SUMMARY
While we have considerable understanding of the transcriptional networks controlling mammalian cell differentiation, our knowledge of post-transcriptional regulatory events is very limited. Using differentiation of primary erythroid cells as a model, we show that the sequence-specific mRNA-binding protein Cpeb4 is strongly induced by the erythroid important transcription factors Gata1 and Tal1 and is essential for terminal erythropoiesis. By interacting with the translation initiation factor eIF3 Cpeb4 represses the translation of a large set of mRNAs, including its own mRNA. Thus transcriptional induction and translational repression combine to form a negative feedback loop to control Cpeb4 protein levels within a specific range that is required for terminal erythropoiesis. Our study provides an example of how translational control is integrated with transcriptional regulation to precisely control gene expression during mammalian cell differentiation.
INTRODUCTION
Gene regulation during development is regulated at multiple transcriptional and post-transcriptional levels, including control of mRNA translation and degradation. Whereas a great deal is known about the transcriptional regulatory networks that control cell type specificity and differentiation, much less is understood about the post-transcriptional regulatory circuits that are essential for mammalian development, particularly somatic cell differentiation. Here we use terminal erythropoiesis as a system to explore post-transcriptional events that control an important terminal cell differentiation pathway.
During the final stage of erythropoiesis, the erythropoietin (Epo)-responsive erythroid CFU-E (colony forming unit erythroids) progenitors undergo dramatic changes in morphology and in protein expression. In the presence of Epo, CFU-Es divide 4–5 times and undergo dramatic decreases in both nuclear and cell sizes, chromatin condensation, hemoglobinization, and ultimately extrusion of the nuclei, forming enucleated reticulocytes. These changes are accompanied by significant transcriptome reprogramming; ~600 genes are induced and ~6000 genes are repressed at the RNA level during terminal erythroid differentiation (Wong et al., 2011). These transcriptomic changes are mediated by several key erythroid-important transcription factors, including Gata1, Tal1, and EKLF, as well as by the Epo-Epo receptor-Jak2 -Stat5 signaling pathway (Hattangadi et al., 2011; Kerenyi and Orkin, 2010). Much less is known concerning post-transcriptional regulation of gene expression.
Here we used primary mouse fetal liver cells to explore the post-transcriptional regulatory events in terminal erythroid differentiation. From embryonic days 12–16 (E12–16), mouse fetal liver is the primary site of erythropoiesis. Most (90%) fetal liver cells are in the erythroid lineage, providing us with a relatively pure source of erythroid cells. Erythroid cells at different developmental stages (BFU-Es, CFU-Es, and mature Ter119+ cells) can be purified using different combinations of cell surface markers (Flygare et al., 2011). Cultured late erythroid progenitor cells, predominantly CFU-Es, undergo terminal proliferation and differentiation into enucleated reticulocytes in vitro in a fashion that recapitulates terminal erythropoiesis in vivo (Ji et al., 2008; Zhang et al., 2003). Critically, transcriptomes, chromatin modifications, and genomic occupancies by erythroid important transcription factors have been well documented in mouse fetal liver erythroid cells at different stages of differentiation (Alvarez-Dominguez et al., 2013; Pilon et al., 2011; Wong et al., 2011). These methods and resources make terminal differentiation of mouse fetal liver erythroid progenitors an ideal system to investigate the interrelationships between transcriptional and post-transcriptional regulatory circuits in mammalian cell development.
Here, using genomic approaches, we identified a sequence-specific RNA-binding protein, Cpeb4, which is dramatically induced in terminal erythroid differentiation by two erythroid important transcription factors, Gata1 and Tal1. Cpeb4 belongs to the cytoplasmic polyadenylation element binding (CPEB) protein family that in mammals has four members, Cpeb1–4. All CPEB proteins in mammals have RNA-binding domains in their carboxy-termini that are responsible for binding to their substrate mRNAs via recognition of specific sequences in the 3’ untranslated region (3’UTR) (Fernandez-Miranda and Mendez, 2012; Huang et al., 2006). Mechanistically, CPEB proteins are best characterized as translational activators through elongating poly(A)-tails of target mRNAs via recruiting cytoplasmic poly(A) polymerases, although CPEB proteins can also repress translation (D’Ambrogio et al., 2013; Fernandez-Miranda and Mendez, 2012). Functionally, despite involvement in many biological processes (e.g. embryo development, neuronal activity, cancer) (D’Ambrogio et al., 2013; Fernandez-Miranda and Mendez, 2012), CPEB proteins’ roles in somatic cell differentiation still remain to be explored.
Here we show that Cpeb4 is induced by the erythroid important transcription factors Gata1 and Tal1, is strongly upregulated during terminal erythroid development, and is essential for terminal erythropoiesis. By binding directly to the translation initiation factor eIF3 complex, Cpeb4 represses the translation of a large set of mRNAs, most of which are normally downregulated during terminal erythroid development. Cpeb4 also binds to its own mRNA to repress its translation, and ectopic expression of Cpeb4 blocks erythroid differentiation. Thus transcriptional induction and translational repression combine to form a negative feedback loop to control Cpeb4 protein levels within a specific range that is required for terminal erythropoiesis. Our study provides an example of how translational control is integrated with transcriptional regulation to precisely control gene expression during mammalian cell differentiation.
RESULTS
Identification of RNA-binding Proteins Enriched in Terminal Differentiating Erythroblasts
RNA-binding proteins (RBPs) play important roles in post-transcriptional regulation of gene expression. Interestingly, ENCODE expression profiles revealed that mammalian RBPs are differentially expressed among different cells and tissues and that many RBPs are highly cell-type specific (Figure S1A) (Stamatoyannopoulos et al., 2012). These expression patterns suggest that some RBPs are required for formation of specific cell types, but the roles of most RBPs in mammalian cell differentiation are unknown. From the ENCODE data we identified 49 RBPs that are enriched in fetal liver erythroblasts. Among these 49 RBPs, six are highly induced during terminal erythroid differentiation from the CFU-E stage (Figure S1B). Here we focused on Cpeb4 because it is highly expressed in terminal differentiating erythroblasts and is dramatically induced during terminal differentiation. Among the four family members of CPEB proteins, only Cpeb4 is highly expressed in mouse fetal liver terminally differentiating erythroblasts (Figure S1C), allowing us to specifically characterize the function of this protein in erythroid development.
During terminal erythropoiesis Cpeb4 is Dramatically Induced by Gata1 and Tal1
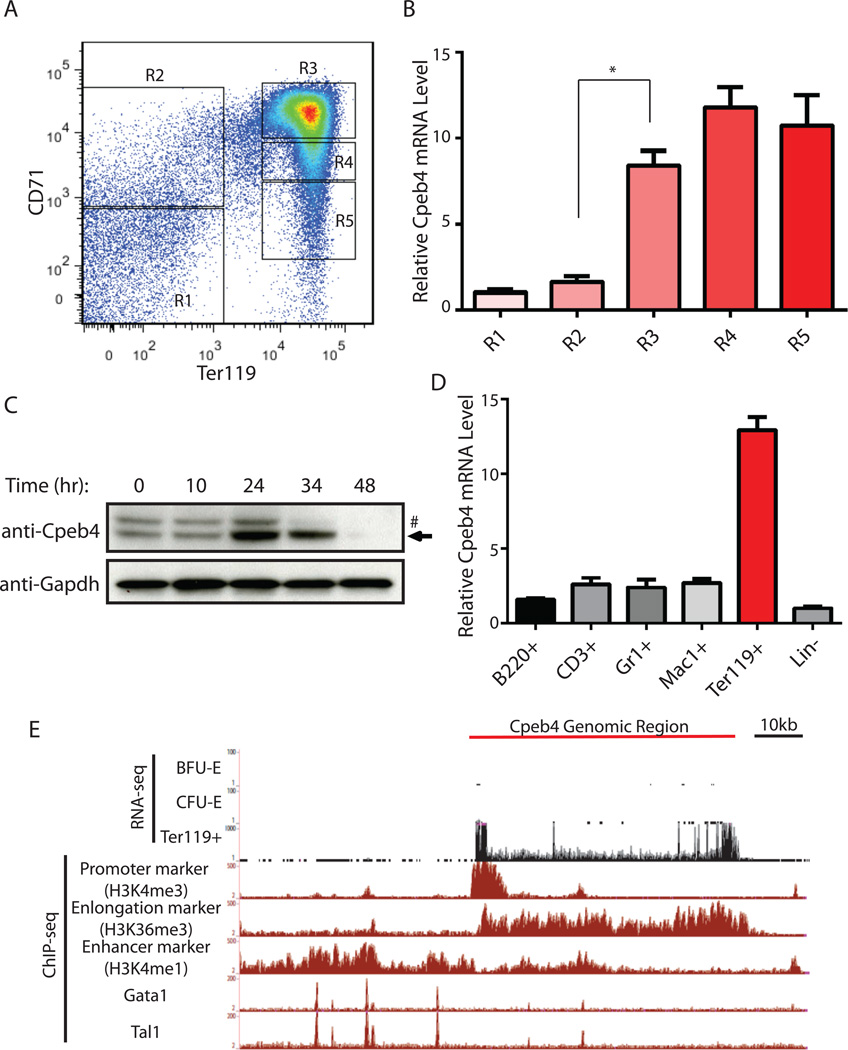
To determine the expression pattern of Cpeb4 in erythropoiesis, we first fractionated total E14 mouse fetal liver into five populations (R1-R5) using the cell surface markers CD71 (transferrin receptor) and Ter119 (glycophorin) (Figure 1A). R1 and R2 fractions are enriched for erythroid progenitors, and R3-R5 contain successively differentiated erythroblasts with R5 containing mostly enucleated reticulocytes (Zhang et al., 2003). Real-time RT-PCR revealed that Cpeb4 mRNA is dramatically induced during the transition from R2 to R3 (Figure 1B), congruent with the period when most of the ~600 erythroid-important genes are upregulated (Wong et al., 2011). In our erythroid culture system (Zhang et al., 2003), Cpeb4 protein levels peak at 24 hrs (Figure 1C), correlating well with the induction of Cpeb4 mRNA and the appearance of hemoglobinized erythroblasts (Ji et al., 2008; Zhang et al., 2003). Collectively, these results indicate that Cpeb4 mRNA and protein are dramatically induced during terminal erythropoiesis.
Figure 1. Cpeb4 is specifically induced in terminal erythroid differentiation.
(A–B) E14 mouse fetal liver cells were fractionated into R1-R5 based on the cell surface markers CD71 and Ter119. Cpeb4 mRNA was quantified in R1-R5 cells. (C) Different lineages of terminal differentiated hematopoietic cells and progenitor cells (Lin-) were isolated by magnetic assisted sorting from E14.5 mouse fetal liver followed by Cpeb4 mRNA quantification (mean ± SD; n = 3; * indicates p < 0.01 using two-tailed Student’s t test). (D) Lin- cells isolated from E14.5 mouse fetal liver were cultured in erythroid differentiation media and Cpeb4 protein levels were determined by Western blot at each indicated time point. The arrow indicates CPEB4 protein, the upper band (#) is a nonspecific band. (E) The RNA-seq data of erythroid cells at different developmental stages and the ChIP-seq datasets from mouse fetal liver Ter119+ cells at the Cpeb4 genomic region. See also Figures S1 and S2.
To further determine whether Cpeb4 is specific to erythroid cells, we fractionated total mouse fetal liver cells into different cell lineages and progenitors using cell surface markers and then quantified the Cpeb4 mRNA level using real-time RT-PCR. Clearly, Cpeb4 is highly enriched in terminal differentiating erythroblasts (Ter119+) compared to other cell types (Figure 1D).
Two lines of evidence indicated that Cpeb4 mRNA is induced by Gata1 and Tal1, two erythroid-important transcription factors. First, in published ChIP-seq (chromatin immunoprecipitation with high throughput DNA sequencing) datasets from primary mouse fetal liver erythroblasts (Wong et al., 2011; Wu et al., 2011), there are several discrete Gata1/Tal1 binding signals within 40kb of the Cpeb4 transcription start site (Figure 1E). Interestingly, these Gata1/Tal1 occupancy sites correlate well with the transcription enhancer marker H3K4me1 (Figure 1E), suggesting that Cpeb4 expression is regulated by Gata1 and Tal1. Second, Gata1 activation in G1E cells is a well-established model for studying Gata1-mediated regulation in erythropoiesis (Weiss et al., 1997); in this system Gata1 binding to the Cpeb4 enhancer regions is temporally congruent with transcriptional activation of Cpeb4 (Figure S2). Together, these observations indicate that Cpeb4 is a target of Gata1/Tal1 in terminal erythropoiesis.
Cpeb4 is Required for Terminal Erythropoiesis
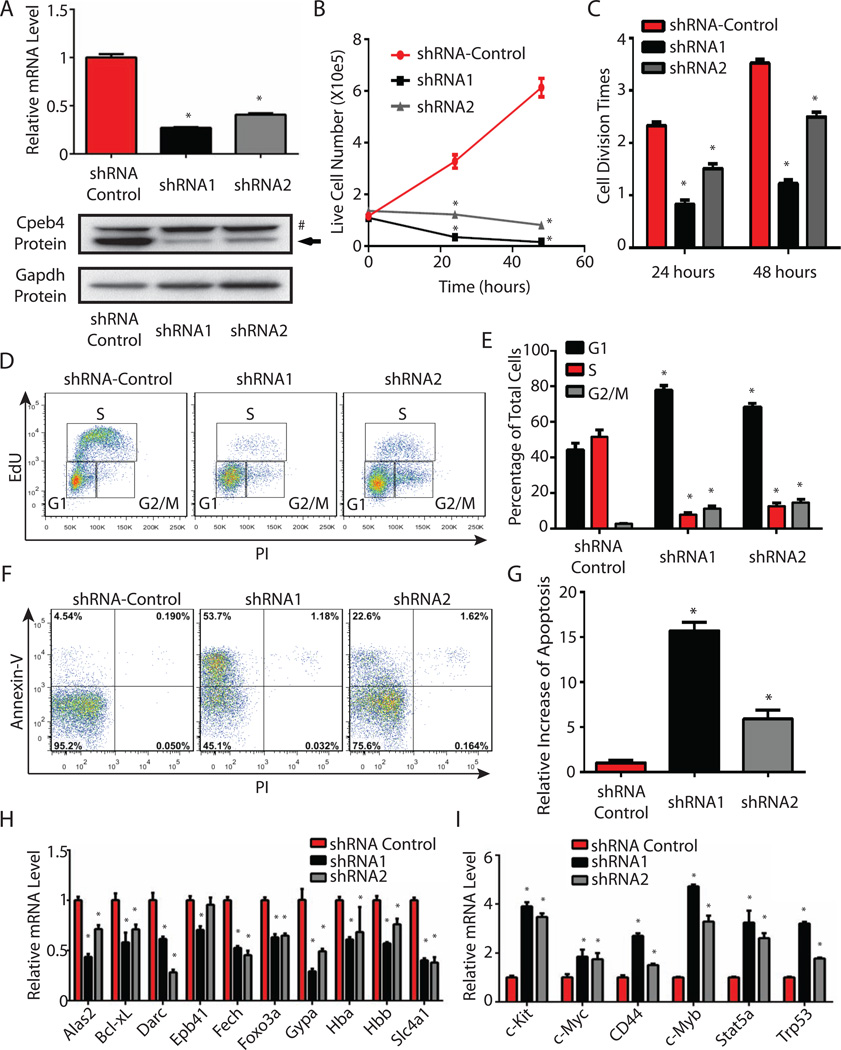
shRNA-mediated loss-of-function studies revealed that Cpeb4 is required for terminal erythroid differentiation. Specifically, two shRNAs targeting different regions of Cpeb4 were introduced into Lin- cells via retroviral transduction. Most of these Lin- cells are CFU-Es (Flygare et al., 2011). The transduced cells were first cultured in the maintenance medium for 24 hours to allow shRNA expression and then switched to differentiation medium. Both shRNAs efficiently knock down both Cpeb4 mRNA and protein levels in differentiating erythroblasts, as compared to a scrambled shRNA control (Figure 2A).
Figure 2. Cpeb4 is required for terminal erythroid differentiation.
(A) Cpeb4 mRNA and Cpeb4 and Gapdh protein levels in shRNA knockdown cells and the shRNA control cells. The arrow indicates Cpeb4 protein, and # indicates a non-specific band. (B) Growth curves of Cpeb4 knockdown cells and the shRNA control cells in the differentiation medium. (C) Measurement of the number of cell divisions at 24 hours and 48 hours in the differentiation medium. (D–E) The cell cycle profiles of Cpeb4 knockdown cells and the shRNA control cells. (F–G) Apoptotic status of Cpeb4 knockdown cells and the shRNA control cells was measured by annexin-V staining. (H–I) Quantification of genes induced and repressed in terminal erythroid differentiation by real-time RT-PCR. All the results are from 3 independent measurements and presented as mean ± SD. * indicates p < 0.01 compared to the corresponding control (two-tailed Student’s t test). See also Figure S3.
Inhibition of Cpeb4 expression caused a marked inhibition of terminal cell proliferation, as determined by counting the numbers of live cells (Figure 2B). To quantify the number of cell divisions in Cpeb4 knockdown cells, we labeled cell membranes with an inert fluorescence dye and then used flow cytometry to determine the fluorescence intensity of the labeled cells at 24 hours and 48 hours in the differentiation medium. During each cell division, the fluorescence dye is diluted by half; thus by comparing the fluorescence intensity at each time point to the original fluorescence intensity, we can calculate how many cell divisions have occurred. Clearly, the number of cell divisions in the Cpeb4 knockdown cells is significantly reduced (Figure 2C), suggesting that cell cycle progression is comprised. To specifically examine the cell cycle, we pulse-labeled differentiating erythroblasts with a reactive deoxynucleotide analog (EdU) to label S-phase cells, and used a DNA dye to discriminate the cells in the G1 and G2/M phases (Figure 2D). Knockdown of Cpeb4 caused cells to accumulate in the G1 phase, accompanied by a dramatic decrease in the numbers of S phase cells (Figure 2D, 2E), indicating that cell proliferation was severely compromised. In addition, there was a large increase of apoptotic cells (annexin-V positive) following Cpeb4 knockdown (Figure 2F, 2G). Collectively, these results indicated that Cpeb4 is required for proper cell cycle progression in terminal differentiating erythroblasts.
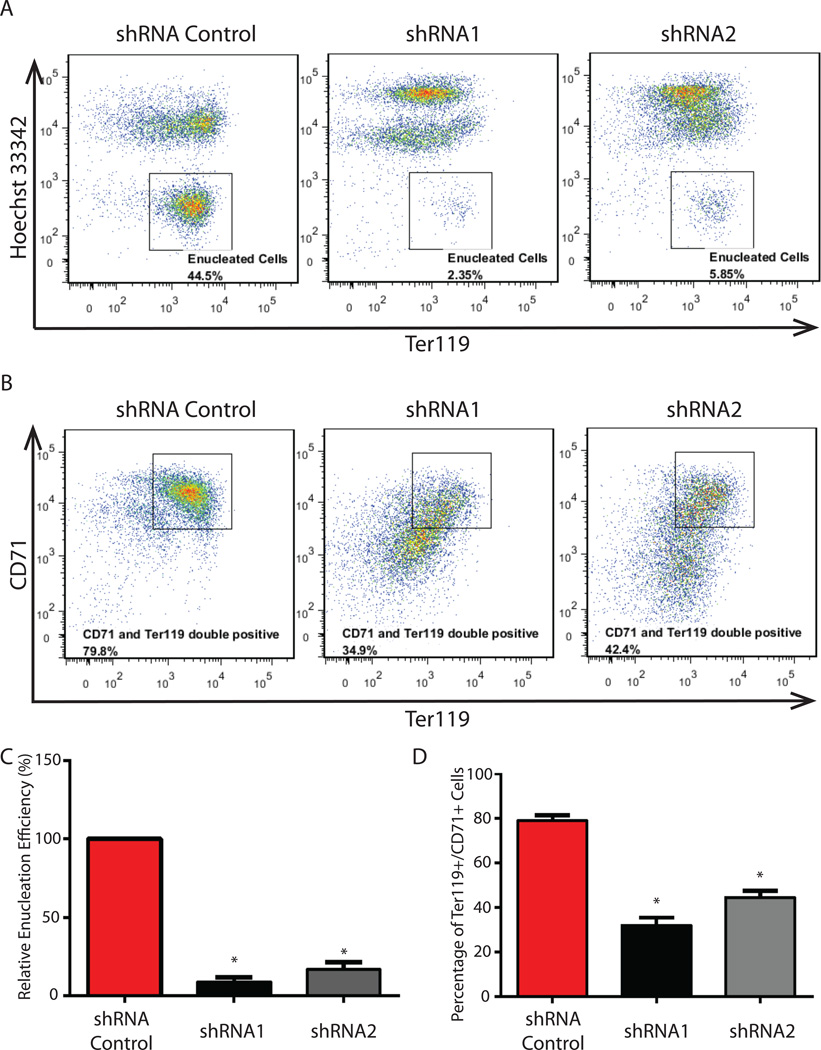
To further determine the changes of gene expression and the downstream differentiation process in Cpeb4 knockdown erythroid cells, we used real-time RT-PCR to quantify the expression of a group of erythroid-important genes induced during terminal differentiation. Induction of these genes was blocked following Cpeb4 knockdown (Figure 2H). Concomitantly, Cpeb4 knockdown cells contained much higher-than-normal levels of key mRNAs normally downregulated during terminal differentiation (Figure 2I). These results revealed that terminal erythroid differentiation in Cpeb4 knockdown cells was compromised. To directly assay the products of terminal erythropoiesis, we measured the generation of enucleated reticulocytes by using a DNA dye to fractionate the cells with nuclei and the enucleated cells. Simultaneously, the maturation of terminal differentiating erythroblasts was monitored by Ter119 and CD71 staining. When Cpeb4 was inhibited, the generation of enucleated reticulocytes was dramatically inhibited (Figure 3A, 3C). In addition, the maturation of differentiating erythroblasts, as indicated by the CD71 and Ter119 double position cell population, was also severely compromised (Figure 3B, 3C). Collectively, these results indicate that induction of Cpeb4 is required for terminal erythroid differentiation.
Figure 3. Cpeb4 knockdown blocks the generation of enucleated reticulocytes.
(A) The generation of enucleated reticulocytes was analyzed in the Cpeb4 knockdown cells and the shRNA control cells. Staining was with Ter119 and the Hoechst DNA- binding dye. (B) The expression of cell surface markers Ter119 and CD71 was analyzed in Cpeb4 knockdown cells and the shRNA control cells. (C–D) Quantifications of relative efficiencies of enucleation and the percentages of Ter119 and CD71 double positive cells in the Cpeb4 knockdown cells and the shRNA control cells. All the results are from 3 independent measurements and presented as mean ± SD. * indicates p < 0.01 using two-tailed Student’s t test.
We believe that these phenotypes do not result from nonspecific effects of the Cpeb4 shRNAs and thus are specific to Cpeb4, because the severity of all these phenotypes correlates well with the Cpeb4 knockdown efficiencies (Figure 2). Furthermore, when the same two shRNAs were introduced into NIH3T3-L1 cells, where Cpeb4 is not expressed, no phenotypic changes in the cells were observed (Figure S3).
Cpeb4 Interacts with eIF3 to Repress mRNA Translation in Erythroid Cells
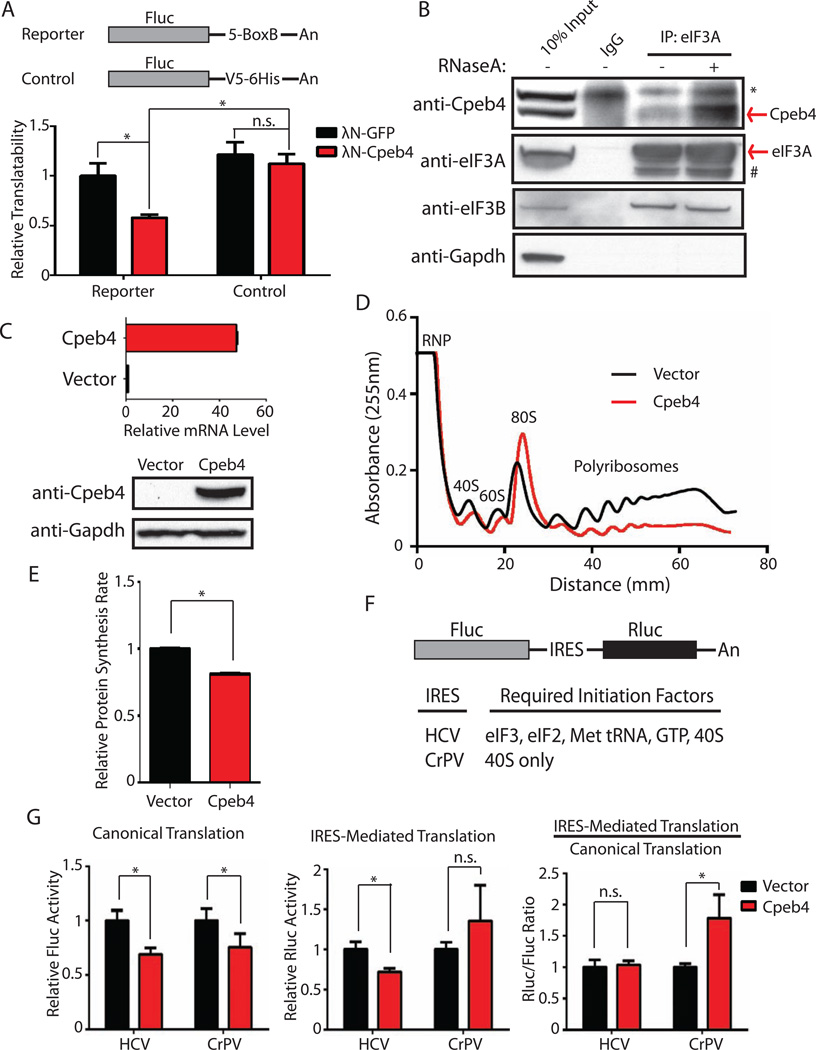
CPEB proteins can both repress and activate target mRNA translation (Fernandez-Miranda and Mendez, 2012). To determine how Cpeb4 regulates gene expression in erythroid cells, we employed a tethering assay (Coller and Wickens, 2007). Using the strong and specific interaction between the bacteriophage λN polypeptide and the BoxB RNA motif, we tethered the λN-Cpeb4 fusion protein to the 3’UTR of a firefly luciferase reporter mRNA (Figure 4A). The translatability of the reporter, defined as luciferase activity normalized to luciferase mRNA level, was assayed in erythroleukemia cells (K562 cells). Interestingly, tethering Cpeb4 significantly reduced the reporter’s translatability compared to tethering a green fluorescence protein (GFP) to the 3’UTR of the reporter mRNA (Figure 4A). In addition, there was no change in the luciferase reporter mRNA level (Figure S4A) when Cpeb4 or GFP was tethered, indicating that the effect was predominantly accomplished by translational inhibition. Importantly, translational repression was dependent on the BoxB sites, since there was no change in the translatability of the control reporter lacking these sites when the λN-Cpeb4 fusion protein was introduced (Figure 4A).This observation indicates that inhibition of translatability requires the binding of Cpeb4 to the reporter mRNA. These results revealed that Cpeb4 is a translation repressor in erythroid cells; consistent with this notion, the two cytoplasmic poly(A) polymerases, Gld2 and Gld4, which are required for CPEB protein-mediated translational activation (D’Ambrogio et al., 2013), are not expressed in terminally differentiating erythroblasts (Figure S4B).
Figure 4. Cpeb4 represses mRNA translation in erythroid cells by interacting with eIF3.
(A) The translatability of the reporters in the presence and absence of λN-Cpeb4 was determined by luciferase assay in K562 cells. (B) Co-immunoprecipitation of endogenous eIF3 complex was performed in mouse fetal liver Ter119+ cells, followed by SDS-PAGE and Western blotting to detect the proteins as indicated. (^) indicates a non-specific band; (#) is likely to be degradation product(s) from eIF3A. (C) Quantification of Cpeb4 mRNA and protein levels in MEL cells transduced by a Cpeb4 expressing retrovirus and control retrovirus. (D–E) The polyribosome profiles (D) and the translation rates (E) of Cpeb4-expressing MEL cells were compared to those of the control cells. (F–G) IRES-based bicistronic luciferase reporters (F) were transfected into K562 cells, and then luciferase assays were performed in the presence and absence of ectopic expression of Cpeb4 to measure the translation status of these reporters. All the results are from 3 independent measurements and presented as mean ± SD. * indicates p < 0.01 and n.s. indicates p > 0.05 using two-tailed Student’s t test. See also Figure S4, Table S1.
To identify the proteins that interact with Cpeb4 in erythroid cells, we performed affinity purification followed by mass spectrometry. Specifically, we immunoprecipitated both endogenous Cpeb4 from primary mouse fetal liver erythroblasts and an epitope-tagged Cpeb4 expressed in K562 cells. This experiment was performed in the presence and absence of RNaseA to discriminate RNA-independent and RNA-mediated interactions. In both types of cells, mass spectrometry analyses identified several eIF3 subunits as the major translation factors that directly associate with Cpeb4 (Figure S4D S4E, Table S1). Interestingly, Maskin (Tacc3 in mouse) and eIF4E, which are used by Cpeb1 in Xenopus oocytes to repress mRNA translation (D’Ambrogio et al., 2013), were not detected, suggesting that Cpeb4 uses a different mechanism to repress mRNA translation in erythroid cells.
The eIF3 complex is the largest and the most complicated translation initiation factor in mammals with 13 non-identical subunits (eIF3a to eIF3m) that can regulate translation initiation at multiple steps (Hinnebusch, 2006, 2014; Sonenberg and Hinnebusch, 2009). The several eIF3 subunits (eIF3a, eIF3b, eIF3c, eIF3l) identified in our mass spectrometry analysis suggested that Cpeb4 directly associates with the eIF3 complex in erythroid cells. To further confirm this interaction, we used an antibody against eIF3a subunit to immunoprecipitate the endogenous eIF3 complex from primary mouse fetal liver erythroblasts (Figure 4B). Indeed, a different eIF3 subunit, eIF3b, was found to co-immunoprecipitate with eIF3a (Figure 4B), indicating that this approach can isolate the eIF3 complex (or part of the eIF3 complex). Western blot analysis revealed that Cpeb4 specifically co-immunoprecipitated with the eIF3 complex and that this interaction is independent of RNA (Figure 4B), indicating that Cpeb4 directly interacts with the eIF3 complex.
We hypothesized that since Cpeb4 interacts with a general translation initiation factor, if expressed at high level Cpeb4 may repress global mRNA translation in erythroid cells. To test this, we created a mouse erythroleukemia cell line (MEL) that overexpresses Cpeb4 (Figure 4C). We first examined global mRNA-ribosome associations by sucrose-gradient mediated polyribosome profiling (Figure 4D). Interestingly, Cpeb4 overexpression resulted in an obvious decrease of polyribosomes with a corresponding increase of 80S ribosomes (Figure 4D), indicating an inhibition of translation initiation. Importantly, overexpression just of the C-terminal segment of Cpeb4, which contains all of the RNA-binding domains (Huang et al., 2006), does not influence global mRNA-ribosome association (Figure S4C). This indicates that Cpeb4-mediated translation inhibition is not caused by general RNA binding. To directly measure the global translation rate, we pulse-labeled cells with a non-radioactive methionine analog and measured how fast it can be incorporated into polypeptides in the MEL cells. We found that Cpeb4 overexpression reduced the global translation rate to 80% of that in the control cells (Figure 4E). The different extents of translation inhibition we observed from polyribosome profiling and the methionine incorporation assay are probably due to the different aspects of translation these two assays measure: mRNA-ribosome binding in the polysome profile vs. translation kinetics in the methionine incorporation assay. Nonetheless, these results indicate that Cpeb4 can repress mRNA translation in erythroid cells and suggest that the Cpeb4-eIF3 interaction is functional.
We next showed that Cpeb4-mediated translation repression requires the eIF3 complex by using an assay that measures translation from an internal ribosome entry site (IRES). IRES elements are structured RNAs that are usually encoded by viral genomes to position and activate eukaryotic translational initiation (Fraser and Doudna, 2007). Importantly, different viral IRES elements require different sets of translation factors (Fraser and Doudna, 2007). To study Cpeb4-mediated translation repression, we used the eIF3 dependent HCV IRES and the eIF3 independent CrPV IRES (Fraser and Doudna, 2007). Specifically, a bicistronic reporter was employed in which firefly luciferase (Fluc) is translated by cap-dependent translation and translation of renilla luciferase (Rluc) is controlled either by HCV-IRES or CrPV-IRES mediated translation (Figure 4F). When these two reporters were transfected into K562 cells (human erythroleukemia cells), as expected cap-dependent translation from both reporters was repressed following Cpeb4 expression (Figure 4G), as determined by the firefly luciferase activity. However, Cpeb4 only repressed HCV-IRES mediated translation but did not affect CrPV-IRES mediated translation (Figure 4G). Since HCV-IRES requires the eIF3 complex to initiate translation while CrPV-IRES does not, these results revealed that Cpeb4-mediated translation repression is dependent on eIF3, indicating that the repressive Cpeb4-eIF3 interaction is operative in erythroid cells.
Collectively, these experiments revealed that Cpeb4 is a translation repressor in erythroid cells; it interacts with the generation translation initiation factor eIF3, and this interaction is functionally relevant for Cpeb4-mediated translation repression.
Cpeb4 Represses the Translation of a Set of mRNAs, Including Its Own mRNA, during Terminal Erythropoiesis
To identify the endogenous mRNA substrates that Cpeb4 regulates, we performed RNA immunoprecipitation (RIP) followed by microarray (RIP-Chip) studies targeting endogenous Cpeb4 in mouse primary fetal liver erythroblasts (Jain et al., 2011) (GEO: GSE57004). We used three criteria to identify Cpeb4 substrate mRNAs from the microarray data. First, the substrate mRNAs should be enriched at least 2.5 fold in the RIP sample versus the input. Second, the substrate mRNAs must have higher signal intensity in the RIP sample compared to that in the IgG control. Third, the substrates should be reproducibly detected in two independent Cpeb4 RIPs. These criteria resulted in 227 putative Cpeb4 target transcripts (Table S2).
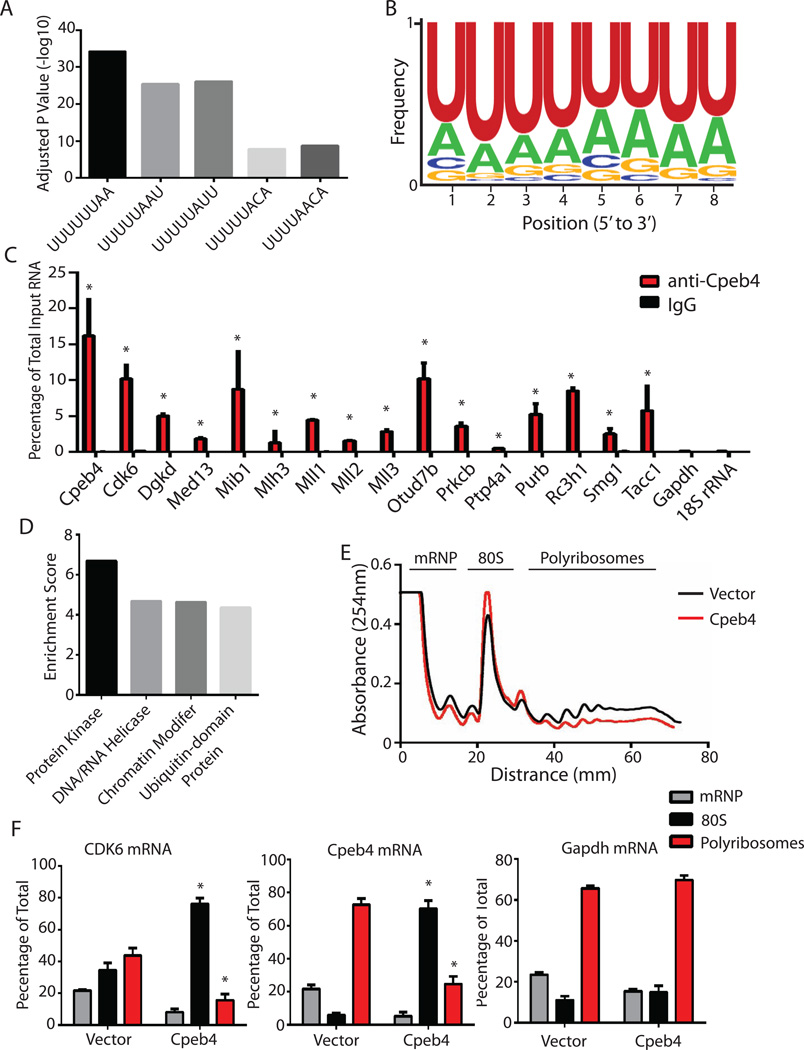
Three lines of evidence indicate that the 227 mRNAs identified by RIP-Chip are Cpeb4 substrates in differentiating erythroblasts. First, all of the five known cytoplasmic polyadenylation elements to which Cpeb4 can bind (Ortiz-Zapater et al., 2012) were significantly enriched in the 3’UTRs of the 227 enriched mRNAs, compared to those of 503 control transcripts that were depleted in the Cpeb4 RIP-Chip (Figure 5A). Second, a uridine-rich (U-rich) 8-nucleotide motif was statistically over-represented in the 3’UTRs of the 227 enriched mRNA compared to those in randomly selected mRNAs (Figure 5B). Critically, this U-rich motif resembles the consensus Cpeb4 binding sequences identified from in vitro binding studies (Huang et al., 2006; Ray et al., 2013). Third, we selected 16 mRNAs from these 227 mRNAs for experimental verification. RIP followed by real-time RT-PCR revealed that all 16 mRNAs were immunoprecipitated in significant amounts by an anti-Cpeb4 antibody but not by the IgG control (Figure 5C). Importantly, Cpeb4 did not bind two abundant control RNAs, Gapdh mRNA and 18S rRNA (Figure 5C), indicating that the Cpeb4-mRNA interaction is specific. Collectively these results indicated that the 227 mRNAs are Cpeb4 substrates in differentiating erythroblasts. Gene ontology analysis indicated that Cpeb4 substrates are enriched for mRNAs encoding protein kinases, nucleic acid helicases, chromatin modifiers, and ubiquitin-domain containing proteins (Figure 5D), suggesting that Cpeb4 can modulate expression of these factors in differentiating erythroblasts.
Figure 5. Cpeb4 represses target mRNA translation in differentiating erythroblasts.
(A–B) The 3’UTRs from Cpeb4 RIP enriched mRNAs were compared to those from depleted mRNAs to determine the enrichment of known CPEs (A), and then 8-nucleotide motifs in the 3’UTRs from the enriched mRNAs was compared to randomly selected mRNAs to identify the most over-represented motifs in the mRNAs associated with Cpeb4 (B). (C–D) RIP followed by real-time PCR was used to verify 16 Cpeb4-associated mRNAs (C), and then gene ontology analysis was performed on all the Cpeb4-assocaited transcripts identified from the Cpeb4 RIP-Chip (D). (E) The polyribosome profiles of Cpeb4 overexpressing MEL cells and control cells. (F) The distribution of two Cpeb4 target mRNAs, the endogenous Cpeb4 mRNA, and Cdk6 mRNA, and a control mRNA on a sucrose-density gradient from MEL cells was determined by real-time RT-PCR in the presence and absence of Cpeb4. All the results are from 3 independent measurements and presented as mean ± SD. * indicates p < 0.01 using two-tailed Student’s t test. See also Figure S5, Table S2.
To determine the functional consequence of Cpeb4 binding to the target mRNAs, we analyzed the distribution of Cpeb4 target mRNAs in MEL cells across a polyribosome gradient both in the presence and absence of Cpeb4 expression (Figure 5E). When Cpeb4 was expressed, both endogenous Cpeb4 mRNA, as determined by real-time RT-PCR targeting the 3’UTR, and Cdk6 mRNA shifted from the polyribosome to the 80S region (Figure 5F), indicating translation inhibition of these two Cpeb4 substrate mRNAs. Importantly, Cpeb4 expression did not change the distribution of Gapdh mRNA on the gradient (Figure 5F), indicating the specificity of Cpeb4-mediated translational repression. These results indicated that Cpeb4 binding results in translational inhibition of multiple substrate mRNAs
Two lines of observations indicated that inhibiting the expression of Cpeb4 substrate mRNAs is important for terminal erythropoiesis. First, downregulation of Cdk6 is known to be required for terminal erythroid differentiation; while constitutive expression of Cdk6 blocks this differentiation process (Matushansky et al., 2000, 2003). Since Cdk6 mRNA is translationally repressed by Cpeb4 in terminal differentiating erythroblasts, these results partially explain why Cpeb4-mediated translation repression is important for terminal erythropoiesis.
Second, we constitutively expressed 7 Cpeb4 substrate mRNAs, including Cdk6 mRNA, in primary fetal liver erythroblasts, and then assayed terminal erythroid differentiation by monitoring the formation of enucleated reticulocytes. We found that constitutive expression of each of these Cpeb4 substrates blocks terminal erythroid differentiation (Figure S5A, S5B). Importantly, constitutive expression of a control protein, renilla luciferase, had no effect on this differentiation process (Figure S5A, S5B). These results suggest that down-regulation of these Cpeb4 target genes is functionally important for terminal erythropoiesis. Interestingly, the levels of most of the 227 Cpeb4 substrate mRNAs decrease monotonically during terminal erythroid differentiation (Figure S5C), suggesting that Cpeb4-mediated translation repression further ensures that the proteins encoded by these mRNAs are not produced in terminally differentiating erythroblasts.
Maintaining Cpeb4 Protein Level within a Specific Range is Required for Terminal Erythropoiesis
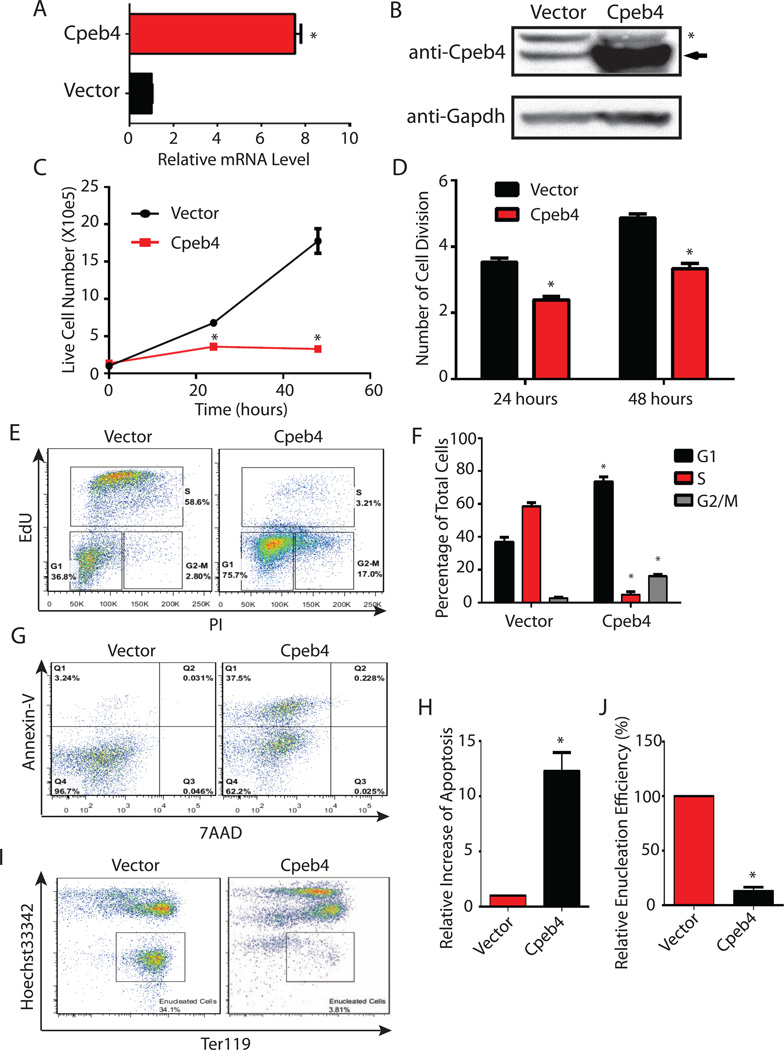
One striking finding is that Cpeb4 can bind to a large amount of its own mRNA (Figure 5C), suggesting that Cpeb4-mediated negative translational regulation functions in a circuit with Gata1/Tal1-mediated transcriptional induction to control Cpeb4 protein levels within a specific range during terminal erythropoiesis. Our loss-of-function studies indicated that decreasing Cpeb4 protein level below this range is detrimental to terminal erythropoiesis (Figure 2, Figure 3). To further test the functional importance of maintaining Cpeb4 protein levels within a specific range, we used retroviral transduction to constitutively express Cpeb4 during terminal erythropoiesis, increasing both Cpeb4 mRNA and protein above their normal levels in differentiating erythroblasts (Figure 6A, 6B). This resulted in a very similar phenotype to Cpeb4 knockdown, as indicated by inhibition of cell proliferation (Figure 6C) and cell division (Figure 6D), disappearance of cells in the S phase (Figure 6E, 6F), increase of apoptotic cells (Figure 6G, 6H), and inability to form enucleated reticulocytes (Figure 6I, 6J). Importantly, constitutive expression of only the C-terminal segment of Cpeb4 that contains all the RNA-binding domains (Huang et al., 2006) does not affect terminal erythroid differentiation, as indicated by the generation of enucleated reticulocytes (Figure S6A–C). This indicates that the phenotypes we observed following Cpeb4 constitutive expression are not caused by general RNA binding. In addition, these phenotypes are specific to Cpeb4, because constitutive expression of another mRNA-binding protein, Carhsp1, which is also dramatically induced during terminal erythropoiesis (Figure S1B), did not inhibit terminal erythroid differentiation (Figure S6D–F). Collectively, these observations revealed that maintaining Cpeb4 protein levels within a specific range is important for terminal erythropoiesis.
Figure 6. Constitutive expression of Cpeb4 inhibits terminal erythroid differentiation.
(A–B) The Cpeb4 mRNA (A) and protein (B) levels in mouse primary fetal liver erythroblasts constitutively expressing ectopic Cpeb4 and control cells. The arrow indicates Cpeb4 protein, and # indicates a non-specific band. (C) Growth curves of Cpeb4 constitutive expressing cells and control cells in the differentiation medium. (D) Measurement of the number of cell divisions at 24 hours and 48 hours in the differentiation medium. (E–F) The cell cycle profiles of Cpeb4 constitutive expressing cells and the control cells. (G–H) Apoptotic status of Cpeb4 constitutive expressing cells and the shRNA control cells was measured by annexin-V staining. (I–J) Enucleation of Cpeb4 constitutive expressing cells and the control cells. All the results are from 3 independent measurements and presented as mean ± SD. * indicates p < 0.01 using two-tailed Student’s t test. See also Figure S6.
Cpeb4 is Functionally Important in vivo
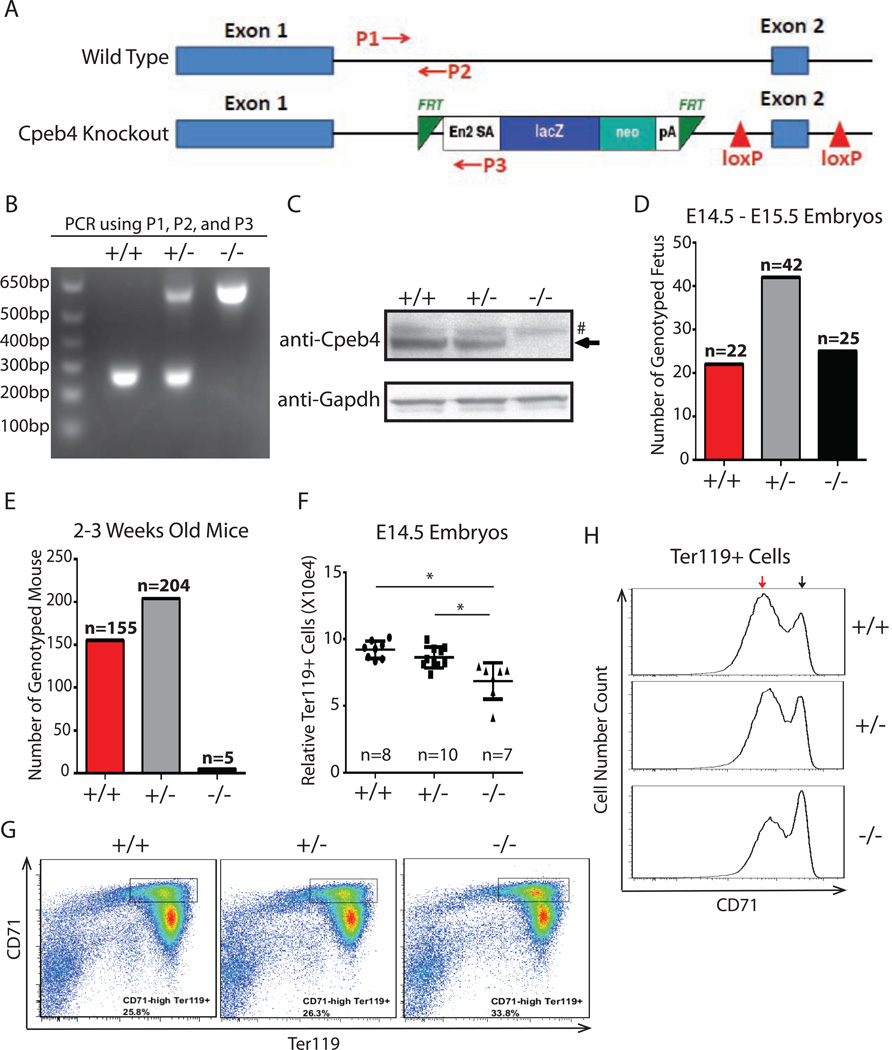
To explore the in vivo functional significance of Cpeb4, we obtained a Cpeb4 heterozygous mouse from The International Knockout Mouse Consortium. The mouse was constructed by inserting after the first exon of Cpeb4 a reporter-tagged sequence that contains a strong transcriptional termination signal (pA signal) (Figure 7A) (Skarnes et al., 2011). We verified the genotype of the knockout mouse by PCR (Figure 7B) and showed by Western blotting that the level of Cpeb4 protein in homozygous knockout E14.5 fetal liver cells is below the detection limit (Figure 7C). In breeding Cpeb4 heterozygous mice, wild-type, heterozygous, and homozygous embryos were present at E14.5-E15.5 in the expected Mendelian ratio (Figure 7D). However, of the 364 mice resulting from matings of the heterozygous Cpeb4 mice we genotyped at 2–3 weeks of age, only 5 were Cpeb4 homozygotes (Figure 7E), while wild type and Cpeb4 heterozygous mice were born in the predicted Mendelian ratio. Thus Cpeb4 homozygous mice die in utero after E15 or within 2 weeks of birth, strongly arguing that Cpeb4 is required for normal development in vivo.
Figure 7. Cpeb4 is functionally important in vivo.
(A) Structures of Cpeb4 genomic region in wild type and Cpeb4 knockout mouse. P1, P2, and P3 are PCR primers for genotyping. (B) Genotyping of wildtype (+/+), Cpeb4 heterozygous (+/−), and Cpeb4 knockout (−/−) mice by PCR. (C) Western blot of E14.5 total fetal liver cells isolated from wildtype, Cpeb4 heterozygous, and Cpeb4 homozygous knockout embryos. (D) Genotyping of E14.5-E15.5 embryos from a mating of Cpeb4 heterozygotes. (E) Genotyping of 2–3 week old mice from matings of Cpeb4 heterozygotes. (F) The quantification of Ter119+ cells in fetal livers from E14.5 embryos. The y axle is the scale relative to a known amount of added beads. * indicates p < 0.05 using two-tailed Student’s t test. (G) CD71 and Ter119 staining of total E14.5 fetal liver cells. (H) CD71 expression levels in Ter119+ cells from E14.5 mouse fetal liver. The black arrow indicates high CD71 level; the red arrow indicates low CD71 level. The results of (G, H) are representative of E14.5 embryos from 4 sets of matings of Cpeb4 heterozygotes.
Two lines of evidence indicate that terminal erythropoiesis is impaired in the Cpeb4 knockout embryo, which may contribute to the death phenotype. First, Cpeb4 homozygous knockout E14.5 fetal liver has significantly fewer terminally differentiating Ter119+ erythroblasts compared to those of wild type and the heterozygous embryos (Figure 7F). Second, during terminal differentiation, Ter119+ erythroblasts cells gradually lose the transferrin receptor (CD71); thus the relative level of CD71 on Ter119+ cells is a measure of their progression during differentiation. We found that E14.5 Cpeb4 −/− Ter119+ cells express higher levels of CD71 compared to Cpeb4 +/− and wild type Ter119+ cells (Figure 7G, 7H), indicating that the differentiation is compromised. Collectively, these observations reveal that a deficiency of Cpeb4 in vivo causes a block in terminal erythroid differentiation.
DISCUSSION
Here we characterized a Cpeb4-mediated negative translational regulatory circuit that is required for terminal erythropoiesis. This translational regulatory circuit is transcriptionally induced by Gata1/Tal1 and is translationally negatively auto-regulated by Cpeb4 itself. We argue that this regulatory circuit limits the level of Cpeb4 expression to within a specific range required for this cell differentiation process.
A previously reported Cpeb4 homozygous knockout mouse was reported to be normal (Tsai et al., 2013); while our Cpeb4 homozygous knockout mouse has a clear late embryonic early neonatal lethal phenotype. This phenotypic difference could arise from different methods used to generate these knockout mice. Specifically, our mouse contains a strong transcription termination signal after the first exon of Cpeb4, thereby, inhibiting the generation of full length Cpeb4 mRNA. In contrast, Tsai et al deleted exon 2 of Cpeb4, resulting in Cpeb4 transcripts with premature stop codons (PTCs). Since truncated proteins can be generated from the PTCs and potential translational read-through at PTCs can generate the full length protein (or longer truncated proteins), the Cpeb4 allele Tsai et al generated may be hypomorphic, which can potentially explain the negative results of their Cpeb4 knockout mouse. Our Cpeb4 −/− mice are lethal sometime after E14.5 and before 2 weeks after birth. Considering that Cpeb4 is expressed in multiple tissues and cells (e.g. neurons, fetal liver erythroblasts, etc.) and Cpeb4 has been implicated in regulating meiosis and mitotic cell-cycle progression (Igea and Mendez, 2010; Novoa et al., 2010), we speculate that the lethal phenotype we observed may be due to defects in multiple tissues. Nonetheless, the defects in terminal erythropoeisis we observed at E14.5 indicate that Cpeb4 is indeed required for proper terminal erythroid differentiation in vivo, and also suggest that these erythropoiesis defects contribute to the lethal phenotype.
CPEB proteins are important translational regulators in diverse biological processes (D’Ambrogio et al., 2013). Among the four family members, Cpeb4 has been implicated in regulating neuronal activities, mitosis, meiosis, and cancer metastasis (Huang et al., 2006; Igea and Mendez, 2010; Novoa et al., 2010; Ortiz-Zapater et al., 2012; Theis et al., 2003). Interestingly, under these biological settings, Cpeb4 predominantly functions as a translational activator by recruiting cytoplasmic poly(A) polymerase(s) to extend the poly(A) tails on the 3’ ends of target mRNAs. For example, Cpeb4 activates target mRNA translation in the activated oocyte and in pancreatic tumors (Ortiz-Zapater et al., 2012). Here, however, we found that in terminal differentiating erythroblasts, the two known cytoplasmic poly(A) polymerases are not expressed and that Cpeb4 functions as a translation repressor. These observations are consistent with the notion that CPEB proteins can both repress and activate mRNA translation.
Then what triggers the functional switch of Cpeb4 from a translation activator to a repressor? We speculate that under different biological contexts post-translational modifications and/or association with different protein complexes may mediate this functional switch for Cpeb4. Such has been observed for Cpeb1, the best characterized CPEB protein family member. Specifically, in the Xenopus oocyte, phosphorylation of Cpeb1 by the Aurora kinase results in disassociation of Cpeb1 from a translation repression complex and formation of a translation activation complex that recruits cytoplasmic polyadenylases (D’Ambrogio et al., 2013; Fernandez-Miranda and Mendez, 2012). Although Aurora kinase phosphorylation sites are absence in Cpeb4 (Theis et al., 2003), it would be very interesting to explore the post-translational modification status of Cpeb4 and the factors with which it associates in terminal erythroid cells as well as in other biological settings.
Mechanistically, we found that Cpeb4 interacts with the eIF3 complex to repress mRNA translation in erythroid cells. This is different from the translational repression mechanism employed by Cpeb1. In the Xenopus oocyte, Cpeb1 interacts with Maskin to interfere with eIF4E’s binding to the 5’ end cap structure of mRNA, thereby inhibiting translation initiation. Although Maskin (Tacc3 in mouse) is abundantly expressed in erythroid cells, we did not observe an interaction with Cpeb4 in our mass spectrometry analyses. In addition, Cpeb4 can inhibit certain IRES (HCV) mediated translational initiation, indicating that Cpeb4 inhibits translation at the step(s) after 5’ end cap recognition. These results indicate that Cpeb4 uses a previously uncharacterized mechanism to repress translation. We believe that eIF3 is important for Cpeb4-mediated translation because Cpeb4 directly associates with eIF3 complex. Furthermore, Cpeb4 represses eIF3-dependent translation (cap-dependent translation and HCV-IRES mediated translation), but not eIF3-indepdent translation (CrPV-IRES mediated translation). These observations indicate that the Cpeb4-eIF3 interaction is functional in translational repression. eIF3 is the most complicated translation initiation factor with 13 non-identical subunits in mammals that can regulate translation initiation at multiple steps, including facilitation of 48S complex formation and recycling of posttermination complexes (Hinnebusch, 2006, 2014; Pisarev et al., 2007). Although we showed the Cpeb4 directly associates with the eIF3 complex, our data does not indicate to which subunit(s) of eIF3 complex Cpeb4 directly binds. Thus it would be of great interest in the future to characterize this Cpeb4-eIF3 interaction in detail, which will shed light into the detailed molecular mechanisms of Cpeb4-mediated translation repression.
In terminal erythropoiesis, differentiating erythroblasts undergo dramatic morphological changes that include chromatin condensation, decrease of nuclei size, and eventually exit of the cell cycle and extrusion of the nucleus, resulting in enucleated reticulocytes. These morphological changes require coordinated control of many factors involved in diverse cellular processes. For example, many histone modifiers and transcription activators that maintain active chromatin status need to be inhibited prior to or during chromatin condensation, and cell cycle activators and DNA replication factors need to be downregulated so that erythroid cells can exit the cell cycle. Interestingly, many Cpeb4 substrate mRNAs encode proteins involved in maintaining active chromatin structures (e.g. Mll1–3 and Med13) and controlling cell cycle and DNA replication (e.g. Cdk6 and nucleic acid helicases). Thus, it is tempting to speculate that Cpeb4 contributes to terminal erythropoiesis by inhibiting the expression of those proteins that need to be downregulated during this terminal cell differentiation process. In addition, we also observed that Cpeb4 binds a group of ubiquitin-domain containing proteins that have decreasing mRNA levels in terminal erythroid differentiation. This observation suggests that down-regulation of these protein degradation regulators may also be important for the generation of enucleated reticulocytes.
As important translational regulators, CPEB proteins can form auto-regulatory loops to control their own expression. In Drosophila oocytes, the Cpeb1 homologue, Orb, binds its own mRNA to stimulate its translation, thereby forming a positive auto-regulatory loop at the translational level to generate large amount of Orb protein that is required for stimulating the translation of developmentally important mRNAs (Tan et al., 2001). In contrast, in erythroid cells, we found that Cpeb4 also binds its own mRNA, but instead Cpeb4 represses translation of its own mRNA, thereby generating a negative auto-regulatory loop to limit Cpeb4 protein levels within a specific range in terminal erythroblasts. Our functional studies indicate that this negative regulatory circuit is important for erythroid differentiation, as disruption of this loop both by loss-of-function and gain-of-function approaches block this developmental process. We speculate that this negative auto-regulation mechanism employed by Cpeb4 ensures that Cpeb4 protein will not accumulate to high levels that may interfere with the translation of other mRNAs that are important for forming enucleated reticulocytes (e.g. hemoglobin mRNA translation). Interestingly, Cpeb4 mRNA is also dramatically induced in human terminal erythropoiesis (Merryweather-Clarke et al., 2011). In addition, there are multiple U-rich motifs in the 3’UTRs from both mouse and human Cpeb4 mRNAs, some of which are conserved between these two species. Thus, we speculate that Cpeb4-mediated translational regulatory circuit is conserved between mouse and human.
In summary, our study characterized a negative translational regulatory circuit that is required for terminal erythropoiesis by inhibiting the translation of many genes normally downregulated during terminal erythropoiesis. Interestingly, this translational regulatory circuit is induced by well-characterized transcriptional regulatory networks, precisely coupling translational and transcription regulation to modulate the expression of multiple genes during terminal erythroid differentiation. We speculate that similar post-transcriptional regulatory circuits exist in other mammalian cell differentiation processes.
Experimental Procedures
Plasmids and oligonucleotides
Mouse strain and fetal liver cell analysis
All the mice used in this study were in a C57BL/6 background. The Cpeb4 heterozygous knockout mouse (C57BL/6NTac-Cpeb4tm1a(EUCOMM)Wtsi/Cnrm) was imported from The International Knockout Mouse Consortium. All animal experiments were in compliance with regulations established by the the Whitehead Institute veterinary staff. To determine the number of Ter119+ cells and the expression of CD71 in E14.5 mouse fetal liver, the cells from a fetal liver were dissociated and resuspended in 300ul PBS. 100ul cells were used for staining with antibodies to Ter119 (APC) and CD71 (FITC) as well as with PI. After staining, the cells were washed and resuspended in 500ul PBS. Then 50ul CountBright Absolute Counting Beads (Life technologies) (0.51 × 10e5 beads/ml) were added as a standard for quantification of the cell number.
Tissue culture and retrovirus transduction
Erythroid progenitor cells were isolated from E14.5 C57BL/6 mouse fetal liver cells and then cultured in maintenance and differentiation medium using the media and methods described previously (Hu et al., 2011). A shRNA-expressing MSCV retroviral vector was used for shRNA-mediated loss-of-function studies (Hu et al., 2011); pRetroX-IRES-DsRedExpress vector from Clontech was used for gain-of-function studies. All the retroviruses were packaged in 293T cells via the pCL-eco packaging vector and then used to transduce erythroid progenitor cells using the methods described previously (Hattangadi et al., 2010). K562 cells were obtained from the ATCC, and MEL cells were a kind gift from Dr. Barry Paw (Harvard Medical School).
RNA and protein analysis
The miRNeasy kit from Qiagen was used to extract total RNA from cells lines or primary cells isolated by fluorescence-activated or magnetic-assisted cell sorting. In all RNA isolation procedures, DNase I on-column digestion (Qiagen) was performed to remove genomic DNA. cDNAs from the isolated RNAs were synthesized using the Superscript II reverse transcriptase (Invitrogen) with random primers. Real-time PCR was performed on an ABI Prism 7900 sequence detection system using SYBR Green PCR Master Mix (Applied Biosystems). Proteins from target cells were extracted by RIPA buffer (50mM Tris-HCl pH7.4, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS, 150mM NaCl, 2mM EDTA, 50mM NaF). Then SDS-PAGE and Western blot were employed to detect target proteins using the following antibodies: Cpeb4 (Abcam: ab83009), Gapdh (Santa Cruz biotechnology, Inc: sc-32233), eIF3A (Cell Signaling: 2538), eIF3B (Abcam: ab133601).
Flow cyotometry analysis
The flow cytometry analyses were performed on a BD LSR ║ flow cytometer. All the antibodies used for flow cytometry were from eBiosciences. Terminal erythroid differentiation and enucleation was analyzed by the same methods as previously described (Ji et al., 2008). Cell proliferation, cell division, cell cycle, and apoptosis were analyzed using the CountBright Absolute Counting Beads (Life Technologies), the CellTrace CFSE Cell Proliferation Kit (Life Technologies), Click-iT EdU Pacific Blue Flow Cytometry Assay Kit (Life Technologies), and Annexin V Pacific Blue conjugate (Life Technologies), respectively, in accordance with the corresponding protocols. For the cell proliferation analysis, 10 uM CFSE dye (carboxyfluorescein diacetate succinimidyl ester) was used to label cells for 10min at 37°C.
Polysome analysis and translation rate measurement
Polysome analysis was performed as described previously (Hu et al., 2009). In brief, 50 million MEL cells were lysed in polysome lysis buffer (10mM Tris-HCl pH7.4, 12mM MgCl2, 100mM KCl, 1% TritonX-100, 100ug/ml cycloheximide). The cell lysate was loaded onto a 10%-50% (w/v) linear sucrose-density gradient and then centrifuged at 37,000 rpm in a SW-41Ti rotor for 2 hours at 4°C. The gradient was then fractionated using a Gradient Station (BioComp) coupled with a UV monitor (Bio-Rad EM-1). The RNA from each fraction was isolated by the method described in Hu et al., 2009 (Hu et al., 2009). The global translation rate was measured by using the Click-iT HPG Alexa Fluor-594 protein synthesis assay kit (Life Technologies).
Luciferase assays
Luciferase assays were performed in accordance with the protocol from the Dual-Luciferase Reporter Assay System (Promega). Specifically, reporters and Cpeb4 expressing plasmids were transfected into K562 cells using the Lipofectamine LTX reagent (Life Technologies), and then luciferase assays were performed at 30 hours after transfection.
Affinity purification and immunoprecipitation
The Cpeb4 complex was purified from 50 million mouse fetal liver Ter119+ cells and 50 million K562 cells stably expressing Cpeb4-GFP using anti-Cpeb4 and anti-GFP (abcam 290) antibodies, respectively. 10ug antibody was used for each purification, and the purification was performed in the presence and absence of RNaseA (200ug/ml) using a magnetic IP and Co-IP kit from Pierce (Cat. 88805). The purified complex was resolved on an SDS-PAGE gel followed by silver staining using reagents compatible with mass spectrometry (Pierce Cat. 24600). After silver staining, each lane on the SDS-PAGE gel was cut into 10 pieces for mass spectrometry to identify proteins at the Biopolymers & Proteomics Lab at the Koch Institute of MIT. 5ug anti-eIF3A antibody was used to immunoprecipitate the eIF3 complex from 20 million mouse fetal liver Ter119+ cells. The purified complex was then subject to SDS-PAGE followed by Western blotting.
Bioinformatic analysis
RIP-Chip data from Affymetrix Mouse Gene ST Arrays were normalized by RMA using the Affy package from Bioconductor. A custom probeset definition was used for processing the arrays as defined by Dai et al.(Dai et al., 2005) such that one probeset represents one Entrez Gene ID. The microarray results were deposited at the Gene Expression Omnibus with accession number GSE57004.
To identify Cpeb4 binding motifs, refGene 3’UTR sequences were downloaded from the UCSC Table Browser in Jan 2014, with repeat sequences masked to N. For each gene, all the possible distinct 8mers were identified with the wordcount function in EMBOSS (Rice et al., 2000). Fisher’s exact test was used to identify enriched 8mers in the Cpeb4 enriched mRNAs relative to either the depleted mRNAs in the Cpeb4 RIP-Chip or a background set of 17791 coding genes in both RNA-seq and RIP-Chip excluding the Cpeb4 enriched mRNAs. To identify new motifs in the 227 enriched mRNAs in the Cpeb4 RIP-Chip, we randomly selected the same number of enriched mRNAs from the 17791 background genes 1000 times. Each time, we ran Fisher’s exact test and looked for the over represented 8mer motifs (FDR-corrected p value < 0.05) in the Cpeb4 bound mRNAs. Only the enriched motifs in all 1000 replicates were used to draw the sequence logo with WebLogo (http://weblogo.berkeley.edu).
The gene ontology analysis was performed using the DAVID Functional Annotation Bioinformatics Microarray Analysis (http://david.abcc.ncifcrf.gov/)
Statistical analysis
Two-tailed Student’s t test was used to determine whether two sets of data are statistically different from each other. All statistical analyses and the resulting graphs were performed and generated using GraphPad Prism version 6. Data are presented as mean ± standard deviation (SD).
Supplementary Material
HIGHLIGHTS.
Cpeb4 is induced by Gata1/Tal1 and is required for terminal erythropoiesis
Cpeb4 interacts with eIF3 to repress mRNA translation in erythroid cells
Cpeb4 binds a large group of mRNAs, including its own, in erythroid cells
Maintaining Cpeb4 level within a range is required for terminal erythropoiesis
Acknowledgements
We thank Drs. Juan Alvarez, Jiahai Shi, Stephen Eichhorn, and Barry Paw for assistance on experiments and reagents, Dr. George Bell for critical reading of this manuscript, and the flow cytometry core at the Whitehead Institute and the Biopolymers & Proteomics Lab at MIT for technical support. Wenqian Hu is a Merck Fellow of the Life Sciences Research Foundation and is supported by a Pathway to Independence Award (1K99HL118157) from the NIH. This research was supported by NIH grants R01DK068348 and 5P01HL066105 to H.F.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez-Dominguez JR, Hu W, Yuan B, Shi J, Park SS, Gromatzky AA, van Oudenaarden A, Lodish HF. Global discovery of erythroid long non-coding RNAs reveals novel regulators of red cell maturation. Blood. 2013 doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Wickens M. Tethered function assays: an adaptable approach to study RNA regulatory proteins. Methods Enzymol. 2007;429:299–321. doi: 10.1016/S0076-6879(07)29014-7. [DOI] [PubMed] [Google Scholar]
- D’Ambrogio A, Nagaoka K, Richter JD. Translational control of cell growth and malignancy by the CPEBs. Nat Rev Cancer. 2013;13:283–290. doi: 10.1038/nrc3485. [DOI] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Miranda G, Mendez R. The CPEB-family of proteins, translational control in senescence and cancer. Ageing Res Rev. 2012;11:460–472. doi: 10.1016/j.arr.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Flygare J, Rayon Estrada V, Shin C, Gupta S, Lodish HF. HIF1alpha synergizes with glucocorticoids to promote BFU-E progenitor self-renewal. Blood. 2011;117:3435–3444. doi: 10.1182/blood-2010-07-295550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- Hattangadi SM, Burke KA, Lodish HF. Homeodomain-interacting protein kinase 2 plays an important role in normal terminal erythroid differentiation. Blood. 2010;115:4853–4861. doi: 10.1182/blood-2009-07-235093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattangadi SM, Wong P, Zhang L, Flygare J, Lodish HF. From stem cell to red cell: regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood. 2011;118:6258–6268. doi: 10.1182/blood-2011-07-356006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. The Scanning Mechanism of Eukaryotic Translation Initiation. Annu Rev Biochem. 2014 doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25:2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Kan MC, Lin CL, Richter JD. CPEB3 and CPEB4 in neurons: analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J. 2006;25:4865–4876. doi: 10.1038/sj.emboj.7601322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igea A, Mendez R. Meiosis requires a translational positive loop where CPEB1 ensues its replacement by CPEB4. EMBO J. 2010;29:2182–2193. doi: 10.1038/emboj.2010.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Devine T, George AD, Chittur SV, Baroni TE, Penalva LO, Tenenbaum SA. RIP-Chip analysis: RNA-Binding Protein Immunoprecipitation-Microarray (Chip) Profiling. Methods Mol Biol. 2011;703:247–263. doi: 10.1007/978-1-59745-248-9_17. [DOI] [PubMed] [Google Scholar]
- Ji P, Jayapal SR, Lodish HF. Enucleation of cultured mouse fetal erythroblasts requires Rac GTPases and mDia2. Nat Cell Biol. 2008;10:314–321. doi: 10.1038/ncb1693. [DOI] [PubMed] [Google Scholar]
- Kerenyi MA, Orkin SH. Networking erythropoiesis. J Exp Med. 2010;207:2537–2541. doi: 10.1084/jem.20102260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matushansky I, Radparvar F, Skoultchi AI. Reprogramming leukemic cells to terminal differentiation by inhibiting specific cyclin-dependent kinases in G1. Proc Natl Acad Sci U S A. 2000;97:14317–14322. doi: 10.1073/pnas.250488697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matushansky I, Radparvar F, Skoultchi AI. CDK6 blocks differentiation: coupling cell proliferation to the block to differentiation in leukemic cells. Oncogene. 2003;22:4143–4149. doi: 10.1038/sj.onc.1206484. [DOI] [PubMed] [Google Scholar]
- Merryweather-Clarke AT, Atzberger A, Soneji S, Gray N, Clark K, Waugh C, McGowan SJ, Taylor S, Nandi AK, Wood WG, et al. Global gene expression analysis of human erythroid progenitors. Blood. 2011;117:e96–108. doi: 10.1182/blood-2010-07-290825. [DOI] [PubMed] [Google Scholar]
- Novoa I, Gallego J, Ferreira PG, Mendez R. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat Cell Biol. 2010;12:447–456. doi: 10.1038/ncb2046. [DOI] [PubMed] [Google Scholar]
- Ortiz-Zapater E, Pineda D, Martinez-Bosch N, Fernandez-Miranda G, Iglesias M, Alameda F, Moreno M, Eliscovich C, Eyras E, Real FX, et al. Key contribution of CPEB4-mediated translational control to cancer progression. Nat Med. 2012;18:83–90. doi: 10.1038/nm.2540. [DOI] [PubMed] [Google Scholar]
- Pilon AM, Ajay SS, Kumar SA, Steiner LA, Cherukuri PF, Wincovitch S, Anderson SM, Mullikin JC, Gallagher PG, Hardison RC, et al. Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood. 2011;118:e139–148. doi: 10.1182/blood-2011-05-355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, Canfield T, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Chang JS, Costa A, Schedl P. An autoregulatory feedback loop directs the localized expression of the Drosophila CPEB protein Orb in the developing oocyte. Development. 2001;128:1159–1169. doi: 10.1242/dev.128.7.1159. [DOI] [PubMed] [Google Scholar]
- Theis M, Si K, Kandel ER. Two previously undescribed members of the mouse CPEB family of genes and their inducible expression in the principal cell layers of the hippocampus. Proc Natl Acad Sci U S A. 2003;100:9602–9607. doi: 10.1073/pnas.1133424100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LY, Chang YW, Lin PY, Chou HJ, Liu TJ, Lee PT, Huang WH, Tsou YL, Huang YS. CPEB4 knockout mice exhibit normal hippocampus-related synaptic plasticity and memory. PLoS One. 2013;8:e84978. doi: 10.1371/journal.pone.0084978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P, Hattangadi SM, Cheng AW, Frampton GM, Young RA, Lodish HF. Gene induction and repression during terminal erythropoiesis are mediated by distinct epigenetic changes. Blood. 2011;118:e128–138. doi: 10.1182/blood-2011-03-341404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Cheng Y, Keller CA, Ernst J, Kumar SA, Mishra T, Morrissey C, Dorman CM, Chen KB, Drautz D, et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 2011;21:1659–1671. doi: 10.1101/gr.125088.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Socolovsky M, Gross AW, Lodish HF. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood. 2003;102:3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.