Abstract
DNA phosphorothioate (PT) modification is a recently identified epigenetic modification that occurs in the sugar-phosphate backbone of prokaryotic DNA. Previous studies have demonstrated that DNA PT modification is governed by the five DndABCDE proteins in a sequence-selective and R P stereo-specific manner. Bacteria may have acquired this physiological modification along with dndFGH as a restriction-modification system. However, little is known about the biological function of Dnd proteins, especially the smallest protein, DndE, in the PT modification pathway. DndE was reported to be a DNA-binding protein with a preference for nicked dsDNA in vitro; the binding of DndE to DNA occurs via six positively charged lysine residues on its surface. The substitution of these key lysine residues significantly decreased the DNA binding affinities of DndE proteins to undetectable levels. In this study, we conducted site-directed mutagenesis of dndE on a plasmid and measured DNA PT modifications under physiological conditions by mass spectrometry. We observed distinctive differences from the in vitro binding assays. Several mutants with lysine residues mutated to alanine decreased the total frequency of PT modifications, but none of the mutants completely eliminated PT modification. Our results suggest that the nicked dsDNA-binding capacity of DndE may not be crucial for PT modification and/or that DndE may have other biological functions in addition to binding to dsDNA.
Introduction
Phosphorothioate linkage was originally developed as an artificial means to stabilize oligonucleotides against nuclease degradation [1]. However, physiological DNA PT modification was recently discovered as a new type of epigenetic modification in which the nonbridging oxygen is replaced with sulfur [2]. Previous studies have shown that PT modification is governed by the products of the five clustered dndABCDE genes [3]. PT linkages are susceptible to Tris peroxide, which accumulates on the anode during conventional or pulsed-field gel electrophoresis, resulting in DNA degradation [3], [4]. DNA PT modification was first discovered in Streptomyces lividans, and it was later found to be widespread across a range of species due to the horizontal transfer of the dnd genes [3], [5], [6], [7]. The genomic mapping of PT sites across bacterial genomes revealed short consensus sequences and partial modifications at given sites [8]. Moreover, a three-gene cluster, dndFGH, which is located adjacent to the dndBCDE in approximately 86 bacterial species constitutes a restriction-modification system to restrict foreign DNA. This system displays similarities to methylation-based R-M systems [9], [10], [11], [12]. However, more than 100 strains possess only dndBCDE and lack dndFGH, suggesting that these genes may encode proteins with functions other than those of a typical R-M system.
An emerging model of Dnd protein functions hypothesizes that DndA is a cysteine desulfurase that catalyzes the removal of sulfur from L-cysteine and assembles DndC as a 4Fe-4S cluster protein. DndC is predicted to have PAPS reductase activity and possesses ATP pyrophosphatase activity [13]. DndB shows homology to a group of transcriptional regulators, whereas DndD has ATPase activity that is potentially related to DNA nicking during sulfur incorporation [3], [14]. DndE is the smallest Dnd protein and consists of 117 amino acids in both Salmonella enterica serovar Cerro 87 and Escherichia coli B7A. It is believed that the DNA phosphodiester linkage within the consensus sequence is nicked by DndD prior to sulfur replacement [14].
DndE is a tetrameric protein that shows a stronger binding affinity for dsDNA with a phosphorylated nick than for intact dsDNA [15]. Six positively charged lysine residues on the surface of DndE are predicted to be involved in the selective binding to negatively charged phosphorylated gaps. Hu et al. replaced the positively charged lysine residues on the DndE surface (K17, K18, K20, K53, K87 and K91) with alanine individually and then tested by binding affinity of the DndE mutants for DNA substrates in vitro. The DndE variants DndEK17A, DndEK20A, DndEK53A and DndEK91A showed 3- to 8-fold decreased binding affinities to intact dsDNA and displayed non-detectable binding to nicked dsDNA [15]. In particular, the mutagenesis of the K18 residue reduced the binding of DndE to both intact and nicked dsDNA below detectable levels [15].
To investigate the impact of the key lysine residues on DNA PT modifications under physiological conditions, we conducted site-directed mutagenesis of dndE in the plasmid pJTU1238, which contains the dndBCDE cluster from S. enterica and confers PT-modified d(GPSA) and d(GPST) to E. coli hosts [6]. The plasmid pJTU1238 and its derivatives were then transformed into E. coli DH10B and XTG102, the S. enterica dndBCDE deletion mutant. The abundance of DNA PT modifications was measured by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). We found that mutating the individual lysines in DndE altered the DNA PT modifications in several ways. However, none of the mutations completely blocked the PT modification pathway. Our results reveal that the disruption of the nicked dsDNA-binding activity of DndE may not be sufficient to abolish DNA PT modifications. Meanwhile, DndE may not be uniquely designed to interact with nicked dsDNA, and therefore may have other functions.
Materials and Methods
Bacterial strains, plasmids and primers
The bacterial strains, plasmids, and primers used in this study are listed in Table 1.
Table 1. Strains, plasmids and primers used in this study.
The in-frame deletion of dndE in pJTU1238
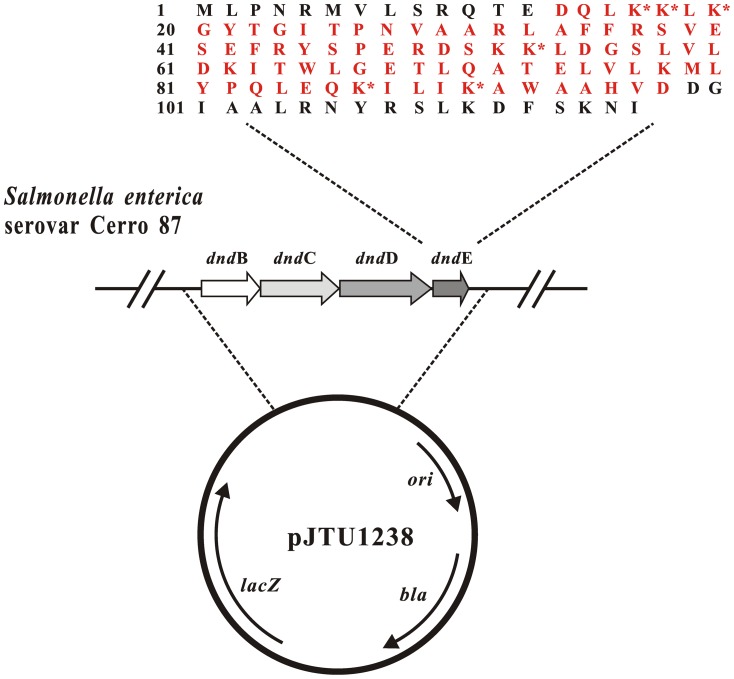
The in-frame deletion of dndE in pJTU1238 was constructed by two-step PCR. In the first step, the upstream and downstream fragments of dndE were generated by PCR using the primer pairs ΔdndE-1/ΔdndE-2 and ΔdndE-3/ΔdndE-4, respectively (Table 1). ΔdndE-2 and ΔdndE-3 are chimeric primers. A mixture of two purified PCR products, which overlapped by 40 bp, served as the template for a ligation PCR using primers ΔdndE-1 and ΔdndE-4. An 1,877-bp product containing the dndE deletion fragment was generated and cloned into the pEASY-Blunt Zero vector (TransGen Biotech), yielding plasmid pWHU675. This plasmid was confirmed by sequencing analysis. The NsiI-XhoI fragment of pWHU675 containing an in-frame deletion of dndE (corresponding to amino acid nos. 14 to 98; red in Figure 1) was inserted into the corresponding sites in pJTU1238 to generate the plasmid pWHU676.
Figure 1. Site-directed mutagenesis of the lysine residues of the dndE gene in pJTU1238.
The indicated mutations of key lysine residues (K17, K18, K20, K53, K87 and K91) were constructed in pJTU1238, a plasmid harboring the dndBCDE genes of S. enterica. The asterisks denote the selected lysine residues that were replaced with alanines. The residues marked in red indicate the amino acids that are deleted in the dndE in-frame deletion mutant.
Site-directed mutagenesis of dndE
The site-directed mutagenesis of the dndE gene on plasmid pJTU1238 was performed using the Muta-direct Site Directed Mutagenesis Kit (SBS Genetech). The primers were designed according to the manufacturer's instructions. The six positively charged lysine residues on the DndE surface (K17, K18, K20, K53, K87 and K91) were individually replaced with alanine (Figure 1). The mutations on pJTU1238 were verified by DNA sequencing using the primers dndE-Mu-F and dndE-Mu-R (Table 1). The derived plasmids were then transformed into XTG102, a S. enterica mutant strain with a deletion of dndBCDE, and E. coli DH10B to detect PT modifications by LC-MS/MS.
DNA digestion conditions
All strains were grown in LB medium to an OD600 of 0.8 at 28°C for Salmonella and 37°C for E. coli. DNA was isolated using the QIAGEN Genomic-tip 100/G kit. After isolation, 20 µg of DNA was hydrolyzed with 2 U of nuclease P1 (USBiological) in 30 mM sodium acetate, pH 5.3, and 0.5 mM ZnCl2 in a total volume of 100 µL at 50°C for 2 h. Following the addition of 10 µL of 1 M Tris-Cl (pH 8.0) to adjust the pH, dephosphorylation was performed by 4 U of alkaline phosphatase (Sigma) at 37°C for another 4 h. The enzymes were subsequently removed by passing the reaction mixture over a PALL Nanosep 10 K OMEGA centrifugal column, and the reaction was concentrated under a vacuum. Prior to the quantitative analysis, the external calibration curves containing varying amounts of d(GPSA) R P and d(GPST) R P standards (5, 10, 20, 40, 60, and 80 pmol) were plotted with 20 pmol of d(GPSA) S P as a reference. The correlation coefficients (r2) were greater than 0.999.
LC-MS/MS analysis of DNA PT modifications
The digested DNA samples were resolved by a Thermo Hypersil GOLD aQ column (150×2.1 mm, 3 µm) by elution at 35°C. The flow rate was set to 200 µL/min using a gradient starting at 97% buffer A (0.1% acetic acid in water) and 3% buffer B (0.1% acetic acid in acetonitrile) for 5 min, followed by a linear gradient to 6% buffer B over 30 min and 6% buffer B to 98% buffer B over 1 min. The buffer was held at 98% buffer B for another 15 min and then changed back to the starting concentration of 3% buffer B over 1 min, followed by a 15 min period to allow the column to re-equilibrate to the 3% buffer B. The column was coupled to a Thermo TSQ Quantum Access MAX mass spectrometer with an electrospray ionization source in positive mode. The selected reaction-monitoring (SRM) scan mode in MS/MS was employed to detect the following ions: d(GPSA) R P [M + H]+ m/z 597 → 136, d(GPST) R P, [M + H]+ m/z 588 → 152 and d(GPSA) S P [M + H]+ m/z 597 → 136. The following parameters were optimized for maximal sensitivity: gas flow, 10 L/min; nebulizer pressure, 25 psi; drying gas temperature, 300°C; capillary temperature, 320°C; source spray voltage, 3.5 kV; sheath gas setting, 40; aux gas setting, 8; capillary voltage, 35 V; and tube lens voltage, 110 V. The collision energy and collision gas pressure were chosen to achieve the lowest background noise.
Quantitative RT-PCR
To assess the expression levels of the dndE genes, total RNA was isolated using a Qiagen RNeasy Protect Bacteria Mini Kit. Samples of 1 µg total RNA were reverse transcribed to synthesize cDNA using RevertAid First Stand cDNA Synthesis kits (Thermo Scientific). Then, 20 ng of cDNA was used as a template for qualitative real-time PCR performed with the SsoFast EvaGreen Supermix with Low ROX Kit (Bio-Rad) and a 7900 HT Fast Real-Time PCR System (Applied Biosystems). An initial incubation at 95°C for 30 s was followed by 40 cycles of 95°C for 10 s and 58.5°C for 30 s. The housekeeping gene gapA, which encodes D-GAPDH, was used as a reference. The primers 5′- TGCTCCCGAATCGAATGGTA-3′ and 5′-AGCGAAACTCACTCTCCACT-3′ were used to amplify dndE, primers 5′-GAAGGCCAGGACATCGTTTC-3′ and 5′-AGTCGCGTGAACAGTAGTCA-3′ were used to amplify gapA from XTG102, and primers 5′-CACGCTACTACCGCTACTCA -3′ and 5′- AGGACGGGATGATGTTCTGG-3′ were used to amplify gapA from E. coli DH10B. The RT-PCR data analysis was performed according to the comparative threshold cycle method (also known as 2−ΔΔCT) between the different strains.
Results
Expression of pJTU1238 and its derivatives in E. coli
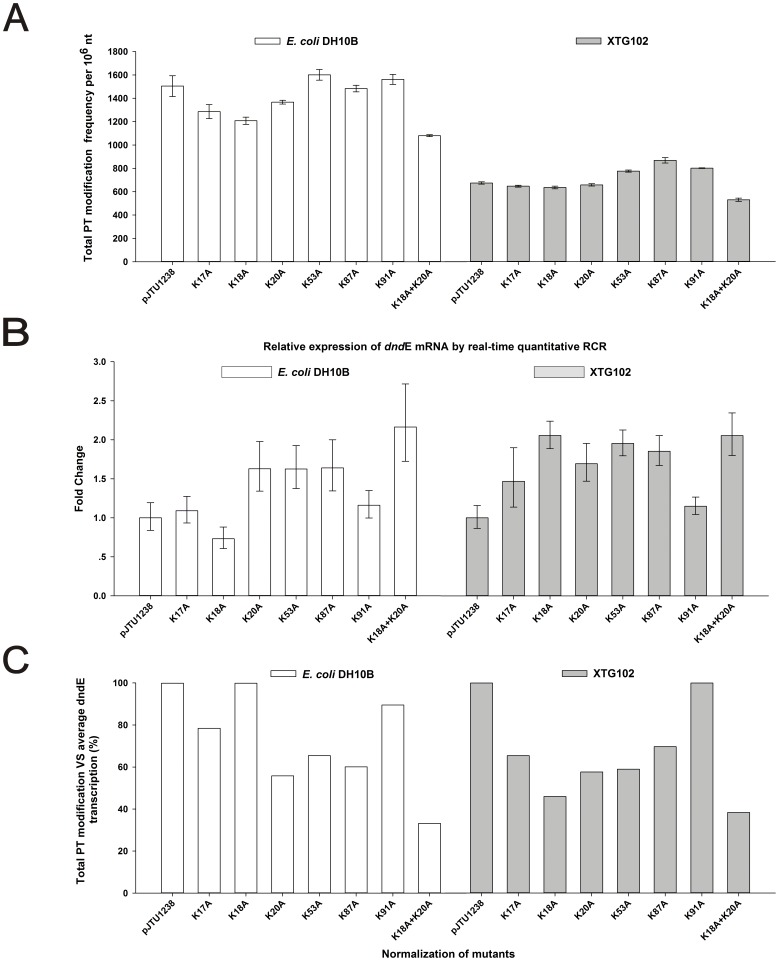
We previously reported that the plasmid pJTU1238 containing the dndBCDE gene cluster conferred the PT modifications of d(GPSA) R P and d(GPST) R P to E. coli DH10B [6]. As a high copy number plasmid, pJTU1238 increased the expression of the dndBCDE genes, leading to a 2-fold increase in the total number of PT modifications compared to wild-type S. enterica [6]. The in-frame deletion of dndE in pJTU1238, however, completely abolished DNA PT modifications, suggesting that dndE plays an essential role in the modification pathway (Figure 1; Table 2). To evaluate the influence of the positively charged lysine residues on DNA PT modification under physiological conditions, mutations were made in pJTU1238 to generate a set of plasmids, pWHU668 to pWHU674, carrying the dnd gene cluster and encoding DndE derivatives (Table 1). The individual replacement of each lysine residue with alanine did not alter the PT sequence specificity of d(GPSA) and d(GPST) (Table 2). The DNA PT frequencies were normalized to dndE transcript levels, and we found that the individual K17A, K20A, K53A and K87A mutations decreased the total PT by 22% to 44%. The K53A mutation slightly increased the frequency of PT modifications by 10%, while the K91A mutation slightly decreased the total frequency of PT modifications in E. coli DH10B by 11%. None of the mutations resulted in a complete loss of PT modifications (Table 2 & 3).
Table 2. PT modifications in E. coli DH10B harboring pJTU1238 and its derivatives.
| Mutations | Strains | PT modifications per 106 nt | ||
| d(GPSA) | d(GPST) | Total PT | ||
| wild type | DH10B(pJTU1238) | 778±29 | 727±60 | 1505±89 |
| K17A | DH10B(pWHU668) | 662±34 | 624±27 | 1286±60 |
| K18A | DH10B(pWHU669) | 629±13 | 579±25 | 1208±31 |
| K20A | DH10B(pWHU670) | 720±12 | 647±5 | 1367±16 |
| K53A | DH10B(pWHU672) | 821±12 | 780±35 | 1601±46 |
| K87A | DH10B(pWHU673) | 765±10 | 718±17 | 1483±28 |
| K91A | DH10B(pWHU674) | 818±17 | 744±29 | 1562±43 |
| K18A+K20A | DH10B(pWHU671) | 569±8 | 510±4 | 1080±8 |
| ΔdndE | DH10B(pWHU676) | ND | ND | ND |
Values represent the mean ± SD for biological triplicates; ND, not detected.
Table 3. Total PT modifications normalized to dndE transcript levels.
| Mutations | Strains | Normalized PT (%) | Strains | Normalized PT (%) |
| wild-type | DH10B(pJTU1238) | 100 | XTG102 (pJTU1238) | 100 |
| K17A | DH10B(pWHU668) | 78.4 | XTG102 (pWHU668) | 65.3 |
| K18A | DH10B(pWHU669) | 110.0 | XTG102 (pWHU669) | 45.9 |
| K20A | DH10B(pWHU670) | 55.8 | XTG102 (pWHU670) | 57.6 |
| K53A | DH10B(pWHU672) | 65.4 | XTG102 (pWHU672) | 58.9 |
| K87A | DH10B(pWHU673) | 60.1 | XTG102 (pWHU673) | 69.6 |
| K91A | DH10B(pWHU674) | 89.5 | XTG102 (pWHU674) | 103.5 |
| K18A+K20A | DH10B(pWHU671) | 33.2 | XTG102 (pWHU671) | 38.3 |
| ΔdndE | DH10B(pWHU676) | ND | XTG102 (pWHU676) | ND |
ND, not detected.
Expression of pJTU1238 and derivatives in S. enterica mutant
To validate the role of mutated DndE in the native host, pJTU1238 and its derivatives were transformed into XTG102, the dndBCDE in-frame deletion mutant of S. enterica (Table 3 & 4). The K17A, K18A, K20A, K53A and K87A mutations in XTG102 reduced the normalized PT by 30% to 54%, while K91A slightly increased the number of PT modifications by 4%. As shown in Figure 2, no lysine replacement completely abolished PT modification. We previously reported that the overexpression of dndBCDE on the high-copy pBluescript SK+ plasmid or the low-copy pACYC184 plasmid caused PT modifications to increase by 2- and 1.5-fold, respectively [6]. However, the overexpression of the dnd cluster on pJTU1238 did not cause a significant increase in the total PT frequency in XTG102, suggesting that the frequency of DNA PT modification in Salmonella is regulated.
Table 4. PT modifications in XTG102 harboring pJTU1238 and its derivatives.
| Mutations | Strains | PT modifications per 106 nt | ||
| d(GPSA) | d(GPST) | Total PT | ||
| wild type | XTG102(pJTU1238) | 374±5 | 299±5 | 673±10 |
| K17A | XTG102 (pWHU668) | 383±3 | 264±5 | 646±8 |
| K18A | XTG102 (pWHU669) | 383±8 | 251±4 | 635±11 |
| K20A | XTG102 (pWHU670) | 392±4 | 265±12 | 657±10 |
| K53A | XTG102 (pWHU672) | 398±8 | 377±16 | 775±9 |
| K87A | XTG102 (pWHU673) | 451±11 | 417±14 | 868±24 |
| K91A | XTG102 (pWHU674) | 410±8 | 390±8 | 800±3 |
| K18A+K20A | XTG102 (pWHU671) | 297±5 | 233±9 | 530±14 |
| ΔdndE | XTG102 (pWHU676) | ND | ND | ND |
Values represent the mean ± SD for biological triplicates; ND, not detected.
Figure 2. The impact of the mutagenesis of the lysine residues on the total PT modifications.
A. The plasmids pWHU668-pWHU674 possessing mutated lysine residues were expressed in E. coli DH10B and XTG102, and the DNA PT frequencies were measured by LC-MS/MS. The columns and error bars represent the mean ± SD; n = 3, biological triplicates. B. qRT-PCR expression analysis of the dndE genes in E. coli DH10B (white bars) and XTG102 (grey bars). The expression levels were normalized to the transcript levels of GAPDH. The columns and error bars represent the mean ± SD; n = 3, biological triplicates. C. PT modifications were normalized to the average dndE expression levels.
Double-site mutagenesis of dndE
X-ray crystallography analysis of DndE revealed that K20 was involved in the formation of hydrogen bonds that produce a positively charged hole [15]. The K20 residue was also proposed as one of the two key residues in DndE based on a DNA-binding affinity assay [15]. Here, we performed double-site mutagenesis, yielding the mutated plasmid pWHU671 encoding DndE with both K18 and K20 replaced with alanine residues. The double-site mutation led to the most significant decrease in total PT modifications, 67% and 62% in DH10B and XTG102, respectively. Although DndEK18A and DndEK20A displayed the most significant decreases in binding affinities to intact dsDNA (native DndE, KD ∼88.3 µM; DndEK20A, KD ∼690 µM; DndEK18A, non-detectable) and nicked dsDNA (DndEK18A and DndEK20A, non-detectable) in the study by Hu et al. [15], the results of the present study showed that the double-site mutation had a remarkable effect on the Dnd modifying efficiency but did not inactivate the effect of the physiological DndE in the PT modification pathway.
Discussion
There are five Dnd proteins involved in the biological pathway of sequence-selective and Rp configuration-specific DNA PT modification. Protein-protein interactions were detected between DndA and DndC, as well as between DndA and DndE, indicating the formation of a Dnd complex [16]. The smallest Dnd protein, DndE, was previously believed to be a sulfotransferase or a phosphoribosylaminoimidazole carboxylase analogue by homology analysis [3]. However, the crystal structure proposed that DndE was a nicked dsDNA-binding protein. The six positively charged lysines on the DndE surface were speculated to be involved in the interaction with nicked dsDNA. The individual mutation of the six lysine residues was later found to significantly decrease the dsDNA-binding affinity in vitro [15]. The mutagenesis of pJTU1238 and the sensitive LC-MS/MS used in this study allowed us to monitor the physiological impact of these positively charged lysine residues on DNA PT modification.
In addition to the non-detectable affinities of DndEK17A, DndEK18A, DndEK20A, DndEK53A and DndEK91A, the DndEK87A mutation showed a decreased KD of 323.5 µM in comparison to 30.6 µM of native DndE in vitro [15]. Our resulted showed that the mutations of K17A, K20A, K53A and K87A on pJTU1238 reduced the total PT frequencies in both E. coli DH10B and XTG102. However, none of the mutations abolished PT modification under physiological conditions. K18 and K20 were characterized as the two most essential lysine residues in binding nicked dsDNA in vitro. However, the K18 and K20 double mutant DndE protein encoded by pWHU671 was still able to generate PT modifications in E. coli DH10B and XTG102, albeit at significantly reduced levels of 33% and 38% compared to pJTU1238, respectively. Surprisingly, we observed an increased level of PT modification after mutating K18A in E. coli DH10B and K91A in XTG102. The comparison of DNA PT modifications in vivo and the DNA-binding affinity assay in vitro revealed that the disruption of the DNA-binding activity of DndE was not sufficient to abolish DNA PT modifications.
It is possible that E. coli may harbor unknown proteins having similar functions with dndE, which may complement the function of dndE when it is mutated. However, the complete loss of PT modification in the deletion mutant of dndE rules out this possibility. It is also possible that the mutation of lysine residues on DndE surface interfered the interaction of Dnd proteins to form complex, which resulted in the different efficiencies of DNA PT modification. Our findings suggest that the dsDNA-binding ability of DndE may not be crucial for its PT modification activity. Whether the perspective role of DndE is a sulfotransferase or a phosphoribosylaminoimidazole carboxylase remains to be validated. Our results also suggest that DndE may have other functions in the DNA PT modification pathway, and therefore may not be uniquely designed to interact with nicked dsDNA.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the 973 programs of the Ministry of Science and Technology of China (2012CB721004, 2013CB734003), National Natural Science Foundation of China (31170049, 31300038), Foundation for the Author of National Excellent Doctoral Dissertation of China, the Program for New Century Excellent Talents in University, Hubei Province's Outstanding Medical Academic Leader program, the Scientific Research Funds of the Education Department of Jiangxi Province, China (GJJ13257). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Eckstein F, Gish G (1989) Phosphorothioates in molecular biology. Trends Biochem Sci 14: 97–100. [DOI] [PubMed] [Google Scholar]
- 2. Wang L, Chen S, Xu T, Taghizadeh K, Wishnok JS, et al. (2007) Phosphorothioation of DNA in bacteria by dnd genes. Nat Chem Biol 3: 709–710. [DOI] [PubMed] [Google Scholar]
- 3. Zhou X, He X, Liang J, Li A, Xu T, et al. (2005) A novel DNA modification by sulphur. Mol Microbiol 57: 1428–1438. [DOI] [PubMed] [Google Scholar]
- 4. Dyson P, Evans M (1998) Novel post-replicative DNA modification in Streptomyces: analysis of the preferred modification site of plasmid pIJ101. Nucleic Acids Res 26: 1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Chen S, Deng Z (2011) Phosphorothioation: An unusual post-replicative modification on the DNA backbone. In: Seligmann H, editor. DNA Replication - Current Advances. Rijeka, Croatia: Intech.
- 6. Wang L, Chen S, Vergin KL, Giovannoni SJ, Chan SW, et al. (2011) DNA phosphorothioation is widespread and quantized in bacterial genomes. Proc Natl Acad Sci U S A 108: 2963–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He X, Ou HY, Yu Q, Zhou X, Wu J, et al. (2007) Analysis of a genomic island housing genes for DNA S-modification system in Streptomyces lividans 66 and its counterparts in other distantly related bacteria. Mol Microbiol 65: 1034–1048. [DOI] [PubMed] [Google Scholar]
- 8. Cao B, Chen C, DeMott MS, Cheng Q, Clark TA, et al. (2014) Genomic mapping of phosphorothioates reveals partial modification of short consensus sequences. Nat Commun 5: 3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu T, Yao F, Zhou X, Deng Z, You D (2010) A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res 38: 7133–7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tock MR, Dryden DT (2005) The biology of restriction and anti-restriction. Curr Opin Microbiol 8: 466–472. [DOI] [PubMed] [Google Scholar]
- 11. Wilson GG, Murray NE (1991) Restriction and modification systems. Annu Rev Genet 25: 585–627. [DOI] [PubMed] [Google Scholar]
- 12. Chen S, Wang L, Deng Z (2010) Twenty years hunting for sulfur in DNA. Protein Cell 1: 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. You D, Wang L, Yao F, Zhou X, Deng Z (2007) A novel DNA modification by sulfur: DndA is a NifS-like cysteine desulfurase capable of assembling DndC as an iron-sulfur cluster protein in Streptomyces lividans. Biochemistry 46: 6126–6133. [DOI] [PubMed] [Google Scholar]
- 14. Yao F, Xu T, Zhou X, Deng Z, You D (2009) Functional analysis of spfD gene involved in DNA phosphorothioation in Pseudomonas fluorescens Pf0-1. FEBS Lett 583: 729–733. [DOI] [PubMed] [Google Scholar]
- 15. Hu W, Wang C, Liang J, Zhang T, Hu Z, et al. (2012) Structural insights into DndE from Escherichia coli B7A involved in DNA phosphorothioation modification. Cell Res 22: 1203–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. An X, Xiong W, Yang Y, Li F, Zhou X, et al. (2012) A novel target of IscS in Escherichia coli: participating in DNA phosphorothioation. PLoS One 7: e51265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Durfee T, Nelson R, Baldwin S, Plunkett G 3rd, Burland V, et al. (2008) The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J Bacteriol 190: 2597–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alting-Mees MA, Short JM (1989) pBluescript II: gene mapping vectors. Nucleic Acids Res 17: 9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.