Abstract
The LRIG (leucine-rich repeats and immunoglobulin-like domains) family of transmembrane proteins contains three vertebrate members (LRIG1, LRIG2, LRIG3) and one member each in flies (Lambik) and worms (Sma-10). LRIGs have stepped into the spotlight as essential regulators of growth factor receptors, including receptor tyrosine and serine/threonine kinases. LRIGs have been found to both negatively (LRIG1, LRIG3) and positively (Sma-10, LRIG3) regulate growth factor receptor expression and signaling, although the precise molecular mechanisms by which LRIGs function are not yet understood. The most is known about LRIG1, which was recently demonstrated to be a tumor suppressor. Indeed, in vivo experiments reinforce the essential link between LRIG1 and repression of its targets for tissue homeostasis. LRIG1 has also been identified as a stem cell marker and regulator of stem cell quiescence in a variety of tissues, discussed within. Comparably less is known about LRIG2 and LRIG3 although studies to date suggest that their functions are largely distinct from LRIG1 and that they likely do not serve as growth/tumor suppressors. Finally, the translational applications of expressing soluble forms of LRIG1 in LRIG1-deficient tumors are being explored and hold tremendous promise.
Keywords: LRIG1, LRIG2, LRIG3, growth factor receptor, tumor suppressor, stem cell
Introduction
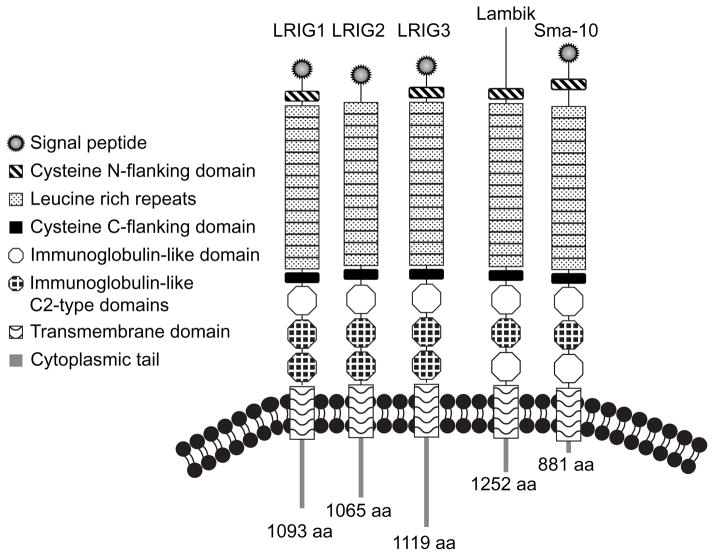
Members of the leucine-rich and immunoglobulin-like domains family of proteins (LRIGs) are single-pass transmembrane proteins implicated in the regulation of growth factor receptors. There is one LRIG gene in flies (Lambik, www.flybase.org) and worms (Sma-10, Gumienny et al., 2010) and three in vertebrates (LRIG1, LRIG2 and LRIG3). The LRIG proteins contain tandem leucine-rich repeats followed by three immunoglobulin-like domains, a transmembrane domain and a large cytoplasmic tail (Figure 1). LRIGs show homology in their extracellular domains and also in the juxtamembrane portion of their cytoplasmic domains. The remainder of the cytoplasmic domain diverges significantly amongst LRIGs (Abraira et al., 2010) and may impart unique functions.
Figure 1. LRIG structure.
The five members of the LRIG family are depicted: Human LRIG1, LRIG2 and LRIG3, Drosophila Lambik and C-elegans SMA-10. Total number of amino acids are listed at the C-terminus of each protein. Domains are color coded as shown in the key. Structure based on SMART (smart.embl-heidelberg.de).
LRIGs are broadly expressed in healthy human tissue and LRIG transcripts have been detected in at least twenty six distinct tissue types with significant variation in relative abundance amongst the LRIGs (Guo et al., 2004). The human LRIG1 gene is located at chromosome 3p14.3 (Nilsson et al., 2001), human LRIG2 at chromosome 1p13 (Holmlund et al., 2004) and human LRIG3 at 12q13.2 (Guo et al., 2004). 3p14 and 1p13 are chromosomal regions known to be frequently deleted in cancer while 12q13 has been reported to be amplified in glioblastoma (Reifenberger et al., 1996, Mischel et al., 2003). 12q13 is also identified as a “region of increased tumor expression” in brain, breast, liver and lung tumors (Zhou et al., 2003).
LRIG1 has been identified as a negative regulator of several receptor tyrosine kinases including ErbBs 1-4 (Laederich et al., 2004, Gur et al., 2004), EGFRvIII (Stutz et al., 2008), Met (Shattuck et al., 2007), Ret (Ledda et al., 2008) and PDGFR-α (Rondahl et al., 2013). With the exception of the Ret receptor and the PDGFR-α (for which it was not examined), LRIG1 has been found to increase the ubiquitination and decrease the half –life of its targets. With the Ret receptor, LRIG1 restricts Ret recruitment to lipid rafts and inhibits binding of the ligand GDNF (glial cell line-derived neurotrophic factor), preventing Ret activation (Ledda et al., 2008). Less is known regarding the specific molecular functions of LRIG2 and LRIG3 although they do not appear to have the same global ability to decrease growth factor receptor expression. LRIG2 has no significant effect on the expression of EGFR, ErbB2 (Rafidi et al., 2013) or PDGFRα/β (Rondahl et al., 2013) while LRIG3 has been found to oppose LRIG1 (Rafidi et al., 2013) and increase expression of ErbBs 1-4 (Rafidi et al., 2013). LRIG1 limits LRIG3 action by promoting its proteolytic degradation, highlighting a complex cross-talk amongst LRIG family members (Rafidi et al., 2013).
The recent discovery of LRIG2 mutations in Urofacial Syndrome is likely to open up new avenues of research and provide unique insight into LRIG function (Stuart et al., 2013). Urofacial Syndrome (UFS) is a congenital autosomal recessive disorder characterized by a severe dysfunction in urinary bladder voiding. Patients also display a telltale facial grimace when attempting to smile (Bacchetta and Cochat, 2010). In 2010, biallelic loss of function mutations in Heparanase-2 (HPSE2) were identified as causative for UFS (Daly et al., 2010, Pang et al., 2010). Heparanase-2 inhibits Heparanase-1 (HPSE1) which in turn, regulates the availability and signaling of growth factors through processing of heparan sulfate proteoglycans (which sequester growth factors). Since HPSE2-linked UFS and LRIG2-linked UFS are reported to be clinically indistinguishable, it has been proposed that HPSE2 and LRIG2 function overlap and that their functional loss may lead to aberrant growth factor receptor signaling (Roberts et al., 2014). Of note, LRIG2 and HPSE2 proteins are co-expressed in human fetal bladder and have been immuno-localized to nerve fascicles growing into the bladder wall, raising the possibility of peripheral nerve involvement in UFS (Daly et al., 2010, Stuart et al., 2013). In direct support of this, recent studies in Xenopus revealed that xHpse2 depletion leads to up-regulated FGF2 signaling and perturbed motor neuron development. (Roberts et al., 2014).
Lrig3 is required for neural crest formation in Xenopus, a process driven by a balance of FGF, BMP and canonical Wnt signaling (Zhao et al., 2008). In experiments using antisense oligos to reduce expression of endogenous Lrig3, Lrig3 was found to be required for the induction of neural crest markers including Slug, Twist, FoxD3 and Sox9. At the receptor level, Lrig3 was found to enhance canonical Wnt signaling but inhibit FGF signaling, in both animal caps and human HEK-293T cells. Lrig3 interacted with and decreased the expression of Xenopus FGFR1, providing a plausible mechanism by which Lrig3 attenuates FGF signaling, but the mechanism by which Lrig3 potentiates Wnt signaling is not known.
Sma-10, the sole C-elegans LRIG, is required for body size regulation, with Sma-10 mutant worms smaller than wild type worms (Gumienny et al., 2010). Lambik, the sole Drosophila LRIG, rescues the body size phenotype in Sma-10 mutants, demonstrating evolutionary conservation of function. However, nothing else is known regarding Lambik. In reporter assays in human HepG2 hepatocellular carcinoma cells, Sma-10 was found to promote BMP (bone morphogenetic protein) signaling. BMPs signal through receptor serine/threonine kinases and ligands and receptors belong to the TGFβ/TGFβ-receptor superfamily. In co-precipitation assays in human embryonic kidney 293T cells, Sma-10 was found to interact with the BMP receptors Sma-6 and Daf-4, Type I and Type II receptors respectively. LRIG1 was also found to interact with Type I receptors including ALK6, ALK1, ALK2 and ALK3 and ACTRIB and Type II receptors including ACTRII and ACTRIIB. However, the functional impact of LRIG1 (positive or negative?) on BMP signaling is unknown. Since LRIG3 is the only mammalian LRIG shown (thus far) to enhance expression of some of its interacting partners, it is tempting to speculate that LRIG3, rather than LRIG1, is the mammalian LRIG which regulates BMP signaling.
LRIG expression in cancer
LRIG1 was cloned from mouse brain in 1996 (Suzuki et al., 1996) and from human brain in 2001 (Nilsson et al., 2001). Its expression in cancer has been a topic of interest for more than a decade, since Hedman and colleagues first proposed that LRIG1 is a tumor suppressor and negative regulator of EGF receptor signaling (Hedman et al., 2002). LRIG1 expression in cancer has been widely examined and of note, LRIG1 recently emerged as one of four genes with prognostic impact in melanoma, glioma, and cancers of breast, bladder and lung. In eight independent cohorts representing these cancers, a decrease in LRIG1 expression was associated with worse patient prognosis (Rouam et al., 2010).
LRIG1 expression is decreased relative to normal tissue in most, but not all, cancer types examined to date (reviewed in Wang et al., 2013 and Hedman & Henriksson, 2007). With recent advancements in genome-wide analysis of human tumors, tumor subtype has become an accessible variable to consider in the analysis of cancer relevant genes. With respect to LRIG1, differences in expression amongst tumor subtypes, both conventional histological subtypes and molecular-based subtypes, are evident. For example, LRIG1 mRNA expression is decreased in clear cell renal cell carcinoma (relative to normal kidney cortex) but not in papillary or chromophobe renal cell carcinomas (Thomasson et al., 2012).
In prostate and breast cancer, LRIG1 expression is induced by androgen and estrogen respectively. High LRIG1 expression observed in hormone receptor-positive disease is likely a reflection of transcriptional activation of the LRIG1 gene by androgen/estrogen receptors (Thomasson et al., 2011, Krig et al., 2011). Indeed, LRIG1 mRNA is enriched in the luminal subtypes of breast cancer (estrogen receptor- positive) but is weakly expressed in the Her2-enriched, basal-like and claudin-low subtypes (mostly estrogen receptor-negative) (Miller et al., 2008, Wang et al., 2013). Despite high expression, in vitro experiments have demonstrated that LRIG1 plays a growth inhibitory role in both prostate and breast cancer cells (Laederich et al., 2004, Thomasson et al., 2011, Krig et al., 2011). In agreement with this, intermediate/high LRIG1 mRNA expression was found to correlate with prolonged relapse-free survival in estrogen receptor-positive breast cancer (Krig et al., 2011). More recently, LRIG1 gene copy number was examined in a large cohort of early stage (I/II) breast cancers (Thompson et al., 2014). Copy number losses were found to be more common in triple-negative and Her2-positive breast cancer when compared to luminal breast cancer, in agreement with weak expression of LRIG1 in these subtypes. Importantly, LRIG1 copy number loss was strongly associated with an elevated and persistent risk of recurrence, distant metastasis and overall mortality. This study also examined LRIG1 mRNA expression across 18 publicly available datasets and revealed that, as with copy number loss, low LRIG1 mRNA expression is significantly and persistently correlated with shorter distant metastasis-free and overall survival. These notable findings highlight the potential utility of clinically assessing LRIG1 expression for patient risk stratification and also implicate LRIG1 in restricting cellular behaviors fundamental to breast cancer metastasis, such as motility and invasion.
In prostate cancer, the relationship between LRIG1 expression and patient outcome may be more complex. In a US cohort of patients who underwent radical prostatectomy, high LRIG1 expression was found to correlate with prolonged overall survival. However, the authors noted that prostate cancer-specific survival information was not available for this cohort and that most of the deaths annotated by overall survival were not likely due to prostate cancer. Conversely, in an independent Swedish cohort who were followed by watchful waiting, high LRIG1 expression was found to correlate with decreased prostate cancer-specific and overall survival. Given the discussed limitations of assessing overall survival, more confidence may be placed in the results from the Swedish cohort although further study is necessary. While LRIG1 inhibits the growth of cultured prostate cancer cells, as a transcriptional target of androgen receptor, its expression in prostate tumors may reflect robust androgen receptor signaling and portend poor prognosis (Thomasson et al., 2011).
In addition to the tumor types/subtypes discussed above, high LRIG1 expression has been linked to good prognosis in squamous cell carcinoma of the skin (Tanemura et al., 2005), cervical adenocarcinoma (Muller et al., 2013), oropharyngeal cancer (Lindquist et al., 2014) and nasopharyngeal cancer (Sheu et al., 2014). In nasopharyngeal cancer (NPC), LRIG1 and phospho-EGFR staining were inversely correlated in primary tumor specimens and ectopic expression of LRIG1 in cultured NPC cells led to the transcriptional down-regulation of EGFR/ErbB activating ligands, providing one mechanism by which LRIG1 inhibits receptor phosphorylation. In both cervical adenocarcinoma and oropharyngeal cancer, LRIG1 expression correlates with positive HPV status and in oropharyngeal cancer, HPV positive tumors with high LRIG1 expression show particularly good prognosis. In oral squamous cell carcinomas, LRIG1 expression is decreased relative to healthy oral mucosa, particularly in poorly differentiated tumors (Jensen et al., 2008, Sheu et al., 2014). This link to tumor differentiation was also observed in squamous cell carcinoma of the skin (Tanemura et al., 2005) and led the authors to propose that LRIG1 deletion may lead to tumor de-differentiation. In astrocytoma, LRIG1 mRNA is decreased compared to non-neoplastic tissue (Ye et al., 2009). In lung cancer, LRIG1 mRNA and protein was found to be decreased in 100% (10/10) of carcinoma in situ (CIS) specimens compared to matched normal airway, with an associated increase in EGFR expression (Lu et al., 2013). Four of these ten CIS specimens showed loss of heterozygosity (LOH) of the LRIG1 locus. Moreover, 75% of 138 lung cancer cell lines and 76% of 37 squamous cell carcinoma cell lines showed loss of heterozygosity of the LRIG1 locus. In a small cohort of malignant ocular surface squamous neoplasias, LRIG1 protein expression was found to be decreased compared to normal and benign tissue (where LRIG1 was over-expressed) and was found to inversely correlate with EGFR expression (Nagata et al., 2014). LRIG1 down-regulation may have potential as a marker of malignancy in a tumor type which completely lacks molecular markers.
Much less is known regarding LRIG2 and LRIG3 expression in cancer and available evidence indicates tissue-specific differences as well as differences dependent upon LRIG subcellular localization. In cervical adenocarcinoma, a high fraction of LRIG3-positive cells correlates with prolonged survival, as observed for LRIG1 (Muller et al., 2013). Peri-nuclear staining of LRIG2 and LRIG3 in diffusely infiltrating astrocytomas was found to correlate with better survival, with LRIG3 emerging as an independent prognostic factor (Guo et al., 2006). The significance of LRIG peri-nuclear localization is not presently understood and the identities of the cellular compartments to which LRIGs localize are unknown although it has been postulated that they may represent endosomes and/or trans-Golgi vesicles (Hedman & Henriksson, 2007). It should be noted that the localization patterns of LRIGs vary significantly, in a cell- and tissue-specific manner, and nuclear (Lindquist et al., 2014, Muller et al., 2013, Karlsson et al., 2008), peri-nuclear (Guo et al., 2006), cytoplasmic (Holmlund et al., 2009, Muller et al., 2013) and cell surface localization has been observed (Laederich et al., 2004). Interestingly, cytoplasmic LRIG2 expression was found to be an independent prognostic factor associated with decreased survival in an oligodendroglioma cohort (Holmlund et al., 2009). Similarly, in early stage squamous cervical carcinoma, cytoplasmic LRIG2 staining was linked to poor survival and identified as an independent prognostic factor while LRIG1 expression was associated with favorable outcome (Hedman et al., 2010). In non-small cell lung cancer, LRIG2 mRNA and protein expression were found to be decreased compared to adjacent tissue. Here again, high cytoplasmic staining of LRIG2 was found to be an independent predictor of poor five year survival (Wang et al., 2014).
The publicly accessible cBioPortal for Cancer Genomics (cbioportal.org, Cerami et al., 2012, Gao et al., 2013) contains 69 cancer genomics studies derived from 17,087 (to date) unique tumor samples. Analysis of LRIG profiles in cBioPortal reveals that LRIG genes are altered in cancer (mutation, deletion, amplification) with distinct overall patterns. For example, gene amplification makes up a greater percentage of LRIG3 alterations, relative to LRIG1 and LRIG2, while most of the LRIG1 and LRIG2 alterations are mutations. Mutation Assessor (available at cBioPortal, Reva et al., 2011) categorizes mutations into those predicted to have high, medium, low and no functional impact based on evolutionary conservation patterns. LRIG mutations with a high impact score according to Mutation Assessor are shown in in Table 1. High impact mutations are localized exclusively to the extracellular domain which is not unexpected given the conservation of this region of LRIGs and the requirement of the extracellular domain for recognition of interacting partners (Gur et al., 2004, Rafidi et al., 2013).
Table 1. LRIG mutations in human malignancy.
LRIG mutations identified in human tumors which are predicted to have a high functional impact. Mutations collated at cbioportal.org and functional impact according to Mutation Assessor, also available at cbioportal.org.
| LRIG1 | LRIG2 | LRIG3 |
|---|---|---|
| N102I, Cutaneous Melanoma, LRRs | N297S, Lung Adenocarcinoma, LRRs | N585Y, Hepatocellular Carcinoma, Ig-like |
| L104S, Endometrial Carcinoma, LRRs | N584D, Stomach Adenocarcinoma, Ig-like | P726R, Bladder Urothelial Carcinoma, Ig-like |
| L146R, Stomach Adenocarcinoma, LRRs | S591Y, Endometrial Carcinoma, Ig-like | |
| N198H, Endometrial Carcinoma, LRRs | C623Y, Cutaneous Melanoma, Ig-like | |
| L213P, Stomach Adenocarcinoma, LRRs | P723T, Pancreatic Adenocarcinoma, Ig-like | |
| L389P, Lung squamous cell carcinoma, LRRs | ||
| L432F, Cutaneous Melanoma, LRRs | ||
| V595M, Endometrial Carcinoma, Ig-like | ||
| P626Q, Lung Adenocarcinoma, Ig-like | ||
| G718R, Cutaneous Melanoma, Ig-like | ||
| G770D, Colorectal, Ig-like |
Knowledge gained from LRIG mouse models
Lrig1 in the skin
The first study of a Lrig knockout (KO) mouse was published in 2002 by Itami’s group at Osaka University (Suzuki et al., 2002). On a mixed (129S7/SvEV and C57BL/6) background, Lrig1 KO mice (neo/neo) developed psoriasiform lesions with epidermal hyperplasia on the tail, face and ears. Lrig1 null keratinocytes were hyper-proliferative and demonstrated perturbed terminal differentiation. This study also revealed that LRIG1 is down-regulated in psoriatic human skin. However, a subsequent study found no down-regulation of LRIG1 mRNA or protein but did reveal changes in cellular and subcellular distribution of the LRIG proteins in psoriatic skin compared to normal skin (Karlsson et al., 2008). Thus, changes in subcellular localization of LRIGs have been observed in both cancer and psoriasis, although as discussed, the significance of these changes is not understood.
The work by Suzuki and colleagues set the stage for subsequent studies from Jensen, Watt and colleagues which identified LRIG1 as an epidermal stem cell marker and regulator of stem cell quiescence. Over-expression of LRIG1 was found to reduce human keratinocyte proliferation and EGF responsiveness through down-regulation of EGFR while keratinocytes in which LRIG1 was depleted had elevated surface EGFR, reinforcing the role of LRIG1 as an essential ErbB negative regulator. Furthemore, LRIG1 depletion resulted in epidermal stem cell expansion (Jensen & Watt, 2006). Analysis of Lrig1 KO mice has provided unique insight into the role of Lrig1 in epidermal homeostasis (Jensen et al., 2009). Paradoxically, Lrig1-positive epidermal stem cells are highly proliferative (Page et al., 2013). Interestingly, this high proliferation index is also observed in one study of Lrig1-positive intestinal stem cells (Wong et al., 2012). Lrig1-positive epidermal stem cells are responsible for the maintenance of the upper pilosebaceous unit and contribute separately to the infundibulum and the sebaceous gland, such that any one Lrig1-positive stem cell contributes to either compartment but not both. Expression of oncogenic K-Ras (G12D) in Lrig1-positive cells is sufficient to drive infundibula and sebaceous gland hyperplasia but tumors are only observed when K-Ras expression is combined with full-thickness wounding.
Lrig1 in the intestine
A major advance in the LRIG field came with the first in vivo evidence that Lrig1 is a tumor suppressor (Powell et al., 2012). At five to six months of age, Lrig1 null mice (CreERT2/CreERT2) were shown to develop highly penetrant duodenal adenomas with increased expression of EGFR, ErbB2 and ErbB3, providing significant in vivo evidence of Lrig1’s role as a negative regulator of ErbB receptors. This study also examined the expression of Lrig1 in intestinal stem cells. Lineage tracing in Lrig1 “reporter mice” (in which β-galactosidase activity reports where endogenous Lrig1 is transcribed) revealed labeling of the base of crypts in both small intestine and colon. Over time, crypts turned completely (and persistently) blue and were populated with an array of differentiated cells, providing unequivocal evidence that Lrig1 marks intestinal stem cells. Endogenous Lrig1 protein expression was also detected with immunostaining and FACS. Lrig1-positive stem cells were relatively quiescent with a lower proliferative index than Lgr5 (leucine rich repeat containing G-protein coupled receptor-5)-positive stem cells. Lrig1-and Lgr5-positive stem cells also diverged at the level of their transcriptomes with Lrig1-positive cells showing evidence of cell cycle repression. However, upon irradiation injury, Lrig1-positive stem cells proliferated and repopulated damaged crypts.
In a simultaneously published study from Jensen’s group, Lrig1 was also found highly enriched in intestinal and colonic stem cells (Wong et al., 2012). However, in this study, Lrig1 and Lgr5 positivity overlapped and most Lrig1-positive cells were proliferative as measured by BrdU (5-bromodeoxyuridine) uptake. Notably, Lrig1 null mice (Suzuki et al., 2002, neo/neo) crossed into the FvB background displayed grossly enlarged abdomens necessitating sacrifice at post-natal day ten. The size of intestinal crypts in these mice was dramatically increased and crypts contained more proliferating cells. Flow cytometry revealed the expansion of the CD24low/mid/UEA-1-negative stem/progenitor population. In Lrig1-null intestinal epithelium, there was a substantial increase in the protein (but not transcript) levels of EGFR, ErbB2, ErbB3 and the Met receptor as well as an increase in the phosphorylation of EGFR/ErbB2/ErbB3, here again emphasizing the essential in vivo role of Lrig1 as a negative regulator of growth factor receptors. In support of this, daily administration of Gefitinib, an EGFR inhibitor, decreased EGFR phosphorylation and restored crypt size and proliferation to wild-type levels. Furthermore, the Lrig1 null phenotype was rescued (in approximately 40% of mice) by crossing with mice heterozygous for Egfrwa-2, an Egfr hypomorph (Luetteke et al., 1994).
Recently, Powell and colleagues (Powell et al., 2014) have reported the development of a superior mouse model of familial adenomatous polyposis (FAP). FAP arises from germline mutations in the APC (adenomatous polyposis coli) tumor suppressor with multiple colonic tumors arising early in life, subsequent to loss of the remaining APC allele. The most widely used mouse model of FAP is the ApcMin mouse (Moser et al., 1990) which also carries a germline mutation in Apc. However, unlike FAP patients, the ApcMin mouse develops mostly intestinal (rather than colonic) tumors of low grade, subsequent to loss of the remaining APC allele. In the model developed by Powell and colleagues (Lrig1-CreERT2/+;Apcfl/+), one copy of Apc is inducibly eliminated in Lrig1-positive (presumably stem/progenitor) cells, precipitating rapid pre-neoplastic changes, with multiple, high grade distal colonic adenomas developing subsequent to Apc loss of heterozygosity (as early as 50 days). In other available models, including the Lgr5-positive model (Barker et al., 2009), simultaneous rather than stepwise loss of Apc is required for tumorigenesis, which does not reflect the human disease. Moreover, the Lrig1-CreERT2/+;Apcfl/+ model shows some of the additional features of FAP, including highly penetrant periampullary tumors, gastric dysplasia and hyperplasia and lesions reminiscent of congenital hypertrophy of the retinal pigment epithelium (CHRPE), present in the majority of FAP patients.
Lrig1 in the lung
Given the prominent role of EGFR in human lung cancer (Siegelin & Borczuk, 2014), Lu and colleagues examined the impact of Lrig1 deletion on airway homeostasis (Lu et al., 2013). In the mouse, Lrig1 is expressed in the epithelium of the trachea and the first few bronchial divisions. In Lrig1 null mice (Suzuki et al., 2002, neo/neo), proliferation of tracheal and bronchial epithelium was increased, accompanied by increased phospho-EGFR staining, providing another in vivo link between Lrig1 and Egfr. This reciprocal relationship between LRIG1 and EGFR was also observed in human lung cancer specimens, as discussed above (LRIG expression in cancer). In murine tracheal epithelial cell (MTEC) air-liquid interface cultures, proliferation of wild type and Lrig1 null cells was similar in pre-confluent cultures but post confluence, Lrig1 null cells had a significant growth advantage, again accompanied by increased EGFR activation. Ectopic expression of LRIG1 in human A549 and H357 lung cancer cell lines, which express little endogenous LRIG1, significantly decreased cell proliferation post-confluence but not pre-confluence. LRIG1-mediated growth inhibition of H357 tumor cells was recapitulated in vivo, in subcutaneous xenografts, in which only one of eleven mice (9%) grew tumors from LRIG1 transduced cells while nine of ten mice (90%) grew tumors from the control group. The mechanism by which LRIG1 regulates cell contact-mediated growth inhibition is likely to be related to its ability to promote interaction between EGFR and E-cadherin. This EGFR/E-cadherin interaction was previously found to inhibit ligand-dependent activation of EGFR (Qian et al., 2004), and the present work from Lu and colleagues reveals a ternary complex containing EGFR/LRIG1/E-cadherin. The presumption is that LRIG1 is an essential mediator of the EGFR/E-cadherin interaction at sites of cell-cell contact but further study is necessary.
Lrig1 in the eye
Recently, gene expression profiling of human corneal keratinocytes demonstrated that LRIG1 is highly expressed in corneal epithelial stem cells (Nakamura et al., 2014), adding to an expanding list of LRIG1-positive stem cells. Analysis of Lrig1 null mice (Suzuki et al., 2002, neo/neo) revealed thickened and keratinized corneal epithelium with rampant inflammation of the corneal stroma, beginning at six months of age and manifesting in 70% of mice by twenty-four months of age. Corneal epithelial cells in Lrig1 null mice were more proliferative and showed evidence of a cell fate change, from corneal to keratinized epithelium. Time-dependent analysis of recovery from corneal wounding (which depends upon the integrity of corneal stem cells) demonstrated a deficit in Lrig1 null mice with incomplete wound healing. Comparison of wild type and null corneas revealed elevated Stat3 protein levels in null tissue, revealing a novel negative regulation of the Stat3 pathway by Lrig1. Upon wounding, phospho-Stat3 (which represents transcriptionally active Stat3) was detected in nuclei of Lrig1 null cornea while no phospho-Stat3 was observed in wild type tissue. Furthermore, a number of inflammatory cytokines were up-regulated in Lrig1 null tissue and this was exacerbated by corneal wounding. Finally, transgenic expression of constitutively active Stat3 in the basal layer of the epithelium recapitulated the Lrig1 null phenotype while inhibition of Stat3 with the small molecule STA21 prevented wound-induced corneal opacity in Lrig1 null mice, cementing the Stat3 pathway as a target of Lrig1. Bone marrow transplantation experiments demonstrated that wild type bone marrow reduced the severity of the wound-induced phenotype, demonstrating a cooperation between Lrig1 null bone marrow and corneal tissue. The mechanism by which Lrig1 regulates the Stat3 pathway is presently unknown. Analysis of other tissue types in Lrig1 null mice may reveal whether Stat3 regulation by Lrig1 is restricted to the cornea or is a more widespread mechanism.
Lrig2 null mice
Recently, the generation of Lrig2 null mice was reported (Rondahl et al., 2013). Lrig2 null mice showed increased mortality compared with heterozygous and wild type mice although the underlying mechanisms are not yet understood. Although embryo and birth weights were not different, null mice were found to be significantly smaller than heterozygous and wild type mice by five days of age and these differences persisted until twelve and fifteen weeks of age for females and males respectively. Given the prior finding that LRIG2 is associated with decreased survival in human oligodendroglioma (Holmlund et al., 2009), the impact of Lrig2 status on oligodendroglioma genesis in vivo was examined. In this study, a PDGF-B (platelet-derived growth factor-B)-driven model was used. Briefly, Ntv-a transgenic mice (which express the Tv-a avian retrovirus receptor under the control of the neural-specific nestin promoter) were generated on a wild type, heterozygous or Lrig2 null background. DF-1 cells producing PDGF-B-encoding retrovirus were then injected intracranially and tumor formation was analyzed. In the wild type setting, 100% of injected mice developed brain tumors by twelve weeks of age (82% low grade, 18% high grade). In the Lrig2 heterozygous setting, 100% of injected mice developed tumors (92% low grade, 8 % high grade) while in the Lrig2 null setting, only 77% of injected mice developed tumors by twelve weeks of age (100% low grade). Both tumor incidence and grade depended on Lrig2 genotype, uncovering a novel role for Lrig2 in promoting PDGFB-driven brain tumor formation. Intriguingly, LRIG2 had no apparent effect on expression or phosphorylation of PDGF receptors (α or β) while ectopic expression of LRIG1 was found to decrease PDGFRα expression. In addition, no effect of LRIG2 was observed on phosphorylation of Akt or Erk1/2, signaling cascades which lie downstream from PDGFR activation. However, the kinetics of immediate early gene expression following PDGF-B stimulation were altered in Lrig2 null MEFs (mouse embryo fibroblasts); expression of c-Fos and Egr2 peaked early and showed a more transient expression pattern. Given the complex interplay between signaling magnitude and duration in specifying cellular outcome (Kao et al., 2001), these changes in signaling kinetics could be functionally important. However, the mechanism by which LRIG2 impacts PDGF signaling is unknown and does not appear to be at the level of the PDGFRs.
LRIG2 has also been found to impact EGFR signaling in human glioma cells (Wang et al., 2009) implying that it may make multifactorial contributions to brain tumor malignancy. Depletion of LRIG2 in the GL15 glioblastoma cell line led to increased EGF-dependent EGFR turnover, decreased EGF-dependent EGFR phosphorylation, decreased cell growth and increased apoptosis, suggesting that LRIG2 plays a supportive role in EGFR signaling. However, LRIG2 depletion also increased GL15 cell invasion in vitro which was unexpected given its effects on EGFR. Since invasion is the major barrier to complete surgical resection of brain tumors (which in theory, would be curative), this suggests that any therapeutic strategy involving LRIGs must be approached carefully. On the other hand, LRIG1 depletion in GL15 cells led to increased EGFR and Akt phosphorylation, increased expression of c-Myc, increased cell proliferation and invasion and decreased apoptosis (Xie et al., 2013), suggesting that LRIG1 may play a more “straightforward” tumor suppressor role in brain malignancy. Indeed, overexpression of LRIG1 in U87MG-EGFRvIII (Stutz et al., 2008) and U251 (Ye et al., 2009, Mao et al., 2013) glioblastoma cells and H4 glioma cells (Ye et al., 2009) inhibited EGFR signaling and decreased cell proliferation and invasion. In U251 cells, overexpression of LRIG1 inhibited growth of tumor cells as subcutaneous xenografts (Mao et al., 2013).
Lrig3 null mice
In a search for regulators of inner ear morphogenesis using gene trap technology, Lrig3 was identified as essential for lateral canal development by Abraira and colleagues in 2008 (Abraira et al., 2008). A precisely timed and appropriately localized fusion process drives canal development and in Lrig3 null mice (Lrig3flox, C57Bl6 background), fusion occurs earlier and beyond normal boundaries compared to wild type mice, leading to lateral canal truncation. This phenotype is also observed in mice in which Lrig3 is specifically deleted in the otic epithelium, further refining Lrig3’s role in canal development (Abraira et al., 2010). Expression of Netrin1, a secreted protein related to laminin and an axon guidance molecule (Arakawa, 2004), is spatially expanded in Lrig3 null mice, demonstrating that one function of Lrig3 is to restrict Netrin1 expression to fusion plate cells. In agreement with this, lowering of the gene dosage of Netrin1 rescued the Lrig3 null phenotype, restoring normal canal development in Lrig3-null/Netrin1-heterozygous mice. The mechanisms by which Lrig3 restricts Netrin1 expression are not presently known although the authors hypothesize that Lrig3 “titrates” the activity of a receptor tyrosine kinase (such as Fgfr, known to be involved in inner ear development) which in turn, fine tunes Netrin1 expression. ErbB receptors do not appear to be involved in the Lrig3 phenotype as disruption of Neuregulin signaling with a dominant-negative ErbB4 has no effect on inner ear development in chick embryos, despite strong expression of ErbB2/ErbB3 in otic epithelium (Abraira et al., 2010).
Double-null mice
Unlike Lrig3 null mice, inner ear development is morphologically normal in Lrig1 and Lrig2 null mice (del Rio et al., 2013). However, in Lrig1/Lrig3 double null mice (Lrig1−/−; Lrig3−/−, C57Bl6 background), inner ear development is more severely impacted than when Lrig3 is deleted alone. Loss of Lrig1 in Lrig3 null mice does not worsen the lateral canal phenotype discussed above (and indeed, Lrig1 is not co-expressed with Lrig3 in lateral canal epithelium) but does lead to worsened phenotypes in sites of Lrig1/Lrig3 co-expression, suggesting a level of functional redundancy for these two Lrigs in the inner ear. For example, the utricle and saccule do not separate in double null mice and the posterior canal is smaller. Of note, Lrig1/Lrig3 double null mice die in utero or at birth and display defects in multiple tissues, sugge sting Lrig1/Lrig3 redundancy in other tissues. Conversely, Lrig1/Lrig2 double null mice (Lrig1−/−; Lrig2−/−) show no deficits in inner ear development despite overlapping expression, demonstrating that Lrig3 is sufficient to orchestrate proper inner ear development.
With respect to auditory sensitivity, Lrig1 single null mice (Lrig1−/−) required sounds thirty-fold more intense than wild type animals to elicit a response while Lrig2 single null (Lrig2−/−) and Lrig3 single null (Lrig3−/−) mice responded normally (del Rio et al., 2013). However, loss of Lrig2 exacerbated the Lrig1 auditory phenotype and raised the threshold for response. Functionally, Lrig1 was shown to be essential for the initial detection of sound while Lrig2 is required for the neuronal response to sound. Interestingly, cochlear morphology was normal in Lrig1/Lrig2 double null mice but innervation was abnormal with sparser efferent innervation. Analysis of Lrig1/Lrig2 double null mice was also complicated by early mortality although a small percentage survived to adulthood.
Soluble LRIG1 as a therapeutic
Restoration of tumor suppressors to deficient tumor cells may represent a promising therapeutic strategy in cancer. For example, the p53 tumor suppressor is the most commonly mutated gene in cancer and as such, there is intense interest in strategies which would enable restoration of wild type p53 function. A variety of approaches are being employed to target the p53 axis and some are already in clinical trials, highlighting the potential of this approach (Hong et al., 2014). Delivery of tumor suppressor microRNAs (as mimetics) or suppression of oncogenic microRNAs (with antagomirs) represents another highly feasible strategy and holds great promise for personalized cancer therapy (Sethi et al., 2014).
Ectopic expression of full length LRIG1 has been shown to inhibit the growth, motility and/or invasion of a variety of tumor cells, in vitro and in vivo (Laederich et al., 2004, Shattuck et al., 2007, Stutz et al., 2008, Miller et al., 2008, Ye et al., 2009, Thomasson et al., 2011, Li et al., 2011, Krig et al., 2011, Wang et al., 2012, Lu et al., 2013, Chang et al., 2013, Qi et al., 2013), making its potential restoration to LRIG1-deficient tumors of great interest. However, the translational applications of expressing full length LRIG1 are limited at this time due to its large size and membrane-bound nature. Nevertheless, the soluble extracellular domain of LRIG1 (sLRIG1), containing only the leucine-rich repeats (with their flanking caps) or a decorin fusion thereof, was found to inhibit the growth of EGFR-expressing A431 squamous carcinoma cells (high expressors) and HeLa cervical adenocarcinoma cells (low expressors) (Goldoni et al., 2007). sLRIG1 was demonstrated to specifically bind to the EGFR and inhibit both ligand-independent (basal) and ligand-dependent EGFR phosphorylation. However, unlike full length LRIG1, sLRIG1 did not stimulate receptor degradation.
Expression of a larger version of sLRIG1, containing the entire extracellular domain, was found to inhibit EGF-dependent Fos induction and cell proliferation in COS-7 cells (Yi et al., 2011). In this study, full length LRIG1 was revealed to undergo proteolytic processing in COS-7 cells, yielding a predominant 100 kD species, likely corresponding to the entire extracellular domain, and a less prominent 60 kD species. Production of sLRIG1 in COS7 cells was largely dependent upon the ADAM17 metalloprotease as it was significantly inhibited by the specific ADAM17 inhibitor, TAPI-2. Expression of full length LRIG1 in HPB-ALL leukemia cells also led to the production of a 100 kD sLRIG1 species and in co-culture experiments, LRIG1-expressing HPB-ALL cells inhibited EGF signaling in COS-7 cells, in a manner dependent upon metalloprotease activity (i.e. LRIG1 shedding from HPB-ALL cells). Analysis of mouse (tail skin) and human (prostate, ileum, stomach and skin) tissues detected the expression of a 60kD Lrig1 species, strongly suggesting that Lrig1 processing occurs in vivo. As reported in the Goldoni et al. study, sLRIG1 did not promote EGFR degradation.
A key study highlighting the translational utility of sLRIG1 was recently published by Johansson and colleagues (Johansson. et al, 2013). Using cell encapsulation technology which allows for stable and prolonged expression of recombinant proteins by cell-based “bioreactors”, the impact of sLRIG1 (entire extracellular domain) on growth of patient-derived, orthotopic glioblastoma xenografts was assessed. sLRIG1 significantly inhibited tumor growth in vivo, in a manner independent of EGFR expression level. sLRIG1 had a similar inhibitory effect on two patient-derived xenografts which differed greatly in their EGFR expression level (one with EGFR gene amplification, one without EGFR amplification). This independence from EGFR expression level was also observed with xenografts of U87 glioblastoma cells; sLRIG1 had a similar growth inhibitory effect on U87, U87-EGFR and U87-EGFRvIII (an oncogenic form of EGFR frequently expressed in glioblastoma) cells. This is in contrast to Goldoni et al. who reported that EGFR expression was necessary for sLRIG1’s inhibitory activity. In co-culture experiments of U87 cells with sLRIG1-expressing cells, sLRIG1 had no effect on total EGFR expression or EGFR cell surface expression (as expected) but also, did not decrease EGF-stimulated EGFR phosphorylation (somewhat unexpected given prior studies). However, sLRIG1 did inhibit EGF-stimulated MAP kinase activity, suggesting that it was impacting EGFR signaling, and slowed cell cycle progression. Most notably, sLRIG1 delivered to patient-derived tumors, which were first allowed to establish for two weeks, was effective in significantly prolonging survival (32% increase in survival for mice implanted with sLRIG1 producing cells). This finding is particularly important because any potential therapy must be effective in the setting of established tumors. These findings emphasize the potential of sLRIG1 as a therapeutic and also extend its likely utility beyond EGFR-overexpressing malignancies.
Beyond sLRIG1, therapeutic possibilities for LRIG1-deficient tumors include micro-RNA based re-expression of LRIG1 and inhibition of pathways that are activated upon LRIG1 depletion. With respect to microRNAs which regulate LRIG1, essentially nothing is known. LRIG1 was found to be under-expressed in chronic lymphocytic leukemia (CLL) patients who expressed low levels of MIR15a/16-1 although there was no molecular analysis conducted (Hanlon et al., 2009). With respect to pathways activated upon LRIG1 depletion, the MAP kinase inhibitor U1026 or the Sphingosine kinase-1 inhibitor SK1-I was found to significantly inhibit the survival, migration and invasion advantages of head and neck cancer cells in which LRIG1 was knocked-down (Sheu et al., 2014). An obvious caveat to this approach is that LRIG1 depletion/loss may lead to the activation of multiple pathways which could not all be effectively inhibited clinically. Indeed, knock-down of LRIG1 in MDA-MB-231 breast cancer cells and SKOV3 ovarian cancer cells leads to increased expression of multiple receptor tyrosine kinases, including Met and Ron (Bai et al., 2012). On the other hand, tumors which retain LRIG1 expression may be particularly sensitive to certain therapeutics, opening a window of opportunity. For example, Smac mimetics, small molecules which are under clinical development, mimic endogenous Smac/DIABLO and inhibit the inhibitors of apoptosis (IAP) proteins cIAP1/2 and XIAP, triggering TNF-α-dependent apoptosis. The requirement of LRIG1 expression for SM-164 (Smac mimetic)-induced TNFα expression and growth inhibition was shown in knock-down experiments (Bai et al., 2012). SM-164 resistant cells expressed low levels of LRIG1 and increased levels of receptor tyrosine kinases. However, the mechanism by which LRIG1 regulates SM-164-induced TNFα expression is unknown. Sensitivity to SM-164 was restored by tyrosine kinase inhibitors which target multiple kinases, including GSK1363089 and PF2341066, suggesting that activation of these kinase pathways (which would happen in a LRIG1-depleted setting) may suppress SM-164-mediated TNFα expression.
LRIG mechanism of action
A persistent area of controversy in the LRIG field regards the molecular mechanism(s) by which the LRIG proteins function. The first studies implicating LRIG1 as a negative regulator of ErbB receptor tyrosine kinases both found that LRIG1 enhanced receptor ubiquitination and proteolytic degradation (Laederich et al., 2004, Gur et al., 2004). Gur et al. delved deeper into the molecular mechanism and reported that LRIG1 recruits the c-Cbl ubiquitin ligase to the EGFR/LRIG1 complex (via interaction of c-Cbl with the LRIG1 cytoplasmic domain) and that c-Cbl is functionally essential for LRIG1-mediated EGFR degradation. The authors proposed that LRIG1 evolved as a surrogate means of c-Cbl recruitment. However, Cbl-70Z, a well characterized dominant negative form of c-Cbl, was found to have no effect on LRIG1-mediated down-regulation of EGFR and EGFRvIII (Stutz et al., 2008), suggesting that regulation of EGFR by LRIG1 is c-Cbl independent. A subsequent study found that LRIG1-mediated Met destabilization is c-Cbl independent (Shattuck et al., 2007), despite Met being a well-known target of c-Cbl (Peschard et al., 2001). In support of this, it was recently reported that the SAIT301 antibody promotes Met receptor degradation, in a LRIG1-dependent, c-Cbl independent manner (Lee et al., 2014). LRIG1 itself becomes ubiquitinated following SAIT301 treatment, promoting its interaction with Hepatocyte growth factor-regulated tyrosine kinase substrate (HRS), which recognizes ubiquitinated cargo and facilitates sorting into multivesicular bodies for eventual lysosomal degradation (Oh et al., 2014). In this study, LRIG1 was found to promote lysosomal degradation of Met through interaction of the LRIG1-Met complex with HRS. Furthermore, LRIG1 was revealed to be a target of the deubiquitinating enzyme UPS8 which was shown to counteract LRIG1 ubiquitination and degradation. Interestingly, SAIT301 triggers LRIG1/Met degradation by interfering with LRIG1-USP8 interaction.
Recently, deletion analysis of LRIG1 led to the finding that Δ-cyto LRIG1 (lacking the entire cytoplasmic domain which contains the c-Cbl binding site) is fully active in mediating ErbB2 degradation (Rafidi et al., 2013), underscoring the existence of a c-Cbl independent mechanism for LRIG1. This is supported by the prior finding that Δ-cyto LRIG1 and wild type LRIG1 have similar abilities to inhibit the growth of glioma cells (Yi et al., 2011).
In light of these recent findings, the model of LRIG1 mechanism of action must be revised. Mounting evidence suggests that it is unlikely that LRIG1 functions in a c-Cbl-dependent manner. If this is the case, the question lingers as to how LRIG1 promotes the degradation of its targets. Knockdown of LRIG1 has been found to increase the cell surface expression of EGFR (Jensen & Watt, 2006) while ectopic expression of LRIG1 has been found to enhance the EGF-stimulated internalization of EGFR (Rafidi et al., 2013), strongly suggesting that LRIG1 does engage the receptor internalization machinery. Co-expression of LRIG1 with ErbB4 leads to their co-localization in intracellular vesicles but the identity of these vesicles is unknown (Abraira et al., 2010). Furthermore, Sma-10, the C-elegans LRIG ortholog, is localized to intracellular puncta, possibly endocytic vesicles (Gumienny et al., 2010). The ability of LRIG1 to enhance EGFR internalization depends upon its extracellular domain as the Δ-ecto mutant of LRIG1 was ineffective in stimulating EGFR internalization (Rafidi et al., 2013). This is not surprising as the extracellular domain is necessary for physical interaction with EGFR and other targets (Gur et al., 2004, Rafidi et al., 2013). However, the LRIG1 extracellular domain could conceivably interact with a third component which spans the membrane and engages the cytosolic receptor degradation machinery (Rafidi et al., 2013). This model does not contradict with the finding that Δ-cyto LRIG1 is as active as full length LRIG1 while the c-Cbl dependent mechanism does not explain the activity of Δ-cyto LRIG1. Recruitment of HRS to LRIG1 (Oh et al., 2014), which presumably occurs through HRS-mediated recognition of ubiquitinated residues in the LRIG1 cyto domain, also does not explain the activity of Δ-cyto LRIG1.
Conclusion
The study of LRIG proteins has spanned nearly two decades, with the initial cloning of mouse LRIG1 in 1996 (Suzuki et al., 1996). Tremendous knowledge has been gained through the concerted efforts of LRIG researchers, using in vitro and in vivo approaches. Most notably, LRIG1 has “arrived” on the cancer biology scene as a tumor suppressor. The seminal hypothesis that LRIG1 is a functional homolog of Kekkon-1, a negative regulator of Egfr signaling in flies (Nilsson et al., 2001, Suzuki et al., 2002, Hedman et al., 2002), spurred the field forward and provided a molecular rationale for the pursuit of LRIG1 (Laederich et al., 2004, Gur et al., 2004). While Lambik (rather than Kekkon-1) is now recognized as the Lrig ortholog in flies, LRIG1 and Kekkon-1 share functional attributes, including the ability to inhibit EGFR phosphorylation and signaling. In this sense, we have come full circle with LRIG1, from candidate regulator of EGFR/ErbB receptors to in vivo validated negative regulator of growth factor receptors, including EGFR/ErbB receptors. Learning more about Lambik function, of which nothing is known beyond its ability to rescue Sma-10 mutant animals (Gumienny et al., 2010), should provide deeper insight into LRIG function. Many incredibly important questions linger in the LRIG field, both in developmental and cancer biology, and we look forward to their resolution in the years to come.
Acknowledgments
Funding
Salary support for the authors was provided by UC-MEXUS (MECP) and by NIH grant CA118384 (CS and CS).
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the review.
References
- Abraira VE, Del Rio T, Tucker AF, Slonimsky J, Keirnes HL, Goodrich LV. Cross-repressive interactions between Lrig3 and netrin 1 shape the architecture of the inner ear. Development. 2008;135:4091–4099. doi: 10.1242/dev.029330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraira VE, Satoh T, Fekete DM, Goodrich LV. Vertebrate Lrig3-ErbB interactions occur in vitro but are unlikely to play a role in Lrig3-dependent inner ear morphogenesis. PLoS One. 2010;5:e8981. doi: 10.1371/journal.pone.0008981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H. Netrin-1 and its receptors in tumorigenesis. Nat Rev Cancer. 2004;4:978–987. doi: 10.1038/nrc1504. [DOI] [PubMed] [Google Scholar]
- Bai L, McEachern D, Yang CY, Lu J, Sun H, Wang S. LRIG1 modulates cancer cell sensitivity to Smac mimetics by regulating TNFalpha expression and receptor tyrosine kinase signaling. Cancer Res. 2012;72:1229–1238. doi: 10.1158/0008-5472.CAN-11-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchetta J, Cochat P. Severe voiding dysfunction: ask the child to smile. Kidney Int. 2010;78:225–226. doi: 10.1038/ki.2010.144. [DOI] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Shi R, Yang T, Li F, Li G, Guo Y, Lang B, Yang W, Zhuang Q, Xu H. Restoration of LRIG1 suppresses bladder cancer cell growth by directly targeting EGFR activity. J Exp Clin Cancer Res. 2013;32:101. doi: 10.1186/1756-9966-32-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly SB, Urquhart JE, Hilton E, McKenzie EA, Kammerer RA, Lewis M, Kerr B, Stuart H, Donnai D, Long DA, et al. Mutations in HPSE2 cause urofacial syndrome. Am J Hum Genet. 2010;86:963–969. doi: 10.1016/j.ajhg.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio T, Nishitani AM, Yu WM, Goodrich LV. In vivo analysis of Lrig genes reveals redundant and independent functions in the inner ear. PLoS Genet. 2013;9:e1003824. doi: 10.1371/journal.pgen.1003824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldoni S, Iozzo RA, Kay P, Campbell S, McQuillan A, Agnew C, Zhu JX, Keene DR, Reed CC, Iozzo RV. A soluble ectodomain of LRIG1 inhibits cancer cell growth by attenuating basal and ligand-dependent EGFR activity. Oncogene. 2007;26:368–381. doi: 10.1038/sj.onc.1209803. [DOI] [PubMed] [Google Scholar]
- Gumienny TL, Macneil L, Zimmerman CM, Wang H, Chin L, Wrana JL, Padgett RW. Caenorhabditis elegans SMA-10/LRIG is a conserved transmembrane protein that enhances bone morphogenetic protein signaling. PLoS Genet. 2010;6:e1000963. doi: 10.1371/journal.pgen.1000963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Holmlund C, Henriksson R, Hedman H. The LRIG gene family has three vertebrate paralogs widely expressed in human and mouse tissues and a homolog in Ascidiacea. Genomics. 2004;84:157–165. doi: 10.1016/j.ygeno.2004.01.013. [DOI] [PubMed] [Google Scholar]
- Guo D, Nilsson J, Haapasalo H, Raheem O, Bergenheim T, Hedman H, Henriksson R. Perinuclear leucine-rich repeats and immunoglobulin-like domain proteins (LRIG1-3) as prognostic indicators in astrocytic tumors. Acta Neuropathol. 2006;111:238–246. doi: 10.1007/s00401-006-0032-5. [DOI] [PubMed] [Google Scholar]
- Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, Amariglio N, Henriksson R, Rechavi G, Hedman H, et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004;23:3270–3281. doi: 10.1038/sj.emboj.7600342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon K, Rudin CE, Harries LW. Investigating the targets of MIR-15a and MIR-16-1 in patients with chronic lymphocytic leukemia (CLL) PLoS One. 2009;4:e7169. doi: 10.1371/journal.pone.0007169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedman H, Nilsson J, Guo D, Henriksson R. Is LRIG1 a tumour suppressor gene at chromosome 3p14.3? Acta Oncol. 2002;41:352–354. doi: 10.1080/028418602760169398. [DOI] [PubMed] [Google Scholar]
- Hedman H, Henriksson R. LRIG inhibitors of growth factor signalling - double-edged swords in human cancer? Eur J Cancer. 2007;43:676–682. doi: 10.1016/j.ejca.2006.10.021. [DOI] [PubMed] [Google Scholar]
- Hedman H, Lindstrom AK, Tot T, Stendahl U, Henriksson R, Hellberg D. LRIG2 in contrast to LRIG1 predicts poor survival in early-stage squamous cell carcinoma of the uterine cervix. Acta Oncol. 2010;49:812–815. doi: 10.3109/0284186X.2010.492789. [DOI] [PubMed] [Google Scholar]
- Holmlund C, Nilsson J, Guo D, Starefeldt A, Golovleva I, Henriksson R, Hedman H. Characterization and tissue-specific expression of human LRIG2. Gene. 2004;332:35–43. doi: 10.1016/j.gene.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Holmlund C, Haapasalo H, Yi W, Raheem O, Brannstrom T, Bragge H, Henriksson R, Hedman H. Cytoplasmic LRIG2 expression is associated with poor oligodendroglioma patient survival. Neuropathology. 2009;29:242–247. doi: 10.1111/j.1440-1789.2008.00970.x. [DOI] [PubMed] [Google Scholar]
- Hong B, van den Heuvel AP, Prabhu VV, Zhang S, El-Deiry WS. Targeting tumor suppressor p53 for cancer therapy: strategies, challenges and opportunities. Curr Drug Targets. 2014;15:80–89. doi: 10.2174/1389450114666140106101412. [DOI] [PubMed] [Google Scholar]
- Jensen KB, Watt FM. Single-cell expression profiling of human epidermal stem and transit-amplifying cells: Lrig1 is a regulator of stem cell quiescence. Proc Natl Acad Sci U S A. 2006;103:11958–11963. doi: 10.1073/pnas.0601886103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Jones J, Watt FM. A stem cell gene expression profile of human squamous cell carcinomas. Cancer Lett. 2008;272:23–31. doi: 10.1016/j.canlet.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Collins CA, Nascimento E, Tan DW, Frye M, Itami S, Watt FM. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell. 2009;4:427–439. doi: 10.1016/j.stem.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Oudin A, Tiemann K, Bernard A, Golebiewska A, Keunen O, Fack F, Stieber D, Wang B, Hedman H, et al. The soluble form of the tumor suppressor Lrig1 potently inhibits in vivo glioma growth irrespective of EGF receptor status. Neuro Oncol. 2013;15:1200–1211. doi: 10.1093/neuonc/not054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S, Jaiswal RK, Kolch W, Landreth GE. Identification of the mechanisms regulating the differential activation of the mapk cascade by epidermal growth factor and nerve growth factor in PC12 cells. J Biol Chem. 2001;276:18169–18177. doi: 10.1074/jbc.M008870200. [DOI] [PubMed] [Google Scholar]
- Karlsson T, Mark EB, Henriksson R, Hedman H. Redistribution of LRIG proteins in psoriasis. J Invest Dermatol. 2008;128:1192–1195. doi: 10.1038/sj.jid.5701175. [DOI] [PubMed] [Google Scholar]
- Krig SR, Frietze S, Simion C, Miller JK, Fry WH, Rafidi H, Kotelawala L, Qi L, Griffith OL, Gray JW, et al. Lrig1 is an estrogen-regulated growth suppressor and correlates with longer relapse-free survival in ERalpha-positive breast cancer. Mol Cancer Res. 2011;9:1406–1417. doi: 10.1158/1541-7786.MCR-11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL, 3rd, Sweeney C. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem. 2004;279:47050–47056. doi: 10.1074/jbc.M409703200. [DOI] [PubMed] [Google Scholar]
- Ledda F, Bieraugel O, Fard SS, Vilar M, Paratcha G. Lrig1 is an endogenous inhibitor of Ret receptor tyrosine kinase activation, downstream signaling, and biological responses to GDNF. J Neurosci. 2008;28:39–49. doi: 10.1523/JNEUROSCI.2196-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Kim B, Lee SB, Jeong Y, Oh YM, Song YJ, Jung S, Choi J, Lee S, Cheong KH, et al. Cbl-independent degradation of Met: ways to avoid agonism of bivalent Met-targeting antibody. Oncogene. 2014;33:34–43. doi: 10.1038/onc.2012.551. [DOI] [PubMed] [Google Scholar]
- Li F, Ye ZQ, Guo DS, Yang WM. Suppression of bladder cancer cell tumorigenicity in an athymic mouse model by adenoviral vector-mediated transfer of LRIG1. Oncol Rep. 2011;26:439–446. doi: 10.3892/or.2011.1304. [DOI] [PubMed] [Google Scholar]
- Lindquist D, Nasman A, Tarjan M, Henriksson R, Tot T, Dalianis T, Hedman H. Expression of LRIG1 is associated with good prognosis and human papillomavirus status in oropharyngeal cancer. Br J Cancer. 2014;110:1793–1800. doi: 10.1038/bjc.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Teixeira VH, Yuan Z, Graham TA, Endesfelder D, Kolluri K, Al-Juffali N, Hamilton N, Nicholson AG, Falzon M, et al. LRIG1 regulates cadherin-dependent contact inhibition directing epithelial homeostasis and pre-invasive squamous cell carcinoma development. J Pathol. 2013;229:608–620. doi: 10.1002/path.4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luetteke NC, Phillips HK, Qiu TH, Copeland NG, Earp HS, Jenkins NA, Lee DC. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes Dev. 1994;8:399–413. doi: 10.1101/gad.8.4.399. [DOI] [PubMed] [Google Scholar]
- Mao F, Wang B, Xiao Q, Xi G, Sun W, Zhang H, Ye F, Wan F, Guo D, Lei T, et al. A role for LRIG1 in the regulation of malignant glioma aggressiveness. Int J Oncol. 2013;42:1081–1087. doi: 10.3892/ijo.2013.1776. [DOI] [PubMed] [Google Scholar]
- Miller JK, Shattuck DL, Ingalla EQ, Yen L, Borowsky AD, Young LJ, Cardiff RD, Carraway KL, 3rd, Sweeney C. Suppression of the negative regulator LRIG1 contributes to ErbB2 overexpression in breast cancer. Cancer Res. 2008;68:8286–8294. doi: 10.1158/0008-5472.CAN-07-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischel PS, Shai R, Shi T, Horvath S, Lu KV, Choe G, Seligson D, Kremen TJ, Palotie A, Liau LM, et al. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22:2361–2373. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- Muller S, Lindquist D, Kanter L, Flores-Staino C, Henriksson R, Hedman H, Andersson S. Expression of LRIG1 and LRIG3 correlates with human papillomavirus status and patient survival in cervical adenocarcinoma. Int J Oncol. 2013;42:247–252. doi: 10.3892/ijo.2012.1702. [DOI] [PubMed] [Google Scholar]
- Nagata M, Nakamura T, Sotozono C, Inatomi T, Yokoi N, Kinoshita S. LRIG1 as a potential novel marker for neoplastic transformation in ocular surface squamous neoplasia. PLoS One. 2014;9:e93164. doi: 10.1371/journal.pone.0093164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Hamuro J, Takaishi M, Simmons S, Maruyama K, Zaffalon A, Bentley AJ, Kawasaki S, Nagata-Takaoka M, Fullwood NJ, et al. LRIG1 inhibits STAT3-dependent inflammation to maintain corneal homeostasis. J Clin Invest. 2014;124:385–397. doi: 10.1172/JCI71488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Vallbo C, Guo D, Golovleva I, Hallberg B, Henriksson R, Hedman H. Cloning, characterization, and expression of human LIG1. Biochem Biophys Res Commun. 2001;284:1155–1161. doi: 10.1006/bbrc.2001.5092. [DOI] [PubMed] [Google Scholar]
- Oh YM, Lee SB, Choi J, Suh HY, Shim S, Song YJ, Kim B, Lee JM, Oh SJ, Jeong Y, et al. USP8 modulates ubiquitination of LRIG1 for Met degradation. Sci Rep. 2014;4:4980. doi: 10.1038/srep04980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page ME, Lombard P, Ng F, Gottgens B, Jensen KB. The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell Stem Cell. 2013;13:471–482. doi: 10.1016/j.stem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang J, Zhang S, Yang P, Hawkins-Lee B, Zhong J, Zhang Y, Ochoa B, Agundez JA, Voelckel MA, Fisher RB, et al. Loss-of-function mutations in HPSE2 cause the autosomal recessive urofacial syndrome. Am J Hum Genet. 2010;86:957–962. doi: 10.1016/j.ajhg.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschard P, Fournier TM, Lamorte L, Naujokas MA, Band H, Langdon WY, Park M. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol Cell. 2001;8:995–1004. doi: 10.1016/s1097-2765(01)00378-1. [DOI] [PubMed] [Google Scholar]
- Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AE, Vlacich G, Zhao ZY, McKinley ET, Washington MK, Manning HC, Coffey RJ. Inducible loss of one Apc allele in Lrig1-expressing progenitor cells results in multiple distal colonic tumors with features of familial adenomatous polyposis. Am J Physiol Gastrointest Liver Physiol. 2014;307:G16–23. doi: 10.1152/ajpgi.00358.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi XC, Xie DJ, Yan QF, Wang YR, Zhu YX, Qian C, Yang SX. LRIG1 dictates the chemo-sensitivity of temozolomide (TMZ) in U251 glioblastoma cells via down-regulation of EGFR/topoisomerase-2/Bcl-2. Biochem Biophys Res Commun. 2013;437:565–572. doi: 10.1016/j.bbrc.2013.06.116. [DOI] [PubMed] [Google Scholar]
- Qian X, Karpova T, Sheppard AM, McNally J, Lowy DR. E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. EMBO J. 2004;23:1739–1748. doi: 10.1038/sj.emboj.7600136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafidi H, Mercado F, 3rd, Astudillo M, Fry WH, Saldana M, Carraway KL, 3rd, Sweeney C. Leucine-rich repeat and immunoglobulin domain-containing protein-1 (Lrig1) negative regulatory action toward ErbB receptor tyrosine kinases is opposed by leucine-rich repeat and immunoglobulin domain-containing protein 3 (Lrig3) J Biol Chem. 2013;288:21593–21605. doi: 10.1074/jbc.M113.486050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenberger G, Ichimura K, Reifenberger J, Elkahloun AG, Meltzer PS, Collins VP. Refined mapping of 12q13-q15 amplicons in human malignant gliomas suggests CDK4/SAS and MDM2 as independent amplification targets. Cancer Res. 1996;56:5141–5145. [PubMed] [Google Scholar]
- Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts NA, Woolf AS, Stuart HM, Thuret R, McKenzie EA, Newman WG, Hilton EN. Heparanase 2, mutated in urofacial syndrome, mediates peripheral neural development in Xenopus. Hum Mol Genet. 2014;23:4302–4314. doi: 10.1093/hmg/ddu147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondahl V, Holmlund C, Karlsson T, Wang B, Faraz M, Henriksson R, Hedman H. Lrig2-deficient mice are protected against PDGFB-induced glioma. PLoS One. 2013;8:e73635. doi: 10.1371/journal.pone.0073635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouam S, Moreau T, Broet P. Identifying common prognostic factors in genomic cancer studies: a novel index for censored outcomes. BMC Bioinformatics. 2010;11:150. doi: 10.1186/1471-2105-11-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi S, Ali S, Sarkar FH. MicroRNAs in personalized cancer therapy. Clin Genet. 2014 doi: 10.1111/cge.12362. [DOI] [PubMed] [Google Scholar]
- Shattuck DL, Miller JK, Laederich M, Funes M, Petersen H, Carraway KL, 3rd, Sweeney C. LRIG1 is a novel negative regulator of the Met receptor and opposes Met and Her2 synergy. Mol Cell Biol. 2007;27:1934–1946. doi: 10.1128/MCB.00757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu JJ, Lee CC, Hua CH, Li CI, Lai MT, Lee SC, Cheng J, Chen CM, Chan C, Chao SC, et al. LRIG1 modulates aggressiveness of head and neck cancers by regulating EGFR-MAPK-SPHK1 signaling and extracellular matrix remodeling. Oncogene. 2014;33:1375–1384. doi: 10.1038/onc.2013.98. [DOI] [PubMed] [Google Scholar]
- Siegelin MD, Borczuk AC. Epidermal growth factor receptor mutations in lung adenocarcinoma. Lab Invest. 2014;94:129–137. doi: 10.1038/labinvest.2013.147. [DOI] [PubMed] [Google Scholar]
- Stuart HM, Roberts NA, Burgu B, Daly SB, Urquhart JE, Bhaskar S, Dickerson JE, Mermerkaya M, Silay MS, Lewis MA, et al. LRIG2 mutations cause urofacial syndrome. Am J Hum Genet. 2013;92:259–264. doi: 10.1016/j.ajhg.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz MA, Shattuck DL, Laederich MB, Carraway KL, 3rd, Sweeney C. LRIG1 negatively regulates the oncogenic EGF receptor mutant EGFRvIII. Oncogene. 2008;27:5741–5752. doi: 10.1038/onc.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Sato N, Tohyama M, Wanaka A, Takagi T. cDNA cloning of a novel membrane glycoprotein that is expressed specifically in glial cells in the mouse brain. LIG-1, a protein with leucine-rich repeats and immunoglobulin-like domains. J Biol Chem. 1996;271:22522–22527. doi: 10.1074/jbc.271.37.22522. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Miura H, Tanemura A, Kobayashi K, Kondoh G, Sano S, Ozawa K, Inui S, Nakata A, Takagi T, et al. Targeted disruption of LIG-1 gene results in psoriasiform epidermal hyperplasia. FEBS Lett. 2002;521:67–71. doi: 10.1016/s0014-5793(02)02824-7. [DOI] [PubMed] [Google Scholar]
- Tanemura A, Nagasawa T, Inui S, Itami S. LRIG-1 provides a novel prognostic predictor in squamous cell carcinoma of the skin: immunohistochemical analysis for 38 cases. Dermatol Surg. 2005;31:423–430. doi: 10.1111/j.1524-4725.2005.31108. [DOI] [PubMed] [Google Scholar]
- Thomasson M, Wang B, Hammarsten P, Dahlman A, Persson JL, Josefsson A, Stattin P, Granfors T, Egevad L, Henriksson R, et al. LRIG1 and the liar paradox in prostate cancer: a study of the expression and clinical significance of LRIG1 in prostate cancer. Int J Cancer. 2011;128:2843–2852. doi: 10.1002/ijc.25820. [DOI] [PubMed] [Google Scholar]
- Thomasson M, Hedman H, Ljungberg B, Henriksson R. Gene expression pattern of the epidermal growth factor receptor family and LRIG1 in renal cell carcinoma. BMC Res Notes. 2012;5:216. doi: 10.1186/1756-0500-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PA, Ljuslinder I, Tsavachidis S, Brewster A, Sahin A, Hedman H, Henriksson R, Bondy ML, Melin BS. Loss of LRIG1 Locus Increases Risk of Early and Late Relapse of Stage I/II Breast Cancer. Cancer Res. 2014;74:2928–2935. doi: 10.1158/0008-5472.CAN-13-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Han L, Chen R, Cai M, Han F, Lei T, Guo D. Downregulation of LRIG2 expression by RNA interference inhibits glioblastoma cell (GL15) growth, causes cell cycle redistribution, increases cell apoptosis and enhances cell adhesion and invasion in vitro. Cancer Biol Ther. 2009;8:1018–1023. doi: 10.4161/cbt.8.11.8375. [DOI] [PubMed] [Google Scholar]
- Wang X, Xiao Q, Xing X, Tian C, Zhang H, Ye F, Wan F, Wang B, Guo D, Lei T. LRIG1 enhances cisplatin sensitivity of glioma cell lines. Oncol Res. 2012;20:205–211. doi: 10.3727/096504013x13589503482770. [DOI] [PubMed] [Google Scholar]
- Wang Y, Poulin EJ, Coffey RJ. LRIG1 is a triple threat: ERBB negative regulator, intestinal stem cell marker and tumour suppressor. Br J Cancer. 2013;108:1765–1770. doi: 10.1038/bjc.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Wu J, Song H. LRIG2 expression and prognosis in non-small cell lung cancer. Oncol Lett. 2014;8:667–672. doi: 10.3892/ol.2014.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong VW, Stange DE, Page ME, Buczacki S, Wabik A, Itami S, van de Wetering M, Poulsom R, Wright NA, Trotter MW, et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol. 2012;14:401–408. doi: 10.1038/ncb2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R, Yang H, Xiao Q, Mao F, Zhang S, Ye F, Wan F, Wang B, Lei T, Guo D. Downregulation of LRIG1 expression by RNA interference promotes the aggressive properties of glioma cells via EGFR/Akt/c-Myc activation. Oncol Rep. 2013;29:177–184. doi: 10.3892/or.2012.2102. [DOI] [PubMed] [Google Scholar]
- Ye F, Gao Q, Xu T, Zeng L, Ou Y, Mao F, Wang H, He Y, Wang B, Yang Z, et al. Upregulation of LRIG1 suppresses malignant glioma cell growth by attenuating EGFR activity. J Neurooncol. 2009;94:183–194. doi: 10.1007/s11060-009-9836-1. [DOI] [PubMed] [Google Scholar]
- Yi W, Holmlund C, Nilsson J, Inui S, Lei T, Itami S, Henriksson R, Hedman H. Paracrine regulation of growth factor signaling by shed leucine-rich repeats and immunoglobulin-like domains 1. Exp Cell Res. 2011;317:504–512. doi: 10.1016/j.yexcr.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Zhao H, Tanegashima K, Ro H, Dawid IB. Lrig3 regulates neural crest formation in Xenopus by modulating Fgf and Wnt signaling pathways. Development. 2008;135:1283–1293. doi: 10.1242/dev.015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Luoh SM, Zhang Y, Watanabe C, Wu TD, Ostland M, Wood WI, Zhang Z. Genome-wide identification of chromosomal regions of increased tumor expression by transcriptome analysis. Cancer Res. 2003;63:5781–5784. [PubMed] [Google Scholar]