Abstract
The recently sequenced Daphnia pulex genome revealed the NR1L nuclear receptor group consisting of three novel receptors. Phylogenetic studies show that this group is related to the NR1I group (CAR/PXR/VDR) and the NR1J group (HR96), and were subsequently named HR97a/b/g. Each of the HR97 paralogs from Daphnia magna, a commonly used crustacean in toxicity testing, was cloned, sequenced, and partially characterized. Phylogenetic analysis indicates that the HR97 receptors are present in primitive arthropods such as the chelicerates but lost in insects. qPCR and immunohistochemistry demonstrate that each of the receptors is expressed near or at reproductive maturity, and that HR97g, the most ancient of the HR97 receptors, is primarily expressed in the gastrointestinal tract, mandibular region, and ovaries, consistent with a role in reproduction. Transactivation assays using an HR97a/b/g-GAL4 chimera indicate that unlike Daphnia HR96 that is promiscuous with respect to ligand recognition, the HR97 receptors do not respond to many of the ligands that activate CAR/PXR/HR96 nuclear receptors. Only three putative ligands of HR97 receptors were identified in this study: pyriproxyfen, methyl farnesoate, and arachidonic acid. Only arachidonic acid, which acts as an inverse agonist, alters HR97g activity at concentrations that would be considered within physiologically relevant ranges. Overall, this study demonstrates that, although closely related to the promiscuous receptors in the NR1I and NR1J groups, the HR97 receptors are mostly likely not multi-xenobiotic sensors, but rather may perform physiological functions, potentially in reproduction, unique to crustaceans and other non-insect arthropod groups.
Keywords: unsaturated fatty acids, energy, reproduction, xenobiotic
INTRODUCTION
As a critical component in the multi-level regulation of gene expression, nuclear receptors (NR) have been considered the Rosetta Stone of physiology (Evans, 2005), as they recognize hormonal, environmental, and dietary signals, and elicit individual physiological responses by altering transcription (Hernandez et al., 2009). NRs constitute a large superfamily of transcriptional regulators involved in a vast array of diverse physiological functions such as reproduction, control of embryonic development, regulation of cell differentiation, and homeostasis (Germain et al., 2006; Thomson et al., 2009; Zollner et al., 2006).
Most NRs contain five domains, of which the DNA binding domain (C domain) is highly conserved, and the ligand binding domain (LBD; E domain) shows several conserved features (Buzón et al., 2012). Ligand binding to the E domain can cause conformational changes that stimulate the dissociation of co-repressors, and generate the association of co-activators and DNA response elements with NRs, thereby increasing transcriptional activity (Mangelsdorf et al., 1995). NRs can function as monomers, homodimers, or heterodimers; many NRs interact with RXR, a common heterodimer partner (Amoutzias et al., 2007).
Since the cloning of the glucocorticoid receptor in 1985 (Hollenberg et al., 1985), new members have been constantly added to the NR superfamily with the various sequencing and annotation projects, providing valuable information on transcriptional regulation and evolution of the animal kingdom (Bridgham et al., 2010; Evans, 2005). Today, there are seven subfamilies (NR0–NR6) of NRs known (Thomson et al., 2009). The recent use of multiple biochemical and phylogenetic analyses has shown that modern NRs evolved through subtle tinkering of an ancestral ligand-dependent receptor related to HNF4 (Bridgham et al., 2010). Studies on NR phylogenetics are crucial to understanding metazoan evolution, as NRs have gone through numerous duplication events during evolution.
Daphnia magna, commonly known as the water flea, is a crustacean in the Branchiopod class. It is a crucial component of many aquatic food webs and is extensively used in aquatic toxicity testing. Although the ecology of Daphnia magna is well understood, genomic studies on this organism have only recently begun (http://server7.wfleabase.org/genome/Daphnia_magna_prerelease/). The related cladoceran, Daphnia pulex, is the first aquatic arthropod and crustacean genome to be fully sequenced (Colbourne et al., 2011), providing biologists with genomic information on the unique mechanisms required for the survival of aquatic arthropods, and greater tools for studying arthropod evolution. Twenty-six NR genes have been identified in D. pulex (Thomson et al., 2009) including an unpublished NR2E6 member. In comparison, Homo sapiens have 48 NRs, Mus musculus 46, Drosophila melanogaster 21, and Caenorhabditis elegans have over 270 NRs (Adams et al., 2000; Maglich et al., 2001; Robinson-Rechavi et al., 2003; Robinson-Rechavi et al., 2005; Robinson-Rechavi et al., 2001). The interspecies diversity is likely to reveal the time and roles of gene duplications in evolution and facilitate phylogenetic reconstruction (Garcia et al., 2003).
A new NR group (HR97/NR1L) consisting of three members has been discovered in Daphnia (Thomson et al., 2009). All three members are orphan receptors as their ligands are not known. The receptors were assigned the names HR97a, HR97b, and HR97g because of their similarity to the NR1J group of NRs that includes HR96 found in Daphnia pulex and Drosophila melanogaster (Fisk and Thummel, 1995; Karimullina et al., 2012). Phylogenetic analyses suggest that HR97g is the evolutionary precursor to the other two HR97 receptors that are found in tandem repeat (Thomson et al., 2009).
In this study, we describe the evolution and partial characterization of these three HR97s in the the cladoceran D. magna. All three HR97s were cloned by 5′- and 3′-RACE, sequenced, and compared genomically and phylogenetically with similar receptors. GAL4-HR97_DEF chimeric plasmids were constructed, and transactivation assays were conducted with several chemicals, including multiple xenobiotics, as we hypothesized that HR97 would be promiscuous similar to its relative HR96 (Karimullina et al., 2012), and other related NRs such as the vertebrate orthologs CAR and PXR (Kliewer et al., 1998; Krasowski et al., 2010; Kretschmer and Baldwin, 2005; Staudinger et al., 2001). In addition, the age- and organ-dependent expression of HR97 receptors was investigated. This is the first study that investigates the evolution, function and expression of the HR97/NR1L group of NRs.
MATERIALS and METHODS
D. magna
Our colony of D. magna has been cultured at Clemson University for over 15 years and was initially provided by Dr. Steve Klaine (Clemson University)(Baldwin et al., 2001). Daphnids were maintained at 15 individuals/L in 22°C moderately hard water under a 16:8 light:dark photoperiod in an environmental chamber. Each adult daphnid received approximately 6 × 106 cells of Pseudokirchneriella subcapitata (formerly Selenastrum capricornutum) daily supplemented with a suspension of Tetrafinn fish food (Tetra Holding Inc, VA) as described previously (Baldwin et al., 1995; Karimullina et al., 2012).
RNA extraction
RNA was extracted from fresh female Daphnia magna of a variety of ages (2–14 days old) with PureZol (BioRad, Hercules, CA) according to the manufacturer’s directions followed by digestion of residual genomic DNA with DNAse (Promega, Madison, WI). cDNA was synthesized from 2μg of RNA with Moloney Murine Leukemia Virus-Reverse Transcriptase (MMLV-RT), a dNTP mixture, and random hexamers (Promega).
Cloning Daphnia magna HR97a (Clem-magnaHR97a)
Each of the HR97 nuclear receptors was cloned from the D. magna housed at Clemson. This Clemson strain and its associated nuclear receptors are called the Clem-magnaHR97s to distinguish that they are D. magna HR97s from the Clemson strain. In contrast, the genome was cloned from strain Xlnb3 from Finland in turn is termed the Fin-magnaHR97s. To clone Clem-magnaHR97a, a small segment of the magnaHR97a gene was isolated using the following primers designed based on the D. pulex HR97a genome sequence (DappuHR97a) (Thomson et al., 2009): F-97a-GL: 5′-CCAACTCGAGTGGAGGAGAG-3′; R-97a-GL: 5′-GCCACCTTTGAGCAGGATAC-3′. The PCR product was ligated into pCR 2.1 by TA cloning (Invitrogen, Carlsbad, CA). DNA sequencing was performed by MacrogenUSA (Rockville, MD). The Invitrogen 3′ RACE kit (Invitrogen, Carlsbad, CA) was used to amplify the 3′ half of the gene. The forward primer used in the first 3′ RACE PCR is F-97a-GL. F-GSP-97a (5′-TAC GCT CGC TTG ATG GCC GAC -3′) was used in the nested PCR. After that, PCR was performed using primer pair 97a-F2/R-97a-GL. 97a-F2 (5′-CAAGAGTTACCATTTCGGCG -3′)was designed based on D. pulex’s sequence. The cloning of HR97a was completed with the Invitrogen 5′ RACE kit (Invitrogen, Carlsbad, CA). The steps of 5′ RACE include first strand cDNA synthesis, TdT tailing of cDNA and PCR of the dT-tailed cDNA. Two reverse primers were designed based on the known magnaHR97a sequence determined from the work described above. Primer sequence information: R-GSP6-97a: 5′-CACACTGGCCACGGTGACAGCAC –3′; R-GSP7-97a: 5′ CGGAACGACGAAAGAAAGCCTTGC –3′. The former was used in the first 5′ RACE PCR and the latter was used in the nested PCR. DNA sequencing was performed by MacrogenUSA (Rockville, MD). The Clem-magnaHR97a sequence has been submitted to GenBank (JQ678702) as HR97a (NR1L1).
Cloning D. magna HR97b (Clem-magnaHR97b)
A small segment of the Clem-magnaHR97b (magnaHR97b) gene was isolated using primers designed based on the D. pulex HR97b (DappuHR97b) sequence (Thomson et al., 2009): F-97b-GL (5′-GAG CTG CCT TCT GAA AGG TG-3′) and R-97b-GL (5′-GCG TGA ACA GAA CGA TCA AG-3′) primers. The 3′-end of the magnaHR97b gene was cloned via 3′-RACE (Invitrogen). The forward primer used in the first 3′-RACE PCR is F-97b-GL. F-GSP-97b (5′-GGG AGT CGA CGA ACC GAC CAT CAT -3′) was used in the nested PCR. Two rounds of 5′-RACE (Invitrogen) was performed to isolate and determine the 5′-sequence of magnaHR97b with gene specific and nested primers (5′ –GGACATGTTGGCGTTTGGCCAGCG –3′; nested:5′ – ACGGGTCGTAGACTAGCGCTCCTC – 3′) (5′ – GCTCGAAAAGTCGGCCATCAGCCG – 3′; nested: 5′ – GGCGAAACGACGAATCAGAGTCCC -3′). DNA sequencing was performed by MacrogenUSA (Rockville, MD). The Clem-magnaHR97b sequence has been submitted to GenBank (JQ678703) as HR97b (NR1L2).
Cloning D. magna HR97g (Clem-magnaHR97g)
Clem-magnaHR97g (magnaHR97g) was cloned using primer sets (F = forward; R = reverse) F5/R4, F1/R1, and F97g-GL/R-97g-GL, which were designed based on the D. pulex HR97g sequence (Thomson et al., 2009). F5: 5′-GAAGATGTCCAGCGTCTTCT -3′; R4: 5′-TCAATTTGCGCCAGTTC-3′; F1:5′-ATGGATGACAGCAACAGTTCT-3′; R1:5′-CGAAGCCATCCTTTCTCCAT-3′; F97g-GL: 5′-ACATGGCCAAACATGTGTCA-3′; R97g-GL: 5′-TGTCTTCAAAGCTTGGTTCG-3′. PCR products were ligated into pCR 2.1 by TA cloning (Invitrogen). Fm2/Rm2 primer pair (Fm2: 5′-CCAATTGGTGCAACACTCCTAG-3′; Rm2: 5′-GCTCGATCGGGCGTAAACAT-3′) was designed after sequencing the PCR products from the previous reactions and used to determine the unknown areas within the middle of the gene. The 3′ end of magnaHR97g was cloned following 3′-RACE (Invitrogen) with the following primers: F-GSP1: 5′-CCTGGAAGATGTCCAGCGCCTTCT-3′; nested primer F-GSP2: 5′-GCCCGATCGAGCAGATCTTGTTGCTCGT-3′. DNA sequencing was performed by MacrogenUSA (Rockville, MD). The Clem-magnaHR97g sequence has been submitted to GenBank (JF792806) as HR97g (NR1L3).
Gene annotations and genomic structure
Annotation of the D. pulex’s NRs was performed previously (Thomson et al., 2009). Identification of D. magna orthologs was performed by degenerative PCR and RACE as described above, and later using the Basic Local Alignment Search Tool (BLAST) available on the D. magna genome project’s webpage (http://server7.wfleabase.org/genome/Daphnia_magna_prerelease/) in order to identify the gene structure (position, length, exon, introns, and phase of each intron) of the three HR97 genes in clone Xlnb3 (Fin-magnaHR97a/b/g). Xlnb3 is the strain of D. magna isolated from a pond in Finland from which the D. magna genome is being sequenced. We were able to find the corresponding HR97a, HR97b, and HR97g sequences and use this information to construct the genomic model of each of the HR97 receptors. Percent protein sequence identities were also compared between D. magna and D. pulex HR97 receptors, and between other distinct NR groups using clustalw (http://www.ebi.ac.uk/Tools/clustalw2/).
Phylogenetics
Phylogenetic analyses of the HR97 receptors was performed using methods described previously (Hannas et al., 2010; Thomson et al., 2009) with some modification. HR97a/b/g from Fin-D. magna was compared to D. pulex and to similar NRs of other species available in GenBank such as Drosophila melanogaster, Xenopus laevis, Danio rerio, Homo sapiens, Ciona intestinalis, Ixodes scapularis, and Caenorhabditis elegans. NCBI accession numbers for the receptors used in the phylogenetic analysis are available in Additional File 1. The DNA binding domain (DBD) and the ligand binding domain (LBD) of each receptor were identified using the conserved domain database (CDD) (Marchler-Bauer et al., 2007). Zf-C4 (pfam00105) was used to identify the DBD, and Hormone recep (pfam00104) was used to identify the LBD. ClustalX default parameters were used to align the domains (Thompson et al., 1997).
Trees were constructed using Bayesian Inference with MrBayes software version 3.1.2 (Ronquist and Huelsenbeck, 2003) on the Computational Biology Service Unit of Cornell University (http://cbsuapps.tc.cornell.edu/mrbayes.aspx). Phylogenetic trees were constructed using the “mixed-model” approach in which the Markov chain Monte Carlo sampler explores nine different fixed-rate amino acid substitution models implemented in MrBayes. We used 4 chains with runs of 5 million generations, chains sampled every 100 generations, a burnin of 10,000 trees with the WAG model (Whelan and Goldman, 2001). The C. elegans NHR-1/NR1K1 receptor was used as the outgroup.
Maximum parsimony and distance parameters were used to provide additional support for the phylogenetic relationships observed. Distance parameters were measured using PAUP 4.0b10 with default characteristics (mean character difference and among site rate variation), and full heuristic searches. Branch support was measured by bootstrap analysis with 1000 replicates. Parsimony was constructed using PAUP version 4.0b10 with heuristic searches, tree-bisection-reconnection, topological constraints not enforced, and multiple tree option in effect with an initial maximum tree setting at 100,000. Branch support was measured by bootstrapping with 10,000 replicates. Trees were visualized with FigTree (http://tree.bio.ed.ac.uk/software).
Expression plasmids for Transactivation Assays
Chimeric genes containing the D, E (LBD), and F domains of the magnaHR97a (GAL4-97aDEF), magnaHR97b (GAL4-97bDEF), or magnaHR97g (GAL4-97gDEF) receptors were attached to the DNA binding domain (DBD) of Gal4 for the purpose of performing transactivation assays. The chimeras were constructed using the Clontech In-Fusion Dry-Down PCR Cloning Kit according to the manufacturer’s directions (Clontech Laboratories, Mountain View, CA). The D, E and F domains of each receptor were amplified using primer pairs F-clontech-97a-BamHI (5′-GAA TTC CCG GGG ATC GAT GAA CGC AAA GCC CTG ATG AAA GCA CGT -3′), R-clontech-97a-XbaI (5′-CTG CGG CCG CTC TAGA TCA TTG GAG CTT GTT GGT ATC TTT GGC TGG TCG -3′) for HR97a, primer pairs F-clontech-97b-BamHI (5′- GAA TTC CCG GGG ATC GAT GAA CGC AAA GCT TTA ATG AAA GCT CGA GC -3′), and R-clontech-97b-XbaI (5′-CTG CGG CCG CTC TAGA TCA GTG GGC GTC GTA AAG CTC TGA ATA TTC TTC -3′) for HR97b, and primer pairs F-clontech-97g-BamHI (5′-GAA TTC CCG GGG ATC GAA GAA AAT GTG AAA ATG AGA GAG GCC AAG-3′) and R-clontech-97g-XbaI (5′-CTG CGG CCG CTC TAGATTA AAC TGG GTT ATC TGT TTC CAT TTG TTG ACT-3′) for HR97g. The PCR products were inserted into the pBIND vector (Promega CheckMate Mammalian Two-hybrid system; Promega, Madison, WI) in frame after the GAL4 DNA binding domain.
Transactivation Assays
HEPG2 human hepatoma cells (ATCC, Rockville MD) were cultured in phenol red free Dulbecco’s Modified Eagle’s Medium (DMEM) (Mediatech, Manassas, VA) supplemented with10% fetal bovine serum (HyClone, Logan, UT), 1% L-glutamine (Invitrogen, Carlsbad, CA) and 1% Penicillin/Streptomycin (Invitrogen, Carlsbad, CA) at 37°C under 5% CO2.
HepG2 cells were plated in 12-well plates at 200,000 cells per well. To determine chemical activation of HR97 receptors, cells were transfected with 0.02 μg of the GAL4-97a/b/gDEF chimeric plasmids, and 0.1 μg the reporter plasmid pG5luc that contains five GAL4 binding sites upstream of the firefly luciferase gene (Promega CheckMate Mammalian Two-hybrid system) using Effectene Transfection Reagent according to the manufacturer’s recommendations(Qiagen, Valencia, CA). Cells were either left untreated or treated with the chemicals of interest dissolved in 0.1% DMSO. Untreated wells also received 0.1% DMSO. Luciferase activity was measured 24 hours after chemical treatment with the Steady-Glo Luciferase Assay System (Promega). Several hormones, fatty acids, and xenobiotics were used to test whether they activated the different HR97 receptors. These included 4-nonylphenol (CAS 84852-15-3), bisphenol A (80-05-7), chlorpyrifos (2921-88-2), cortisol (50-23-7), dihyroandrosterone (1852-53-5), ethyinyl estradiol (57-63-6), β-estradiol (50-28-2), endosulfan (115-29-7), parathion (56-38-2), methyl farnesoate (10485-70-8), pyriproxyfen (95737-68-1), palmitic acid (57-10-3), stearic acid (57-11-4), oleic acid (112-80-1), linoleic acid (60-33-3), and arachidonic acid (506-32-1). None of the chemicals are toxic at the concentrations tested as determined by MTS assays with the exception of 30 μM pyriproxyfen (Promega).
Typically, data are presented as the mean of triplicate assays ± standard error. Statistical significance was determined by ANOVA followed by Dunnett’s test as the post hoc test when comparing samples to control samples (following chemical treatment or no treatment), or Tukey’s multiple comparison test as the post-hoc test when comparing multiple treatments (following the mammalian two-hybrid assays). The GraphPad Prizm 4.0 statistical and graphing package (La Jolla, CA) was used for statistical analysis and graphing. For the concentration-response studies, activities were normalized as a percent of the maximal activation and concentrations were log transformed to determine EC50 values from the sigmoidal concentration-response curves generated by GrapPad Prizm 4.0 as described previously (Baldwin and Roling, 2009; Karimullina et al., 2012).
Quantitative Real-time PCR (qPCR)
Daphnia magna age 2, 4, 7, and 14 days were collected from four 1 L culture beakers per age group. RNA was extracted and cDNA made as described above. qPCR was performed as described by us (Mota et al., 2010; Roling et al., 2006) and others (Muller et al., 2002) using βactin (Heckmann et al., 2006; Kim et al., 2009) (Forward 5′-CCA CAC TGT CCC CAT TTA TGA AG-3′-3′; Reverse 5′-CGC GAC CAG CCA AAT CC-3′, annealing temperature 52.2°C) as the reference gene. PCR efficiency was determined from standard curves developed from 1:1, 1:5, 1:25, 1:125, 1:625, and 1:3125 dilutions of a composite cDNA sample. Samples were diluted 1:5 and amplifications were performed in triplicate with a 96-well iQ5 real-time PCR (Bio-Rad) with SYBRGreen PCR mastermix (SABiosciences, Frederick, MD) to quantify gene expression. qPCR results were normalized to the expression of β-actin (Heckmann et al., 2006). Samples were analyzed by taking the efficiency curve of the qPCR reaction to the power of the threshold cycle divided by the reference gene as described previously (Muller et al., 2002).
The sequence and annealing temperatures of the HR97 qPCR primers are: magnaHR97a: Forward 5′-ACA GTG GTG CTA CAG CCG CTA ATA-3′; Reverse 5′-AGA GTC GCA AGA TAA TCC GCC GAA-3′, annealing temperature 56°C. magnaHR97b: Forward 5′-TAA-CCG-ACG-CCG-CTC-AAT-CCT-ATT-3′; Reverse 5′-ATG-TTG-AAC-CGG-CGG-GAA-ATG-ATG-3′, annealing temperature 56°C. magnaHR97g: Forward 5′-GCG TAC ATG GCC AAA CAT GTG TCA-3′; Reverse 5′-TGC CTA GCT TGG CCT CTC TCA TTT-3′ at annealing temperature of 57°C. The sequence and annealing temperatures of the vitellogenin 2 (Vtg2) qPCR primers are: Vtg2: Forward 5′-GCA CTC TTT CTC GTT ATT GCT G; Reverse 5′-GAT CTT GAC TGG GCT ATT GAT T-3′ at annealing temperature of 60°C (Tokishita et al., 2006). Data are presented as mean ± standard error (n = 4–5). Statistical significance was determined by ANOVA followed by Dunnett’s test as the post hoc test using the GraphPad Prizm 4.0 statistical and graphing package (La Jolla, CA).
Immunohistochemistry of HR97g
Organ specific expression of HR97g was evaluated by immunohistochemistry. Daphnia magna were preserved in 10% formaldehyde (Fisher Scientific), processed in a LEICA ASP300S automatic sample processer (Leica Microsystems, Bannockburn, IL), and embedded in paraffin blocks using the LEICA EG1150C (Leica). Five micron sections were cut with a LEICA RM2165 rotary microtome, transferred onto a slide coated with 3-aminopropyltriethoxysilane and fixed by drying on a hot platform. Samples were deparaffinized and rehydrated using a standard step wise protocol with a xylene, alcohol, PBS gradient prior to antigen retrieval. Immunohistochemistry was performed as described as previously performed at the Clemson University Animal Histology facility (Johnstone et al., 2008).
An antigen affinity purified Rabbit anti-Daphnia HR97g antibody was used to determine organ specific expression of HR97g by immunohistochemistry. The antibody was prepared against a KLH-conjugated N-terminal (A/B domain) sequence (GSSNEENAVPENKSC) of HR97g (Genscript, Piscataway, NJ). Specificity was demonstrated by ELISA and confirmed by peptide competition IHC assays with 10 and 100-fold peptide dilutions (Additional File 2). Slices of whole daphnids were incubated with the HR97g primary antibody (40 mg/ml) in blocking solution at room temperature in a humidified chamber for 60 minutes followed by three washings with PBS for 5 minutes each. Samples were incubated with a 1:200 dilution of goat anti-rabbit secondary antibody (Invitrogen) tagged with Alexa fluor green fluorescent dye in blocking solution for 60 minutes, washed three times in PBS for 5 minutes each, and mounted with a cover slip prior to imaging. Images were obtained with an Optishot 2 under a DAPI green fluorescent filter (Nikon, Melville, NY), and captured with a Spot camera model 1.1.0 (Diagnostic Instruments, Sterling Heights, MI).
RESULTS
Phylogenetics
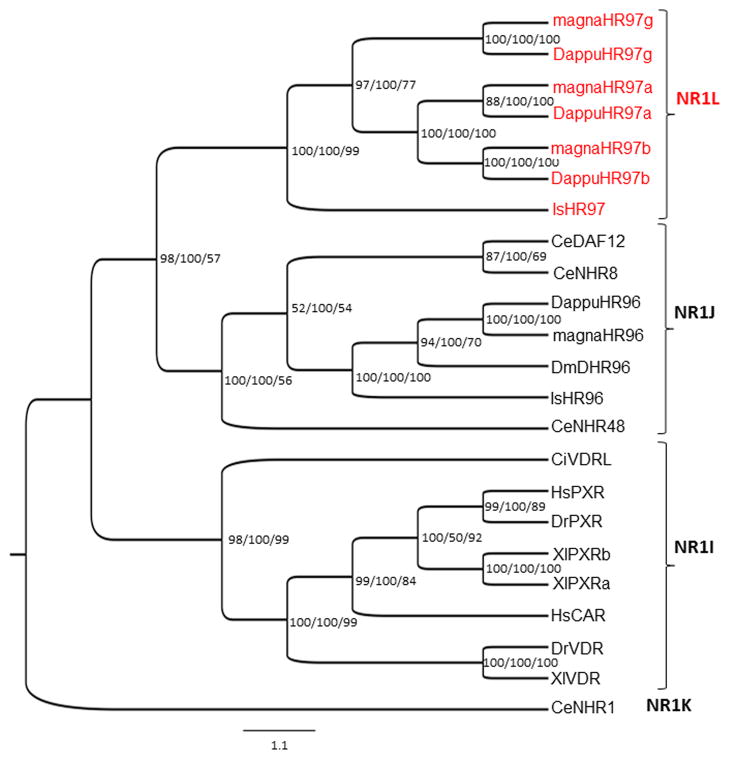
The combined C (DBD) and E (LBD) domains of the three NR1L group members (Clem-magnaHR97a/b/g; Clemson University strain of D. magna) were phylogenetically compared to the related NR1J group that contains HR96 (Thomson et al., 2009) and its relatives, the NR1I group that contains CAR, PXR, and VDR (Karimullina et al., 2012; Lin et al., 2011). The phylogenetic tree separates the receptors into three clades: a NR1L clade that contains the HR97s, a NR1J clade that contains the HR96s, CeNHR48, CeNHR8 and CeDAF12, and a NR1I clade that contains CAR, PXR and VDR (Fig. 1). There are no disagreements between the three programs; Bayesian Inference, Maximum Parsimony and Neighbor-Joining. The HR97s are more closely related to the HR96s than to their vertebrate relatives, CAR, PXR and VDR. Differentiation among the three HR97 paralogs occurred before the speciation of D. magna and D. pulex, and is in agreement with previous studies that HR97g is the precursor of HR97a and HR97b (Thomson et al., 2009). Overall, the phylogenetic tree further confirms that the HR97s constitute a distinct NR group related to HR96.
Fig. 1. Phylogenetic analysis of the NR1L nuclear receptor group.
Bayesian Inference, Maximum Parsimony, and Neighbor-Joining were used to determine the relationship of the NR1L (HR97) group of NRs to the related NR1J and NR1I groups. NRs from several different species were analyzed following alignment of their conserved C (DBD) and E (LBD) domains using Bayesian Inference, Maximum parsimony and Neighbor-Joining methods. The Bayesian tree is shown with posterior probabilities from the Bayesian tree, and bootstrap support values (frequency of occurrence) from the Neighbor-Joining and Maximum Parsimony trees provided in order from left to right, respectively as confirmatory analysis of the Bayesian analysis at each node. CeNHR1 (NR1K1) was chosen as the outgroup. Abbreviation of species names are as follows: magna=Daphnia magna, Dappu=Daphnia pulex, Is=Ixodes scapularis, Ce=Caenorhabditis elegans, Dm=Drosophila melanogaster, Ci=Ciona intestinalis, Dr=Danio rerio, Hs=Homo sapiens, Xl=Xenopus laevis. Accession numbers of the analyzed NRs are provided in Additional File 1.
Genomic Structure and Nuclear Receptor Domain Comparisons
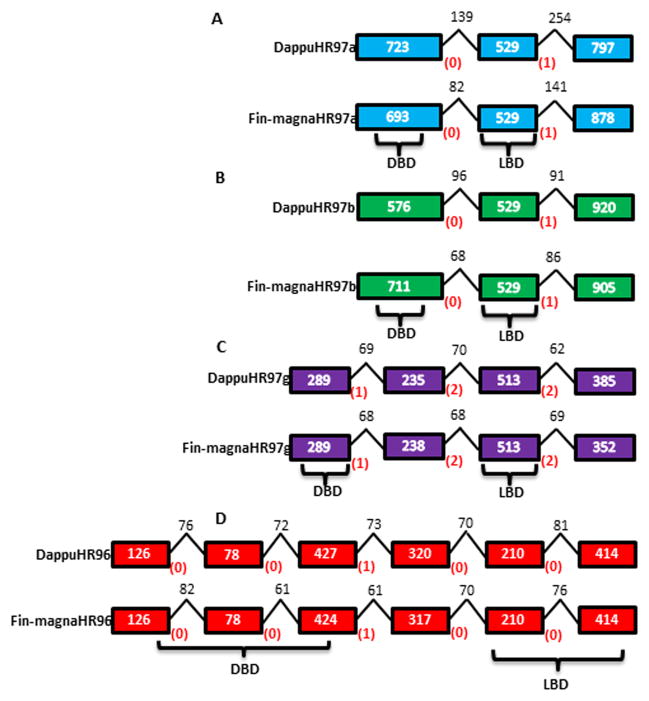
The position, length, and phase of each HR97a/b/g intron (0, 1, or 2 base pairs into a codon) from the genomic sequenced D. magna clone, Xlnb3 (Fin-magna)(https://wiki.cgb.indiana.edu/display/magna/Daphnia+magna+Genome) was determined following D. magna genome BLAST searches. Similar to DappuHR97a and HR97b, the Fin-magnaHR97a and HR97b genes are found in tandem repeat within the genome (Thomson et al., 2009). The intron phases of each HR97 receptor are identical between each Fin-magna receptor and its D. pulex homolog; however, HR97a/b differs from HR7g in both the number of introns and intron phases (Fig. 2).
Fig. 2. The gene structure of the NR1L nuclear receptors is identical between D. magna and D. pulex.<.
br>The blocks represent exons, the numbers inside the blocks indicate exon size, the numbers above the lines indicate intron size, and the numbers in parentheses represent the intron phases. The gene structures of HR97a (A), HR97b (B), HR97g (C), and HR96 (D).
The five NR domains of Clem-magnaHR97a/b/g were also compared to their corresponding domains in Fin-magnaHR97a/b/g and DappuHR97a/b/g (Table 1). ClustalX alignments of HR97a, HR97b, and HR97g between the different strains and species are available in Additional Files 3–5. The Clem-magna sequences agree well with the Fin-magna strain of D. magna with only minor variances. In general, the five domains, especially the C and E domains, are also very similar between D. magna and D. pulex with identity scores greater than 90. The A/B and F domains showed the lowest scores. These differences in scores are primarily due to differences in the size of the corresponding domains from D. magna to D. pulex. However, there are exceptions as the identity score between D. magna and D. pulex HR97a’s A/B domain is 68 due to differences in both size and amino acid composition, and the identity score of 70 between D. magna and D. pulex HR97g’s D domain is almost exclusively caused by a region of this domain with little similarity. Overall, the HR97 receptors do not show as high of sequence or genomic structure conservation as the related HR96 receptors (Fig. 2e).
Table 1.
The DNA-binding domains (C domain) and ligand binding domains (E domain) of the HR97 receptors are well conserved between species and strains. The HR97 NRs from the Clemson strain of D. magna (Clem-magna) were aligned to the corresponding HR97 NRs from the Finland D. magna (Fin-magna) strain and D. pulex (Dappu), and identity scores were determined.
| A. HR97a | A/B | C | D | E | F |
|---|---|---|---|---|---|
| Clem-magnaHR97a | 100 | 100 | 100 | 100 | 100 |
| Fin-magnaHR97a | 97 | 100 | 100 | 97 | 99 |
| DappuHR97a | 68 | 100 | 84 | 90 | 47 |
| B. HR97b | A/B | C | D | E | F |
|---|---|---|---|---|---|
| Clem-magnaHR97b | 100 | 100 | 100 | 100 | 100 |
| Fin-magnaHR97b | 98 | 98 | 98 | 100 | 99 |
| DappuHR97b | 68 | 98 | 92 | 97 | 60 |
| C. HR97g | A/B | C | D | E | F |
|---|---|---|---|---|---|
| Clem-magnaHR97g | 100 | 100 | 100 | 100 | 100 |
| Fin-magnaHR97g | 100 | 97 | 99 | 99 | 100 |
| DappuHR97g | 83 | 84 | 70 | 90 | 76 |
In addition, the five domains of Clem-magnaHR97a were aligned with the corresponding domains of several other NRs (Table 2). MagnaHR97a is highly similar to magnaHR97b in the C and E domains, but not as similar to other receptors, including HR97g. This confirms that HR97g is substantially different from the duplicated HR97a and HR97b forms, and suggests that HR97g has unique ligand and DNA binding characteristics. HR97g also contains the same DBD base-contact residues unique to NR1J members such as HR96 (Antebi et al., 2000; Thomson et al., 2009), while magnaHR97a, magnaHR97b, and IsHR97 contain slightly different base-contact residues (Additional File 6).
Table 2.
Percent identity of Clem-magnaHR97a to other NR1I/J/L subfamily members.
| Nuclear Receptor | A/B | C | D | E | F |
|---|---|---|---|---|---|
| magnaHR97a | 100 | 100 | 100 | 100 | 100 |
| magnaHR97b | 24 | 98 | 74 | 76 | 21 |
| magnaHR97g | 20 | 63 | 31 | 40 | 8 |
| Fin-magnaHR96 | 12 | 48 | 20 | 29 | 33 |
| DappuHR96 | 12 | 48 | 23 | 29 | 33 |
| DmDHR96 | 25 | 44 | 14 | 25 | 33 |
| HsCAR | 12 | 42 | 11 | 20 | 50 |
| magnaRXR | 15 | 42 | 8 | 20 | 0 |
Data shown as identity scores to magnaHR97a derived from ClustalW.
Hs = Homo sapiens, Dm = Drosophila melanogaster, Dappu = Daphnia pulex, magna = D. magna
Age and Tissue-Dependent Expression of HR97g
To determine age-related expression of HR97a/b/g in D. magna, groups of organisms were cultured and collected at 2, 4, 7, and 14-days of age, and expression analyzed by qPCR (Fig. 3). HR97a, HR97b, and HR97g mRNA levels generally increase with age with the highest expression at day 7 or 14. Both HR97a and HR97b, which are found in tandem repeat, show incredibly similar expression profiles with the greatest expression during adolescence (Fig. 3AB). HR97g shows a similar pattern of expression as HR97a/b, but HR97g mRNA expression shows a steady increase with increasing age into adulthood (Fig. 3C). Vitellogenin 2 (Vtg2), the ovarian produced egg lipoprotein (Hannas et al., 2011), was used to confirm ovarian maturation in these stages (Fig. 3D). Vtg2 expression demonstrates that the ovaries are mature or maturing by 7-days of age. Vtg2 also follows a similar pattern of preferential expression to HR97s receptors in the 7-day and older daphnids; however, instead of an increase of expression of approximately 5X, expression was increased 31–33X.
Fig. 3. Age-dependent expression of the HR97 receptors.<.
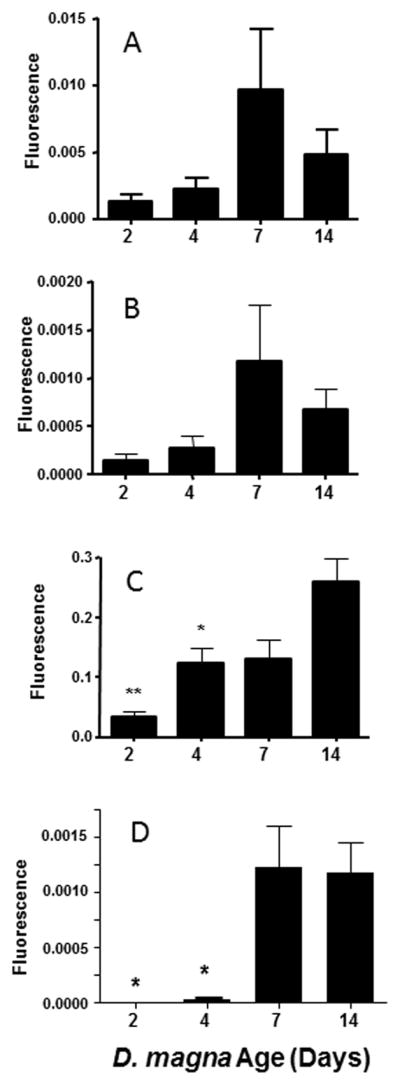
br>QPCR was performed with primers specific to HR97a (A), HR97b (B), or HR97g (C) with daphnids at age 2, 4, 7, and 14-days old. Data is expressed as mean + SEM. An asterisk (p< 0.05) or two asterisks (p < 0.001) indicate significance compared to 14-day old daphnids by ANOVA followed by Tukey’s multiple comparision test (n = 4–5).
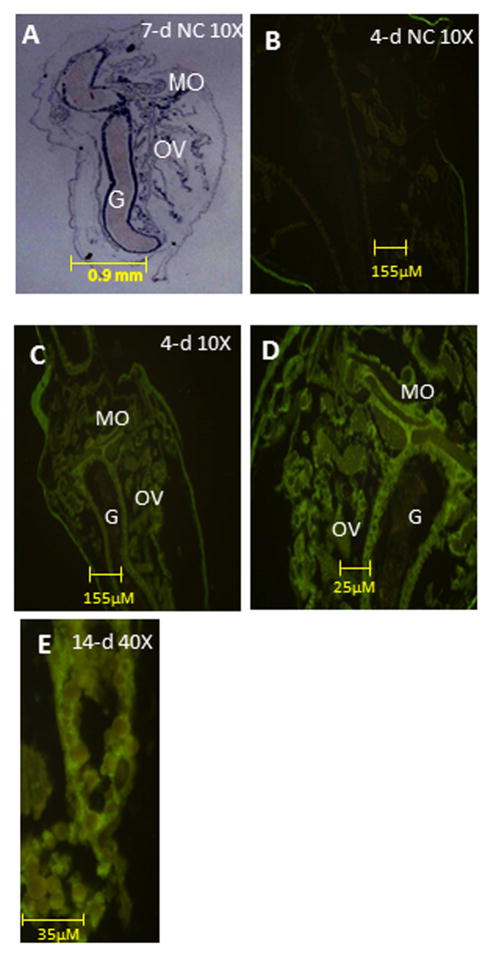
Immunohistochemistry (IHC) of HR97g from 4 and 14-day old daphnids indicates greater fluorescence in the mandibular region, ovary (OV), and gastrointestinal tract (G) than other tissues of D. magna (Fig. 4) with a strong increase in expression in the ovaries by day 14 (Fig. 4E). The localization of HR97g implies a role in diet, reproduction, or potentially juvenoid hormone function as the mandibular organ is the site of methyl farnesoate synthesis—methyl farnesoate is the crustacean juvenile hormone (Laufer and Biggers, 2001).
Fig. 4. Organ-dependent expression of HR97g.<.
br>HR97g is primarily expressed in the mandibular organ (MO), ovaries (OV), and gastrointestinal tract (G). Immunohistochemical analysis of HR97g expression was performed as noted in the Materials and Methods. Hematoxylin and eosin staining (H&E) of D. magna (A). IHC fluorescence in the absence of the primary antibody in a 4-day old juvenile daphnid (10X magnification)(B). IHC fluorescence in a juvenile daphnid (4-days old) at 10X and (C) 40X magnification (D). IHC from an ovary of a 14-day old daphnid (E).
Constitutive HR97 Transactivation
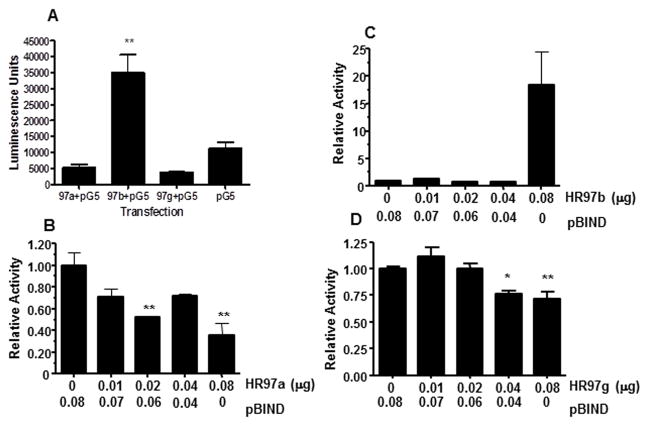
Cells were transfected with Gal4 chimeric receptors containing Clem-magnaHR97a, Clem-magnaHR97b, or Clem-magnaHR97g (GAL4-97a/b/gDEF) and the pG5luc reporter gene to determine if the magnaHR97 receptors are ligand-independent activators of gene expression. HR97a and HR97g repressed constitutive transcription of the luciferase reporter in the absence of added ligand, while HR97b significantly increased transcription under the same conditions (Fig. 5A). We subsequently transfected cells with increasing concentrations of the GAL4-97a/b/g-DEF chimeric plasmids in the presence of the unaltered pBIND-GAL4 vector to ensure equal plasmid loading and further characterized the constitutive activity of the HR97 receptors. HR97a and HR97g exerted weak, but discernable concentration-dependent repression of pBIND-induced transcriptional activity in the absence of ligand (Fig. 5B, Fig. 5D), suggesting that these receptors are transcriptional repressors in the absence of ligand. Other NRs, such as PXR, have also been found to repress gene expression in the absence of ligands because of their recruitment of co-repressors (Ourlin et al., 2003; Takeshita et al., 2002). In contrast, HR97b activity was inhibited in the presence of the empty pBIND-GAL4 plasmid, and HR97b alone significantly increased transcriptional activity (Fig. 5C). These data may also indicate that there is competition for binding to the GAL4 DNA binding site on the pG5luc plasmid between the GAL4-97a/b/g-DEF chimeric plasmids and the empty pBIND-GAL4 with GAL4-HR97b showing greater activity and GAL4-HR97a and g showing repressive activity.
Fig. 5. Constitutive transcriptional activity of HR97 receptors.<.
br>A) HepG2 cells were transfected with 0.02 mg of pBIND-GAL4-97a/b/g-DEF chimeric plasmid and 0.1 μg of the reporter plasmid, pG5luc. Control cells were transfected with pG5luc only and transactivation activity was measured as described in the Materials and Methods. Only HR97b increases constitutive activity. (B–D) Cells were transfected with 0, 0.01, 0.02, 0.04 or 0.08 μg pBIND-GAL4-97a/b/g-DEF chimeric plasmid supplemented with empty pBIND-GAL4 vector to ensure all wells contained 0.08μg of total expression plasmid and luciferase activity measured. Assays were performed two times. An asterisk (p < 0.05) or two asterisks (p < 0.01) indicates statistical significance as determine by ANOVA followed by Dunnett’s multiple comparison test with GraphPad Prizm 4.0 (n = 3).
Ligand-dependent HR97 transactivation
Transactivation assays were also performed in the presence of several different chemicals to test whether the magnaHR97 receptors act as promiscuous xenobiotic/endobiotic sensors similar to some of the related NR1I (CAR/PXR) and NR1J (HR96, C. elegans-nhr-8) receptors (Hernandez et al., 2009; Kliewer et al., 1998; Lindblom et al., 2001), including Daphnia HR96 (Karimullina et al., 2012). Of the chemicals tested at 10 μM, only the juvenile hormone analog, pyriproxyfen significantly activated transcription (Fig. 6). Pyriproxyfen increased magnaHR97g-mediated transcriptional activity by 54%, but not magnaHR97a or HR97b. Arachidonic acid (AA), an n-6 polyunsaturated fatty acid, decreased magnaHR97a, HR97b, and HR97g activity (Fig. 6). Overall, the data suggests the HR97 receptors are not promiscuous NRs at least when compared to related receptors (Baldwin and Roling, 2009; Hernandez et al., 2009; Karimullina et al., 2012).
Fig. 6. Ligand-dependent activation of HR97g.
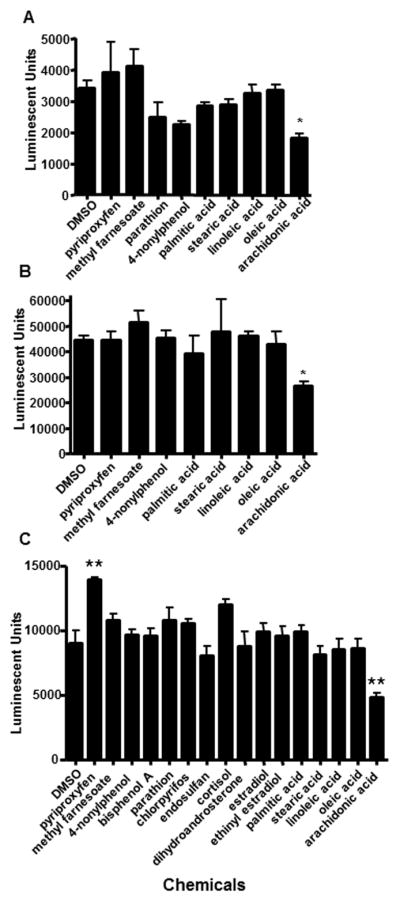
HepG2 cells transfected with 0.02 mg of GAL4-97a-DEF (A) GAL4-97b-DEF (B), or Gal4-97g-DEF (C) and 0.1 mg of the pG5luc reporter plasmid were treated with several chemicals, and luciferase activity measured. All chemicals were tested at 10 mM except oleic, linoleic and arachidonic acid, which were tested at 100 mM. Statistical significance was determined by ANOVA followed by Dunnett’s multiple comparison test as the post-hoc test. An asterisk indicates p < 0.05 and two asterisks indicate p < 0.01 (n = 3–4).
Methyl farnesoate, a juvenile hormone, and its analog pyriproxyfen initiate the production of males for sexual reproduction in cyclic parthenogenic Daphnia species (Olmstead and LeBlanc, 2002; Wang et al., 2005). Because pyriproxyfen activates HR97g suggests the possibility that HR97g contributes to the regulation of male sex determination, therefore, further concentration-dependent assays were performed with both pyriproxyfen and methyl farnesoate, the endogenous juvenoid hormone produced by crustaceans (Eads et al., 2008). Pyriproxyfen did not activate HR97a or HR97b (Fig. 7). Pyriproxyfen did increase transcriptional activation of HR97g in a concentration-dependent fashion with peak activation at 10 μM and a significant decrease at 30 μM, presumably because of noticeable cell toxicity. Concentration-response curves indicate that the EC50 of pyriproxyfen is 3.36 μM, which is quite high and its activation of about 1.5X is quite small. Methyl farnesoate also activates HR97g, but not HR97a or HR97b (Fig. 7d). HR97g-mediated luciferase activity is increased in the presence of methyl farnesoate in a concentration-dependent manner with 30 μM exerting the maximal transcriptional effect (Fig. 7e).
Fig. 7. Pyriproxyfen and methyl farnesoate activate HR97g but not HR97a and HR97b in transactivation assays.<.
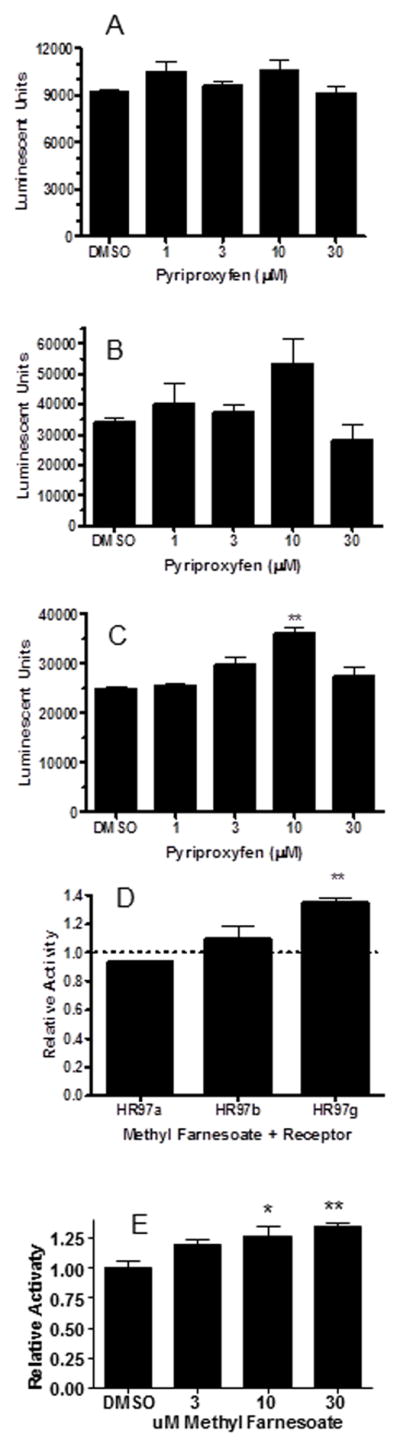
br>Cells transfected with GAL4-97aDEF (A), GAL4-97bDEF (B), or GAL4-97gDEF (C), and the pG5luc reporter were exposed to pyriproxyfen at concentrations from 3–30 mM and luciferase activity measured as described in the Materials and Methods. (D) Activation of HR97a, HR97b, and HR97b by methyl farnesoate at 30 mM. (E) Activation of HR97g by methyl farnesoate. An asterisk indicates significance at p < 0.05, and two asterisks indicate significance at p < 0.01 by ANOVA followed by Dunnett’s multiple comparison test using GrapPad Prizm 4.0 (n = 3).
AA was the only chemical tested that repressed constitutive HR97a, HR97b, and HR97g transactivation activity (Fig. 6), and therefore AA is considered an inverse agonist (Baldwin and Roling, 2009; Forman et al., 1998; Huang et al., 2004). The EC50 of AA was lower for HR97g than HR97a or HR97b. Overall, AA was more efficacious and potent at inhibiting HR97g activity than HR97a or HR97b (Fig. 8). Concentration-response analysis with HR97g revealed an EC50 of 26.8μM with a 95% Confidence interval of 12 – 60 μM (Fig. 8A). These values are within physiological ranges, which vary from approximately 0.5 – 15 μM in uninvolved or inactive tissues to 35 – 100 μM in active or inflamed tissues that recently unesterified arachidonic acid (Brash, 2001). In other words, HR97g is inhibited by arachidonic acid at concentrations found in cells that have recently undergone physiological changes or stress that caused arachidonic acid release.
Fig. 8. Inhibition of HR97 paralogs by arachidonic acid.<.
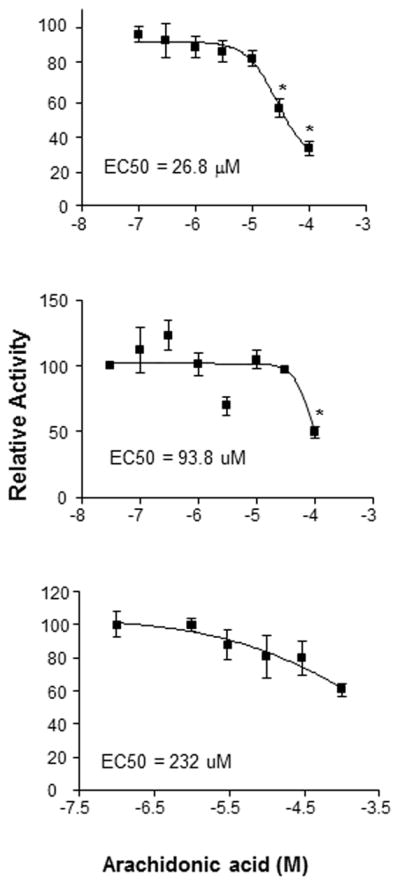
br>HepG2 cells transfected with GAL4-97gDEF (A), GAL4-97aDEF (B), or GAL4-97bDEF, and the pG5luc reporter were exposed to arachidonic acid at concentrations from 0.1–100 mM and luciferase activity measured as described in the Materials and Methods. Concentration-response curves and EC50s were determined with GrapPad Prizm 4.0 (n = 3).
DISCUSSION
The HR97 receptors are a newly discovered clade of receptors that were placed in the NR1L group (Thomson et al., 2009). D. magna and D. pulex both express three distinct HR97 receptors of which HR97a and HR97b are found in tandem duplicate in the genome. Here we confirm that they are a novel group; separate but related to the invertebrate HR96 receptors that are involved in lipid metabolism and chemical detoxification (Bujold et al., 2010; Karimullina et al., 2012; King-Jones et al., 2006; Sieber and Thummel, 2009).
While discovered in Daphnia species, the HR97 receptors are not specific to Daphnia. Phylogenetic analyses confirm that the Ixodes scapularis (deer tick) genome contains a member of the NR1L group (IsHR97) (Fig. 1), as does Metaseiulus occidentalis (western predatory mite; GenBank accession number XP_003748267; data not shown). Ixodes and Metaseiulus are arachnids and found within the Chelicerata, an ancient order of Arthropoda (Regier et al., 2010). Nevertheless, the HR97 receptors have not been observed in any of the insect genomes sequenced (Adams et al., 2000; Colbourne et al., 2011; Consortium, 2008; Cruz et al., 2009; Emes et al., 2003; Maglich et al., 2003). The arthropods are typically separated into different clades based on the Pancrustacea or Mandibulata (Pancrustacea + Myriapoda) hypothesis described previously (Giribet and Ribera, 2000; Regier et al., 2010; Rota-Stabelli et al., 2011) (see Figure 9 for a graphical representation of arthropod evolution). Therefore, HR97 receptors could be present in several other arthropod genomes, including spiders, centipedes, ostracods, and potentially decapod crustaceans. HR97 receptors may also be present in arthropod ancestors. Based on this data, we conclude that HR97 is found in ancient arthropods, was amplified within the genome sometime during the evolution of arthropods, but lost in the insects (Hexapoda; a modern lineage).
Fig. 9. Cladogram designed based on the pancrustacean hypothesis.<.
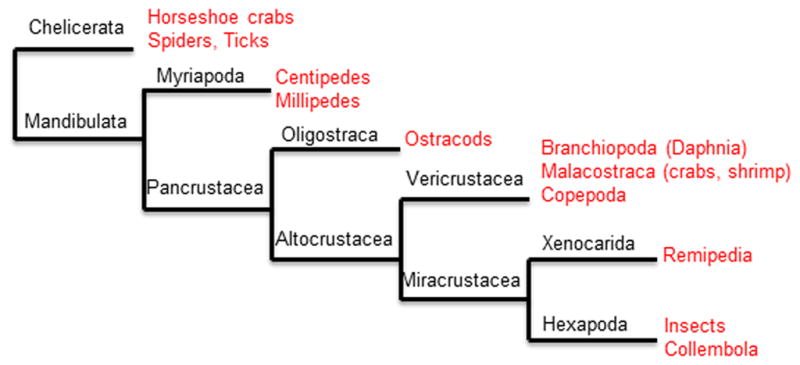
br>The different clades are shown in black, and examples are in red. Daphnia are members of the Vericrustacea, which also includes crabs, shrimp, lobsters, copepods, and amphipods.
We hypothesized that D. magna HR97s may be promiscuous toxicant receptors activated by a plethora of environmental chemicals because they are related to CAR, PXR, and HR96 (King-Jones et al., 2006; Kretschmer and Baldwin, 2005). However, our data does not support this hypothesis as few of the chemicals we tested activated HR97 (Fig. 5); many of these chemicals activated Daphnia HR96 (Karimullina et al., 2012). Not all receptors related to the HR97s are promiscuous chemical sensors. The NR1J member DAF-12 in C. elegans, for example, has been shown to regulate dauer diapause and development (Antebi et al., 2000). The VDR is primarily activated by the steroid hormone, vitamin D (Haussler and McCain, 1977; Umesono et al., 1991). Furthermore, many of the promiscuous xenobiotic receptors such as HR96, CAR, and PXR have other functions; most likely as regulators of lipid homeostasis (Dong et al., 2009; Finn et al., 2009; Horner et al., 2009; Sieber and Thummel, 2009).
The n-6 unsaturated fatty acid, AA has been identified as a putative ligand for each of the HR97 receptors with HR97g showing the greatest response (Fig. 6, 8); although AA does not completely block HR97g activity. Interestingly, AA is one of the few unsaturated fatty acids tested that does not alter HR96 activity. Six different fatty acids were previously tested as Daphnia HR96 transcriptional modulators (arachidonic, oleic, linoleic, α-linolenic, DHA, and eicosapentaenoic (EPA) acids). The n-3 fatty acids were inhibitors and the n-6/9 fatty acids were activators with the exception of AA, which had no activity towards HR96 (Karimullina et al., 2012). Instead, AA inhibits HR97 activity. This suggests a division of function between HR96 and the HR97s potentially associated with AA metabolism or use in Daphnia.
AA is the only chemical we have found at this time that can modulate HR97 receptors at physiological concentrations as HR97g has an EC50 of 26.8μM. In inactive tissues, AA concentrations are typically below 30μM; however, concentrations can reach much higher during inflammatory reaction or stimulated tissues where concentrations have been reported as high as 100 μM (Brash, 2001). An example of another receptor that is activated by endogenous ligands at μM concentrations is RXR. The EC50 of 9-cis-retinoic acid for RXR is 0.5μM (Schwimmer et al., 2004), while the EC50 of the putative endogenous RXR ligand docosahexaenoic acid (DHA) is > 50 μM (Calderon and Kim, 2007). However, it is also possible that AA metabolites such as the prostaglandins, leukotrienes, and epoxyeicosatrienoic acids are HR97 ligands (Du et al., 2005; Hurtado et al., 2009; Sears and Ricordi, 2012).
Daphnia incorporate and utilize the fatty acids they eat, and their fatty acid composition reflects their primarily algal diet (Brett et al., 2006) with the rapid conversion of DHA to EPA and the preferential retention of two key fatty acids; AA (n-6), and EPA (n-3) (Taipale et al., 2011; von Elert, 2002). Given HR96’s proposed role in triglyceride absorption and energy metabolism(Sieber and Thummel, 2009), and the potential for it to be involved in unsaturated fatty acid signaling through the n-3 fatty acids, DHA and EPA (Karimullina et al., 2012), it is possible that other putative NRs (i.e. HR97s) may recognize the retained n-6 fatty acid, AA. Studies indicate that AA is not involved in growth, and instead is (Becker and Boersma, 2005); associated with early oocyte maturation in crustaceans (Chansela et al., 2012).
The preferential expression of the HR97s in adults (Fig. 3), and specifically within the ovaries (Fig. 4) suggests a role for the HR97s in reproduction. Under the temperature and culture conditions used, daphnids typically bear eggs in their brood chamber at approximately day 7 and release their first brood at approximately 9–10 days of age. Vtg2 expression indicates a mature ovary by day 7 (Fig. 3). Our data indicates that HR97a and HR97b is preferentially expressed as daphnids transition from juveniles to reproductively competent adults, and HR97g is preferentially expressed in reproductively competent adults. Thus, HR97s may help a rapidly reproducing R species such as D. magna best allocate energy resources towards reproduction.
Free fatty acids and polar lipids have recently been found to bind or activate a number of NRs. For example, triacylglycerols, cholesterol, and unsaturated fatty acids have been shown to activate HR96 (Karimullina et al., 2012; Sieber and Thummel, 2012). Linoleic acid is found within the binding pocket of HNF4α (Yuan et al., 2009), LRH-1 is sensitive to DLPC, a phosphatidylcholine (Lee et al., 2011), phosphatidyl insositols bind SF-1 (Krylova et al., 2005), and the PPARs respond to a suite of unsaturated and saturated fatty acids (Desvergne and Wahli, 1999). Taken together, it is not unique that AA, a critically important unsaturated fat in Daphnia and other species, could be a NR ligand.
However, it cannot be ruled out that there are other HR97 ligands or each HR97 has its own distinct ligand. The genomic structure and amino acid identity of the HR97 receptors vary considerably between the different HR97s (Table 2) and between different daphnid species (Table 1). In constrast, HR96’s amino acid identity (Karimullina et al., 2012) and genomic structure (Fig. 2) are extremely similar when comparing between the different daphnid species. These differences in the C and E domains suggest that HR97a/b have similar DNA binding properties and therefore potentially similar transcriptional functions, but HR97g’s probably differ. All three HR97 receptors show differences in their LBDs with HR97g showing the greatest disparity compared to the other receptors (Table 2). In turn, pyriproxyfen and methyl farnesoate activate HR97g but not HR97a and HR97b (Fig. 6, 7). However, activation is weak (about 1.5X) and probably only at supraphysiological concentrations. Therefore HR97g is mostly likely not a juvenoid receptor. Furthermore, two separate studies have demonstrated a key role for MetR (methoprene resistance receptor) as the methyl farnesoate receptor involved in male production (LeBlanc et al., 2013; Miyakawa et al., 2013) further reducing the likelihood that HR97g is a juvenoid receptor.
The amplification and alteration of the HR97 NRs in the Daphnia genome is intriguing. A recent study, using genomic, biochemical, structural, and phylogenetic analyses, has shown that modern NRs evolved through subtle tinkering of a ligand-dependent ancestral receptor related to HNF4 (Bridgham et al., 2010). Differences in the constitutive regulatory functions of HR97a and HR97b provides further evidence that closely related NRs could develop different roles through subtle changes in amino acid composition during evolution. The relationship between HR97a and HR97b is similar to CAR and PXR, two closely related receptors in the NR1I group. PXR, like HR97a, represses gene expression in the absence of ligand because of its recruitment of co-repressors such as NCoR (nuclear receptor co-repressor) and SMRT (silencing mediator for retinoid and thyroid hormone receptors)(Hernandez et al., 2009; Takeshita et al., 2002). CAR, like HR97b, has significant constitutive activity in the absence of ligands (Baes et al., 1994).
Interestingly, the constitutive activity of CAR is attributed to the active orientation of the AF (activation function)-2 helix within AF-2 motif of the LBD (Xu et al., 2004). There are two primary motifs in a NR that are involved in the recruitment of cofactors; AF-1 in the A/B domain and AF-2 in the LBD (Hernandez et al., 2009; King-Jones and Thummel, 2005). Because the AF-1 domain of HR97a/b was not included in our chimeric plasmids, the difference in the constitutive activity of the NRs in the transactivation assays is most likely attributed to the AF-2 motif within the LBD. This study provides additional evidence that the LBD of a NR is crucial for functions beyond ligand-binding, including its distinct activation functions, and that these LBD functions are conserved in a crustacean species.
In summary, this study provides a partial characterization of the newly discovered HR97 (NR1L) group of nuclear receptors in D. magna (Fig. 1). Phylogenetic comparison to other model organisms demonstrates that the HR97 receptors exist in early arthropod species such as chelicerates (tick). Therefore based on the Pancrustacean/Mandibulata hypothesis (Regier et al., 2010; Rota-Stabelli et al., 2011), we hypothesize that the HR97 receptors may be found in several distinct arthropod groups that evolved after the chelicerates and before the insects (Fig. 9). However, unlike their NR relative HR96/CAR/PXR, the HR97 receptors do not appear to be promiscuous. Similar to many other NR1 family members (Bujold et al., 2010; Dong et al., 2009; Finn et al., 2009; Horner et al., 2009; Sieber and Thummel, 2009), the HR97 receptors appear to respond to dietary cues, specifically the dietary fatty acid, AA or its metabolites. Last, the expression patterns of the HR97 receptors suggest a role in reproduction and because AA is one of two unsaturated fatty acids specifically retained by crustaceans and concentrated in the ovaries (Chansela et al., 2012). Future studies will investigate the role of AA in Daphnia reproduction. In conclusion, the HR97 group is a novel NR group that we propose based on its expression and the limited data we have for putative ligands, can sense dietary changes and in turn alter reproduction.
Supplementary Material
Highlights.
The NR1L (HR97) group of nuclear receptors (NRs) are newly discovered in Daphnia
The HR97s are also present in early arthropods such as the chelicerates but lost in insects
They are related to CAR, PXR, and HR96, but are not promiscuous
Arachidonic acid inhibits HR97g activity at physiologically relevant concentrations
HR97 NRs are primarily expressed in adults as they enter reproductive maturity
Acknowledgments
Funds for this project were provided by Clemson University start-up, a George R. MacDonald Fellowship to Gautam Ginjupalli, a Public Service Agency Fellowship to Yangchun Li, and National Institutes of Environmental Health Sciences grant R15-ES017321 to William Baldwin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Amoutzias GD, et al. A protein interaction atlas for the nuclear receptors: Properties and quality of a hub-based dimerisation network. BMC Systems Biology. 2007;1:34. doi: 10.1186/1752-0509-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A, et al. Daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Baes M, et al. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol. 1994;14:1544–1552. doi: 10.1128/mcb.14.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin WS, et al. Incomplete ecdysis is an indicator of ecdysteroid exposure in daphnia magna. Environ Toxicol Chem. 2001;20:1564–1569. [PubMed] [Google Scholar]
- Baldwin WS, et al. Physiological and biochemical perturbations in daphnia magna following exposure to the model environmental estrogen diethylstilbestrol. Environ Toxicol Chem. 1995;14:945–952. [Google Scholar]
- Baldwin WS, Roling JA. A concentration addition model for the activation of the constitutive androstane receptor by xenobiotic mixtures. Toxicol Sci. 2009;107:93–105. doi: 10.1093/toxsci/kfn206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker C, Boersma M. Differential effects of phosphorus and fatty acids on daphnia magna growth and reproduction. Limnol Oceanogr. 2005;50:388–397. [Google Scholar]
- Brash AR. Arachidonic acid as a bioactive molecule. J Clin Invest. 2001;107:1339–1345. doi: 10.1172/JCI13210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett MT, et al. Daphnia fatty acid composition reflects that of their diet. Limnol Oceanogr. 2006;51:2428–2437. [Google Scholar]
- Bridgham JT, et al. Protein evolution by molecular tinkering: Diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biology. 2010;8:e1000497. doi: 10.1371/journal.pbio.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujold M, et al. Nuclear receptor DHR96 acts as a sentinel for low cholesterol concentrations in Drosophila melanogaster. Mol Cell Biol. 2010;30:793–805. doi: 10.1128/MCB.01327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzón V, et al. A conserved surface on the ligand binding domain of nuclear receptors for allosteric control. Mol Cell Endocrinol. 2012;348:394–402. doi: 10.1016/j.mce.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Calderon F, Kim H-Y. Role of RXR in neurite outgrowth induced by docosahexaenoic acid. Prostaglandins, Leukotrienes and Essential Fatty Acids. 2007;77:227–232. doi: 10.1016/j.plefa.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chansela P, et al. Composition and localization of lipids in penaeus merguiensis ovaries during the ovarian maturation cycle as revealed by imaging mass spectrometry. PLoS One. 2012;7:e33154. doi: 10.1371/journal.pone.0033154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz J, et al. Nuclear receptors in the mosquito Aedes aegypti: Annotation, hormonal regulation and expression profiling. FEBS J. 2009;276:1233–1254. doi: 10.1111/j.1742-4658.2008.06860.x. [DOI] [PubMed] [Google Scholar]
- Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: Nuclear control of metabolism. Endocrine Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- Dong B, et al. Activation of nuclear receptor car ameliorates diabetes and fatty liver disease. Proc Natl Acad Sci U S A. 2009;106:18831–18836. doi: 10.1073/pnas.0909731106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, et al. Evidence that cytochrome P450 Cyp2b19 is the major source of epoxyeicosatrienoic acids in mouse skin. Arch Biochem Biophys. 2005;435:125–133. doi: 10.1016/j.abb.2004.11.023. [DOI] [PubMed] [Google Scholar]
- Eads BD, et al. Ecological genomics in Daphnia: Stress responses and environmental sex determination. Heredity. 2008;100:184–190. doi: 10.1038/sj.hdy.6800999. [DOI] [PubMed] [Google Scholar]
- Emes RD, et al. Comparison of the genomes of human and mouse lays the foundation of genome zoology. Human Mol Genetics. 2003;12:701–709. doi: 10.1093/hmg/ddg078. [DOI] [PubMed] [Google Scholar]
- Evans RM. The nuclear receptor superfamily: A rosetta stone for physiology. Mol Endocrinol. 2005;19:1429–1434. doi: 10.1210/me.2005-0046. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. Unsaturated fatty acid regulation of cytochrome p450 expression via a CAR-dependent pathway. Biochem J. 2009;417:43–54. doi: 10.1042/BJ20080740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk GJ, Thummel CS. Isolation, regulation, and DNA-binding properties of three drosophila nuclear hormone receptor superfamily members. Proc Natl Acad Sci U S A. 1995;92:10604–10608. doi: 10.1073/pnas.92.23.10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, et al. Androstane metabolites bind to and deactivate the nuclear receptor car-beta. Nature. 1998;395:612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- Garcia HE, et al. Nuclear receptors are markers of animal genome evolution. J Struct Funct Genom. 2003;3:177–184. [PubMed] [Google Scholar]
- Germain P, et al. Overview of nomenclature of nuclear receptors. Pharmacol Rev. 2006;58:685–704. doi: 10.1124/pr.58.4.2. [DOI] [PubMed] [Google Scholar]
- Giribet G, Ribera C. A review of arthropod phylogeny: New data based on ribosomal DNA sequences and direct character optimization. Cladistics. 2000;16:204–231. doi: 10.1111/j.1096-0031.2000.tb00353.x. [DOI] [PubMed] [Google Scholar]
- Hannas BR, et al. Interactions of the crustacean nuclear receptors HR3 and E75 in the regulation of gene transcription. Gen Comp Endocrinol. 2010;167:268–278. doi: 10.1016/j.ygcen.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannas BR, et al. Regulation and dysregulation of vitellogenin mRNA accumulation in daphnids (Daphnia magna) Aquat Toxicol. 2011;101:351–357. doi: 10.1016/j.aquatox.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussler MR, McCain TA. Basic and clinical concepts related to vitamin d metabolism and action. N Engl J Med. 1977;297:974–983. doi: 10.1056/NEJM197711032971804. [DOI] [PubMed] [Google Scholar]
- Heckmann L-H, et al. Expression of target and reference genes in daphnia magna exposed to ibuprofen. BMC Genomics. 2006;7:175. doi: 10.1186/1471-2164-7-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JP, et al. Activation of CAR and PXR by dietary, environmental and occupational chemicals alters drug metabolism, intermediary metabolism, and cell proliferation. Curr Pharmacog Personal Med. 2009;7:81–105. doi: 10.2174/187569209788654005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg SM, et al. Primary structure and expression of a functional human glucocorticoid receptor cdna. Nature. 1985;318:635–641. doi: 10.1038/318635a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner MA, et al. The Drosophila DHR96 nuclear receptor binds cholesterol and regulates cholesterol homeostasis. Genes Dev. 2009;23:2711–2716. doi: 10.1101/gad.1833609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, et al. Meclizine is an agonist ligand for mouse constitutive androstane receptor (car) and an inverse agonist for human car. Mol Endocrinol. 2004;18:2402–2408. doi: 10.1210/me.2004-0046. [DOI] [PubMed] [Google Scholar]
- Hurtado MA, et al. Arachidonic acid (20:4n-6) effect on reproduction, immunology, and prostaglandin e2 levels in crassostrea corteziensis (hertlein, 1951) Aquaculture. 2009;294:300–305. [Google Scholar]
- Johnstone MB, et al. Visualization of shell matrix proteins in hemocytes and tissues of the eastern oyster, Crassostrea virginica. J Exp Zool B Mol Dev Evol. 2008;310:227–239. doi: 10.1002/jez.b.21206. [DOI] [PubMed] [Google Scholar]
- Karimullina E, et al. Hr96 is a promiscuous endo- and xeno-sensing nuclear receptor. Aquat Toxicol. 2012;116–117:69–78. doi: 10.1016/j.aquatox.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, et al. Phototoxicity and oxidative stress responses in Daphnia magna under exposure to sulfathiazole and environmental level ultraviolet b irradiation. Aquat Toxicol. 2009;91:87–94. doi: 10.1016/j.aquatox.2008.10.006. [DOI] [PubMed] [Google Scholar]
- King-Jones K, et al. The dhr96 nuclear receptor regulates xenobiotic responses in drosophila. Cell Metab. 2006;4:37–48. doi: 10.1016/j.cmet.2006.06.006. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Thummel CS. Nuclear receptors: A perspective from Drosophila. Nature Rev Genetics. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Krasowski MD, et al. Evolution of promiscuous nuclear hormone receptors: LXR, FXR, VDR, PXR, and CAR. Mol Cell Endocrinol. 2011;334:39–48. doi: 10.1016/j.mce.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer XC, Baldwin WS. CAR and PXR: Xenosensors of endocrine disrupters? Chem-Biol Interac. 2005;155:111–128. doi: 10.1016/j.cbi.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Krylova IN, et al. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Laufer H, Biggers WJ. Unifying concepts learned from methyl farnesoate for invertebrate reproduction and post-embryonic development. Amer Zool. 2001;41:442–457. [Google Scholar]
- LeBlanc GA, et al. A transgenerational endocrine signaling pathway in crustacea. PLoS One. 2013;8:e61715. doi: 10.1371/journal.pone.0061715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, et al. A nuclear-receptor-dependent phosphatidylcholine pathway with antidiabetic effects. Nature. 2011;474:506–510. doi: 10.1038/nature10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin GG, et al. Hormone receptor-like in 96 and broad-complex modulate phenobarbital induced transcription of cytochrome P450 cyp6d1 in Drosophila s2 cells. Insect Mol Biol. 2011;20:87–95. doi: 10.1111/j.1365-2583.2010.01047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblom TH, et al. A C. elegans orphan nuclear receptor contributes to xenobiotic resistance. Curr Biol. 2001;11:864–868. doi: 10.1016/s0960-9822(01)00236-6. [DOI] [PubMed] [Google Scholar]
- Maglich JM, et al. The first complete genome sequence from a teleost fish (Fugu rubripes) adds significant diversity to the nuclear receptor superfamily. Nucleic Acids Res. 2003;31:4051–4058. doi: 10.1093/nar/gkg444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich JM, et al. Comparison of complete nuclear receptor sets from the human, Caenorhabditis elegans and Drosophila genomes. Genome Biol. 2001;2:research0029.1–0029.7. doi: 10.1186/gb-2001-2-8-research0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, et al. The nuclear receptor superfamily: The second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. Cdd: A conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:237–240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa H, et al. A mutation in the receptor methoprene-tolerant alters juvenile hormone response in insects and crustaceans. Nature Commun. 2013;4:1856. doi: 10.1038/ncomms2868. [DOI] [PubMed] [Google Scholar]
- Mota LC, et al. CAR-null mice are sensitive to the toxic effects of parathion: Association with reduced cytochrome p450-mediated parathion metabolism. Drug Metab Dispos. 2010;38:1582–1588. doi: 10.1124/dmd.110.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PY, et al. Processing of gene expression data generated by quantitative real-time rt-pcr. Biotechniques. 2002;32:1372–1379. [PubMed] [Google Scholar]
- Olmstead AW, LeBlanc GA. Juvenoid hormone methyl farnesoate is a sex determinant in the crustacean Daphnia magna. J Exp Zool. 2002;293:736–739. doi: 10.1002/jez.10162. [DOI] [PubMed] [Google Scholar]
- Ourlin JC, et al. The small heterodimer partner interacts with the pregnane x receptor and represses its transcriptional activity. Mol Endocrinol. 2003;17:1693–1703. doi: 10.1210/me.2002-0383. [DOI] [PubMed] [Google Scholar]
- Regier JC, et al. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature. 2010;463:1079–1083. doi: 10.1038/nature08742. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, et al. The nuclear receptor superfamily. J Cell Sci. 2003;116:585–586. doi: 10.1242/jcs.00247. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, et al. Explosive lineage-specific expansion of the orphan nuclear receptor HNF4 in nematodes. J Mol Evol. 2005;60:577–586. doi: 10.1007/s00239-004-0175-8. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, et al. Euteleost fish genomes are characterized by expansion of gene families. Genome Res. 2001;11:781–788. doi: 10.1101/gr.165601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roling JA, et al. Hexavalent chromium reduces juvenile growth and alters gene expression in Fundulus heteroclitus. Environ Toxicol Chem. 2006;25:62–70. doi: 10.1897/05-659r.1. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rota-Stabelli O, et al. A congruent solution to arthropod phylogeny: Phylogenomics, microRNAs, and morphology support monophyletic mandibulata. Proc R Soc B. 2011;278:298–306. doi: 10.1098/rspb.2010.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwimmer LJ, et al. Creation and discovery of ligand receptor pairs for transcriptional control with small molecules. Proc Natl Acad Sci USA. 2004;1–1:14707–14712. doi: 10.1073/pnas.0400884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears B, Ricordi C. Role of fatty acids and polyphenols in inflammatory gene transcription and their impact on obesity, metabolic syndrome and diabetes. Eur Rev Med Pharmacol Sci. 2012;16:1137–1154. [PubMed] [Google Scholar]
- Sieber MH, Thummel CS. The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell Metab. 2009;10:481–490. doi: 10.1016/j.cmet.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber MH, Thummel CS. Coordination of triacylglycerol and cholesterol homeostasis by dhr96 and the Drosophila LipA homolog magro. Cell Metab. 2012;15:122–127. doi: 10.1016/j.cmet.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger J, et al. Coordinate regulation of xenobiotic and bile acid homeostasis by pregnane x receptor. Drug Metab Dispos. 2001;29:1467–1472. [PubMed] [Google Scholar]
- Taipale SJ, et al. Diet-switching experiments show rapid accumulation and preferential retention of highly unsaturated fatty acids in Daphnia. Oikos. 2011;120:1674–1682. [Google Scholar]
- Takeshita A, et al. Putative role of the orphan nuclear receptor sxr (steroid and xenobiotics receptor) in the mechanism of cyp3a4 inhibition by xenobiotics. J Biol Chem. 2002;277:32453–32458. doi: 10.1074/jbc.M111245200. [DOI] [PubMed] [Google Scholar]
- Thompson JD, et al. The clustalx windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson SA, et al. Annotation, phylogenetics, and expression of the nuclear receptors in Daphnia pulex. BMC Genomics. 2009;10:500. doi: 10.1186/1471-2164-10-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokishita S-i, et al. Organization and repression by juvenile hormone of a vitellogenin gene cluster in the crustacean, Daphnia magna. Biochem Biophys Res Comm. 2006;345:362–370. doi: 10.1016/j.bbrc.2006.04.102. [DOI] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Umesono K, et al. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin d3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Elert E. Determination of limiting polyunsaturated fatty acids in daphnia galeata using a new method to enrich food algae with single fatty acids. Limnol Oceanogr. 2002;47:1764–1773. [Google Scholar]
- Wang HY, et al. The screening of chemicals for juvenoid-related endocrine activity using the water flea Daphnia magna. Aquat Toxicol. 2005;74:193–204. doi: 10.1016/j.aquatox.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Xu RX, et al. A structural basis for constitutive activity in the human CAR/RXRα heterodimer. Mol Cell. 2004;16:919–926. doi: 10.1016/j.molcel.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Yuan X, et al. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS One. 2009;4:e5609. doi: 10.1371/journal.pone.0005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollner G, et al. Role of nuclear receptors in the adaptive response to bile acids and cholestasis: Pathogenetic and therapeutic considerations. Mol Pharm. 2006;3:231–251. doi: 10.1021/mp060010s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.