Abstract
The nitroheterocyclic classes of drugs have a long history of use in treating anaerobic infections, as exemplified by metronidazole as a first-line treatment for mild-to-moderate Clostridium difficile infection (CDI). Since direct comparisons of the three major classes of nitroheterocyclic drugs (i.e. nitroimidazole, nitazoxanide and nitrofurans) and nitrosating agents against C. difficile are under-examined, in this study their actions against C. difficile were compared. Results show that whilst transient resistance occurs to metronidazole and nitazoxanide, stable resistance arises to nitrofurans upon serial passage. All compounds killed C. difficile at high concentrations in addition to the host defence nitrosating agent S-nitrosoglutathione (GSNO). This suggests that GSNO killing of C. difficile contributes to its efficacy in murine CDI. Although nitric oxide production could not be detected for the nitroheterocyclic drugs, the cellular response to metronidazole and nitrofurans has some overlap with the response to GSNO, causing significant upregulation of the hybrid-cluster protein Hcp that responds to nitrosative stress. These findings provide new insights into the action of nitroheterocyclic drugs against C. difficile.
Keywords: Nitroimidazole, Nitrofurans, Nitazoxanide, Nitrosative stress, Anti-Clostridium difficile action
1. Introduction
Clostridium difficile is the main cause of hospital-acquired diarrhoea in developed countries such as the USA and Europe. Each year in the USA alone, there are >400 000 cases of C. difficile infection (CDI), with >14 000 deaths [1]. Since the 1980s, metronidazole, a 5-nitroimidazole prodrug, has been established as a first-line therapy for mild-to-moderate CDI [2]. Despite its long history of use for treating CDI, the cellular action of metronidazole against C. difficile is not well characterised [3]. However, based on studies in other organisms, metronidazole is bioreductively activated by cellular oxidoreductases (e.g. nitroreductases), whereby its nitro group is reduced by an electron to produce a highly reactive and unstable nitroimidazole anion that can have several fates [4]. The unstable nitroimidazole anion may be further reduced to nitroso and hydroxylamine intermediates or may undergo decomposition yielding additional reactive species in the form of an imidazole radical and a nitrite anion from which nitric oxide (NO) is derived [4]. These nitroimidazole reactive derivatives and NO cause damage to cellular targets, namely proteins and DNA, leading to cell death [4,5]. If NO is produced upon metronidazole bioreduction in C. difficile, this might indicate that it imposes nitrosative stress in C. difficile in a manner similar to the innate immune system [6]. However, the genetic response of C. difficile both to nitroheterocyclic drugs, including metronidazole, and host-derived NO-generating molecules such as S-nitrosoglutathione (GSNO) is relatively undercharacterised [7].
Interestingly, there are only a few reports of metronidazole resistance in C. difficile [3,8]. This extremely low incidence of metronidazole resistance in C. difficile is confounded by the instability of the metronidazole-resistant phenotype, with resistance being lost during freezer storage or following brief passage in microbiological media [3,9]. The rarity of metronidazole resistance in C. difficile is unusual considering that resistance to metronidazole occurs by several different mechanisms in other bacteria [10]. This prompted us to question whether the lack of metronidazole resistance in C. difficile is also displayed by other nitroheterocyclic drugs. Besides metronidazole, other members of the nitroheterocyclic drug class are also important treatments for other anaerobic infections, namely the 5-nitrofuran and nitrothiazolyl drugs [11,12].
Furthermore, the nitrothiazolyl nitazoxanide is considered an alternate treatment for CDI and has been successfully modified to produce improved analogues [12]. A key difference in these three nitroheterocyclic drug types arises from their redox potential, which dictates the spectrum of activity, mechanism of bioreduction and cellular effects [13]. Interestingly, nitazoxanide acts as a non-competitive inhibitor of pyruvate:ferredoxin oxidoreductase (PFOR) in anaerobes (e.g. C. difficile), protozoa and Helicobacter pylori, which is distinct to the cellular action of 5-nitroimidazoles and 5-nitrofurans, involving the formation of reactive species and inhibition of multiple targets [10].
In this study, we sought to better understand the action of these three subclasses of nitroheterocyclic drugs against C. difficile by directly comparing their effects on cell viability, propensity to select for stable resistance, and the cellular responses of DNA damage and nitrosative stress. The results suggest that all three nitroheterocyclic subclasses show characteristic mode of action profiles, with nitroimidazoles and nitrofurans bearing some resemblance to GSNO. We also report for the first time that C. difficile is rapidly killed by GSNO, which now provides an additional basis for the observed efficacy of this molecule in mice with CDI [6].
2. Materials and methods
2.1. Chemicals, bacterial strains and growth conditions
Clostridium difficile strains CD196 (a historic NAP1 strain) and R20291 (a contemporary NAP1 strain) were kindly provided by Dr A.L. Sonenshein (Tufts University, Medford, MA) and strain BAA-1875 (NAP7) was obtained from ATCC (Manassas, VA). All strains were routinely grown in pre-reduced BHITY broth [brain–heart infusion, tryptone (1% w/v) and yeast extract (0.5% w/v)] at 37 °C in a Whitley A35 anaerobic workstation (Don Whitley Scientific, Shipley, UK). Antibiotics were obtained from the following sources: metronidazole, vancomycin, fusidic acid, rifaximin and nitazoxanide were from Sigma-Aldrich (St Louis, MO); ornidazole was from Alfa Aesar (Ward Hill, MA); nitrofurazone was from TCI America (Portland, OR); nifuroxazide and furazolidone were from MP Biomedicals (Santa Ana, CA); and nitrofurantoin was from Acros Organics (Fair Lawn, NJ). GSNO was from Enzo Life Sciences (Farmingdale, NY) and was prepared fresh in culture medium adjusted to pH 6.5 and was tested in medium of pH 6.5. Stock solutions of other compounds were stored at −80 °C in dimethyl sulphoxide (DMSO) at 10 mg/mL.
2.2. Determination of minimum inhibitory concentrations (MICs)
MICs of compounds were determined in 24-well plates, or in 96-well round-bottom microtitre plates (Thermo Scientific, Waltham, MA) for freeze–thawed cultures from serial passage experiments [14]. In 24-well plates, compounds were first two-fold serially diluted in 500 µL of BHITY broth, followed by the addition of 500 µL of 106 CFU/mL of C. difficile, with test concentrations ranging from 0.06–64 µg/mL. Plates were incubated anaerobically at 37 °C for 24 h. MICs were similarly determined in 96-well microtitre plates but using antibiotic dilution volumes of 50 µL with a final volume of 100 µL after addition of the inoculum. There was no difference in the MICs determined in 96-well microtitre plates or 24-well plates.
2.3. Determination of minimum bactericidal concentrations (MBCs)
MBCs were determined against log-phase cells in 24-well plates as described previously [14]. Briefly, 900 µL of mid-logarithmic cells [optical density at 600 nm (OD600) ≈ −0.3] was added to 100 µL of two-fold serially diluted compounds. After 24 h of incubation, viable counts were enumerated on BHITY agar containing 20% w/v activated charcoal [14]. MBCs were determined at least twice and were defined as the lowest concentration of compound killing ≥3 log of the initial inoculum (ca. −107 CFU/mL).
2.4. Time–kill assay
Time–kill assay experiments were performed as described previously [14]. Mid-logarithmic phase cultures (OD600 ≈ 0.3) were exposed to metronidazole and nitazoxanide at their MBC, nitrofurantoin at 8× and 16× its MIC and S-nitrosoglutathione (GSNO) at its MIC. Total viable counts were performed on samples recovered at different points. All experiments were performed at least twice.
2.5. Selection of nitroheterocyclic-resistant mutants
To compare the potential for emergence of de novo resistance in C. difficile, methods for plating of concentrated cultures or serial passage in the presence of subinhibitory concentrations of drug were adopted as described below.
2.5.1. Mutant selection on agar
Overnight cultures in BHITY broth (10 mL) were concentrated 10-fold resulting in 109 CFU/mL. The entire 1 mL of sample was plated (100 µL per plate) onto BHITY agars containing antibiotic at 4× MIC. Following incubation for 48 h, the mutation frequency was obtained from the total number of colonies on selection plates divided by the total number of viable cells.
2.5.2. Selection of stable resistance by serial passage
Serial passage experiments were conducted in 96-DeepWell plates (Thermo Scientific), essentially as described previously [15] except for the use of BHITY broth (1 mL) as the growth medium. Initially, compounds were two-fold serially diluted to yield antibiotic concentrations that were eight-fold above and four-fold below their MICs. Each well was inoculated with C. difficile to yield ca. 106 CFU/well. Following incubation for 24 h at 37 °C, the lowest concentration of compound preventing visible growth was recorded as the MIC. Bacteria growing one dilution below the MIC were then used to inoculate the subsequent round of passaging in fresh broth containing antibiotic. This was repeated for a total of 20 passages and at each stage the MIC obtained was used to define the compound concentration ranges for the subsequent passage. To isolate individual colonies from populations showing growth at elevated concentrations, cultures were plated onto agar with antibiotic at 4× MIC. To determine whether stable resistance to compounds arose during passaging, stocks of bacteria were first stored at −80 °C. For every fourth passage in the series, frozen stocks were plated onto non-selective BHITY agar. After overnight growth, the entire plate was scrapped into fresh broth, incubated overnight and MICs were determined in 96-well microtitre plates.
2.6. Measurement of nitrite and nitric oxide
Two methods were used to detect the production of reactive nitrite or NO in cultures as described below.
2.6.1. Griess reagent assay
Since the initial experiments revealed quenching of Griess reagent assay in BHITY, these experiments were performed in TYG broth (1% w/v tryptone, 0.5% w/v yeast and 1% w/v glucose). There were no differences in the MICs of compounds in TYG or BHITY broth. Aliquots (1 mL) of concentrated C. difficile CD196 (ca. 109 CFU/mL) were exposed to metronidazole and nitrofurantoin at 64 µg/mL (0.37 mM and 0.27 mM, respectively) and to GSNO (50.4 µg/mL; 0.15 mM) for 30 min and 90 min. Cells were centrifuged and supernatants were retained, whereas cell pellets were lysed by bead beating. Both the supernatant and cell lysates (50 µL) were assayed for nitrite according to instructions for the Griess reagent kit from Promega (Madison, WI). Nitrite concentrations were determined from a standard curve of nitrite at 1.5–200 µM; the limit of detection was 2.5 µM.
2.6.2. β-Galactosidase (β-gal) reporter assay
Escherichia coli strain JOEY426 (MG1655 derivative of JOEY19 [16]) carrying a NO-inducible promoter (ytfE) fused to lacZ was used as reporter strain in co-cultures with C. difficile CD196; ytfE is specifically induced by NO. Both cultures were grown separately to OD600 ≈ −0.3 in BHITY and lysogeny broth–Lennox medium, respectively. Cells were centrifuged, washed and re-suspended in pre-reduced defined medium [17]. Clostridium difficile CD196 and E. coli JOEY426 were mixed to yield a final OD600 of 2.5 and 0.3, respectively, in 1 mL, and metronidazole (64 µg/mL; 0.37 mM), nitrofurantoin (64 µg/mL; 0.27 mM) and GSNO (50.4 µg/mL; 0.15 mM) were added for 30 min and 90 min followed by measuring β-gal activity [16].
2.7. Gene expression analysis by quantitative reverse transcription PCR (qRT-PCR)
To compare the effect of compounds and GSNO on gene expression, qRT-PCR was performed on logarithmic cultures exposed to growth inhibitory concentrations (GICs). The GIC is considered more relevant than the MIC to probe primary transcriptional responses [18] since there is a significantly larger population of bacteria compared with what is used for MIC testing; also, the GIC does not cause cell death, unlike the MBC. GICs were determined by adding compounds to cultures (OD600 ≈ 0.3) followed by optical density measurements. The concentration of test compound at which no increase in optical density was observed up to 6 h was designated as the GIC. To probe the transcriptional response to the GIC, cultures were grown in BHITY to OD600 ≈ 0.3 and compounds were added at their GICs as follows: 8 µg/mL metronidazole; 16 µg/mL nitazoxanide; 64 µg/mL nitrofurantoin; and 0.038 mM GSNO. Cultures were then incubated for 20 min at 37 °C and before adding bacterial RNAprotect™ reagent (QIAGEN, Valencia, CA). Total RNA was isolated using a SV Total RNA Isolation System (Promega) and 2 µg was converted to cDNA using M-MLV reverse transcriptase (Promega) with random hexamers. Maxima SYBR Green Master Mix (Thermo Scientific) and gene-specific primers (Supplementary Table S1) were used to amplify gene transcripts in an Applied Biosystems 7300 Real Time PCT System (Applied Biosystems, Grand Island, NY). Results were calculated by the comparative Ct Method (ΔΔCT method) [19] with endogenous 16S rRNA serving as a control to normalise mRNA levels.
3. Results
3.1. Activities of nitroheterocyclic drugs against Clostridium difficile
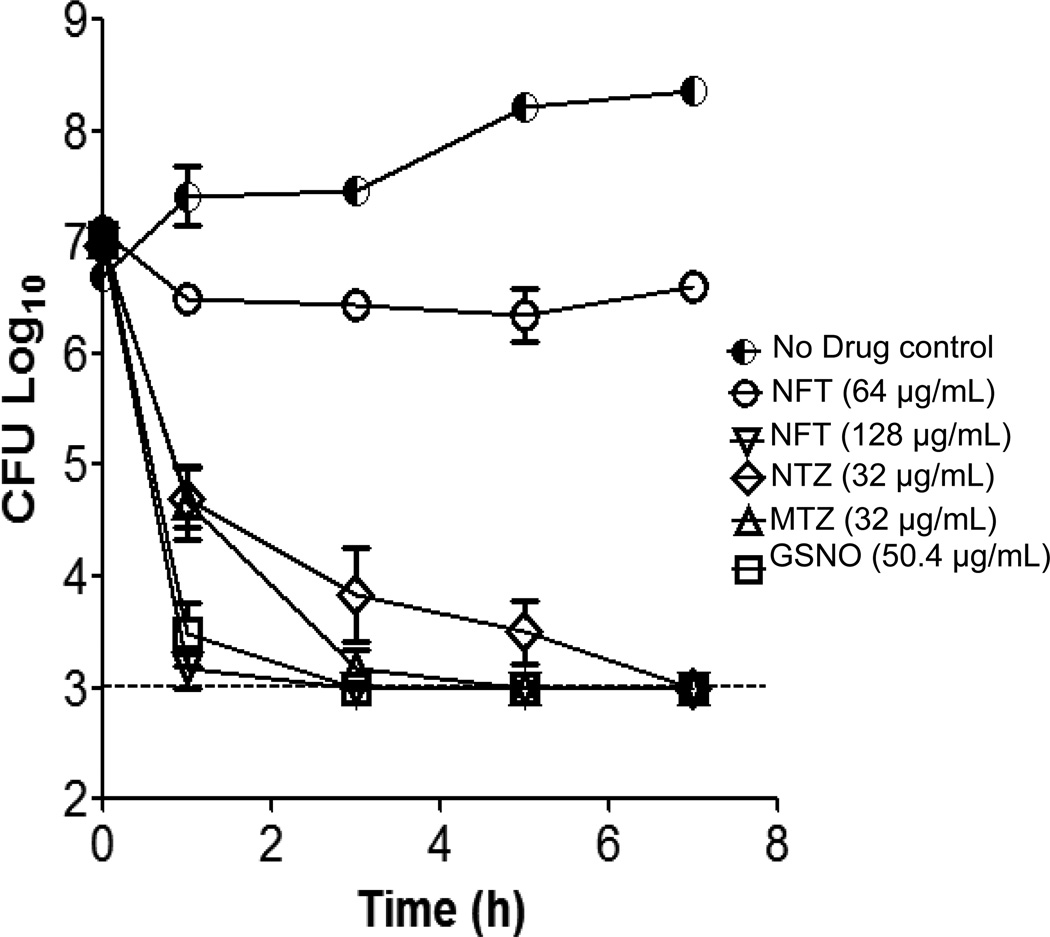
A comparative assessment of the activities of the three main subclasses of nitroheterocyclic drugs against C. difficile, under the same test conditions, is shown in Table 1. As expected, the nitroimidazoles metronidazole and ornidazole were highly active, inhibiting the growth of C. difficile at low concentrations (0.25–0.5 µg/mL). Surprisingly, under the test conditions, nitazoxanide only inhibited growth of the three test strains at high concentrations of 8–16 µg/mL, which was comparable with nitrofurans that inhibited the growth of C. difficile at 2–8 µg/mL. Despite nitazoxanide requiring higher concentrations to inhibit growth, it was found to be as potent as metronidazole in killing logarithmic cultures over a 24-h period (Table 1), as both caused a 3 log reduction in cells at similar test concentrations (i.e. 16–64 µg/mL). Conversely, all four nitrofurans failed to kill more than 2 log of bacteria within a 24-h exposure period at 64 µg/mL. Interestingly, the NO donor GSNO was also bactericidal to all three C. difficile test strains with MBCs (201.8–403.5 µg/mL; 0.6–1.2 mM) that were ≥4-fold its MIC (Table 1). Since MBC measurements are endpoint assays, time–kill experiments were performed against CD196 to compare the rate of kill with metronidazole, nitazoxanide, nitrofurantoin and GSNO. As shown in Fig. 1, GSNO at its MIC (i.e. 50.4 µg/mL; 0.15 mM) was the most rapidly killing agent, since a 1-h exposure reduced the number of viable cells to <103 CFU/mL (i.e. the detection limit of the assay); however, by 24 h the surviving cells regrew to ca. 107 CFU/mL explaining the higher MBCs of 4–8-fold above the MIC. Metronidazole and nitazoxanide at 32 µg/mL killed ca. 3 log of cells in 3 h, but the limit of detection was only reached in 5 h and 7 h, respectively. Consistent with the endpoint MBCs, nitrofurantoin was entirely bacteriostatic at 64 µg/mL (Fig. 1), but a 2-fold increase in concentration (128 µg/mL) resulted in rapid bactericidal activity comparable with metronidazole and nitazoxanide.
Table 1.
Antibacterial activities (µg/mL) of nitroheterocyclic drugs and other test compounds against Clostridium difficile strainsa
| Compound | CD196 | BAA-1875 | R20291 | |||
|---|---|---|---|---|---|---|
| MIC | MBClog | MIC | MBClog | MIC | MBClog | |
| Metronidazole | 0.5 | 32 | 0.25 | 32 | 0.5 | 16 |
| Ornidazole | 0.25 | 32 | 0.25 | 64 | 0.5 | 64 |
| Nitazoxanide | 8 | 32 | 8 | 32 | 16 | 64 |
| Nitrofurantoin | 8 | >64 | 8 | >64 | 8 | >64 |
| Nitrofurazone | 4 | >64 | 4 | >64 | 4 | >64 |
| Nifuroxazide | 8 | >64 | 8 | >64 | 8 | >64 |
| Furazolidone | 4 | >64 | 2 | >64 | 2 | >64 |
| Vancomycin | 1 | >64 | 0.5 | >64 | 1 | >64 |
| Fusidic acid | 0.1 | >64 | 0.05 | >64 | 0.1 | >64 |
| GSNOb | 0.15 | 1.2 | 0.15 | 0.6 | 0.30 | 1.2 |
| (50.4) | (403.5) | (50.4) | (201.8) | (100.8) | (403.5) | |
MIC, minimum inhibitory concentration; MBClog, minimum bactericidal concentration against log-phase cells; GSNL, S-nitrosoglutathione.
Values are the mean of at least three independent experiments.
MICs are in mM (with values in parenthesis in µg/mL). The pH of BHITY broth [brain–heart infusion, tryptone (1% w/v) and yeast extract (0.5% w/v)] was adjusted to 6.5; the MIC for GSNO in broth where the pH was not adjusted (pH 7.3) was >10 mM. A pH of 6.5 had no effect on the MICs of nitroheterocyclic drugs.
Fig. 1.
Time–kill kinetics against log-phase Clostridium difficile CD196. Data are shown for metronidazole (MTZ) and nitazoxanide (NTZ) at their minimum bactericidal concentration (MBC), nitrofurantoin (NFT) at 8× and 16× its minimum inhibitory concentration (MIC) and S-nitrosoglutathione (GSNO) at its MIC. Data are the mean of at least two independent experiments, with error bars showing the standard error of the mean.
3.2. Unlike metronidazole and nitazoxanide, stable resistance to nitrofurans may be obtained by serial passage
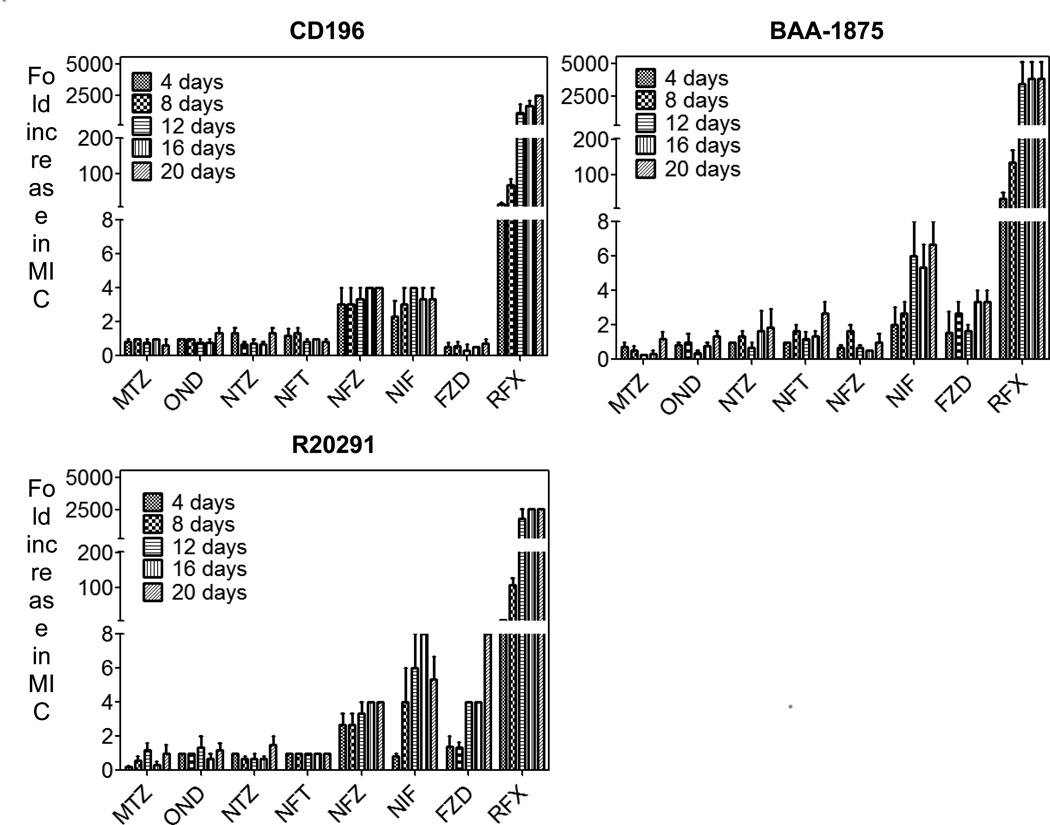
No mutants could be obtained on agar from concentrated stationary-phase cultures (108–109 CFU/mL) of the three C. difficile strains, suggesting that all three classes of nitroheterocyclic drugs have a low propensity for first-step resistance. To further examine the likelihood of resistance development, cultures were serially passaged under increasing concentrations of antibiotics to enrich the mutant population. During passaging, it was observed that C. difficile could be grown at concentrations that were above the MIC for each test compound (Supplementary Table S2) suggesting the presence of subpopulations exhibiting phenotypic resistance or unstable de novo resistant mutants. None the less, the presumptive de novo mutants in cultures exposed to metronidazole, ornidazole or nitazoxanide could not be isolated on agar and the population regained susceptibility after storage at −80 °C (Fig. 2; Supplementary Table S2). Conversely, stable nitrofuran-resistant mutants could be isolated from the populations showing reduced susceptibility to the tested nitrofurans (Fig. 2; Table 2). As shown in Table 2, nitrofuran-resistant mutants were 2–16-fold more resistant to nitrofurans and typically exhibited cross-resistance only within the nitrofuran class. As a control, all test strains yielded stable mutants to rifaximin (Fig. 2).
Fig. 2.
Changes in the susceptibility of Clostridium difficile strains to nitroheterocyclic drugs during serial passage for 20 days. Values represent the mean of three independent experiments, with error bars showing the standard error of the mean. MTZ, metronidazole; OND, ornidazole; NTZ, nitazoxanide; NFT, nitrofurantoin; NFZ, nitrofurazone; NIF, nifuroxazide; FZD, furazolidone; RFX, rifaximin. Mean starting minimum inhibitory concentrations (MICs) against CD196, BAA-1875 and R20291 were 0.01, 0.005 and 0.03 µg/mL, respectively.
Table 2.
Class-specific cross-resistance to nitrofurans in Clostridium difficilea
| Agent | Parent strain | Mutant | MIC (fold increase) | |||
|---|---|---|---|---|---|---|
| NFZ | NIF | FZD | NFT | |||
| NFZ | CD196 | MK-2131 | 4 | Noneb | 16 | None |
| CD196 | MK-2631 | 8 | 8 | 16 | 4 | |
| BAA-1875 | MK-2634 | 4 | 8 | 16 | None | |
| R20291 | MK-2137 | 4 | 4 | 16 | None | |
| NIF | CD196 | MK-2132 | 8 | 4 | 16 | None |
| CD196 | MK-2632 | 8 | 4 | 16 | None | |
| BAA-1875 | MK-2135 | 4 | 4 | 8 | 4 | |
| BAA-1875 | MK-2635 | 4 | 8 | 16 | None | |
| NFT | CD196 | MK-2133 | 8 | 4 | 4 | 2 |
| BAA-1875 | MK- 2636 | 8 | 8 | 8 | 8 | |
MIC, minimum inhibitory concentration; NFZ, nitrofurazone; NIF, nifuroxazide; FZD, furazolidone; NFT, nitrofurantoin.
No cross-resistance to nitazoxanide and metronidazole was observed.
‘None’ indicates that no fold increase was observed.
3.3. Metronidazole and other nitroheterocyclics do not appear to produce measureable nitric oxide
To detect and quantify the production of these nitro radicals, Griess reagent was initially used. With the control GSNO at 0.15 mM, 12.2 ± 0.1.4 µM and 20.2 ± 2.4 µM of nitrite was detected after 30 min and 90 min of incubation, respectively. In contrast, both metronidazole and nitrofurantoin at 64 µg/mL (0.37 mM and 0.27 mM, respectively) failed to generate detectable signals above the detection limit of 2.5 µM (Supplementary Table S3). Similarly, no discernable signal above basal levels could be detected for metronidazole and nitrofurantoin using JOEY426, the NO-responsive β-gal reporter strain (Supplementary Table S4). However, β-gal production was induced in the presence of GSNO even at a low concentration of 0.0015 mM.
3.4. Transcriptional response to nitroheterocyclic drugs and S-nitrosoglutathione
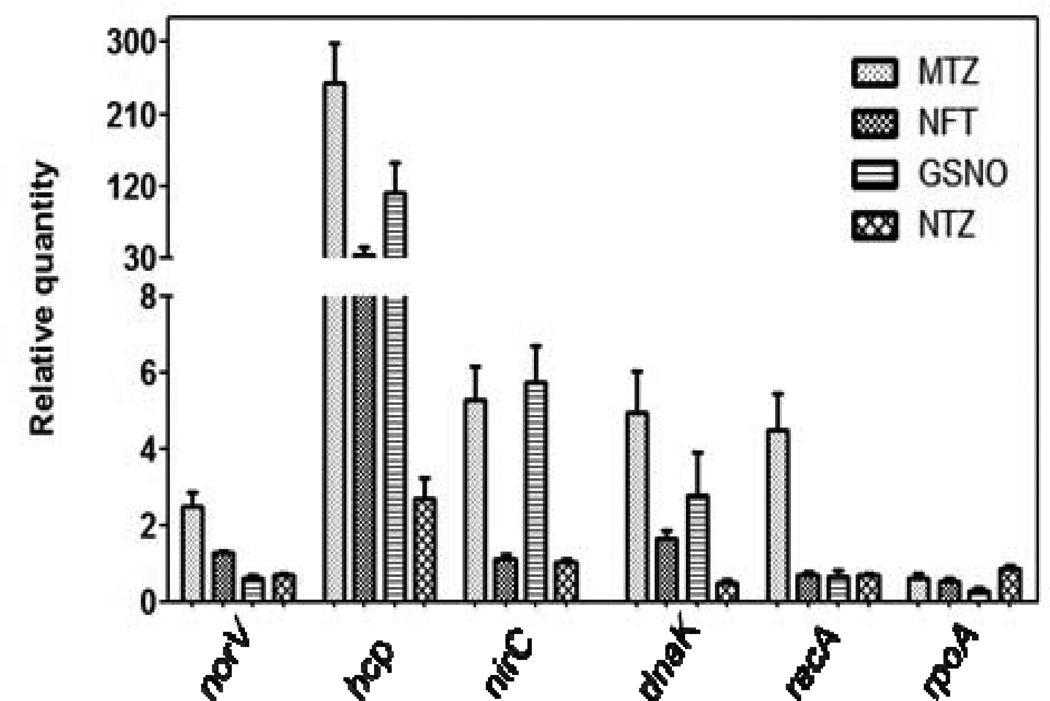
To further understand the cellular action of nitroheterocyclics and GSNO, the expression of genes known to respond to DNA damage [recombinase A (recA)] [20], nitrosative stress [hybrid-cluster protein (hcp), nitric oxide reductase flavorubredoxin (norV) and putative hydrosulphide/nitrite transporter (nirC)] [21] and general stress [Hsp70 molecular chaperone (dnaK)] was analysed. These results are summarised in Fig. 3, with a >2.5-fold change in gene expression considered as significant. Exposure to GSNO, metronidazole and nitrofurantoin dramatically increased expression of the hybrid-cluster protein gene hcp by 112.6 ± 36-, 249.3 ± 49- and 35 ± 8-fold, respectively. However, we were surprised to find such high induction of hcp by metronidazole and nitrofurantoin, as NO/nitrite could not be detected or appear to be produced in only low amounts compared with GSNO (Supplementary Table S3). Consistent with nitazoxanide targeting the activity of PFOR and not undergoing reduction to form reactive species [10], hcp was only induced by three-fold, possibly due to cellular redox changes. With respect to metronidazole, other genes involved in nitrogen metabolism (norV and nirC) were also upregulated by 2.5 ± 0.3-fold and 5.2 ± 0.9-fold, respectively. Induction of recA (4.4 ± 0.9-fold) by only metronidazole is consistent with it triggering the SOS response in C. difficile owing to DNA strand breaks [5]. The test control vancomycin did not affect expression of the studied genes (data not shown).
Fig. 3.
Gene expression analysis in response to metronidazole (MTZ), nitrofurantoin (NFT), nitazoxanide (NTZ) and S-nitrosoglutathione (GSNO). The bar depicts the relative quantities of mRNA for respective genes, measured by quantitative reverse transcription PCR and normalised against the 16S rRNA gene. The mean of three independent experiments is shown, with error bars showing the standard error of the mean.
4. Discussion
Despite being used for more than 30 years to treat CDI, the mode of action of metronidazole in C. difficile is still underexplored, in contrast to other pathogens such as H. pylori and protozoans [10,22]. Thus, we explored the action of metronidazole against C. difficile, comparing it with other clinically relevant nitroheterocyclic drugs. This study advances present knowledge on the potential for de novo resistance and the cellular response of C. difficile not only to metronidazole but to other subclasses of nitroheterocyclic drugs. We also demonstrate that C. difficile is highly sensitive to GSNO, which is found in the gastrointestinal tract [6]. It is therefore likely that the bactericidal activity of GSNO contributes to its efficacy in the murine model of CDI [6]. This observation also adds to the existing literature that C. difficile and its spores are highly sensitive to acidified nitrite [23].
The inability to select spontaneous mutants exhibiting stable resistance to metronidazole might reflect the presence of multiple nitroreductases that can activate metronidazole, the possibility that the nitroreductase(s) is essential (e.g. PFOR), or the likelihood that mutations causing resistance also impose biological fitness costs in C. difficile [3]. Consequently, we were unable to obtain stable mutants showing resistance to metronidazole and the related nitroimidazole ornidazole. These findings are consistent with prior studies which showed that metronidazole-heteroresistant subpopulations obtained through serial passage revert back to wild-type susceptibilities upon freezer storage and thaw [9]. Similarly, we show that spontaneous mutants are also not readily selected with nitazoxanide, even though temporal decreases in the susceptibility of cultures occur during serial passage. This may be due to the distinct mode of action of nitazoxanide, involving inhibition of PFOR by abstracting a proton from the substrate cofactor thiamine pyrophosphate (TPP) [10,12]; thus mutations in PFOR may not be an avenue for nitazoxanide resistance [10]. In contrast, stable resistant mutants were enriched during serial exposure to nitrofurans. Since these nitrofuran-resistant mutants were not cross-resistant to metronidazole or nitazoxanide, this suggests that the use of nitrofurans for other disease indications, including gastrointestinal infections, may not act as a conduit for the emergence of resistance to metronidazole and nitazoxanide in C. difficile. Furthermore, since nitazoxanide retains activity against clinical C. difficile exhibiting resistance to metronidazole [24], it is possible that all three nitroheterocyclic classes could play roles in treating CDI.
The nitroimidazole or nitrofuranyl anion formed upon reduction may either undergo decomposition to produce NO or become further reduced to nitroso and hydroxylamine intermediates [4]. In the present studies, the levels of free NO were either absent or too low to be detected, which leads us to speculate that in C. difficile the nitroheterocyclic anions of metronidazole and nitrofurantoin do not undergo decomposition. However, we cannot negate that even if NO is produced it may react with target proteins, making it unavailable for detection in our assays involving the Griess reagent and an E. coli reporter strain that are only capable of detecting micromolar levels of NO. Hence, more sensitive analytical methods will be required to confirm the current findings and to elucidate the fate of activated nitroheterocyclic drugs in C. difficile.
Interestingly, dramatic induction of the hybrid cluster protein hcp occurred upon exposure to metronidazole, nitrofurantoin and GSNO. Induction of hcp by GSNO is not surprising since it is known to respond to nitrosative stress, where it was originally proposed to act as a hydroxylamine reductase, protecting bacteria from reactive species generated during nitrite or NO reduction [21]. However, we are unaware of reports demonstrating that hcp responds to nitroimidazole and nitrofuran drugs. We believe this suggests that there are some intersections in the types of cellular damage caused or proteins targeted by nitroimidazoles, nitrofurans and nitrosating agents [25]. We speculate this overlap primarily occurs at the proteome level, resulting from their respective reactive species either forming adducts or causing damage to similar cellular proteins [25]. Therefore, Hcp appears to have a broader role in responding to cellular damage caused by reactive species and not just in the detoxification of hydroxylamine radicals. This might support the recent hypothesis by Cole [25] that Hcp may be involved in repairing damaged metalloproteins. Furthermore, low induction of hcp by nitazoxanide supports its distinct mode of action as an inhibitor of PFOR activity, which does not involve reduction to reactive species that occur with other nitroheterocyclic drugs. Hcp may represent a marker for differentiating the actions of future nitroheterocyclic agents, e.g. nitroimidazole versus nitrothiazolyl analogues. This study suggests that activated metronidazole also causes DNA strand breaks in C. difficile as in other bacteria, resulting in the triggering of the SOS response, but this is not the primary mechanism of action for nitazoxanide and nitrofurantoin, which is also evident in H. pylori [13].
Further studies will be needed in C. difficile to determine the cellular mechanism(s) of activation of metronidazole and indeed proteins modified by both metronidazole and GSNO, as this could identify new cellular targets to discover treatments for CDI. This study also provided further insight to the actions of nitroheterocyclic drugs against C. difficile. It also indicates that C. difficile is highly sensitive to GSNO and provides a framework for new hypotheses to probe the action of these agents to control CDI.
Supplementary Material
Highlights.
Unstable resistance arises during serial passage of C. difficile in metronidazole or nitazoxanide
Contrastingly, stable resistance arises to nitrofurans with no cross resistance
Nitrosating agent GSNO rapidly kills C. difficile and is an alternate mechanism of controlling CDI
An overlap in the mechanisms for cellular damage is suggested for nitrofurantoin, metronidazole and GSNO
Acknowledgment
The authors are thankful to Dr Stephen Spiro (The University of Texas at Dallas, Dallas, TX) for providing E. coli JOEY426.
Funding: Funding for this research was provided by the National Center for Complementary and Alternative Medicine, National Institutes of Health [grant 5R01AT006732].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: None declared.
Ethical approval: Not required.
References
- 1.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 2.Kyne L, Hamel MB, Polavaram R, Kelly CP. Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile. Clin Infect Dis. 2002;34:346–353. doi: 10.1086/338260. [DOI] [PubMed] [Google Scholar]
- 3.Lynch T, Chong P, Zhang J, Hizon R, Du T, Graham MR, et al. Characterization of a stable, metronidazole-resistant Clostridium difficile clinical isolate. PLoS One. 2013;8:e53757. doi: 10.1371/journal.pone.0053757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson RJ, Groundwater PW, Todd A, Worsley AJ. Nitroimidazole antibacterial agents. In: Anderson RJ, Groundwater PW, Todd A, Worsley AJ, editors. Antibacterial agents: chemistry, mode of action, mechanisms of resistance and clinical applications. Chichester, UK: John Wiley & Sons, Ltd; 2012. pp. 85–102. [Google Scholar]
- 5.Lofmark S, Edlund C, Nord CE. Metronidazole is still the drug of choice for treatment of anaerobic infections. Clin Infect Dis. 2010;50(Suppl 1):S16–S23. doi: 10.1086/647939. [DOI] [PubMed] [Google Scholar]
- 6.Savidge TC, Urvil P, Oezguen N, Ali K, Choudhury A, Acharya V, et al. Host S-nitrosylation inhibits clostridial small molecule-activated glucosylating toxins. Nat Med. 2011;17:1136–1141. doi: 10.1038/nm.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emerson JE, Stabler RA, Wren BW, Fairweather NF. Microarray analysis of the transcriptional responses of Clostridium difficile to environmental and antibiotic stress. J Med Microbiol. 2008;57:757–764. doi: 10.1099/jmm.0.47657-0. [DOI] [PubMed] [Google Scholar]
- 8.Baines SD, O'Connor R, Freeman J, Fawley WN, Harmanus C, Mastrantonio P, et al. Emergence of reduced susceptibility to metronidazole in Clostridium difficile. J Antimicrob Chemother. 2008;62:1046–1052. doi: 10.1093/jac/dkn313. [DOI] [PubMed] [Google Scholar]
- 9.Peláez T, Cercenado E, Alcalá L, Marín M, Martín-López A, Martínez-Alarcón J, et al. Metronidazole resistance in Clostridium difficile is heterogeneous. J Clin Microbiol. 2008;46:3028–3032. doi: 10.1128/JCM.00524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman PS, Sisson G, Croxen MA, Welch K, Harman WD, Cremades N, et al. Antiparasitic drug nitazoxanide inhibits the pyruvate oxidoreductases of Helicobacter pylori, selected anaerobic bacteria and parasites, and Campylobacter jejuni. Antimicrob Agents Chemother. 2007;51:868–876. doi: 10.1128/AAC.01159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Upcroft P, Upcroft JA. Drug targets and mechanisms of resistance in the anaerobic protozoa. Clin Microbiol Rev. 2001;14:150–164. doi: 10.1128/CMR.14.1.150-164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warren CA, van Opstal E, Ballard TE, Kennedy A, Wang X, Riggins M, et al. Amixicile, a novel inhibitor of pyruvate: ferredoxin oxidoreductase, shows efficacy against Clostridium difficile in a mouse infection model. Antimicrob Agents Chemother. 2012;56:4103–4111. doi: 10.1128/AAC.00360-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sisson G, Goodwin A, Raudonikiene A, Hughes NJ, Mukhopadhyay AK, Berg DE, et al. Enzymes associated with reductive activation and action of nitazoxanide, nitrofurans, and metronidazole in Helicobacter pylori. Antimicrob Agents Chemother. 2002;46:2116–2123. doi: 10.1128/AAC.46.7.2116-2123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu X, Cherian PT, Lee RE, Hurdle JG. The membrane as a target for controlling hypervirulent Clostridium difficile infections. J Antimicrob Chemother. 2013;68:806–815. doi: 10.1093/jac/dks493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leeds JA, Sachdeva M, Mullin S, Barnes SW, Ruzin A. In vitro selection, via serial passage, of Clostridium difficile mutants with reduced susceptibility to fidaxomicin or vancomycin. J Antimicrob Chemother. 2014;69:41–44. doi: 10.1093/jac/dkt302. [DOI] [PubMed] [Google Scholar]
- 16.Bodenmiller DM, Spiro S. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J Bacteriol. 2006;188:874–881. doi: 10.1128/JB.188.3.874-881.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karasawa T, Ikoma S, Yamakawa K, Nakamura S. A defined growth medium for Clostridium difficile. Microbiology. 1995;141:371–375. doi: 10.1099/13500872-141-2-371. [DOI] [PubMed] [Google Scholar]
- 18.Muthaiyan A, Silverman JA, Jayaswal RK, Wilkinson BJ. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob Agents Chemother. 2008;52:980–990. doi: 10.1128/AAC.01121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlacher K, Cox MM, Woodgate R, Goodman MF. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature. 2006;442:883–887. doi: 10.1038/nature05042. [DOI] [PubMed] [Google Scholar]
- 21.Rodionov DA, Dubchak IL, Arkin AP, Alm EJ, Gelfand MS. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput Biol. 2005;1:e55. doi: 10.1371/journal.pcbi.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller J, Schildknecht P, Muller N. Metabolism of nitro drugs metronidazole and nitazoxanide in Giardia lamblia: characterization of a novel nitroreductase (GlNR2) J Antimicrob Chemother. 2013;68:1781–1789. doi: 10.1093/jac/dkt106. [DOI] [PubMed] [Google Scholar]
- 23.Cunningham R, Mustoe E, Spiller L, Lewis S, Benjamin N. Acidified nitrite: a host defence against colonization with C. difficile spores? J Hosp Infect. 2014;86:155–157. doi: 10.1016/j.jhin.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Freeman J, Baines SD, Todhunter SL, Huscroft GS, Wilcox MH. Nitazoxanide is active against Clostridium difficile strains with reduced susceptibility to metronidazole. J Antimicrob Chemother. 2011;66:1407–1408. doi: 10.1093/jac/dkr077. [DOI] [PubMed] [Google Scholar]
- 25.Cole JA. Legless pathogens: how bacterial physiology provides the key to understanding pathogenicity. Microbiology. 2012;158:1402–1413. doi: 10.1099/mic.0.059048-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.