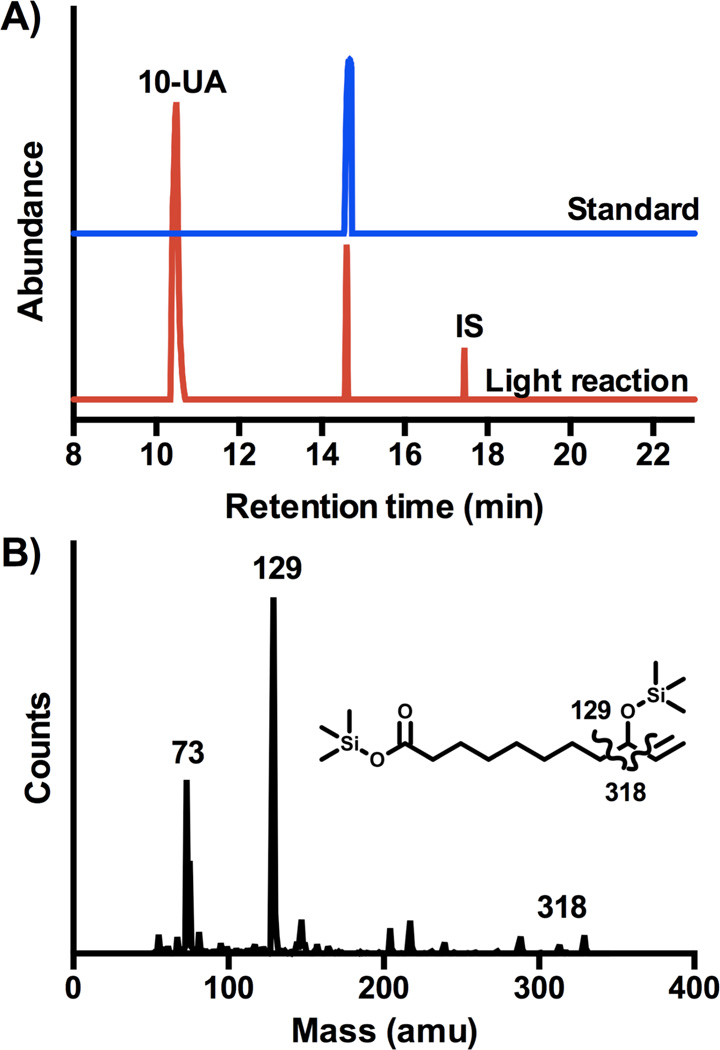
Following the flash quench reductive conditions previously reported,20 we investigated the functionalization of 10-undecenoic acid (10-UA) by the hybrid P450 BM3 WT/L407C-Ru1 enzyme upon light activation. Determination of the binding constant (Kd = 780 µM) reflects poorer substrate binding compared to its saturated counterpart.20 While the oxidation of dodecanoic acid led previously to three mono hydroxylated products at subterminal positions,19 only a sole monohydroxylated product is obtained with 10-UA as substrate (40% yield) in the light-driven reaction as shown in Figure 2. The nature of the hydroxylated product can be assigned to the 9-hydroxy-10-undecenoic acid (1) based on the mass fragmentation pattern after derivatization as established previously for the lauric acid.20 An authentic sample of 1 was also prepared showing the same retention time and fragmentation pattern, unambiguously confirming the nature of the proposed hydroxylated product. The initial reaction rate of 6 mol of product/mol of enzyme/min, determined after 1 min reaction, is much lower than the one observed for the light-driven hydroxylation of lauric acid19 but similar to the one reported for the full-length enzyme using NADPH.25
Figure 2.
A) Gas chromatograms of the trimethylsilylated mixture from the light-driven reaction (blue) and authentic sample (red). 12-hydroxydodecanoic acid was used as internal standard (I.S., 10 nmol) for product quantification. B) Mass fragmentation of the peak at 14.3 min with the corresponding structure indicating the major fragments observed.
3.2. Synthesis of the racemic standard
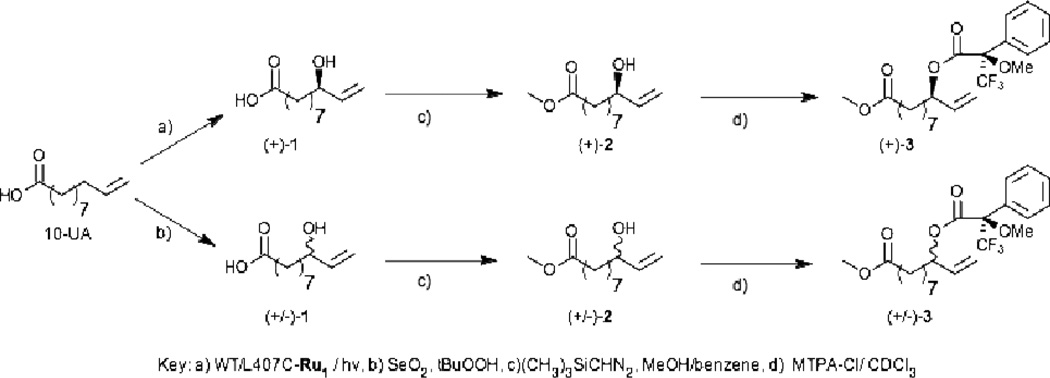
A racemic mixture of the proposed hydroxylated product (1) can also be prepared synthetically from the allylic oxidation of 10-undecenoic acid. The compound is easily obtained using stoichiometric amount of Selenium dioxide and tertbutylhydroperoxide in reasonable yield (40%) after stirring for 48 hours at room temperature.21 Once purified, Compound 1 can be prepared for stereochemical analysis by converting to its methyl ester (2) using trimethylsilyldiazomethane and further to its Mosher ester (3) using (S)-(+)-α-Methoxy-α-trifluoromethylphenylacetyl chloride (MTPA-Cl) in CDCl3 according to Scheme 1.
Scheme 1.
Synthetic routes to convert 9-hydroxy-10-undecenoic acid (1) obtained from either light driven enzymatic reaction (a) or synthetically (b) to its methyl (2) and Mosher (3) esters.
3.3. Investigation of the stereoselectivity of the hydroxylated product
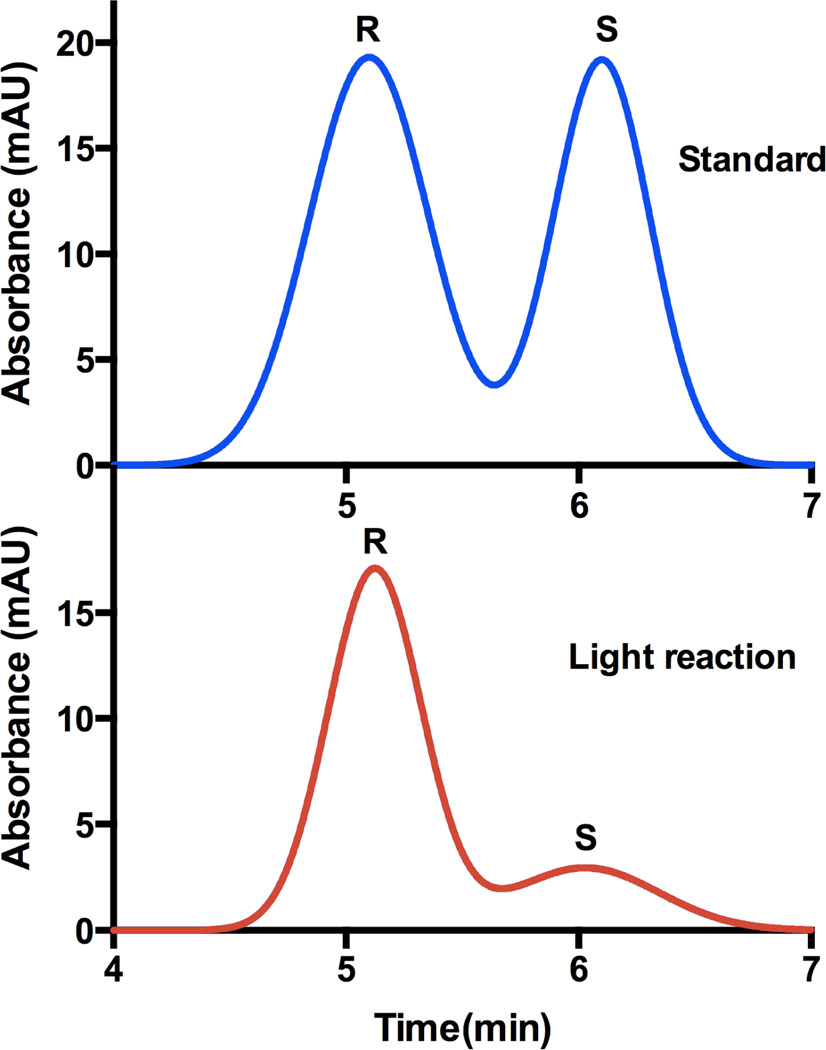
The synthetic racemic Mosher esters (3) from Section 3.2 were then analyzed on a chiral HPLC column leading to the elution of the R enantiomer at 5.1 min and the S enantiomer at 6.1 min as shown in Figure 3. Following similar procedures, we investigated the stereochemistry of the hydroxylated product from the light-activated enzymatic reaction. Mainly the R enantiomer was obtained (85% ee) consistent with the previous report on the saturated fatty acid counterpart.26
Figure 3.
Chromatographic resolution of the enantiomers from the synthetic standard (blue) and the monohydroxylated product generated during the light reaction (red) after conversion to their respective Mosher esters.
4. Discussion
P450 BM3 enzyme is known to catalyze the regio- and stereoselective oxidation of various saturated and unsaturated long chain fatty acids.5 Interestingly, terminally unsaturated long chain fatty acids have not received much attention. Early report indicated that terminal unsaturated substrates of various chain lengths resulted in rapid enzyme inactivation.27 Chen et al. recently reported that allylic hydroxylation of 10-undecenoic acid (10-UA) is preferred under turnover conditions with small amount of epoxidation at the terminal unsaturation.25 Depending on the method of Compound I generation in this study, a change in the product ratio between hydroxylation and oxidation is observed. We have been interested in studying the oxidation of the 10-undecenoic acid using our light-activated P450 BM3 heme domain biocatalyst, WT-L407C-Ru1. We recently demonstrated that this light-activated approach is valuable in the hydroxylation of lauric acid, a C12 fatty acid yielding high photocatalytic activity and initial reaction rate.19 In addition, the ratio of hydroxylated products in the photocatalytic reaction is identical to that observed with the wild-type heme domain.
Under the photocatalytic conditions previously developed, the oxidation of 10-UA by the light activated WT/L407C-Ru1 hybrid enzyme leads to a single monohydroxylated product as shown in the GC chromatogram (Figure 2). From the mass fragmentation pattern, the hydroxylation occurs exclusively at the 9 position as also confirmed with the synthetic analog. It is worth noting that under the photocatalytic conditions, no reduction of the starting material is observed. In addition, no epoxidation product is detected in the GC chromatograms. As shown previously, absence of light or reductive quencher do not show any product formation.
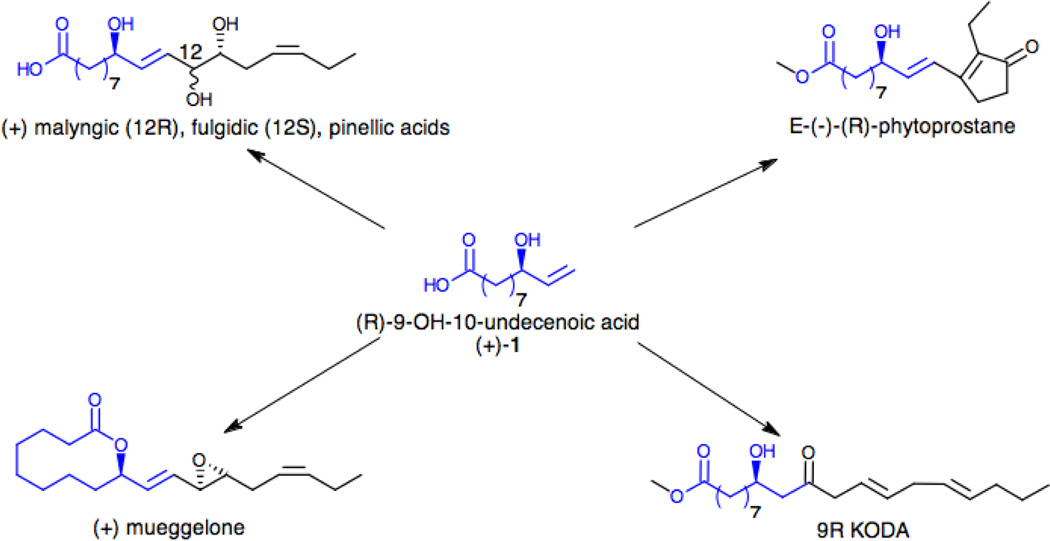
While obtaining a single hydroxylated product is attractive, the possibility of having one of the two enantiomers from the enzymatic reaction would represent a high added value. Besides being a natural product, cochadylonic acid,28 the (R)-9-hydroxy-10-undecenoic acid ((+)-1) is also a useful synthon for the synthesis of various natural products as illustrated in Scheme 2. Its alcohol and carboxylic acid moieties can react under Yamaguchi conditions29 to yield a 10-membered ring lactonization product, while the presence of the terminal double bond has been used for chain elongation via metathesis reaction30 using Grubbs catalyst. Several compounds have been synthesized following these approaches including (+)-mueggelone,31 an inhibitor of fish development as well as malyngic, fulgidic32, pinellic acids33 and the botanical analogues of prostaglandins, phytoprostanes.34
Scheme 2.
Synthetic routes to various natural products using the (R)-9-hydroxy-10-undecenoic acid, (+)-1
The synthesis of one of the two enantiomers has been rather challenging often requiring several synthetic steps31–34 despite recent advances using a chiral palladium catalyst.35 The chirality of the active site of the P450 BM3 enzyme is well documented leading often exclusively to the R enantiomer for the monooxygenated long chain fatty acids.26 We have then investigated the stereochemistry of the enzymatically hydroxylated 10-UA. In order to determine its absolute configuration, the isolated product as well as the synthetic racemic mixture were methylated using trimethylsilyldiazomethane and then converted to their respective Mosher esters using MTPA-Cl. The two enantiomers of the racemic mixture (±)-3 were resolved on a chiral HPLC column as shown in Figure 3 with the R enantiomer eluting first. The enzymatically hydroxylated product shows mainly the R enantiomer eluting at 5.1 min consistent with the stereoselectivity observed for their saturated counterpart. The enantiomeric excess for the reaction was determined to be 85% ee. Obtaining the R enantiomer in high ee renders the hydroxylated product from the light-activated system a valuable synthon and further expanding the light-activated P450 BM3 biocatalysis.
5. Conclusion
Oxidation of 10-undecenoic acid by the light-activated hybrid WT/L407C-Ru1 enzyme leads directly to a highly enantiomerically enriched monohydroxylated product highlighting the advantages of biocatalysis over traditional methods. The resulting (R)-9-hydroxy-10-undecenoic acid is a useful synthon for the synthesis of a wide range of natural products. From the wealth of data available in engineering P450 BM3 enzymes, tighter substrate binding36 could be achieved as well as inversion of the stereochemistry.37
Acknowledgments
The authors would like to thank the National Institute of Health (GM095415) for financial support. This research was also supported by an award from Research Corporation for Science Advancement. L.C. thanks the National Science Foundation (MRI grant 0923573) and San José State University for the use of mass spectrometry facilities in the PROTEIN LAB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.(a) Clouthier CM, Pelletier JN. Chem. Soc. Rev. 2012;41:1585. doi: 10.1039/c2cs15286j. [DOI] [PubMed] [Google Scholar]; (b) Schulz S, Girhard M, Urlacher VB. Chemcatchem. 2012;4:1889. [Google Scholar]; (c) Hall M, Bommarius AS. Chem. Rev. 2011;111:4088. doi: 10.1021/cr200013n. [DOI] [PubMed] [Google Scholar]
- 2.Denisov IG, Makris TM, Sligar SG, Schlichting I. Chem. Rev. 2005;105:2253. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 3.(a) Luthra A, Denisov IG, Sligar SG. Arch. Biochem. Biophys. 2011;507:26. doi: 10.1016/j.abb.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Krest CM, Onderko EL, Yosca TH, Calixto JC, Karp RF, Livada J, Rittle J, Green MT. J. Biol. Chem. 2013;288:17074. doi: 10.1074/jbc.R113.473108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rittle J, Green MT. Science. 2010;330:933. doi: 10.1126/science.1193478. [DOI] [PubMed] [Google Scholar]
- 5.Whitehouse CJC, Bell SG, Wong LL. Chem. Soc. Rev. 2012;41:1218. doi: 10.1039/c1cs15192d. [DOI] [PubMed] [Google Scholar]
- 6.Coelho PS, Brustad EM, Kannan A, Arnold FH. Science. 2013;339:307. doi: 10.1126/science.1231434. [DOI] [PubMed] [Google Scholar]
- 7.(a) McIntosh JA, Coelho PS, Farwell CC, Wang ZJ, Lewis JC, Brown TR, Arnold FH. Angew. Chem. Int. Ed. 2013;52:9309. doi: 10.1002/anie.201304401. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Singh R, Bordeaux M, Fasan R. ACS Catal. 2014;4:546. doi: 10.1021/cs400893n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Urlacher VB, Girhard M. Trends Biotechnol. 2012;30:26. doi: 10.1016/j.tibtech.2011.06.012. [DOI] [PubMed] [Google Scholar]; (b) Fasan R. ACS Catal. 2012;2:647. [Google Scholar]
- 9.Dietrich JA, Yoshikuni Y, Fisher KJ, Woolard FX, Ockey D, McPhee DJ, Renninger NS, Chang MCY, Baker D, Keasling JD. ACS Chem. Biol. 2009;4:261. doi: 10.1021/cb900006h. [DOI] [PubMed] [Google Scholar]
- 10.(a) Zhang KD, Shafer BM, Demars MD, Stern HA, Fasan R. J. Am. Chem. Soc. 2012;134:18695. doi: 10.1021/ja3073462. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sawayama AM, Chen MMY, Kulanthaivel P, Kuo MS, Hemmerle H, Arnold FH. Chem. Eur. J. 2009;15:11723. doi: 10.1002/chem.200900643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Bell SG, Chen XH, Sowden RJ, Xu F, Williams JN, Wong LL, Rao ZH. J. Am. Chem. Soc. 2003;125:705. doi: 10.1021/ja028460a. [DOI] [PubMed] [Google Scholar]; (b) Sowden RJ, Yasmin S, Rees NH, Bell SG, Wong LL. Org. Biomol. Chem. 2005;3:57. doi: 10.1039/b413068e. [DOI] [PubMed] [Google Scholar]
- 12.Girhard M, Machida K, Itoh M, Schmid RD, Arisawa A, Urlacher VB. Microb. Cell Fact. 2009;8:36. doi: 10.1186/1475-2859-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schewe H, Holtmann D, Schrader J. Appl. Microbiol. Biotechnol. 2009;83:849. doi: 10.1007/s00253-009-1917-8. [DOI] [PubMed] [Google Scholar]
- 14.Hollmann F, Witholt B, Schmid A. J. Mol. Catal. B: Enzym. 2002;19:167. [Google Scholar]
- 15.Munro AW, Girvan HM, McLean KJ. Biochim. Biophys. Acta. 2007;1770:345. doi: 10.1016/j.bbagen.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 16.(a) Schwaneberg U, Appel D, Schmitt J, Schmid RD. J. Biotechnol. 2000;84:249–257. doi: 10.1016/s0168-1656(00)00357-6. [DOI] [PubMed] [Google Scholar]; (b) Udit AK, Arnold FH, Gray HB. J. Inorg. Biochem. 2004;98:1547. doi: 10.1016/j.jinorgbio.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Sadeghi SJ, Fantuzzi A, Gilardi G. Biochim. Biophys. Acta. 2011;1814:237. doi: 10.1016/j.bbapap.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 18.(a) Jensen K, Jensen PE, Moller BL. ACS Chem. Biol. 2011;6:533. doi: 10.1021/cb100393j. [DOI] [PubMed] [Google Scholar]; (b) Zilly FE, Taglieber A, Schulz F, Hollmann F, Reetz MT. Chem. Commun. 2009:7152. doi: 10.1039/b913863c. [DOI] [PubMed] [Google Scholar]
- 19.Tran NH, Nguyen D, Dwaraknath S, Mahadevan S, Chavez G, Nguyen A, Dao T, Mullen S, Nguyen TA, Cheruzel LE. J. Am. Chem. Soc. 2013;135:14484. doi: 10.1021/ja409337v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.(a) Tran NH, Huynh N, Chavez G, Nguyen A, Dwaraknath S, Nguyen TA, Nguyen M, Cheruzel L. J. Inorg. Biochem. 2012;115:50. doi: 10.1016/j.jinorgbio.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tran NH, Huynh N, Bui T, Nguyen Y, Cooper ME, Cheruzel LE. Chem. Commun. 2011;47:11936. doi: 10.1039/c1cc15124j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umbreit MA, Sharpless KB. J. Am. Chem. Soc. 1977;99:5526. [Google Scholar]
- 22.Hashimoto N, Aoyama T, Shioiri T. Chem. Pharm. Bull. 1981;29:1475. [Google Scholar]
- 23.Rao AVR, Reddy ER, Purandare AV, Varaprasad CVNS. Tetrahedron. 1987;43:4385. [Google Scholar]
- 24.Hoye TR, Jeffrey CS, Shao F. Nat. Protoc. 2007;2:2451. doi: 10.1038/nprot.2007.354. [DOI] [PubMed] [Google Scholar]
- 25.Chen XH, Su Z, Horner JH, Newcomb M. Org. Biomol. Chem. 2011;9:7427. doi: 10.1039/c1ob06035j. [DOI] [PubMed] [Google Scholar]
- 26.Cryle MJ, Matovic NJ, De Voss JJ. Tetrahedron Lett. 2007;48:133. [Google Scholar]
- 27.Shirane N, Sui ZH, Peterson JA, Ortiz de Montellano PR. Biochemistry. 1993;32:13732. doi: 10.1021/bi00212a044. [DOI] [PubMed] [Google Scholar]
- 28.Yoshikawa M, Murakami T, Shimada H, Yoshizumi S, Saka M, Yamahara J, Matsuda H. Chem. Pharm. Bull. 1998;46:1008. doi: 10.1248/cpb.46.1008. [DOI] [PubMed] [Google Scholar]
- 29.Takimoto S, Inanaga J, Katsuki T, Yamaguchi M. Bull. Chem. Soc. Jpn. 1981;54:1470. [Google Scholar]
- 30.Miura A, Kuwahara S. Tetrahedron. 2009;65:3364. [Google Scholar]
- 31.Yadav JS, Somalah R, Ravindar K, Chandraiah L. Tetrahedron Lett. 2008;49:2848. [Google Scholar]
- 32.Kurashina Y, Miura A, Enomoto M, Kuwahara S. Tetrahedron. 2011;67:1649. [Google Scholar]
- 33.Prasad KR, Swain B. Tetrahedron: Asymmetry. 2008;19:1636. [Google Scholar]
- 34.Perlikowska W, Mikolajczyk M. Tetrahedron: Asymmetry. 2011;22:1767. [Google Scholar]
- 35.Covell DJ, White MC. Tetrahedron. 2013;69:7771. doi: 10.1016/j.tet.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haines DC, Hegde A, Chen BZ, Zhao WQ, Bondlela M, Humphreys JM, Mullin DA, Tomchick DR, Machius M, Peterson JA. Biochemistry. 2011;50:8333. doi: 10.1021/bi201099j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munzer DF, Meinhold P, Peters MW, Feichtenhofer S, Griengl H, Arnold FH, Glieder A, de Raadt A. Chem. Commun. 2005:2597. doi: 10.1039/b501527h. [DOI] [PubMed] [Google Scholar]