Summary
Mitotic spindles are critical for accurate chromosome segregation. Centrosomes, the primary microtubule nucleating centers of animal cells, play key roles in forming and orienting mitotic spindles. However, the survival of Drosophila without centrosomes suggested they are dispensable in somatic cells, challenging the canonical view. We used fly wing disc epithelia as a model to resolve these conflicting hypotheses, revealing that centrosomes play vital roles in spindle assembly, function, and orientation. Many acentrosomal cells exhibit prolonged spindle assembly, chromosome mis-segregation, DNA damage, misoriented divisions, and eventual apoptosis. We found that multiple mechanisms buffer the effects of centrosome loss, including alternative microtubule nucleation pathways and the Spindle Assembly Checkpoint. Apoptosis of acentrosomal cells is mediated by JNK signaling, which also drives compensatory proliferation to maintain tissue integrity and viability. These data reveal the importance of centrosomes in fly epithelia, but also demonstrate the robust compensatory mechanisms at the cellular and organismal level.
Introduction
Evolution has shaped mechanisms ensuring that accurate chromosome segregation occurs with high fidelity via microtubule-based mitotic spindles. Animal cell spindles are bipolar structures formed primarily via microtubule (MT) nucleation by a pair of centrosomes (Walczak and Heald, 2008). They facilitate equal segregation of the genome to the two daughters. Defects in spindle formation or function can lead to chromosome mis-segregation and aneuploidy (Nicholson and Cimini, 2011), a common form of chromosomal instability (CIN) and hallmark of most cancer cells (Hanahan and Weinberg, 2011). Furthermore, many tumors show misregulated centrosome number or function, suggesting centrosomes serve a central role in preventing CIN and cancer (Gordon et al., 2012). Mutations in centrosomal proteins also underlie microcephaly (MCPH), a developmental disorder resulting in reduced brain size (Megraw et al., 2011). However, in both cancer and MCPH, it remains unclear how defects in centrosome function contribute to disease, underscoring the need for in vivo, mechanistic examinations of centrosomes in mitosis and development.
Surprisingly, despite the many important roles of animal centrosomes, fruit flies lacking centrioles, core centrosome components, survive to adulthood (Basto et al., 2006; they die soon after due to the separate role of centrioles in cilia, and thus sensory neurons). This led to the conclusion that fly somatic cells do not need centrosomes to effectively conduct mitosis, suggesting non-centrosomal MT nucleation pathways (chromatin-based Ran and Augmin pathways; Clarke and Zhang, 2008; Goshima and Kimura, 2010; Goshima et al., 2008) are sufficient for mitotic spindle assembly. In normal cells, these pathways function in parallel with centrosomal MT nucleation to form spindles. This suggested an alternate model in which centrosomes are redundant machinery cells employ to enhance spindle formation and ensure high fidelity chromosome segregation. Interestingly, plant cells lack centrosomes and form mitotic spindles via the Ran and Augmin pathways (Hotta et al., 2012; Nakaoka et al., 2012; Zhang and Dawe, 2011), and meiotic spindles of many animal oocytes form via acentrosomal pathways (Dumont and Desai, 2012). We recently explored how cells and animals respond to the removal of another mitotic fidelity regulator, APC2 (Poulton et al., 2013). We found that redundant mechanisms and buffering by checkpoint proteins help cells cope with APC2 loss. We thus wondered whether similar compensatory mechanisms might explain survival of flies without centrosomes.
We used fly wing epithelial cells to study the consequences of centrosome loss in vivo. We found that although the organism can survive without centrosomes, their absence has serious consequences at the cell and tissue levels. Wing disc cells lacking centrosomes have highly elevated apoptosis. Mitosis is highly inefficient and error-prone, leading to significant reliance on the Augmin and Ran pathways for spindle assembly and the Spindle Assembly Checkpoint (SAC) to delay anaphase onset. In addition, we found that centrosomes help properly orient spindles in fly epithelia, which is important to maintain cell viability. Interestingly, although acentrosomal cells had elevated DNA damage, their apoptosis was p53 independent. Instead, cell death was mediated by the JNK pathway. Despite the dramatic increase in cell death, compensatory proliferation of the remaining cells largely, but not perfectly, maintains tissue architecture. Thus, centrosomes do play a vital role in mitosis in fly somatic cells, ultimately impacting cell viability through multiple aspects of mitosis.
Results
Centriole loss leads to highly elevated rates of apoptosis in wing imaginal discs
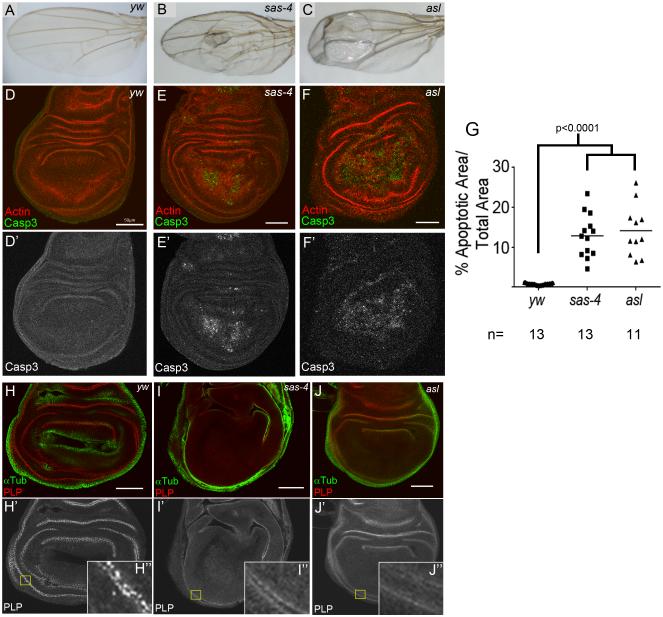
We used as our model Drosophila larval wing imaginal discs, a well characterized epithelium. Flies lacking either Sas-4 or Asl, both essential for centriole duplication, survive to adulthood (Basto et al., 2006; Blachon et al., 2008), but we observed that sas-4 or asl adults often possessed wing defects (vein mis-patterning, blisters, black spots, and curling; Fig 1A-C). These can result from increased cell death during larval/pupal development. We thus compared levels of apoptosis in wildtype (WT) and centriole deficient 3rd instar wing discs, measuring percent area stained for the apoptotic marker cleaved Caspase 3 (Casp3). WT wing discs have very low levels of apoptosis (0.7±2.2% of disc area Casp3 positive; mean±st.dev;Fig 1D), but surprisingly, we found highly elevated levels of Casp3 in sas-4 and asl mutants (12.9±5.4% and 14.2±6.5% of disc area, respectively; Fig 1E-G). We confirmed that discs mutant for sas-4 or asl lacked centrioles, using the centriole-associated protein Pericentrin Like Protein (PLP;Fig 1H-J), as was seen in larval brains (Basto et al., 2006; Blachon et al., 2008). Thus, centriole loss is not without consequence in fly somatic cells, but leads to highly elevated apoptosis.
Fig1. Centrosome loss leads to elevated apoptosis.
(A) WT adult wing. (B-C) Flies mutant for sas-4 or asl show morphological phenotypes. (D,D’,G) WT discs have minimal apoptosis, as indicated by Casp3 staining. (E-G) sas-4 and asl mutant discs display highly elevated levels of apoptosis. (H-H”) PLP labels centrioles in WT wing discs. (I,J) sas-4 and asl mutant discs lack discernible centrioles. Scale bars=50μm in all figures, unless noted. See also Fig S1.
Centrioles Promote Cell Viability Through Roles in Mitotic Spindle Assembly and Chromosome Segregation
Centrioles are multifunctional, generating both cilia and centrosomes. Fly epithelia, however, lack primary cilia (Ma and Jarman, 2011), suggesting apoptosis in sas-4 and asl mutant discs results from centrosome loss, not cilia. To test this we examined apoptosis in wing discs lacking the key centrosomal protein Cnn, which recruits many pericentriolar proteins, promoting MT nucleation, but which is not required for centriole or cilia formation (Megraw et al., 1999; 2001). Consistent with a role for centrosomes in somatic cell viability, cnn mutant wing discs displayed elevated apoptosis similar to centriolar mutants (18.9±6.3% of disc Casp3 positive, n=12; p<0.0001 vs. WT). Thus centrosomes are the centriole-generated structure maintaining cell viability.
In mammalian somatic cells, centrosomes are the primary nucleating centers for both mitotic spindles and interphase MTs. In fly cells, however, centrosomes do not nucleate interphase MTs; separate pathways govern that process (Rogers et al., 2008). Consistent with this, sas-4 wing disc cells had normal interphase MTs and apicobasal cell polarity (Fig S1A-D). We thus hypothesized that centrosome loss in fly epithelia specifically affects mitotic spindle assembly. To characterize this, we first stained WT and sas-4 mutant discs for α-tubulin (αtub) and the mitotic chromatin marker phospho-HistoneH3 (PH3). As expected, in prophase WT cells, MTs nucleated from the centrosomes (Fig 2A) served as focal points of bipolar spindle assembly during prometaphase (Fig 2B). In acentrosomal cells, MT nucleation appeared to occur around the chromatin itself (Fig 2E), yet these cells could establish bipolar spindles (Fig 2F). To observe MT dynamics, we live-imaged WT and sas-4 wing discs expressing the chromatin marker Histone:RFP (His:RFP) and the MT marker Jupiter:GFP. WT centrosomes became active in mitosis, nucleating many MTs that connected to chromosomes, forming a spindle (Fig S2A; Movie S1). By contrast, in sas-4 cells, MTs of nascent spindles originated around the chromosomes in prometaphase and the process was slowed (Fig S2B; Movie S2), as in other acentrosomal cell types (Basto et al., 2006; Lecland et al., 2013). Thus, spindles still assemble in the absence of centrosomes, but do so via a different process.
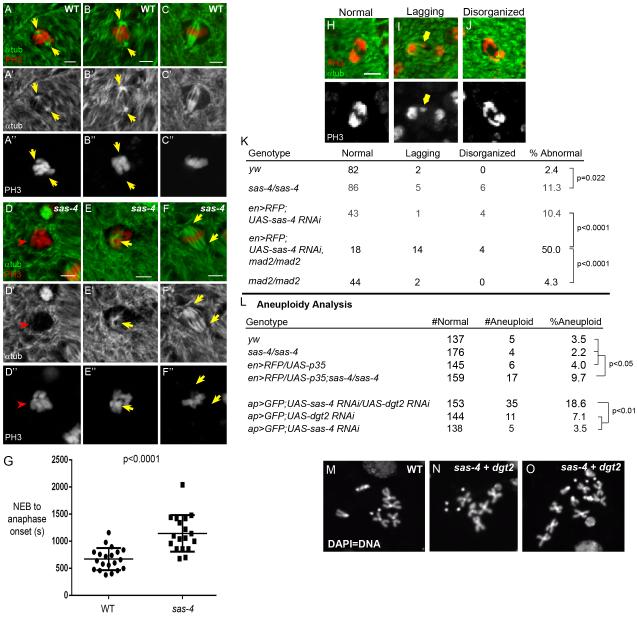
Fig2. Acentrosomal cells assemble spindles by a different pathway.
(A-C) WT wing discs stained for mitotic marker PH3 (red) and αtub (green) show centrosomal MT nucleation at prophase (A; yellow arrows). (B) Prometaphase. Spindle MTs initiate from centrosomes. (C) A fully formed metaphase spindle. (D-F) sas-4 mutant. No obvious MT nucleation is visible at prophase (D; red arrowhead). (E) After NEB, MTs nucleate around chromatin, and form a bipolar spindle (F). (G) Time from NEB to anaphase onset, determined by Polo:GFP (not shown), is 2× as long in acentrosomal cells (n=18) than WT (n=20). (H-J) Example anaphase images used to quantitate segregation defects i.e. lagging or disorganized chromosomes. (K) There are significantly more segregation defects in sas-4 mutant discs. mad2 loss did not increase segregation errors, but mad2 mutants expressing sas-4 RNAi had dramatic increases in segregation defects. (L) Rates of aneuploidy in various genetic backgrounds (see text). (M) normal karyotype, vs aneuploid (N) and near-tetraploid (O) cells frequently observed in Sas-4/Dgt2 codepletion. See also Fig S2. Scale bars=2μm.
Our live imaging of spindle assembly suggested acentrosomal cells were delayed in anaphase initiation. We hypothesized that, in the absence of centrosomes, MT nucleation and spindle formation were less efficient than in WT, and that inefficiency in spindle assembly would leave the SAC unsatisfied, thus delaying anaphase. To determine the time required to satisfy or perhaps by-pass the SAC, we measured time from nuclear envelope breakdown (NEB) to anaphase onset in WT and sas-4 mutants by live-imaging His:RFP and the mitotic kinase Polo (Polo:GFP); Polo invades the nuclear space immediately following NEB and rapidly localizes to kinetochores, making its initial detection on kinetochores an effective proxy for NEB (D’Avino et al., 2007). As predicted, anaphase onset took almost twice as long to initiate in acentrosomal cells (WT=11.2±3.4 min; sas-4=19.1±5.6 min; Fig 2G), consistent with an important role for the SAC in delaying anaphase to permit the inefficient spindle assembly in acentrosomal cells.
Proper, timely spindle assembly ensures accurate chromosome segregation. We thus examined the consequences of prolonged spindle assembly in acentrosomal cells on chromosome segregation. We first characterized the rate of segregation errors by examining anaphase figures from fixed WT and sas-4 mutant wing discs. Consistent with an important role for centrosomes in fidelity, segregation errors significantly increased; 11.3% of anaphase cells had lagging chromosomes, bridges, or disorganized chromosomes versus 2.4% in WT (Fig.2K;p=0.022). Chromosome segregation defects can lead to aneuploidy, which initiates apoptosis in some cells (Dekanty et al., 2012; Sir et al., 2013). Thus, the increased apoptosis in acentrosomal wing cells could result from aneuploidy, although acentrosomal fly neuroblasts (neural progenitors; NBs) did not have a significant increase in aneuploidy (Basto et al., 2006). To determine if centrosomes are important in ensuring accurate segregation of chromosomes in fly epithelia, we used chromosome squashes to measure aneuploidy rates in WT and sas-4 wing cells. Similar to NBs, we did not detect a significant difference in sas-4 wing discs (2.2% aneuploid; 4/180 cells) versus WT (3.5%; 5/142 cells; Fig.2L). Of course, if aneuploidy induced apoptosis, we might not recover aneuploid cells in this assay. To test this, we blocked apoptosis in acentrosomal cells, expressing the anti-apoptotic protein p35 in the posterior half of sas-4 mutant wing discs. After cell death was blocked, aneuploidy significantly increased (9.7%; p<0.05 vs WT, sas-4 mutant, or en>RFP/p35 alone; this probably underestimates aneuploidy in sas-4 mutants when apoptosis is blocked, because p35 was only expressed in the posterior compartment, but, for technical reasons, aneuploidy was measured in the entire wing). Thus, contrary to current models, centrosomes are important for efficient spindle assembly and accurate chromosome segregation in fly somatic cells.
Acentrosomal Cells Depend on Alternative Microtubule Nucleation Pathways
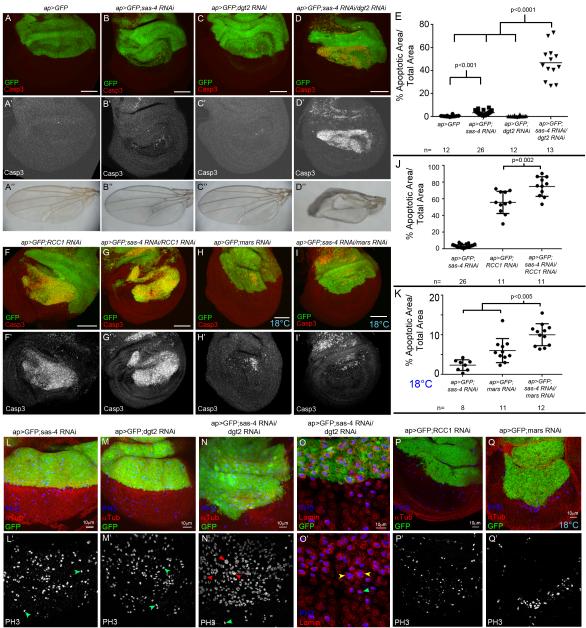
Because acentrosomal wing cells still formed spindles, we hypothesized that their mitosis relies on non-centrosomal spindle assembly pathways. Consistent with this, flies double mutant for cnn and the Augmin protein msd1 are lethal while each single mutant is viable (Wainman et al., 2009). To test this directly we first used ap-Gal4 to drive RNAi against sas-4 alone. sas-4 RNAi significantly increased apoptosis in the Gal4-expressing region of the wing disc relative to controls (Fig 3A,B,E; apoptosis was not as high as sas-4 null mutants, consistent with partial knockdown; we confirmed Sas-4 knockdown; Fig S3A). We then reduced the Augmin complex, via its component Dim gamma tubulin2 (Dgt2;Goshima et al., 2007; 2008). In contrast to sas-4 RNAi, dgt2 RNAi alone did not increase apoptosis (Fig 3C,E). However, double knockdown of both Sas-4 and Dgt2 led to an even more dramatic increase in apoptosis (Fig 3D,E; either single versus double RNAi: p<0.0001). This increase in apoptosis had consequences for wing development—while knocking down either gene alone had little effect on adult wings, Sas-4/Dgt2 double knockdown resulted in severely dysmorphic wings (Fig 3A”-D”). Knockdown of Sas-4 with other Augmin components Dgt3, Dgt4, and Dgt6 had similar effects (Fig S4). Thus, while centrosome loss alone reduces cell viability in a significant fraction of wing cells, many cells can still use the Augmin complex to conduct acentrosomal divisions. However, disrupting both Augmin and centrosomal MT nucleation dramatically elevated cell death and disrupted wing development, suggesting spindle assembly is irreparably compromised.
Fig3. Acentrosomal cells depend on alternative microtubule nucleation pathways.
(A-E, F-K). Apoptosis (Casp3), quantified in (E,J,K), (B“-D”) Adult wings. (A) Control=ap-Gal4 driving UAS-GFP (ap>GFP) in the dorsal compartment. (B) ap>GFP driving sas-4 RNAi. (C) ap>GFP driving Augmin component dgt2 RNAi. (D) Double Sas-4/Dgt2 knockdown. (F) Knockdown of RCC1 alone. (G) RCC1/Sas-4 codepletion (H) Knockdown of Mars at 18°C. (I) Mars/Sas-4 double knockdown at 18°C. (L,M) Sas-4 or Dgt2 knockdown in the dorsal wing. No change in the distribution of prophase, metaphase (green arrows), anaphase, or telophase cells. (N) Codepleting Sas-4 and Dgt2. Obvious enrichment of mitotic cells with virtually all cells stalled in prophase/prometaphase; note apparently polyploid nuclei (red arrows versus green arrow). (O) Sas-4/Dgt2 codepleted disc. Lamin staining (nuclear envelope) confirms that large mitotic nuclei are within a single cell (yellow versus green arrows). (P,Q) RCC1 or Mars knockdown. Fewer mitotic cells in knockdown region. See also Fig S3-S5.
To characterize mitotic fidelity in cells lacking both centrosomes and Augmin function, we attempted to analyze anaphase figures from cells codepleted of Sas-4 and Dgt2. Surprisingly, a majority of the cells in the region of the disc expressing RNAi against both genes appeared stalled in prophase/prometaphase (Fig 3N). There were virtually no anaphase nuclei present, thus precluding analysis of chromosome segregation, but underscoring the dependence of acentrosomal cells on the Augmin complex to promote MT nucleation and build spindles. We could karyotype these cells, and found significantly increased aneuploidy in cells with Sas-4/Dgt2 double knockdown (Fig 2L) relative to either single knockdown; interestingly Dgt2 knockdown alone moderately increased aneuploidy. Importantly, occasional Sas-4/Dgt2 double knockdown cells had dramatically larger mitotic nuclei than normal or single knockdown cells (Fig 3L-O). In fact we occasionally observed near tetraploid karyotypes in our chromosome squashes of wing cells codepleted of Sas-4 and Dgt2 (Fig 2O); such dramatic chromosomal abnormalities were never observed in our other genotypes. Given the difficulty these cells appear to have in forming spindles, we speculate polyploidy may result from a complete mitotic failure, possibly mitotic catastrophe. Further research will be required to determine the precise mechanism by which polyploid cells are generated, and how that contributes to increased apoptosis in cells lacking both centrosomes and Augmin.
To explore alternate mechanisms of acentrosomal spindle assembly, we disrupted chromatin-mediated spindle assembly via Ran-based MT nucleation, using RNAi against the RanGEF Regulator of Condensed Chromatin1 (RCC1; Clarke and Zhang, 2008), or the Ran dependent spindle assembly factor Mars (=HURP;Hayward et al., 2014). Interestingly, knocking down either alone led to very high levels of apoptosis (Fig 3F,J;data not shown), much greater even than those in sas-4 mutants. This suggests chromatin-mediated MT nucleation is more important in spindle assembly than centrosomes, and/or that these genes play additional roles in wing discs; the latter is likely for RCC1, which regulates other Ran-dependent processes like nuclear import. To test the importance of the Ran pathway in acentrosomal cells, we codepleted cells of Sas-4 and either RCC1 or Mars. Knocking down both Sas-4 and RCC1 elevated apoptosis over either single knockdown (Fig 3G,J). Mars knockdown alone led to such high levels of apoptosis that we could not reliably assess increased apoptosis (data not shown). To temper the strong effects of Mars knockdown, we repeated the experiment at 18°C to reduce Gal4 activity and thus knockdown. At 18°C, Mars knockdown alone only modestly reduced cell viability (Fig 3H,K), but codepleting Mars and Sas-4 significantly increased apoptosis (Fig 3I,K; RNAi against sas-4 alone at 18°C had minimal effects; Fig 3K). Interestingly, knocking down RCC1 or Mars also reduced the number of mitotic cells in regions of wing discs expressing RNAi (Fig 3P,Q), as did double knockdown of Sas-4 and RCC1 or Mars (data not shown). ap-Gal4-driven knockdown of Mars or RCC1 led to larval/pupal lethality, precluding analysis of adult wings. We thus used the wing specific MS1096-Gal4 to knockdown RCC1, with or without sas-4 RNAi. Consistent with the apoptosis in these genotypes, RCC1 RNAi alone led to abnormal adult wings, and this was enhanced by sas-4 RNAi (Fig S5). Together, these data suggest that mitotic spindle assembly relies on chromatin-mediated MT nucleation, in both normal and especially acentrosomal fly epithelia.
The Spindle Assembly Checkpoint is Essential for Mitotic Fidelity and Viability of Acentrosomal Cells
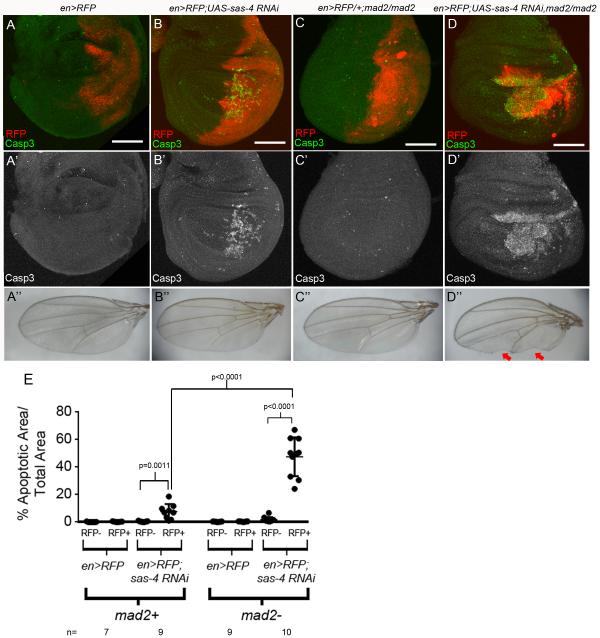
The increased apoptosis and chromosome segregation errors in acentrosomal cells suggested that spindle assembly may be error-prone. Spindle assembly is monitored by the SAC ensuring all kinetochores are attached to MTs before allowing anaphase onset (Foley and Kapoor, 2013). Disrupting the SAC permits entry into anaphase with incomplete kinetochore attachments, which can lead to chromosome mis-segregation. Based on the delayed anaphase onset in acentrosomal cells, we hypothesized disrupting the SAC in acentrosomal cells would increase apoptosis by preventing cells from compensating for slowed or error-prone spindle assembly. To test this, we disrupted the SAC in acentrosomal cells. We first examined flies homozygous mutant for mad2, a key SAC component (Buffin et al., 2007). They are adult viable (Buffin et al., 2007) and have modestly elevated apoptosis in wing discs (Fig 4C,E). We then generated larvae homozygous mutant for both mad2 and sas-4. Strikingly, third instar larvae lacking both centrosomes and the SAC completely lacked any wing, leg or haltere discs, and double mutants died as larvae/pupae. These phenotypes of mad2,sas-4 double mutants demonstrate the importance of the SAC for viability of acentrosomal epithelia in flies.
Fig4. Acentrosomal cells become reliant on the SAC.
(A) en-Gal4 driving UAS-RFP (en>RFP) in the posterior compartment did not increase apoptosis. (B) Expressing sas-4 RNAi by en>RFP elevated apoptosis. (C) While there was a modest, non-significant increase in apoptosis throughout homozygous mad2 mutant discs, en>RFP did not further increase apoptosis. (D) Expressing sas-4 RNAi in a mad2 mutant significantly increased apoptosis, and led to loss of adult posterior wing tissue (D”, arrows). (E) Quantification of apoptosis.
To characterize mitotic fidelity in cells with reduced centrosome function and lacking the SAC, we circumvented the complete lack of discs in mad2,sas-4 mutants by expressing sas-4 RNAi in the posterior compartment of wing discs in flies homozygous mutant for mad2. Whereas loss of Mad2 alone did not significantly elevate apoptosis (Fig 4C,E); there was a dramatic increase in cell death in the region of the disc with Mad2 loss and Sas-4 knockdown (the posterior compartment; Fig 4D), compared to knockdown of Sas-4 alone (Fig 4B). Notably, the posterior compartment was significantly smaller than normal (Fig 4D), which led to loss of adult wing tissue (Fig 4D”). In addition, while each single mutant is viable, almost all mad2 mutant+sas-4 RNAi flies died as pupae (1% of adults had that genotype (1/103); predicted Mendelian=33%). We then analyzed chromosome segregation in mad2 mutant cells with Sas-4 knockdown. Strikingly, while loss of Mad2 alone had little effect on chromosome segregation (4.3% defective) and Sas-4 knockdown alone moderately increased segregation errors (10.4%), in wing discs mutant for mad2 and expressing sas-4 RNAi, 50% of all anaphase figures had chromosome segregation errors (Fig 2K). Thus acentrosomal cells rely heavily on the SAC to delay anaphase, presumably allowing a more functional spindle to form prior to attempted chromosome segregation.
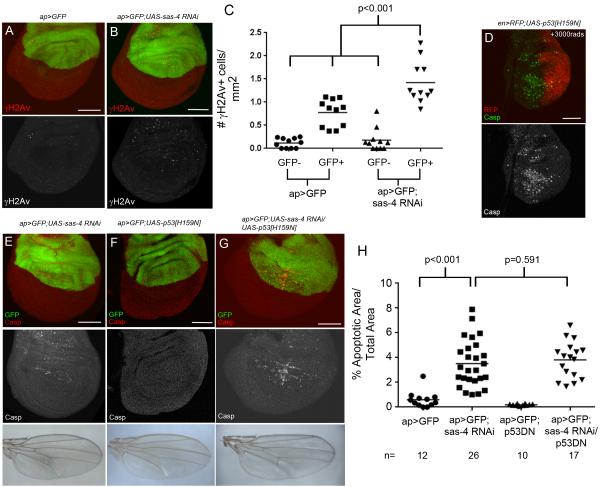
Acentrosomal Cells Have Increased DNA damage but p53 is not essential for apoptosis
DNA damage can activate the p53-mediated DNA damage response, leading to apoptosis (Brodsky et al., 2000; Jin et al., 2000; Ollmann et al., 2000), and DNA damage is generated by many types of mitotic errors. For example, lagging chromosomes were suggested to cause DNA damage through at least two processes, including chromosome pulverization following production of micronuclei or infringement of cytokinetic furrows on lagging chromosomes (Crasta et al., 2012; Janssen et al., 2011). DNA damage can also result from mitotic delay via multiple processes, including telomere deprotection or caspase-induced activation of DNase after mitochondrial disruption (Cenci, 2009; Ganem and Pellman, 2012; Hayashi and Karlseder, 2013). Because acentrosomal cells had increased apoptosis, chromosome segregation errors, and mitotic delay, we hypothesized that acentrosomal mitosis leads to DNA damage. To test this, we stained wing discs expressing sas-4 RNAi with γH2Av, a DNA damage marker (Madigan et al., 2002). sas-4 knockdown significantly increased the number of γH2Av positive cells (Fig 5A-C), though the fraction of cells with DNA damage did not reach the fraction of apoptotic cells in discs of the same genotype (Fig 3B). We then tested whether increased apoptosis in acentrosomal cells was mediated by p53. Interestingly, coexpressing a dominant-negative form of p53 with sas-4 RNAi in wing discs did not decrease apoptosis (Fig 5E-H; p53DN did block irradiation-induced apoptosis; Fig 5D;Ollmann et al., 2000). Thus, although our data suggest centrosomes help maintain genome integrity, the cell death we observed does not appear to result from a p53-mediated DNA damage response.
Fig5. Acentrosomal cells accumulate excess DNA damage but apoptosis does not require p53 activity.
(A,C) Ap>GFP control discs, with a slight increase in DNA damage in the Gal4/GFP expressing compartment, as indicated by γH2Av staining (n=11). (B,C) ap>GFP driving sas-4 RNAi significantly increased γH2Av positive cells (n=11). Expressing p53DN[H159N] represses the apoptotic response to DNA damage induced by irradiation (D), but does not reduce apoptosis associated with sas-4 RNAi (E,G,H). Expressing p53DN alone does not increase apoptosis (F,H).
JNK Signaling Mediates Apoptosis in Acentrosomal Cells
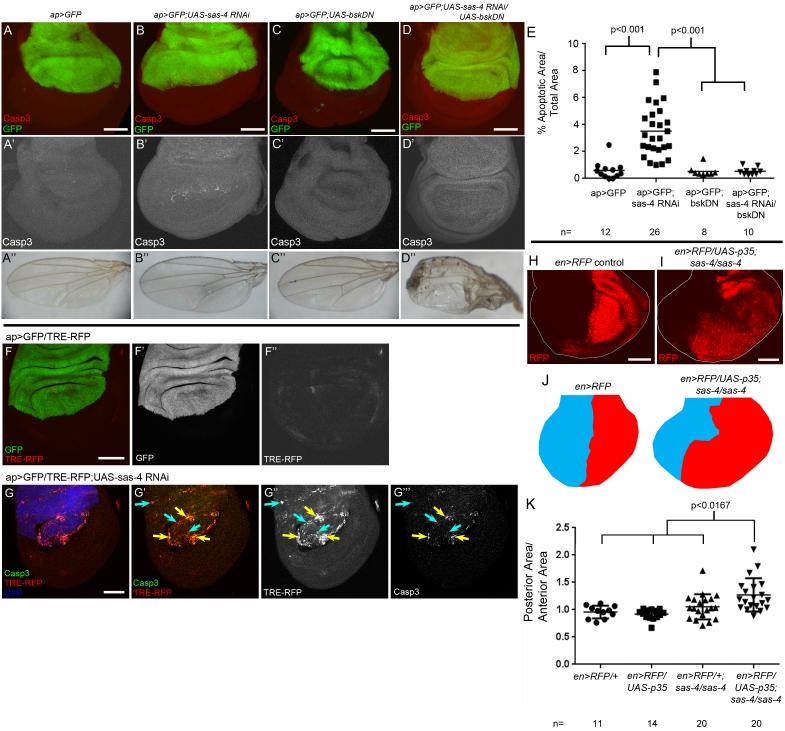
Multiple cellular insults (e.g. aneuploidy, DNA damage, cell stress) can lead to wing disc apoptosis by activating Jun Kinase signaling (JNK; Dekanty et al., 2012; Igaki, 2009; Kanda and Miura, 2004; McEwen and Peifer, 2005; Moreno et al., 2002). To determine if JNK signaling triggers apoptosis in acentrosomal cells, we used ap-Gal4 to coexpress sas-4 RNAi and a dominant negative form of fly JNK (Basket; Bsk). BskDN reduced apoptosis after sas-4 RNAi to WT levels (Fig 6D,E). Interestingly, adult wings of these flies were severely dysmorphic (Fig 6D”), whereas expression of sas-4 RNAi or BskDN alone had negligible effects on adult wings (Fig 6B”,C”). Overexpressing Puckered, a JNK negative regulator (Martin-Blanco et al., 1998), had similar effects (data not shown). We also examined JNK activity in acentrosomal cells by driving sas-4 RNAi in wing discs expressing the JNK reporter TRE-RFP (Chatterjee and Bohmann, 2012). Centrosome loss noticeably elevated JNK reporter expression overall, with highest levels in apoptotic cells (Fig 6F,G). These data indicate JNK signaling is the primary mediator of apoptosis in acentrosomal cells, and suggest that blocking JNK-dependent cell death disrupts wing development.
Fig6. JNK signaling mediates apoptosis in acentrosomal cells but tissue homeostasis is maintained by compensatory proliferation.
(A-E) Apoptosis caused by sas-4 RNAi (B,E) is completely repressed when JNK signaling is blocked via bskDN (D,E). Blocking JNK-induced apoptosis leads to highly defective adult wings (D”). (F) ap>GFP control disc expressing the JNK reporter TRE-RFP. Note minimal RFP expression (F”) and negligible apoptosis (see Fig.3A). (G) sas-4 RNAi dramatically increases TRE-RFP expression throughout the dorsal compartment (G”), however only a subset of cells appear to be undergoing apoptosis (G’,G”’). Blue arrows=JNK activation that does not overlap Casp3, yellow arrows=coincidence of Casp3 and reporter. GFP channel is false-colored blue in G. (H,I,K) Blocking apoptosis in the posterior compartment of sas-4 mutant discs significantly increases posterior compartment size (ratio of posterior relative to anterior area versus control). Quantified in (K) using t-test, Bonferroni correction for multiple comparisons set α=0.0167). (J) Diagrammatic model. Posterior compartment (red) and anterior (blue) for en>RFP (control) and en>RFP/UAS-p35;sas-4/sas-4 discs (based on image I). See also Fig S6.
Apoptosis Caused by Centrosome Loss Triggers Compensatory Proliferation
Despite the significant increase in apoptosis in flies lacking centrioles, they develop to adulthood with only modest defects in adult structures, suggesting developmental processes help buffer flies against the loss of substantial numbers of cells. One candidate for maintaining tissue homeostasis is compensatory proliferation. During this process, dying cells may activate JNK signaling, increasing the proliferation rate of neighboring cells to compensate for the loss of dying cells and maintains tissue integrity (Perez-Garijo et al., 2004; Ryoo et al., 2004). To determine if compensatory proliferation occurs in sas-4 mutant discs, we first examined the proliferative index. Using EdU incorporation as an indicator of S phase, we found that sas-4 mutant discs had significantly more proliferating cells than WT (15.5±2.6 cells/mm2, n=19 discs; p=0.007 vs WT=12.6±2.9 cells/mm2, n=12). In addition, as noted above, JNK signaling is active in acentrosomal cells (Fig 6G), and is at its highest levels in those cells undergoing apoptosis. Compensatory proliferation also causes developmental delay, increasing time to eclosion (Simpson et al., 1980; Stieper et al., 2008). Consistent with this, homozygous sas-4 mutants required ~1 extra day to complete development (mean days to eclosion=13.4 days for sas-4 vs. 12.2 days for heterozygous siblings; it is possible that the sensory defects of sas-4 mutants might also promote developmental delay; Ainsley et al., 2008; Basto et al., 2006). Together these data suggest that wing discs undergo compensatory proliferation to counter the cell death associated with centrosome loss.
Another assay for compensatory proliferation is to block apoptosis, commonly using p35, to determine if hyperplastic growth occurs. p35 blocks the effector caspase Drice but allows the upstream caspase Dronc to remain active (Martin et al., 2009). This effectively traps apoptotic cells in an “undead” state, continuously providing JNK-induced mitogenic signals such as Wg and Dpp to neighboring cells, driving tissue overgrowth (Kondo et al., 2006; Martin et al., 2009; Perez-Garijo et al., 2004; Perez-Garijo et al., 2009; Ryoo et al., 2004). We blocked apoptosis in the posterior compartment of sas-4 homozygous mutant wing discs using en>RFP to drive UAS-p35. If compensatory proliferation occurs in sas-4 mutant discs, we would expect tissue overgrowth. Consistent with this, posterior compartments of sas-4 mutant wing discs underwent hyperplastic growth when apoptosis was blocked (Fig 6I,K). Expressing p35 with en-Gal4 in sas-4 mutants resulted in pupal lethality, although p35 expression alone is not lethal; this precluded examining adult wings. To circumvent lethality, we used MS1096-Gal4 to co-express p35 and sas-4 RNAi. Expression of either alone had little effect on adult wings, but blocking the apoptosis induced by sas-4 RNAi resulted in severely dysmorphic adult wings (Fig S6), indicating that undead sas-4 mutant cells significantly disrupted development. In addition, Wg levels dramatically increased when apoptosis was blocked in sas-4 mutant cells (Fig S6). While these data are consistent with compensatory proliferation, recent work revealed considerable complexity in the response of imaginal disc cells to blocking apoptosis, with compensatory proliferation being one of several distinct responses (Mollereau et al., 2013; Morata et al., 2011; Perez-Garijo et al., 2009), and thus drawing a simple conclusion from the hyperplastic growth is problematic—we address this in the Discussion.
Centrosomes are key players in orienting symmetric divisions in epithelia
Epithelial cells divide symmetrically with a stereotyped orientation, parallel to the plane of the epithelium. Recent work revealed that disrupting spindle orientation in wing discs can lead to apoptosis (Nakajima et al., 2013). Centrosomes play a central role in orienting mitotic spindles during asymmetric divisions of certain stem cells (Yamashita, 2009a), among them fly NBs, one of the best models of this process. Interestingly, ~70% of NBs lacking centrosomes still divide with the correct orientation (Basto et al., 2006), suggesting centrosomes are partially but not completely dispensable for orienting NB spindles. The importance of centrosomes in symmetric divisions of epithelial cells has not been examined.
Wing disc cells divide parallel to the plane of the tissue (Fig 7I,J;Nakajima et al., 2013). Disrupting known players in spindle orientation, e.g. Mud (=human NuMA), results in some cells dividing at more random angles relative to the epithelial sheet (Nakajima et al., 2013), after which the basal daughter cell undergoes apoptosis. Since flies without centrosomes develop to adulthood, this might suggest mechanisms orienting mitotic spindles function efficiently in the absence of centrosomes. Alternatively, the elevated apoptosis in acentrosomal cells could suggest misoriented spindles as a mechanism by which centrosome loss leads to apoptosis.
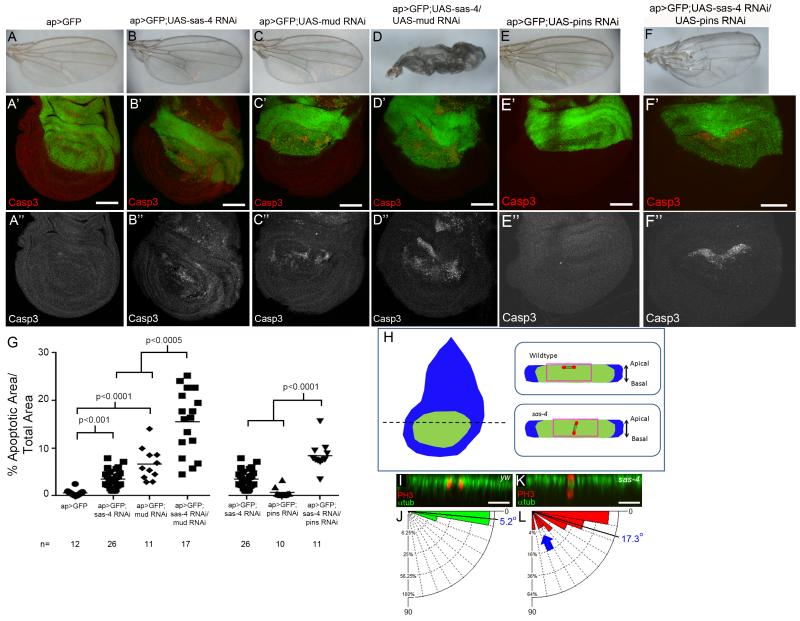
Fig7. Centrosomes and polarity cues both help maintain spindle orientation.
(A,G) Minimal apoptosis in control ap>GFP discs. (B,G) Single RNAi against sas-4 or the spindle orientation pathway component mud (C) elevate apoptosis, but have no appreciable effects on adult wings. (D,G) Co-expressing sas-4 and mud RNAi significantly increased apoptosis and severely disrupted adult wings. (E) RNAi against pins does not increase apoptosis. (F) Apoptosis increased after Sas-4/Pins double knockdown. This also perturbs adult wing development. (H) Model of en face view of wing disc. Spindle orientation analyzed in the pouch (green). Dotted line=Example of location of reconstructed cross-sectional views in I,K. Right=Diagrammatic examples (anaphase nuclei, red; MT in black; Apicobasal polarity indicated). Pink boxes=close-ups in I,K. (I,J) In WT discs, anaphase nuclei (PH3, red) divide in parallel with the epithelial sheet (n=43). (K,L) In sas-4 mutants, we found an increased mean and expanded distribution of division angles (blue arrow; n=34). See also Fig S3,S7. Scale bars A-F=50 μm; I,K=10μm.
To distinguish these hypotheses, we examined genetic interactions between sas-4 and the spindle orientation pathway proteins mud and pins (Morin and Bellaiche, 2011). RNAi against sas-4 alone modestly increased apoptosis (Fig 7B’,G), with no appreciable defects in adult wings (Fig 7B). In agreement with previous work, mud RNAi alone elevated levels of apoptosis (Fig 7C’,G;Nakajima et al., 2013), whereas pins RNAi did not increase apoptosis (Fig 7E’,G; we confirmed knockdown of both targets; Fig S3B,C). Knocking down Mud or Pins alone did not disrupt adult wings (Fig 7C,E). Strikingly, codepleting Sas-4 and either Mud or Pins substantially increased apoptosis and led to dysmorphic wings (Fig 7D,F,G), as did codepleting Mud and either Asl or Sas-6 (both required for centriole duplication; Fig S7A-F;Rodrigues-Martins et al., 2008), or codepleting Sas-4 and Mud using a second mud RNAi line (Fig S7G-K). These data are consistent with the hypothesis that centrosomes help orient mitotic spindles in wing discs. In addition, since JNK signaling mediates apoptosis in acentrosomal wing cells, if defects in spindle orientation underlie this increased apoptosis, then the apoptosis associated with disrupted spindle orientation should also be mediated by JNK. We thus blocked JNK signaling (using bskDN) in cells with compromised spindle orientation (mud RNAi). Consistent with JNK-mediated apoptosis, the apoptosis associated with Mud knockdown was completely blocked by expressing bskDN (Fig S7L).
To directly test the hypothesis that centrosomes help orient spindles, we analyzed spindle orientation in WT and sas-4 wing discs. As in previous work (Nakajima et al., 2013), we found that virtually all WT cells divided in the plane of the sheet (mean=5.2±3.8°, n=43;Fig 7I,J). In sas-4 discs, while most cells divided in the plane, a significant fraction divided more randomly relative to the sheet (Fig 7K,L;mean=17.3±19.6°, n=34; p=0.0002 versus WT); 15% divided at angles >40°, while all WT cells divided at angles <20°. Together these data indicate that centrosomes play an important role in orienting symmetric divisions of wing disc cells, and suggest that at least some of the apoptosis associated with acentrosomal mitosis is attributable to spindle orientation defects.
Discussion
Given the textbook view that centrosomes are critical for efficient spindle assembly, it was a significant surprise to find that flies survived to adulthood without centrosomes (Basto et al., 2006; Megraw et al., 2001; centrosomes do play critical roles in rapid mitotic divisions of early embryos and spermatid meiosis; Rodrigues-Martins et al., 2008; Stevens et al., 2007; Varmark et al., 2007). This led to the conclusion that fly somatic cells do not require centrosomes for cell division. We re-examined how somatic cells and whole animals respond to loss of this key cellular organelle, revealing new insights into the importance of centrosomes and the regulation of mitotic fidelity.
Centrosomes are essential for efficient spindle assembly in fly epithelia
We found that centrosomes promote timely spindle formation, mitotic progression, and spindle orientation in wing epithelia, with an ~15 fold increase in apoptosis in acentriolar cells. Because fly epithelia lack primary cilia, cell death does not result from perturbing ciliogenesis. Furthermore, because centrosomes do not nucleate interphase MTs in fly somatic cells (Rogers et al., 2008), apoptosis likely results from mitotic spindle defects. Indeed, spindles in acentrosomal cells are significantly compromised in efficiency of assembly, fidelity of chromosome segregation, and ability to orient within the tissue.
Spindle assembly defects can lead to aneuploidy, DNA damage, and delayed mitosis (Nicholson and Cimini, 2011), each of which can initiate cell death, as can spindle orientation defects (Nakajima et al., 2013; it will be interesting to determine if spindle misorientation also affects other aspects of mitosis, perhaps activating the SAC). Our data support a model in which centrosome loss has multiple impacts on mitotic fidelity. Anaphase onset is significantly delayed, likely reflecting prolonged SAC activity due to the slower process of chromatin-mediated spindle assembly. Unfortunately, we could not visualize MT-kinetochore connections in the small cells of the wing, but we hope new reagents and microscopy techniques will make this possible. Consistent with an important role for centrosomes in spindle assembly, simultaneously reducing centrosome function and the Augmin or Ran pathways or the SAC further increased cell death. Furthermore, centrosome loss led to significant increases in chromosome segregation errors, both lagging chromosomes and aneuploidy, and codepleting Augmin or the SAC even more dramatically increased mitotic error rates, helping explain the concomitant increases in apoptosis. Thus fly somatic cells have multiple, semi-redundant mechanisms to assemble spindles, and a robust checkpoint to prevent anaphase until this occurs. Cultured chick somatic DT40 cells also depend on centrosomes for proper mitosis—acentriolar DT40 cells had increased apoptosis, prolonged anaphase entry, and increased aneuploidy and lagging chromosomes (Sir et al., 2013), all of which we observed in acentriolar fly cells (though increased aneuploidy was only seen after inhibiting apoptosis). Our in vivo data reinforce these findings from cell culture, demonstrating that centrosomes play critical roles in mitosis in somatic cells of both species.
Centrosome loss triggers JNK dependent apoptosis and tissues respond by compensatory proliferation
What then triggers apoptosis in the absence of centrosomes? We found at least four potential contributing factors: mitotic delay, DNA damage, aneuploidy, and spindle misorientation. Delayed mitosis due to prolonged SAC activity can lead to cell death (Mollinedo and Gajate, 2003; Rieder and Maiato, 2004), potentially driving apoptosis in acentrosomal wing cells. Second, DNA damage is elevated in cells without centrosomes, and this could trigger apoptosis (Roos and Kaina, 2013). However, we do not think DNA damage is the sole contributor since the fraction of cells with obvious DNA damage was substantially lower than those undergoing apoptosis, and apoptosis does not appear to require p53 (JNK signaling may mediate a p53-independent DNA damage response). Interestingly, mouse embryos lacking centrioles have high levels of p53-dependent apoptosis-however this is without obvious DNA damage, suggesting mitotic delay activates p53 (Bazzi and Anderson, 2014). It will be important to determine precisely how centrosome loss leads to increased DNA damage in flies. Mitotic errors can cause DNA damage in many ways, including chromosome segregation errors and delayed mitotic progression (Ganem and Pellman, 2012), both of which occur in acentrosomal wing cells. Third, aneuploidy can also initiate apoptosis in wing discs (Dekanty et al., 2012), and centrosome loss elevated aneuploidy in wing cells. Importantly, this only occurred when we blocked apoptosis, suggesting aneuploidy in acentrosomal cells is probably an important cause of apoptosis. Finally, defects in planar spindle orientation could contribute, though the frequency of substantial orientation errors was again lower than that of apoptosis, suggesting this also is not the sole cause. Thus we hypothesize that multiple factors combine to lead to the elevated rate of apoptosis resulting from centrosome loss. At the level of an individual cell, it is not clear whether apoptosis is induced by a single, strong defect in one process, or the combined effect of errors in multiple events that together push the cell beyond some threshold (presumably of JNK activity). It will be interesting to distinguish these hypotheses.
This range of mitotic defects led to JNK-mediated apoptosis. JNK signaling is activated in response to many cellular insults, including DNA damage, aneuploidy, disrupted cell polarity, and spindle misorientation (Dekanty et al., 2012; Igaki, 2009; Kanda and Miura, 2004;this study). However, it is not known precisely how JNK is activated in response to this wide range of cellular defects. In some contexts (e.g. loss of cell polarity or apoptosis-induced apoptosis;Igaki et al., 2009; Perez-Garijo et al., 2013), this involves the TNF ligand Eiger, while in other circumstances (e.g. morphogen gradient discontinuities or DNA damage) the mechanism of JNK activation remains unclear (Igaki, 2009). We tested the potential role of Eiger in our system by coexpressing sas-4 RNAi and eiger RNAi, using previously characterized eiger lines (Perez-Garijo et al., 2013; Rhiner et al., 2010). We saw no reduction in apoptosis and no adult wing phenotypes (data not shown), in contrast to coexpressing sas-4 RNAi and bskDN. Thus, it does not appear that centrosome loss activates JNK signaling through Eiger. Apoptosis itself can activate JNK signaling (Kuranaga et al., 2002; Shlevkov and Morata, 2012), but we do not believe this is the sole cause in acentrosomal cells, as the JNK reporter is upregulated in most cells expressing sas-4 RNAi, though only subset of these cells undergo apoptosis (Fig 6G). This is consistent with a model in which JNK is activated at lower levels by diverse mitotic errors to promote repair mechanisms, but at higher levels of JNK activation cells enter apoptosis. It is not clear precisely when JNK signaling is activated in response to mitotic errors caused by centrosome loss—some cells may respond to certain mitotic defects by activating JNK well after the defect has actually occurred, perhaps even as a feedback response to initiating apoptosis. It will be important to identify the mechanism(s) by which JNK is activated in response to mitotic defects of acentrosomal cells.
Despite the ability of alternative pathways and checkpoints to ameliorate problems inherent in acentrosomal mitosis, many cells still die, but several days later near-normal flies eclose. In the face of dramatically increased cell death, our data are consistent with the hypothesis that the tissue responds through compensatory proliferation triggered by JNK. We also noted hyperproliferation after blocking cell death, but our p35 experiments are subject to several caveats. First, p35 allows Dronc to remain active, which itself can activate JNK (Kondo et al., 2006; Martin et al., 2009); thus undead cells may lead to continuous JNK activation downstream of Dronc, driving Wg and Dpp expression and tissue growth. This apoptosis-induced proliferation is distinct from compensatory proliferation (Mollereau et al., 2013; Morata et al., 2011; Perez-Garijo et al., 2009). Second, Wg and Dpp are not required for compensatory proliferation in response to radiation-induced apoptosis (Perez-Garijo et al., 2009)—it will be important to test whether Wg is important for the response to centrosome loss. Intriguingly, blocking apoptosis in aneuploid wing cells also led to hyperplastic growth (Dekanty et al., 2012). Importantly, in this situation, blocking cell death through dronc loss, which should prevent apoptosis-induced JNK activation, also led to hyperplastic growth. Thus the hyperplastic growth response to blocking apoptosis associated with aneuploidy may be distinct from apoptosis-induced proliferation. Further, undead aneuploid cells dramatically increased Wg expression and Wg was involved in the hyperplastic growth response (Dekanty et al., 2012). This suggests that the hyperplastic growth in undead sas-4 cells maybe a consequence of the aneuploid cells remaining present in the tissue. Thus, although our data on whole are consistent with compensatory proliferation in sas-4 mutant discs, there are alternative interpretations of the undead-based experiments, including apoptosis-induced proliferation or hyperplastic growth derived from persistence of aneuploid cells.
In epithelia and neuroblasts, centrosomes serve similar roles with dissimilar consequences
Much of the previous in vivo analysis of acentrosomal cells focused on fly NBs as a model. It is of interest to compare and contrast these studies to our findings in epithelia. In both, centrosomes play important but non-essential roles in spindle orientation. In asymmetric NB divisions, disrupting spindle orientation affects cell fate decisions of daughter cells (Yamashita, 2009b). In the symmetric divisions of epithelia, however, our data and others suggest spindle orientation is critical in maintaining cell viability (Nakajima et al., 2013). In both tissues, other polarity cues also contribute to spindle orientation (Basto et al., 2006; Guilgur et al., 2012; Nakajima et al., 2013). How do most acentrosomal spindles find their proper orientation? As was suggested in NBs (Basto et al., 2006), acentrosomal spindle MTs may reach the cortex and interact with the spindle orientation machinery (e.g. Mud, Pins). Alternatively, centrosomes and the spindle orientation pathway may act in parallel, where centrosomes read some inherent cortical polarity to guide spindle positioning (e.g. junctional complexes). If so, it will be important to identify these cues. Regardless, when both the canonical orientation pathway and centrosomes are impaired, the consequences for cell viability are exaggerated, once again providing an example of system robustness.
Both wing discs and NBs rely on centrosomes for timely mitotic spindle formation. Although the cause of the NB delay was not directly tested, we found the mitotic delay in epithelia likely reflects SAC activity. In strong contrast to epithelia, centrosome loss does not increase NB cell death (Basto et al., 2006). Thus, tissues can differ dramatically in their response to centrosome loss, with NBs more refractory to loss, whereas epithelial cells appear very sensitive. This may reflect dramatic differences in the nature of these tissues. Larval brains contain a defined number of identifiable NBs, each giving rise to particular cell types (Doe, 2008)—thus losing a single NB could lead to adult brain defects. In contrast, larval wing epithelial cells are simply part of a pool of cells ultimately giving rise to a wing, with individual cells largely dispensable. Since centrosome dysfunction is implicated in cancer, and many cancers are of epithelial origin, wing disc epithelia may be a relevant model for evaluating centrosome function and its potential links to tumorigenesis. It will be important to continue to investigate differences and similarities of centrosome function in different cell types, as well as how cells and tissues respond to the consequences of centrosome loss.
Methods
Genetics
Fly stocks and their sources are included in the Supplemental Experimental Procedures.
Immunocytochemistry
Antibody staining was as in Roberts et al., (2012). Antibodies: cleaved Caspase3 (1:200, Cell Signaling), PH3 (1:1000, Millipore), αtub (1:2000, Sigma), γH2Av (1:2000 J. Sekelsky), Wg (1:50, DSHB), MMP1 (1:50, DSHB), Dlg (1:100, DSHB), Lamin (1:500, DSHB), aPKC (1:1000, Santa Cruz), Sas-4 (1:100), PLP (1:1000, gifts of N. Rusan), Mud (1:200), and Pins (1:1000, F. Matsuzaki). Alexa secondary antibodies were used at 1:500. Confocal images were acquired on a Zeiss Pascal microscope. PhotoshopCS4 (Adobe) was used to adjust levels so the range of signals spanned the entire output grayscale and to adjust brightness and contrast. Live imaging was performed on a Nikon TE2000-E microscope with Visitech Infinity-Hawk multi-point array scanner with 100× Nikon objective, a Ludl emission filter wheel with Semrock filters, and Hamamatsu ORCA-R2 camera. Movies were assembled in ImageJ.
Quantifying apoptosis
Due to the high levels of apoptosis in many genotypes, it was not possible to distinguish individual dying cells. We therefore measured the area of Casp3 signal (from maximum intensity projections) in the hinge and pouch region of 3rd instar wing discs and standardized that by the total area of hinge and pouch. In experiments with Gal-4, these measures were done separately in Gal-4 expressing and non-expressing areas. For simplicity, we did not present data from non-Gal-4 expressing areas, except where otherwise noted. Statistical comparisons used Student’s t-test (Excel). Exposure to 3000rads of γ-irradiation was used to confirm block of DNA damage-induced apoptosis by p53DN.
Proliferative Index
EdU labeling: Discs were incubated 15’ in Schneider’s media with 100ug/mL EdU, washed in PBS and fixed 20’ in 4% formaldehyde. We completed EdU detection as in the manufacturer’s protocol (Click-It EdU Imaging Kit, Invitrogen). To standardize measurements we drew a circle of 2mm2 in the center of the wing pouch and counted EdU positive cells; dividing by 2 yielded EdU+ cells/mm2. Statistical comparison was based on Student’s t-test (Excel).
Aneuploidy
We directly scored chromosome content of wing disc cells using chromosome squashes following a published protocol (Morais da Silva et al., 2013).
Spindle orientation
We stained wing discs for PH3 and α-tub, and acquired z-stacks at 1μm intervals. Using the Zeiss LSM Browser software, we identified anaphase nuclei in the pouch region by PH3 signal and the presence of bundled midzone MTs, extracted the region of the image stack containing these nuclei, and generated a 3D projection along the Z axis. We rotated the projection to select the frame maximizing the distance between nuclei and imported that frame into ImageJ. We then used the Angle Tool to draw one leg parallel to the plane of the epithelial sheet (using apical accumulation of α-tub as a marker of sheet orientation), with the vertex of the angle in the center of the more apical nucleus, and the other leg drawn through the center of the basal nucleus. Angles were graphed using Oriana4 software and compared by Student’s t-test (Excel).
Supplementary Material
Acknowledgements
We thank N Rusan, D Fox, R Karess, J Sekelsky, Y Tamori, D Bohmann, F Matsuzaki, B Duronio, and T Avidor-Reiss, Bloomington Stock Center, Developmental Studies Hybridoma Bank, and Vienna Drosophila Resource Center for reagents, and T Perdue and S Crews for assistance in microscopy. The work was supported by NIH R01GM067236. JSP was supported by an NIH NRSA and JCC by a UNC SURF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ainsley JA, Kim MJ, Wegman LJ, Pettus JM, Johnson WA. Sensory mechanisms controlling the timing of larval developmental and behavioral transitions require the Drosophila DEG/ENaC subunit, Pickpocket1. Dev Biol. 2008;322:46–55. doi: 10.1016/j.ydbio.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Bazzi H, Anderson KV. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc Natl Acad Sci USA. 2014;111:E1491–1500. doi: 10.1073/pnas.1400568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachon S, Gopalakrishnan J, Omori Y, Polyanovsky A, Church A, Nicastro D, Malicki J, Avidor-Reiss T. Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics. 2008;180:2081–2094. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- Buffin E, Emre D, Karess RE. Flies without a spindle checkpoint. Nat Cell Biol. 2007;9:565–572. doi: 10.1038/ncb1570. [DOI] [PubMed] [Google Scholar]
- Cenci G. Drosophila cell cycle under arrest: uncapped telomeres plead guilty. Cell Cycle. 2009;8:990–995. doi: 10.4161/cc.8.7.7960. [DOI] [PubMed] [Google Scholar]
- Chatterjee N, Bohmann D. A versatile PhiC31 based reporter system for measuring AP-1 and Nrf2 signaling in Drosophila and in tissue culture. PLoS One. 2012;7:e34063. doi: 10.1371/journal.pone.0034063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol. 2008;9:464–477. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino PP, Archambault V, Przewloka MR, Zhang W, Lilley KS, Laue E, Glover DM. Recruitment of Polo kinase to the spindle midzone during cytokinesis requires the Feo/Klp3A complex. PLoS One. 2007;2:e572. doi: 10.1371/journal.pone.0000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekanty A, Barrio L, Muzzopappa M, Auer H, Milan M. Aneuploidy-induced delaminating cells drive tumorigenesis in Drosophila epithelia. Proc Natl Acad Sci USA. 2012;109:20549–20554. doi: 10.1073/pnas.1206675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Dumont J, Desai A. Acentrosomal spindle assembly and chromosome segregation during oocyte meiosis. Trends Cell Biol. 2012;22:241–249. doi: 10.1016/j.tcb.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Pellman D. Linking abnormal mitosis to the acquisition of DNA damage. J Cell Biol. 2012;199:871–881. doi: 10.1083/jcb.201210040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- Goshima G, Kimura A. New look inside the spindle: microtubule-dependent microtubule generation within the spindle. Curr Opin Cell Biol. 2010;22:44–49. doi: 10.1016/j.ceb.2009.11.012. [DOI] [PubMed] [Google Scholar]
- Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilgur LG, Prudencio P, Ferreira T, Pimenta-Marques AR, Martinho RG. Drosophila aPKC is required for mitotic spindle orientation during symmetric division of epithelial cells. Development. 2012;139:503–513. doi: 10.1242/dev.071027. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hayashi MT, Karlseder J. DNA damage associated with mitosis and cytokinesis failure. Oncogene. 2013;32:4593–4601. doi: 10.1038/onc.2012.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward D, Metz J, Pellacani C, Wakefield JG. Synergy between multiple microtubule-generating pathways confers robustness to centrosome-driven mitotic spindle formation. Dev Cell. 2014;28:81–93. doi: 10.1016/j.devcel.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta T, Kong Z, Ho CM, Zeng CJ, Horio T, Fong S, Vuong T, Lee YR, Liu B. Characterization of the Arabidopsis augmin complex uncovers its critical function in the assembly of the acentrosomal spindle and phragmoplast microtubule arrays. Plant Cell. 2012;24:1494–1509. doi: 10.1105/tpc.112.096610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaki T. Correcting developmental errors by apoptosis: lessons from Drosophila JNK signaling. Apoptosis. 2009;14:1021–1028. doi: 10.1007/s10495-009-0361-7. [DOI] [PubMed] [Google Scholar]
- Igaki T, Pastor-Pareja JC, Aonuma H, Miura M, Xu T. Intrinsic tumor suppression and epithelial maintenance by endocytic activation of Eiger/TNF signaling in Drosophila. Dev Cell. 2009;16:458–465. doi: 10.1016/j.devcel.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- Jin S, Martinek S, Joo WS, Wortman JR, Mirkovic N, Sali A, Yandell MD, Pavletich NP, Young MW, Levine AJ. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:7301–7306. doi: 10.1073/pnas.97.13.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda H, Miura M. Regulatory roles of JNK in programmed cell death. J Biochem. 2004;136:1–6. doi: 10.1093/jb/mvh098. [DOI] [PubMed] [Google Scholar]
- Kondo S, Senoo-Matsuda N, Hiromi Y, Miura M. DRONC coordinates cell death and compensatory proliferation. Mol Cell Biol. 2006;26:7258–7268. doi: 10.1128/MCB.00183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuranaga E, Kanuka H, Igaki T, Sawamoto K, Ichijo H, Okano H, Miura M. Reaper-mediated inhibition of DIAP1-induced DTRAF1 degradation results in activation of JNK in Drosophila. Nat Cell Biol. 2002;4:705–710. doi: 10.1038/ncb842. [DOI] [PubMed] [Google Scholar]
- Lecland N, Debec A, Delmas A, Moutinho-Pereira S, Malmanche N, Bouissou A, Dupre C, Jourdan A, Raynaud-Messina B, Maiato H, et al. Establishment and mitotic characterization of new Drosophila acentriolar cell lines from DSas-4 mutant. Biol Open. 2013;2:314–323. doi: 10.1242/bio.20133327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Jarman AP. Dilatory is a Drosophila protein related to AZI1 (CEP131) that is located at the ciliary base and required for cilium formation. J Cell Sci. 2011;124:2622–2630. doi: 10.1242/jcs.084798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan JP, Chotkowski HL, Glaser RL. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 2002;30:3698–3705. doi: 10.1093/nar/gkf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, Martinez-Arias A. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 1998;12:557–570. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FA, Perez-Garijo A, Morata G. Apoptosis in Drosophila: compensatory proliferation and undead cells. Int J Dev Biol. 2009;53:1341–1347. doi: 10.1387/ijdb.072447fm. [DOI] [PubMed] [Google Scholar]
- McEwen DG, Peifer M. Puckered, a Drosophila MAPK phosphatase, ensures cell viability by antagonizing JNK-induced apoptosis. Development. 2005;132:3935–3946. doi: 10.1242/dev.01949. [DOI] [PubMed] [Google Scholar]
- Megraw TL, Kao LR, Kaufman TC. Zygotic development without functional mitotic centrosomes. Curr Biol. 2001;11:116–120. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- Megraw TL, Li K, Kao LR, Kaufman TC. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 1999;126:2829–2839. doi: 10.1242/dev.126.13.2829. [DOI] [PubMed] [Google Scholar]
- Megraw TL, Sharkey JT, Nowakowski RS. Cdk5rap2 exposes the centrosomal root of microcephaly syndromes. Trends Cell Biol. 2011;21:470–480. doi: 10.1016/j.tcb.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollereau B, Perez-Garijo A, Bergmann A, Miura M, Gerlitz O, Ryoo HD, Steller H, Morata G. Compensatory proliferation and apoptosis-induced proliferation: a need for clarification. Cell Death Differ. 2013;20:181. doi: 10.1038/cdd.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinedo F, Gajate C. Microtubules, microtubule-interfering agents and apoptosis. Apoptosis. 2003;8:413–450. doi: 10.1023/a:1025513106330. [DOI] [PubMed] [Google Scholar]
- Morais da Silva S, Moutinho-Santos T, Sunkel CE. A tumor suppressor role of the Bub3 spindle checkpoint protein after apoptosis inhibition. J Cell Biol. 2013;201:385–393. doi: 10.1083/jcb.201210018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morata G, Shlevkov E, Perez-Garijo A. Mitogenic signaling from apoptotic cells in Drosophila. Dev Growth Differ. 2011;53:168–176. doi: 10.1111/j.1440-169X.2010.01225.x. [DOI] [PubMed] [Google Scholar]
- Moreno E, Basler K, Morata G. Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature. 2002;416:755–759. doi: 10.1038/416755a. [DOI] [PubMed] [Google Scholar]
- Morin X, Bellaiche Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell. 2011;21:102–119. doi: 10.1016/j.devcel.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Nakajima Y, Meyer EJ, Kroesen A, McKinney SA, Gibson MC. Epithelial junctions maintain tissue architecture by directing planar spindle orientation. Nature. 2013;500:359–362. doi: 10.1038/nature12335. [DOI] [PubMed] [Google Scholar]
- Nakaoka Y, Miki T, Fujioka R, Uehara R, Tomioka A, Obuse C, Kubo M, Hiwatashi Y, Goshima G. An inducible RNA interference system in Physcomitrella patens reveals a dominant role of augmin in phragmoplast microtubule generation. Plant Cell. 2012;24:1478–1493. doi: 10.1105/tpc.112.098509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JM, Cimini D. How mitotic errors contribute to karyotypic diversity in cancer. Adv Cancer Res. 2011;112:43–75. doi: 10.1016/B978-0-12-387688-1.00003-X. [DOI] [PubMed] [Google Scholar]
- Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, Buchman A, et al. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell. 2000;101:91–101. doi: 10.1016/S0092-8674(00)80626-1. [DOI] [PubMed] [Google Scholar]
- Perez-Garijo A, Fuchs Y, Steller H. Apoptotic cells can induce non-autonomous apoptosis through the TNF pathway. Elife. 2013;2:e01004. doi: 10.7554/eLife.01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Garijo A, Martin FA, Morata G. Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development. 2004;131:5591–5598. doi: 10.1242/dev.01432. [DOI] [PubMed] [Google Scholar]
- Perez-Garijo A, Shlevkov E, Morata G. The role of Dpp and Wg in compensatory proliferation and in the formation of hyperplastic overgrowths caused by apoptotic cells in the Drosophila wing disc. Development. 2009;136:1169–1177. doi: 10.1242/dev.034017. [DOI] [PubMed] [Google Scholar]
- Poulton JS, Mu FW, Roberts DM, Peifer M. APC2 and Axin promote mitotic fidelity by facilitating centrosome separation and cytoskeletal regulation. Development. 2013;140:4226–4236. doi: 10.1242/dev.094425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhiner C, Lopez-Gay JM, Soldini D, Casas-Tinto S, Martin FA, Lombardia L, Moreno E. Flower forms an extracellular code that reveals the fitness of a cell to its neighbors in Drosophila. Dev Cell. 2010;18:985–998. doi: 10.1016/j.devcel.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell. 2004;7:637–651. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Roberts DM, Pronobis MI, Alexandre KM, Rogers GC, Poulton JS, Schneider DE, Jung KC, McKay DJ, Peifer M. Defining components of the ß-catenin destruction complex and exploring its regulation and mechanisms of action during development. PLoS One. 2012;7:e31284. doi: 10.1371/journal.pone.0031284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. From centriole biogenesis to cellular function: centrioles are essential for cell division at critical developmental stages. Cell Cycle. 2008;7:11–16. doi: 10.4161/cc.7.1.5226. [DOI] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Peifer M, Rogers SL. A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol Biol Cell. 2008;19:3163–3178. doi: 10.1091/mbc.E07-10-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos WP, Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013;332:237–248. doi: 10.1016/j.canlet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell. 2004;7:491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Shlevkov E, Morata G. A dp53/JNK-dependant feedback amplification loop is essential for the apoptotic response to stress in Drosophila. Cell Death Differ. 2012;19:451–460. doi: 10.1038/cdd.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P, Berreur P, Berreur-Bonnenfant J. The initiation of pupariation in Drosophila: dependence on growth of the imaginal discs. J Embryol Exp Morphol. 1980;57:155–165. [PubMed] [Google Scholar]
- Sir JH, Putz M, Daly O, Morrison CG, Dunning M, Kilmartin JV, Gergely F. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J Cell Biol. 2013;203:747–756. doi: 10.1083/jcb.201309038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Raposo AA, Basto R, St Johnston D, Raff JW. From stem cell to embryo without centrioles. Curr Biol. 2007;17:1498–1503. doi: 10.1016/j.cub.2007.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieper BC, Kupershtok M, Driscoll MV, Shingleton AW. Imaginal discs regulate developmental timing in Drosophila melanogaster. Dev Biol. 2008;321:18–26. doi: 10.1016/j.ydbio.2008.05.556. [DOI] [PubMed] [Google Scholar]
- Varmark H, Llamazares S, Rebollo E, Lange B, Reina J, Schwarz H, Gonzalez C. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr Biol. 2007;17:1735–1745. doi: 10.1016/j.cub.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Wainman A, Buster DW, Duncan T, Metz J, Ma A, Sharp D, Wakefield JG. A new Augmin subunit, Msd1, demonstrates the importance of mitotic spindle-templated microtubule nucleation in the absence of functioning centrosomes. Genes Dev. 2009;23:1876–1881. doi: 10.1101/gad.532209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Yamashita YM. The centrosome and asymmetric cell division. Prion. 2009a;3:84–88. doi: 10.4161/pri.3.2.8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM. Regulation of asymmetric stem cell division: spindle orientation and the centrosome. Front Biosci (Landmark Ed) 2009b;14:3003–3011. doi: 10.2741/3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Dawe RK. Mechanisms of plant spindle formation. Chromosome Res. 2011;19:335–344. doi: 10.1007/s10577-011-9190-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.