Abstract
An inducible RNA-silencing pathway, involving a single Dicer protein, DCL2, and a single Argonaute protein, AGL2, was recently shown to serve as an effective antiviral defense response in the chestnut blight fungus Cryphonectria parasitica. Eukaryotic RNA-dependent RNA polymerases (RdRPs) are frequently involved in transcriptional and posttranscriptional gene silencing and antiviral defense. We report here the identification and characterization of four RdRP genes (rdr1–4) in the C. parasitica genome. Sequence relationships with other eukaryotic RdRPs indicated that RDR1 and RDR2 were closely related to QDE-1, an RdRP involved in RNA silencing (“quelling”) in Neurospora crassa, whereas RDR3 was more closely related to the meiotic silencing gene SAD-1 in N. crassa. The RdRP domain of RDR4, related to N. crassa RRP-3 of unknown function, was truncated and showed evidence of alternative splicing. Similar to reports for dcl2 and agl2, the expression levels for rdr3 and rdr4 increased after hypovirus CHV-1/EP713 infection, while expression levels of rdr1 and rdr2 were unchanged. The virus-responsive induction patterns for rdr3 and rdr4 were altered in the Δdcl2 and Δagl2 strains, suggesting some level of interaction between rdr3 and rdr4 and the dcl2/agl2 silencing pathway. Single rdr gene knockouts Δrdr1–4, double knockouts Δrdr1/2, Δrdr2/3, Δrdr1/3, and a triple knockout, Δrdr1/2/3, were generated and evaluated for effects on fungal phenotype, the antiviral defense response, viral RNA recombination activity and transposon expression. None of the single or multiple rdr knockout strains displayed any phenotypic differences from the parental strains with or without viral infection or any significant changes in viral RNA accumulation or recombination activity or transposon RNA accumulation, indicating no detectable contribution by the C. parasitica rdr genes to these processes.
Introduction
Mycoviruses and other selfish genetic elements, e.g., transposons, are widely distributed throughout the filamentous fungi. A variety of mechanisms have been identified by which fungi restrict the accumulation and transmission of such elements [Reviewed in 1,2]. The genetic non-self recognition system, vegetative incompatibility, restricts transmission of mycoviruses [3]–[6] and potentially transposable elements [7], [8] between genetically dissimilar individuals in a population. Silencing mechanisms act to control the transcription of transposable elements and accumulation of viruses. Demonstrated fungal transposon silencing mechanisms include repeat-induced point mutations (RIP) [9] and methylation [10]. RNA interference has been shown to restrict both transposon [11]–[15] and virus accumulation in filamentous fungi [16], [17].
The RNA-silencing antiviral pathway is highly conserved among plants, invertebrates and fungi [18], consisting of two core components belonging to the Dicer-like and Argonaute nuclease families [19]. Dicer-like nucleases recognize and, with an associated RNase III-type activity, process viral double-stranded or highly structured RNA into small RNAs of 21–24 nts in length. With the aid of an Argonaute protein, these virus-derived small RNAs (vsRNAs) are subsequently incorporated into an effector complex called the RNA-induced silencing complex (RISC). One strand of the vsRNA is degraded and the remaining strand then serves to guide the effector complex to the cognate viral RNA that is then cleaved by an RNase H-like activity associated with the Argonaute protein. The role of RNA silencing as an antiviral defense mechanism in fungi was first demonstrated for the chestnut blight fungus Cryphonectria parasitica [16]. Although C. parasitica encodes two Dicer-like genes and four Argonaute-like genes, only Dicer 2 (dcl2) and Argonaute 2 (agl2) are required for antiviral defense [16], [20]. DCL2 and AGL2 were also shown to contribute significantly to viral RNA recombination [20], [21].
As clearly demonstrated for plants [14], [22], [23] and the nematode Caenorhabditis elegans [24]–[26], the Dicer/Argonaute-dependent antiviral RNA-silencing response is further amplified by host RNA-dependent RNA polymerases. Fungal-encoded RdRPs have been reported to play a role in transgene silencing [27]–[28], meiotic silencing [29]–[30], production of qiRNA induced by DNA damage [31], sex-induced silencing [32] and restriction of transposon accumulation [11]–[14]. The role of fungal RdRPs in the antiviral defense response is unknown. The primary goal of this study was to identify and characterize the RdRP genes in C. parasitica and examine their contribution to the RNA-silencing antiviral defense response and RNA silencing-mediated promotion of viral RNA recombination.
Materials and Methods
2.1. Fungal strains, growth condition and virulence assay
Wild-type C. parasitica strain EP155 (ATCC 38755) was used for generating rdr1 gene disruption strain Δrdr1. Strain DK80, a mutant of EP155 disrupted in the ku80 gene for non-homologous end-joining DNA repair to promote integration of homologous DNA sequence [33], was used to generate all other RdRP gene-disruption strains. All fungal cultures were maintained on PDA at 22°C. Viral infection of C. parasitica strains was achieved by transfecting viral RNA-coding strand transcripts into fungal spheroplasts by electroporation [34]. The in vitro growth of virus-infected and virus-free fungal mycelium of rdr gene knockouts and parental strains was assessed on PDA plates; colony diameters were measured after 7 days of growth. Asexual sporulation was measured in cultures grown on PDA for 14 days. Conidia were washed from plates with 1% Tween 20, subjected to serial dilution, counted in a hemocytometer and plated on PDA to test for viability.
Virulence assays were performed on dormant American chestnut tree stems as described by Hillman et al. [35], with six duplicate inoculations per fungal strain. Inoculated stems were kept at room temperature in a glass tank to maintain humidity. Cankers were measured after 21 days.
The sensitivity of wild-type and mutant strains to DNA mutagens was measured with a modified spot test [31]. Conidia were harvested with 0.8% NaCl, filtered through Miracloth, counted in a hemocytometer and diluted with 0.8% NaCl to a concentration of 108/ml. Aliquots of the conidial stock solution were incubated on a shaker for 2 hrs at 60 rpm and 30 C with or without the DNA mutagens histidine (1.5 or 3.5 mg/ml) or hydroxyurea (2 or 4 mg/ml). The treated condial suspensions were taken through serial dilution and 10 µl, each containing 106, 5×103, 5×102 or 50 conidia, was spotted in duplicate on PDA plates containing the concentration of histidine or hydroxyurea used in the original treatment. Colony diameter was measured after 3 days of incubation at room temperature. Average values for the two replica plates were recorded.
2.2. Analysis of RdRP genome sequences, gene organization and phylogenetic relationships
The genome sequence assembly of the C. parasitica reference strain EP155, annotated with predicted or experimentally verified gene models and protein functions was retrieved from the U.S. Department of Energy’s Joint Genome Institute (JGI; at http://genomeportal.jgi-psf.org/Crypa2/Crypa2.info.html) and interrogated to identify putative RdRP genes. Additional sequence analyses of PCR-amplified cDNAs were performed by Macrogen (Rockville, MD).
Known or putative RdRP protein sequences from all major eukaryotic taxa were retrieved from GenBank or species genome websites. The sequences were entered into NCBI’s Conserved Domain Search tool (at http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), to identify the protein regions corresponding to the conserved RdRP domain, which were manually extracted and FASTA formatted. Alignments of the RdRP domain sequences were performed with the MUSCLE algorithm (Version 3.8 at http://www.ebi.ac.uk/Tools/msa/muscle/), using the default parameters. The protein alignments were imported into JalView [36] for inspection of alignments and construction of trees with the neighbor-joining method based on percent identity between sequences.
2.3. Generation of RdRP gene knockouts
Identified RdRP gene candidates were disrupted by replacing all or large parts of the rdr coding sequences (CDS) with selectable marker cassettes via homologous recombination, using the DNA transformation protocol developed by Churchill et al [37]. To increase the capacity for producing strains that contain multiple rdr gene disruptions, the Cre-loxP recombination system [38] was adapted to allow recycling of the limited number of selectable marker genes available for C. parasitica transformation [39]. The diagrams in Figs. S1–S3 show the details of the disruption schemes and origins of rdr1, rdr2 and rdr4 gene disruptions. The disruption scheme for rdr3 was previously described in Zhang et al. [39]. Overlap-extension PCR was used to generate the gene-disruption fragments comprising the antibiotic resistance gene cassette and flanking sequences of the targeted rdr genes. Δrdr1 was generated by replacing rdr1 with a cassette consisting of the trpC promoter from Aspergillus nidulans in front of the hygromycin B-phosphotransferase (hph) gene (ProTrpC-hph) (Fig. S1). Δrdr2 was obtained by replacing a 1.5-kb rdr2 fragment with a 3-kb hph cassette (Fig. S2). Δrdr3 was generated with a split marker approach using two fragments of a loxP-protubB-hph-loxP cassette as described in Zhang et al. [39]. Δrdr4 was generated by replacing the rdr4 gene with the intact loxP-ProtubB-hph-loxP cassette (Fig. S3).
To generate double rdr knockout strains, a neomycin (neo) gene cassette in pSK666 [20] was cloned into pKAES183 (provided by Dr. Christopher Schardl, University of Kentucky) by restriction-enzyme digestion and T4 enzyme ligation, to replace ProtubB-phleomycin with the neo gene behind the gpd promoter from A. nidulans, resulting in a loxP-Progpd-neo-loxP construct. The Δrdr1/3 and Δrdr2/3 double mutant strains were generated by disrupting the rdr1 or rdr2 genes with the loxP-Progpd-neo-loxP cassette in the Δrdr3 strain background as described in Zhang et al. [39]. The Δrdr1/2 double mutant strain was generated by disrupting the rdr1 gene with the loxP-Progpd-neo-loxP cassette in the Δrdr2 strain background [described in Fig. 1B of 39]. The Cre-loxP system relies on the Cre recombinase to eliminate marker genes flanked by loxP sites, thus enabling multiple reuse of the same marker genes for generating serial knockouts [40]. As described in Zhang et al. [39] and included here for convenience, for use in recycling selectable marker genes in C. parasitica, a Progpd–Cre gene construct was cloned into pCPX-BSD1 [41], resulting in the construct pCPX-Cre, containing blasticidin as selectable marker. Using the method of Churchill et al. [37], the pCPX-Cre plasmid was transformed into Δrdr1/3 and Δrdr2/3 spheroplasts for Cre expression to eliminate selectable marker genes via recombination at the loxP sites flanking the hph or neo marker genes to produce marker gene-free strains Δrdr1/3-M and Δrdr2/3-M. Blasticidin was used at 600 µg/ml for transformant selection on regeneration media; although untransformed colonies also grew at this concentration, they did so at a slower rate than the pCPX-Cre-containing transformants. Therefore, putative transformants were selected by their larger colony size to screen for the loss of hygromycin and/or neomycin resistance by replica-plating on three different PDA media: medium containing 50 µg/ml hygromycin PDA, medium containing 25 µg/ml neomycin PDA or antibiotic-free medium. The antibiotic-containing plates were screened for transformed colonies that displayed little or no growth, and the corresponding colonies on the antibiotic-free plates were subjected to single-spore isolation on PDA. Excision of the neo and hph genes was confirmed by PCR analysis using the same primer pairs used to confirm rdr gene knockouts.
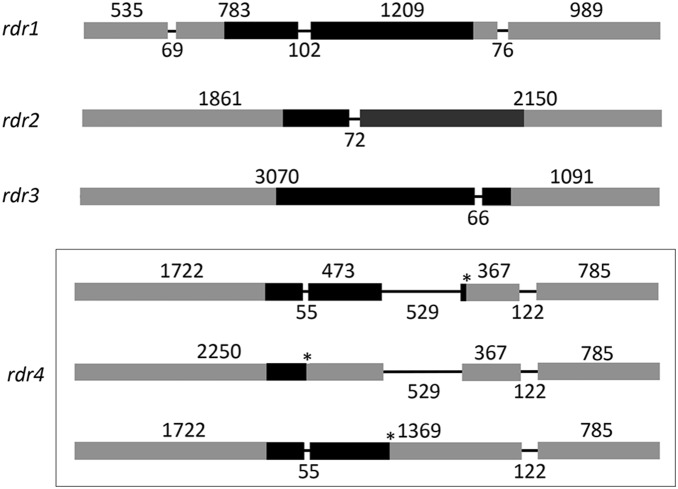
Figure 1. Gene organization of rdr1, rdr2, rdr3 and rdr4.
Positions of exons (boxes) and introns (solid lines) were determined by comparisons of genomic and cDNA sequences for each gene (see text). Black boxes indicate regions corresponding to the RdRP domains in the encoded RDR proteins. Three alternatively spliced sequences were identified in rdr4, each predicted to result in a truncated RdRP domain because of premature stop codons indicated by asterisks. Lengths are indicated above and below exons and introns, respectively.
Δrdr1/2/3 triple knockout strains were engineered in two ways. One strain (F1) was constructed by generating a rdr1-disruption fragment containing ProTrpC-hph by direct PCR amplification from the disrupted rdr1 gene in EP155-based disruption strain Δrdr1, the product of which was then used to transform strain Δrdr2/3-M. A second triple knockout strain, C1, was generated in the Δrdr1/3-M strain background by disrupting the rdr2 gene with a ProtubB-hph cassette without flanking loxP sites from pKAES173 (provided by Dr. Chris Schardl, University of Kentucky). Transformants were single spored and disruptions of all rdr genes were confirmed by PCR.
These extensive disruption studies took advantage of C. parasitic strain DK80 derived from reference strain EP155 by disruption of the ku80 gene for non-homologous end-joining DNA repair to promote homologous DNA recombination [33]. The rdr1 disruption mutant Δrdr1 was generated in strain EP155 prior to the availability of strain DK80. No phenotypic differences have been observed between the DK80 strain and reference strain EP155, whether uninfected or following hypovirus infection. However, strain EP155 was used as the parental control strain for all experiments involving Δrdr1 and DK80 was used as the parental control in experiments involving all other individual and multiple rdr gene disruption mutants. This resulted in the inclusion of both parental strains in many of the experiments described in this report.
2.4. Nucleic acid preparation and analysis
DNA extraction was performed with a small-scale method previously described by Spiering et al. [42]. RNA used for agarose gel analysis of viral full-length and defective interfering (DI) double-stranded RNAs was prepared as described by Sun et al. [20]. Total RNA for reverse-transcription real-time PCR (RT-qPCR) and RNA-seq analysis was extracted with Qiagen’s (Valencia, CA) RNeasy kit. The Monster 1st strand cDNA synthesis kit (Epicentre, Madison, WI) was used for cDNA synthesis from total RNA, following the manufacturer’s protocol, and full-length cDNA copies of each rdr gene were amplified by PCR (Takara Tech Enzyme, Takara) and sequenced.
Transcript accumulation for selected genes was measured by one-step reverse-transcription real-time PCR (RT-qPCR), using AgPath-ID RT-PCR reagents (Ambion USA) and an ABI 7300 real-time PCR system. To guard against potential amplification of contaminating genomic DNA in the RNA samples, at least one primer or probe for each rdr gene was designed to span an intron/exon boundary, identified by comparison of cDNA and genomic DNA sequences. Primer and probe design and general RT-qPCR methods were performed as described by Zhang et al. [43]. Calculations of transcript accumulation values were performed with the comparative threshold cycle method (ΔΔCt) normalized against the β-tubulin gene transcript levels and calibrated against the normalized value in the wild-type strain. The primer/probe sets for the Dicer-2 and Argonaute-2 genes, dcl2 and agl2, respectively, were the same as described by Sun et al. [20]. Other primers used in this study are shown in Table 1.
Table 1. Primers and probes used in this study.
| Primer name | Sequence (5′→ 3′) | Note |
| R1f | AAC CCG CAC GAG ACA CAG TGG GCA GAT | Δrdr1 (ProtrpC-hph) check |
| R1r | GTG TGG AGC AGC AGA TGG AGA AAT ATG | |
| R2f | ATG AAC ACT TCC CAG GGC CA | Δrdr2 (hph cassette) check |
| R2r | TCA ATC GTC CTC GTC CAA CTC | |
| R4f | GTC CTT GTA GAC CAT GTG TGT CCC TT | Δrdr4 (loxP-ProtubB-hph-loxP) check |
| R4r | TCG GAC CCC AGG GTA ACC ACC GCC AAC GTG | |
| TQMrdr1F | AGA TCT CCA AGT TGA GCA CGG GAA | rdr1 gene expression level assay with RT-qPCR |
| TQMrdr1R | TCG CAG ACC CGA TAG CAG TGA TAA | |
| TQMrdr1probe | /56-FAM/AAA GCT CCT/ZEN/CGG ATC CTG TAC ATC GT/3IABkFQ/ | |
| TQMrdr2F | GGT CCC AGC CTG CAT TAA AGT TGT | rdr2 gene expression level assay with RT-qPCR |
| TQMrdr2R | TCA TCT GGC TGG GTT CGA AGA TGA | |
| TQMrdr2probe | /56-FAM/TTG TCC TCA/ZEN/GCA GAA CTA GCC CGA CA/3IABkFQ/ | |
| TQMrdr3F | AAA CGA CTC TCT GGG ACT CAT AGC CA | rdr3 gene expression level assay with RT-qPCR |
| TQMrdr3R | GGA ATT TGG CAA GCT CAA GGC ACT | |
| TQMrdr3probe | /56-FAM/ACA AGG CCT/ZEN/GGG CGG ACA AA/3IABkFQ/ | |
| TQMrdr4F | TGG CGA GGA GCT TGA CCA ACA TAA | rdr4 gene expression level assay with RT-qPCR |
| TQMrdr4R | CAG CGA CTG AAG CGT TGG TTG AAA | |
| TQMrdr4probe | /56-FAM/ACG AAA CTC/ZEN/CAA CAC CGC CAG ATC T/3IABkFQ/ | |
| rdr1d1 | ATG CCG CCG CGA AAA TCA TT | full-length cDNA amplification |
| rdr1u1 | TCA ATC ATC CTC TTG CTG GCT | |
| rdr2d1 | ATG AAC ACT TCC CAG GGC CA | |
| rdr2u1 | TCA ATC GTC CTC GTC CAA CTC | |
| rdr3d1 | ATG ACC ACC CCT ACC AGG GCC CCT AA | |
| rdr3u1 | TTA GTG CTC ATC ACG TAC GCC AGT A | |
| rdr4d1 | ATG GAG GTC CAC ATG CAC | |
| rdr4u1 | TCA TTT ACC ACT GCA GCC |
2.5. Viral recombination assay
All rdr-knockout, DK80 and Δdcl2 strains were transfected with a replication-competent, GFP-containing viral vector Ctp40[2AEGFP] [44] by transfection with spheroplasts [34]. One week after the transfection, each infected strain was transferred to PDA and then sub-cultured to new PDA plates every 5 days for up to 6 transfers. Total RNA was isolated from sub-cultured colonies to monitor for presence of defective interfering viral dsRNAs and to serve as template to synthesize cDNA for PCR analysis with primer pair CHV1-2KF/CHV1-3KR [45] to monitor for maintenance of the EGFP coding domain in the viral vector RNA. GFP accumulation in infected fungal hyphae was also monitored under a fluorescence stereomicroscope (Model MVX10; Olympus) [21].
2.6. RNA-seq analysis of transposon expression
To identify and extract transposon sequences from the C. parasitica genome assembly at JGI (http://genomeportal.jgi-psf.org/Crypa2/Crypa2.home.html), the keywords of all gene models were queried with the following search terms: “transposon,” “retrotransposon,” “transposase,” “transposable,” “transposable element,” “copia,” “FOT,” “Flipper,” “gag,” “gag-pol,” “gypsy,” “LINE,” “pogo” and “SINE.” This search identified >500 gene models whose IDs and associated annotations were downloaded as csv files and sequences as FASTA files. Since these searches frequently resulted in repeated retrieval of the same sequences, the retrieved sequences IDs were manually checked and all duplicate sequences removed. Because many transposon copies in fungal genomes are highly similar in sequence–potentially affecting the read mappings in the RNA-seq analyses–all of the retrieved sequences were aligned with each other in BLASTN. Those sequences having >99% identity along >95% of their total length in the BLAST alignment were then combined as a single sequence consensus in JalView [36] for further analysis. In addition, sequences <500 bp long were excluded from the analysis, resulting in a final set of 104 sequences. Of these, 71 were identified by their annotations in JGI or by BLASTX searches as class I transposable elements (i.e., retroelements or retrotransposons), 21 as class II (DNA) transposons and 12 sequences were similar to RNA-associated or hypothetical proteins (Table S1).
To quantify expression from each of the transposon sequences, total RNA was extracted from the rdr gene knockouts Δrdr1/2/3(C1), Δrdr1/2 and Δrdr4 and the DK80 parental strain from mycelium grown on PDA for 7 days. Three biological replicates were used for Δrdr1/2/3 and Δrdr1/2 and analyzed separately by RNA-seq. One biological sample was used for Δrdr4. The RNA was submitted to the Institute for Bioscience and Biotechnology Research DNA Sequencing Facility for construction of sequencing libraries containing cDNA fragments with an average length of 200 nucleotides. Paired-end reads sequencing data were acquired on an Illumina HiSeq 1000 instrument. On average, >40 million sequence reads were obtained per sample. The reads were uploaded into CLC Genomics Workbench 6.0 (CLCBio, Aarhus, Denmark) and mapped to the transposon sequences for RNA-seq analysis. The following (default) settings were used in the read mappings: a minimum length fraction of 0.9, minimum similarity fraction of 0.8 and maximum number of hits per read of 10; the minimum and maximum distances between paired reads were set at 100 and 250, respectively. Expression was estimated as reads per kb of coding sequence per million mapped reads (RPKM). The RNA-seq expression data from rdr gene knockouts and the DK80 control were used to calculate fold changes in expression by ratio-based analysis to identify differentially expressed transcripts from transposon sequences in each rdr gene knockout strain. Raw RNA-seq data have been deposited in the NCBI sequence read archive (SRA) under BioProject ID PRJNA258639.
Results
3.1. Identification and features of RdRP-like genes in C. parasitica strain EP155
The C. parasitica genome sequence assembly generated by the U.S. Department of Energy’s Joint Genome Institute for reference strain, EP155, was used as a resource for RdRP gene identification in this study. To identify RdRP gene candidates, the keywords in the annotated EP155 genome sequence were queried with “RdRP” as search term. This search yielded four RdRP-like sequences. In addition, a BLASTP search of the EP155 sequence was conducted, using QDE-1, the RdRP required for transgene silencing (Quelling) in Neurospora crassa [27], [46], as query, which did not result in any additional sequences. The RdRP-like genes identified were designated rdr1 (scaffold_11∶693898–697090 in JGI), rdr2 (scaffold_1∶365214–369296), rdr3 (scaffold_5∶3858759–3862568) and rdr4 (scaffold_4∶4500867–4504921). The genomic sequences from each putative RdRP gene were then used to obtain cDNA amplicons by RT-PCR and sequencing. The PCR-amplified cDNAs for rdr1, rdr2 and rdr3 gave a single product as judged by agarose gel analysis, and intron and exon positions in each of the three genes were identified by sequence comparison with the cognate genomic DNA sequences (Fig. 1). rdr1 had three introns and four exons, whereas rdr2 and rdr3 each contained only one intron and two exons. The location within the coding domain of the second intron in rdr1 corresponded to that of the only intron in rdr2 as judged by alignment of the encoded amino acid sequences relative to the exon–intron boundaries in the two genes. No such correspondence was seen for the only intron in rdr3.
Amplification of the full-length ORF in rdr4 gave multiple cDNA bands. To determine the reason for this potential diversity in rdr4 transcripts, the most prominent three bands were individually gel purified and sequenced. This analysis revealed that the variable banding pattern had most likely originated from alternative splicing events in the rdr4 gene. rdr4 contained three introns, and the first two introns were either spliced out or remained unspliced (Fig. 1). The four rdr genes have the following GenBank accession numbers: rdr1 (HF912382); rdr2 (HF912383); rdr3 (HF912384); rdr4 (HF912385).
Searches for conserved protein domains with the CD-Search Tool at NCBI (at http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) revealed that RDR1, RDR2 and RDR3 each contained an RdRP-type conserved domain (pfam ID: pfam05183; E-values <10−80). Interestingly, the rdr4 gene did not contain an intact RdRP domain; the predicted RDR4 protein sequence of the spliced transcripts contained in all cases a disrupted RdRP domain (Fig. 2A) that lacked the DXDGD signature motif of eukaryotic RDRs (Fig. 2B).
Figure 2. Conserved RdRP domains and amino acid sequences of C. parasitica rdr genes.
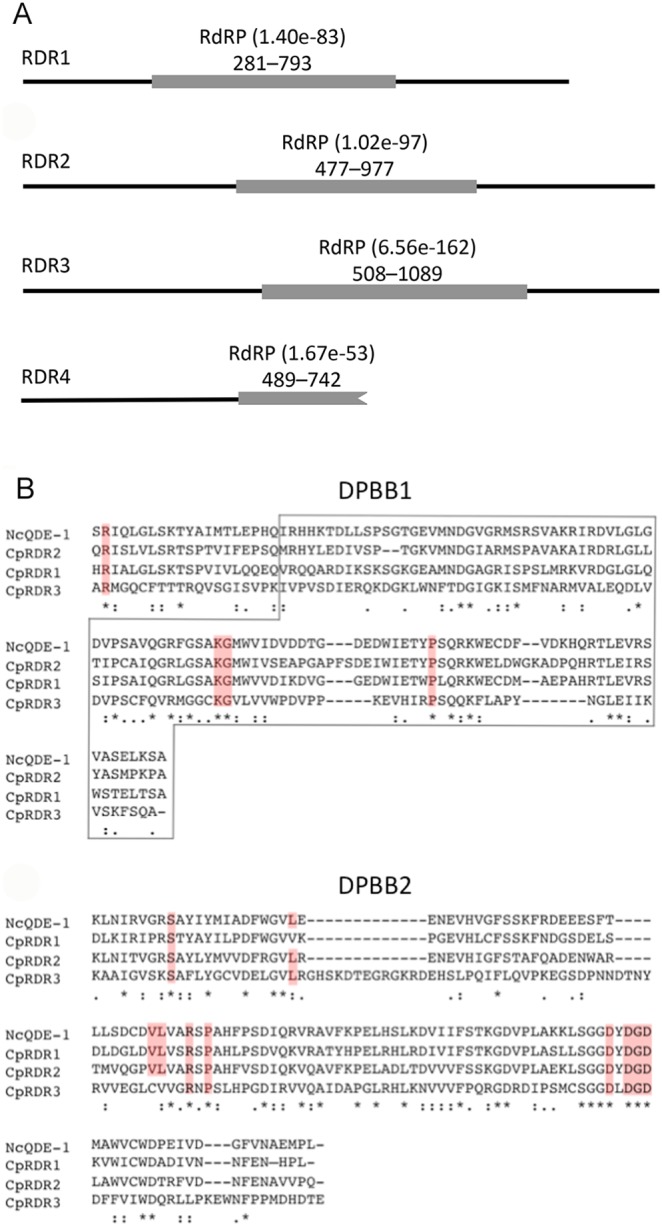
A). The RdRP domains were identified by searching NCBI’s Conserved Domains database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and are indicated by grey boxes; solid lines represent full-length proteins. E-values of each RdRP domain match and the range of amino acids included in the match are indicated next to and below the domain name, respectively. The longest predicted RDR4 protein is indicated here and contains an RdRP domain that is truncated due to a premature stop codon in its gene transcript and that lacks a conserved DPBB domain. B). The three C. parasitica RDRs containing complete RdRP domains were included in an alignment with QDE-1 from N. crassa, an RdRP involved in RNA silencing (Quelling); highly conserved DPBB domains and residues in QDE-1 [47] are indicated by boxes and are highlighted in red, respectively.
QDE-1 from N. crassa is involved in RNA silencing (Quelling) [27] and several key amino acid residues and domains required for its activity have recently been identified [47]. To see if the C. parasitica RdRPs also contained these residues and domains, we conducted comparative sequence analysis of RDR1– RDR3 with QDE-1. Since rdr4 did not appear to encode a functional RDR protein, it was not included in the analysis. All three C. parasitica RDRs contained three invariant aspartic acid residues, corresponding to D1007, D1009 and D1011 in QDE-1 in its catalytic double-psi-β-barrel (DPBB2) domain [47] that constitute the DXDGD motif essential for catalyzing the formation of phosphodiester bonds during RNA synthesis [47]. Furthermore, all three RDRs contained three absolutely conserved residues present in the DPBB1 domain of QDE-1 (Fig. 2B). Including the DXDGD motif, ten highly conserved amino acid residues are present in the DPBB2 domain of QDE-1 [48], all of which were also present in RDR2; RDR1 shared nine and RDR3 eight out of the ten highly conserved residues in QDE-1. This analysis indicated that RDR1– RDR3 possess the active-site residues required for RDR activity; it also revealed that the RDR1 and RDR3 sequences were more divergent from QDE-1 than RDR2 in highly conserved RDR regions.
BLAST sequence alignments (all having E-values <10−19) of the full-length RdRP sequences supported the above pattern of divergence among the three RdRPs: whereas QDE-1 aligned with RDR2 at 42% identity (59% similarity) along 1,125 amino acids, its alignment with RDR1 gave 40% identity (56% similarity) along 1,014 residues, indicating a somewhat greater level of divergence between RDR1 and QDE-1 (alignments not shown). RDR3 failed to give a contiguous alignment with QDE-1, and resulted in only 26% identity (43% similarity) along 340 residues of the longest aligned region. These analyses suggested that out of the three likely functional RdRP-like proteins in C. parasitica, RDR2 appeared to be the one most closely related to QDE-1. However, despite this relatively close relationship, RDR2 displayed only moderate (<60%) similarity to QDE-1, suggesting the possibility of some functional divergence between the two proteins. None of the three C. parasitica RDRs contained an RNA-recognition motif (RRM; pfam ID: cd00590); such RRM domain was identifiable in N. crassa SAD-1 (meiotic silencing), but not in QDE-1, suggesting that the RRM motif is not essential for RNA silencing activity.
3.2. Sequence relationships of RdRP-like proteins from C. parasitica with other RdRP protein sequences
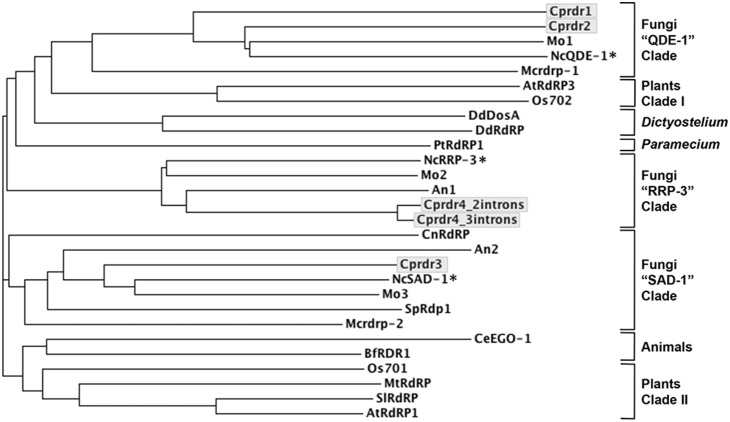
To gain some broader insight into the relationship of the C. parasitica RdRP-like sequences with putative and confirmed RdRP sequences from other species, a multiple sequence alignment of the RdRP-like C. parasitica sequences with those from a range of taxa from plants, protists, animals and other fungi was performed. The alignment was restricted to the predicted RdRP domains in each protein, and used to reconstruct a tree. This yielded several major clades, each represented by RDR sequences originating from a single kingdom (Fig. 3). Consistent with the general topology of RdRP trees in previous reports [49], RdRPs from animals grouped in a single well-defined clade, whereas those from fungi, plants and protists assembled in multiple clades, homogeneous for each organismal group. Interestingly, each of the three fungal RdRP clades contained an RdRP from N. crassa, i.e., QDE-1 (Quelling), SAD-1 (meiotic silencing) or RRP-3 (function unknown), a pattern that was closely tracked by the four RdRP-like sequences from C. parasitica. RDR2 was very closely affiliated with QDE-1, whereas the position of RDR1, although residing in the same (“QDE1”) clade suggested a more distant relationship with both RDR2 and QDE-1. RDR4 (included as two different predicted sequences inferred from alternative splicing and theoretical prediction) was affiliated with RDRs clustering with RRP-3, while RDR3 resided in another fungal clade that included SAD-1. These sequence relationships among the fungal RdRPs were reproducibly reconstructed with additional tree-building methods and evolutionary models (not shown). This analysis suggested distinct biological roles for the three functional C. parasitica RdRPs. For example, the affiliation with QDE-1 suggested involvement of RDR2 and possibly RDR1 in posttranscriptional silencing, whereas clustering of RDR3 with SAD-1, an RDR involved in meiotic silencing [29], might suggest that RDR3 functions in a similar, but as-yet unreported, pathway in C. parasitica. A possible function for RDR4 was less clear, since it lacked a complete RdRP domain and no functional roles have yet been ascribed to its closest relative in N. crassa, RRP3. The analysis thus supported an early split of the four putative C. parasitica RdRP-like sequences, very likely preceding the speciation events that had resulted in the species shown in the tree.
Figure 3. Sequence relationships of C. parasitica RDR-like proteins with putative and known RDRs in other eukaryotes.
RdRP domains were extracted from each sequence and aligned for tree reconstruction as described in the Methods. The four RDR-like C. parasitica sequences are shown in shaded boxes; two alternatively spliced forms of RDR4, identified by cDNA sequencing (resulting in a truncated RdRP domain) are shown. The RDRs in N. crassa are indicated by asterisks and were used to name the three fungal RDR clades, indicated on the right, along with the clades containing RDR sequences in other eukaryotes. Sequence accessions and species names are the following: An1 = AN2717 (Aspergillus nidulans); An2 = AN4790 (A. nidulans); AtRdRP1 = NP_172932.1, RNA-dependent RNA polymerase 1 (Arabidopsis thaliana); AtRdRP3 = NP_179581.2, RNA-dependent RNA polymerase-like protein (A. thaliana); BfRDR1 = AAQ10792.1, RNA-directed RNA polymerase-like protein (Branchiostoma floridae); CeEGO-1 = NP_492132.1, Protein EGO-1 (Caenorhabditis elegans); CnRdRP = ACJ04634.1, RNA-dependent RNA polymerase (Cryptococcus neoformans var. grubii); Cprdr1 = HF912382, putative RNA-dependent RNA polymerase (Cryphonectria parasitica); Cprdr2 = HF912383, putative RNA-dependent RNA polymerase (C. parasitica); Cprdr3 = HF912384, putative RNA-dependent RNA polymerase (C. parasitica); Cprdr4 = HF912385, putative rdr4 pseudogene (C. parasitica); DdDosA = AF117611_1, DosA protein (Dictyostelium discoideum); DdRdRP = EAL62541.1, RNA-directed RNA polymerase (D. discoideum); Mcrdrp-1 = JGI scaffold_05∶2640291–2644233 (Mucor circinelloides); Mcrdrp-2 = JGI scaffold_05∶245898–249911 (M. circinelloides); Mo1 = EHA51817.1, RNA-dependent RNA polymerase (Magnaporthe oryzae); Mo2 = EHA52286.1, RNA-dependent RNA polymerase (M. oryzae); Mo3 = EHA46264.1, RNA-dependent RNA polymerase 1 (M. oryzae); MtRdRP = XP_003608944.1, RNA-dependent RNA polymerase (Medicago truncatula); NcQDE-1 = AJ133528, qde-1 gene (Neurospora crassa); NcRRP-3 = XP_963405, hypothetical protein (N. crassa); NcSAD1 = AAK31733, suppressor of ascus dominance (N. crassa); Os701 = NP_918046.1, putative RNA-directed RNA polymerase (Oryza sativa); Os702 = NP_913147.1 (O. sativa); PtRDRP1 = CAI39057.1, RNA-dependent RNA polymerase (Paramecium tetraurelia); SpRdp1 = CAB11093.1, RNA-directed RNA polymerase Rdp1 (Schizosaccharomyces pombe); SlRdRP = CAA71421.1, RNA-directed RNA polymerase (Solanum lycopersicum).
3.3. Expression of rdr3 and rdr4 is induced upon infection with CHV1-EP713 and suppressed by the viral p29 protein
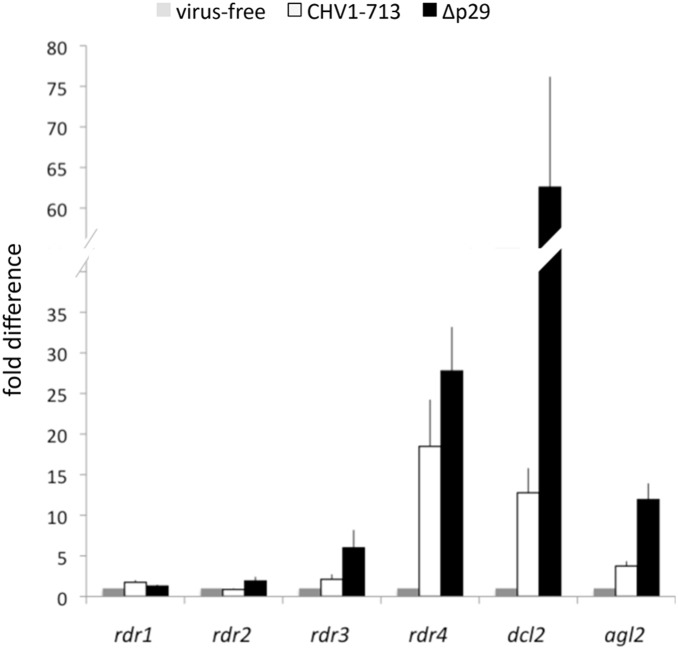
Expression of dcl2 and agl2 involved in the C. parasitica RNA silencing antiviral defense response is induced following hypovirus infection and suppressed by the virus-encoded RNA-silencing suppressor, papain-like protease p29 [20]. Thus, if one or more of the rdr genes were involved in anti-viral defenses, rdr expression would also be expected to be induced in response to viral infection and suppressed by p29. Therefore, the influence of infections by hypovirus CHV-1/EP713 and CHV-1/EP713 in which p29 was deleted (Δp29) on rdr gene expression was examined. Expression of rdr3 and rdr4 was significantly increased upon viral infection, and this response was greater when strain EP155 was infected by Δp29 (Fig. 4). By contrast, rdr1 and rdr2 expression showed only very minor responses to infection with CHV-1/EP713 or Δp29. Expression of rdr4 showed the strongest upregulation, similar to or even exceeding upregulation of dcl2 and agl2 in response to CHV1-EP713. These results indicated that expression of rdr3 and rdr4 was significantly induced by hypovirus infection, a response that was attenuated by the viral p29 RNA-silencing suppressor, thus exhibiting transcriptional responses that were somewhat similar to those of the anti-viral defense genes, dcl2 and agl2.
Figure 4. Accumulation of C. parasitica rdr transcripts in response to infection by hypovirus CHV-1/EP713 and the CHV-1/EP713 mutant Δp29 that lacks the p29 suppressor of RNA silencing.
The relative levels of rdr1–4 gene transcripts were measured by quantitative real-time RT-PCR (2−ΔΔCT method) for uninfected (grey bars), CHV-1/EP713-infected (open bars) and Δp29-infected (black bars) C. parasitica strain EP155. The induction patterns for dcl2 and agl2 transcript accumulation has been previously reported [20], [21] and were also measured in parallel here. The identities of the transcript being measured are indicated at the bottom of the histogram. The fold differences in transcript accumulation are indicated on the Y axis and were calculated using the comparative threshold cycle (ΔΔCT) normalized against tubβ transcript levels and calibrated against the normalized value in the wild-type strain, with standard deviations, based on three independent measurements of two independent RNA preparations, indicated by the error bars. Transcript levels for uninfected C. parasitica were set to a value of one.
3.4. Virus-induced expression patterns for rdr3 and rdr4 are altered in Δagl2 and Δdcl2 strains
To investigate whether the expression of rdr genes may be affected by the deletion of the antiviral genes dcl2 or agl2, the strains Δdcl2 and Δagl2 generated in previous studies [20], [21] were examined for rdr gene expression. Each strain was infected with CHV-1/EP713 or Δp29 and gene expression analyzed by RT-qPCR. Expression of rdr1 and rdr2 in virus-infected and virus-free strains Δdcl2 and Δagl2 (Fig. 5A–B) was largely similar to expression in strain EP155 (Fig. 4), suggesting little or no effect of the anti-viral gene deletions on expression of rdr1 and rdr2. In contrast, rdr3 expression was increased ≥10-fold upon virus infection in both Δdcl2 and Δagl2, much greater than the 2 to 6-fold increase in expression of rdr3 in virus-infected EP155. rdr4 expression was only marginally (≤2-fold) increased in both Δdcl2 and Δagl2 in response to CHV1-EP713 infection, down from ∼20-fold upregulation in CHV1-EP713-infected EP155. The level of upregulation of rdr4 expression in Δp29-infected Δagl2 was largely similar to that in Δp29-infected EP155, whereas deletion of dcl2 markedly decreased rdr4 upregulation to ∼3fold, compared to the almost 30-fold upregulation of rdr4 in Δp29-infected EP155. Expression of rdr1 and rdr2 did not exhibit any detectable changes. These results suggested interactions between rdr3 and rdr4 expression and the induction of the dcl2/agl2 RNA silencing pathway in response to virus infection.
Figure 5. Accumulation of rdr transcripts in response to virus infection in dcl2 and agl2 disruption mutant strains.
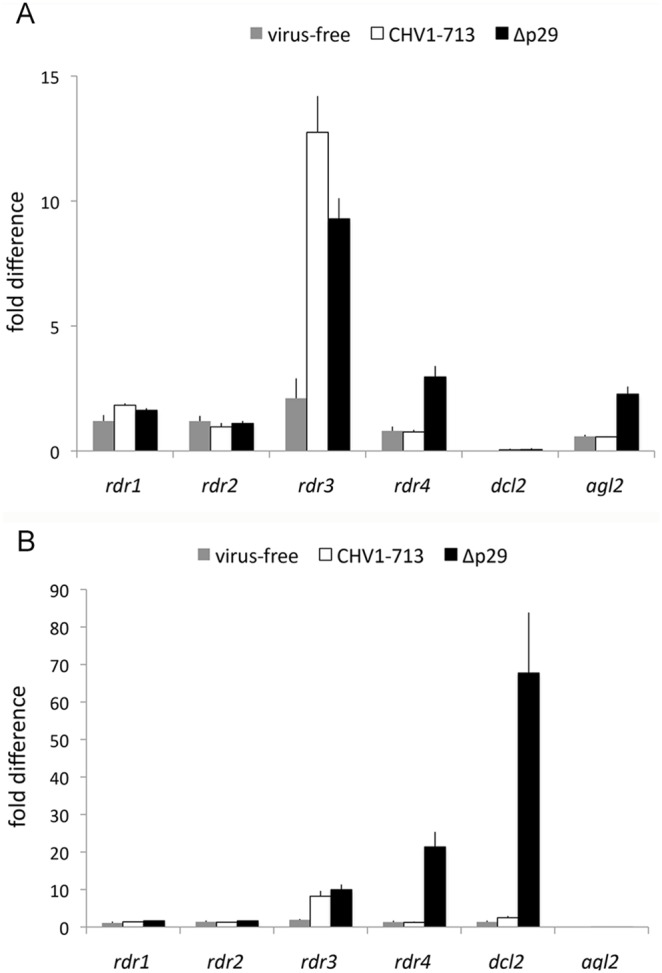
Panel A. Transcript levels for rdr1–4 and agl2 genes were measured in uninfected (grey bars), CHV-1/EP713- (open bars) and Δp29- (black bars) infected Δdcl2 disruption strain as described for Figure 4. Panel B. Transcript levels for rdr1–4 and dcl2 genes were measured in uninfected (grey bars), CHV-1/EP713- (open bars) and Δp29- (black bars) infected Δagl2 disruption strain as described for Figure 4.
3.5. Disruption of RdRP genes and testing of contributions to the RNA silencing antiviral defense response
To further investigate the potential roles of the C. parasitica RdRP genes in the RNA silencing antiviral defense response, all four RdRP genes were disrupted individually and in combination. C. parasitica strains carrying disruptions of the antiviral genes dcl2 and agl2 became severely debilitated following hypovirus infection [16], [20]. So it was a reasonable expectation that disruption of an RdRP gene involved in the RNA silencing antiviral defense pathway would likewise result in similar readily detectable phenotypic changes. The single Δrdr disruption strains each showed growth phenotypes that were essentially identical to the phenotype of the parental strains both in the absence and presence of infecting hypovirus CHV-1/EP713 (Fig. 6).
Figure 6. Effect of rdr gene disruption and hypovirus CHV-1/EP713 on C. parasitica colony growth and morphology.
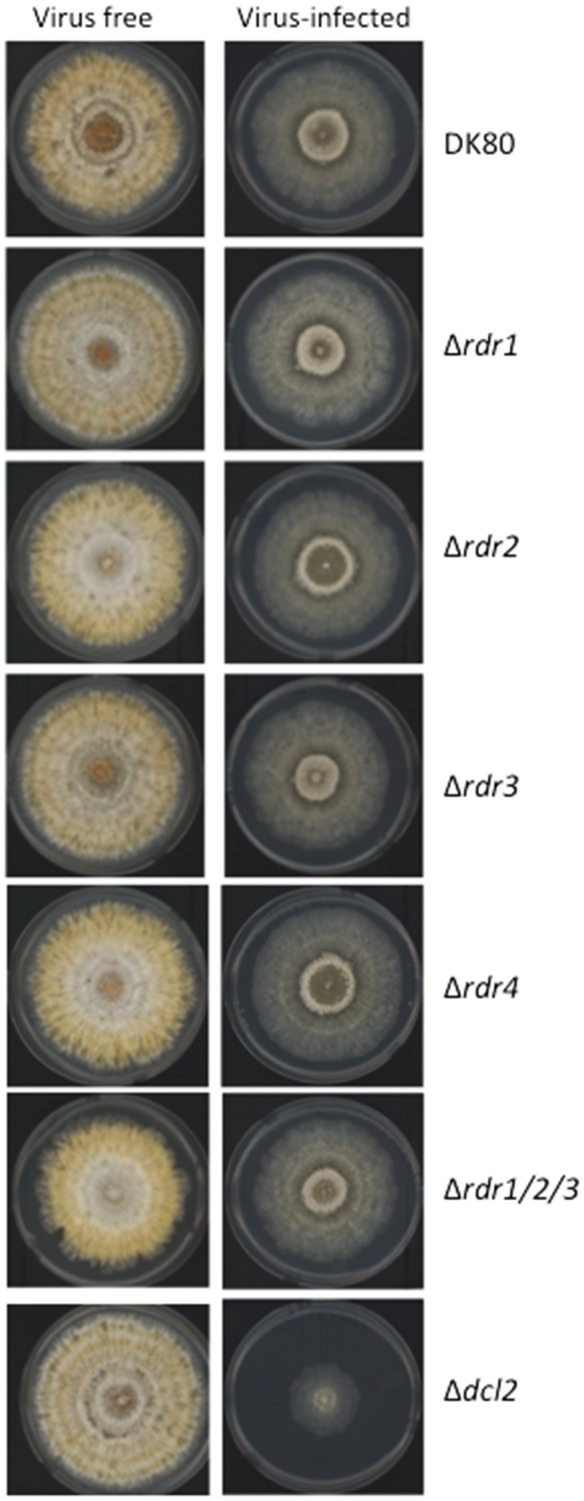
The colonies of uninfected parental strain DK80 and gene disruption mutant strains are shown in the left column while the corresponding hypovirus CHV-1/EP713- infected strains are shown in the right column. The uninfected triple rdr mutant strain Δrdr1/2/3 and uninfected Δdcl2 strains are shown at the bottom of the left column while the corresponding CHV-1/EP713-infected strains are shown in corresponding positions in the right column. Note the severe growth and morphological changes exhibited by the CHV-1/EP713-infected Δdcl2 strain. Cultures were grown for 7 days on PDA.
In view of these results and to account for the possibility that the RdRP genes have some functional redundancy, a series of double- and triple-disruption mutant strains was generated as described in the methods. rdr4 was not included in this analysis, since, as shown above, its predicted protein product lacks a functional RdRP domain. Double and triple Δrdr1/2/3 knockout strains also exhibited normal growth and colony morphology (examples shown in Fig. 6). Additional analysis revealed no reduction in growth of the multiple rdr knockout strains in a virulence assay on inoculated dormant chestnut stems relative to the parental strain or any change in the production of asexual spores (data not shown). N. crassa qde-1 mutant strains showed sensitivity to DNA damaging agents [31]. In contrast, the C. parasitica double and triple rdr mutant strains were as sensitive as the wild-type strain to increasing concentrations of histidine and hydroxyurea (Table 2). The combined results for all single, double and triple rdr knockout strains revealed no essential role for the rdr genes in fungal growth, asexual sporulation, virulence or response to DNA damage-related stress.
Table 2. Colony diameter (cm) in DNA-damage sensitivity assay measured after 3 days of growth.
| Treatment | PDA only | 1.5 mg/ml histidine | 3.5 mg/ml histidine | 2 mg/ml hydroxyurea | 4 mg/ml hydroxyurea | |||||||||||||||
| Spores | 106 | 5×103 | 5×102 | 50 | 106 | 5×103 | 5×102 | 50 | 106 | 5×103 | 5×102 | 50 | 106 | 5×103 | 5×102 | 50 | 106 | 5×103 | 5×102 | 50 |
| EP155 | 3.5 | 4.5 | 4.7 | 4.2 | 4.8 | 4.7 | 4.6 | 4.1 | 3.4 | 3.8 | 3.9 | 3.5 | 2.3 | 2.0 | 1.8 | na+ | 1.3 | na | na | na |
| DK80 | 3.5 | 3.9 | 4.0 | 3.8 | 3.4 | 3.2 | 3.1 | 2.8 | 2.8 | 3.2 | 3.1 | 2.8 | 2.5 | 2.0 | 1.4 | na | 1.6 | na | na | na |
| Δdcl2 | 4.9 | 4.7 | 4.1 | 4.3 | 3.2 | 4.1 | 4.2 | 3.8 | 3.5 | 3.7 | 3.5 | 3.2 | 2.3 | 1.6 | 1.4 | na | 2.0 | na | na | na |
| Δrdr1/2 | 3.8 | 4.4 | 4.5 | 3.9 | 3.4 | 4.1 | 3.7 | 3.3 | 3.2 | 3.4 | 3.3 | 2.9 | 2.4 | 2.0 | 1.7 | na | 1.3 | na | na | na |
| Δrdr1/3 | 3.2 | 4.5 | 3.6 | 3.8 | 3.7 | 3.7 | 3.5 | 3.2 | 3.0 | 3.0 | 3.1 | 3.0 | 2.6 | 2.2 | 1.5 | na | 1.9 | na | na | na |
| Δrdr2/3 | 3.9 | 4.6 | 3.7 | 3.7 | 3.9 | 3.7 | 3.4 | 3.4 | 3.6 | 3.5 | 3.4 | 3.1 | 2.2 | 2.1 | 1.7 | na | 1.6 | na | na | na |
| Δrdr1/2/3 | 3.4 | 4.1 | 4.5 | 4.0 | 3.5 | 3.3 | 3.0 | 3.2 | 3.2 | 3.1 | nd* | nd | 2.1 | 2.2 | 1.5 | na | 1.5 | na | na | na |
| Δrdr4 | 3.7 | 4.6 | 4.3 | 4.4 | 3.7 | 4.3 | 4.0 | 3.6 | 3.0 | 3.5 | 3.3 | 3.0 | 2.1 | 2.2 | 1.6 | na | 1.6 | na | na | na |
*nd, not determined due to contamination on at least one plate.
na, not available due to the poor growth of colonies.
CHV-1/EP713 infection of the double and triple rdr mutant strains resulted in the standard phenotypic changes that were indistinguishable from those of the CHV-1/EP713- infected parental strains (examples shown in Fig. 6). In contrast, growth of the CHV1-EP713-infected Δdcl2 strain used as a control for deletion of antiviral genes in this experiment exhibited a highly debilitated growth phenotype (Fig. 6), as previously described [16]. Moreover, viral RNA accumulation levels were not significantly different between parental and rdr mutant strains (data not shown). Based on the combined results, the C. parasitica rdr genes appear to play no significant role in the RNA silencing antiviral defense response.
3.6. Contribution of host RdRPs to viral recombination
The C. parasitica RNA-silencing antiviral defense pathway was shown to contribute to viral RNA recombination [reviewed in 50]. Defective interfering (DI) RNAs that are readily formed in CHV-1/EP713-infected strain EP155, fail to form in either Δdcl2 or Δagl2 mutant strains [20], [21]. Moreover, non-viral sequences that are rapidly deleted from recombinant hypovirus RNA virus vectors in wild type C. parasitica are stably maintained over prolonged periods of sub-culturing in the Δdcl2 and Δagl2 strains [20], [21]. The collection of rdr gene-disruption mutant strains was tested for their ability to support viral RNA recombination. Mutant and control strains were infected with hypovirus CHV-1/EP713 and independently with recombinant hypovirus Ctp40[2AEGFP] that expresses the enhanced green fluorescent protein (EGFP). The infected strains were sub-cultured every 5 days and monitored for fluorescence, maintainence of the EGFP sequence and production of DI RNAs. Fluorescence disappeared in all independent rdr gene-disruption mutants and in the Δrdr1/2/3 triple mutant within three subcultures, similar to what was observed for the control-infected parental strain. In contrast, fluorescence was retained in the infected Δdcl2 strain for extended periods of sub-culturing. PCR analysis of the extracted viral RNA confirmed the loss of the recombinant viral EGFP gene sequence in the Δrdr mutant strains as early as the first sub-culture and persistence of the EGFP gene sequence in the Δdcl2 strain. Similary, DI RNAs were readily detected in the virus-infected parental strains and in the individual and triple rdr gene disruption strains, but not in the virus-infected Δdcl2 strain (data not shown). Thus, these extensive sets of infection sub-culturing experiments (performed in triplicate) failed to reveal any observable contribution of the rdr genes to RNA silencing-mediated promotion of viral RNA recombination.
3.7. Contribution of RdRPs to transposon silencing
We next investigated whether knocking out one or more of the rdr genes affected the expression of transposable elements. Because the C. parasitica genome contains >100 transposon and transposon-like consensus sequences of >500 bp (see Methods), RNA-seq was performed to investigate whether global transposon expression was altered in the rdr gene knockouts. To identify and retrieve transposon sequences, we used a series of queries to search the annotated C. parasitica genome on the JGI website (see Methods), which identified 104 known or putative transposable elements. Illumina sequencing reads representing the entire transcriptome in Δrdr1/2/3, Δrdr1/2 and Δrdr4 and the parental DK80 strain were then used in RNA-seq mappings to these transposon sequences and expression analysis in CLCBio’s Genomics Workbench. About 0.1% of the total reads in each strain mapped to the transposon sequences (not shown), and RNA-seq detected expression for 60 of the transposon sequences in at least one of the strains. The RNA-seq results indicated that expression of these transposon sequences was very variable: some sequences yielded <10 mapped reads, whereas others resulted in >9,000 mapped reads. However, only ∼20 transposable elements displayed significant expression (i.e., >100 mapped reads per sequence), none of which showed significant (i.e., ≥2 fold) up- or down-regulation in any of the rdr gene knockout strains (results for Δrdr1/2/3 and Δrdr4 are shown in Table S1). Thus, disruption of the rdr genes had no apparent effect on transposon transcript accumulation.
Discussion
Molecular genetic characterization of the RNA silencing components encoded by C. parasitica has clearly demonstrated that the role of RNA silencing as an antiviral defense response observed in plants and invertebrates extends to the fungi. However, only a subset of the RNA silencing apparatus, a single Dicer protein, DCL2, and a single Argonaute protein, AGL2, are required for this important function. The antiviral defense response functions independently of the additional Dicer gene, dcl1, and three additional Argonaute genes, agl1, agl3 and agl4, encoded by this fungus. The extensive analysis of the C. parasitica RdRP genes described here failed to find any evidence for contributions by any of these components to the antiviral defense response.
The four C. parasitica rdr genes fit into the three clades of RdRPs found in most filamentous fungi [51] that correspond to the three N. crassa RdRP genes. The C. parasitica rdr4 gene clustered in the clade containing N. crassa RRP-3, for which no function has been identified. Interestingly, rdr4 displayed alternative splicing, resulting in each case in transcripts whose products lacked an intact conserved RdRP domain, including the essential DXDGD motif. Thus, rdr4 appears to be a non-functional RdRP pseudogene. In this regard, there is considerable evidence for the loss and expansion of RNA silencing genes in fungi [52], e.g., the apparent degeneration of the Aspergillus nidulans QDE-1 ortholog [51] and truncation of A. nidulans Dicer DCL-1 and Argonaute SMS-2 orthologs [17].
Of the three C. parasitica RdRP genes with intact RdRP motifs, rdr3 clustered with N. crassa SAD-1, required for meiotic silencing of unpaired DNA (MSUD), while both rdr1 and rdr2 clustered with N. crassa qde-1, involved in vegetative silencing or Quelling. Since the single Dicer gene dcl2 and single Argonaute gene agl2 required for the antiviral defense response in C. parasitica are orthologs of the N. crassa Dicer (dcl-2) and Argonaute (qde-2) genes involved in Quelling, the rdr1 and rdr2 genes would logically be the most likely candidates to be involved in possibly amplifying the antiviral response.
Examination of RdRP gene expression levels in response to hypovirus infection showed that rdr1 and rdr2 transcript levels did not increase following virus infection as was observed for the dcl2 and agl2 antiviral defense genes. Surprisingly, the accumulation of rdr3 and rdr4 transcripts did significantly increase in response to virus infection (Fig. 4). Moreover, as reported for dcl2 and agl2 [20], [21], rdr3 and rdr4 expression was further increased following infection of the Δp29 virus lacking the suppressor of RNA silencing p29 (Fig. 4). Altered patterns of rdr3 and rdr4 expression were also observed in the Δdcl2 and Δagl2 mutant strains (Fig. 5). Interestingly, expression of the N. crassa rdr4 ortholog rrp3 increased 30-fold in response to dsRNA, leading Choudhary et al. [53] to propose that rrp3 may play a role in the formation of viral or transposon dsRNA.
While the transcript accumulation results indicated some level of interaction at the expression level between the antiviral defense genes and at least rdr3 and rdr4, supporting evidence for a role of the rdr genes in the antiviral defense response was not forthcoming from extensive disruption analysis of the rdr genes. No alterations in either virus-induced symptoms or in virus accumulation levels, as observed for dcl2 and agl2 disruption mutants, were detected in single or multiple rdr gene disruption mutant strain including the triple rdr1, rdr2, rdr3 mutant. Thus, we conclude that C. parasitica is able to mount a robust RNA-silencing antiviral defense response without contributions from the encoded rdr genes. This clearly contrasts with the observations that RdRPs serve to amplify the RNA silencing antiviral defense response in plants [14], [22], [23] and in C. elegans [24]–[26]. However, the very large induction in C. parasitica dcl2 expression in response to CHV1-EP713 infection [20], [21] may preclude the need for RdRP amplification of the antiviral defense signal.
We recently reported that the C. parasitica Dicer and Argonaute proteins involved in antiviral defense also contribute to viral RNA recombination to produce viral defective interfering (DI) RNAs and to delete non-viral RNA sequences from viral-based expression vectors [20], [21]. DI RNA formation and vector RNA stability was indistinguishable in parental, individual rdr mutant and triple rdr mutant strains. Thus, the rdrs also appear not to contribute to this RNA-silencing antiviral defense-related effect.
The accumulation of transposable element transcripts and fragments have been reported to be modulated by RNA-silencing pathways in several filamentous fungi [12], [13], [15], [32], [54], [55], and specific roles for RdRPs in this process have been reported for Cryptococcus neoformans [12], [32], Mucor circinelloides [13] and N. crassa [54]. Illumina sequencing of C. parasitica small RNAs isolated from wild type and Δdcl2/Δagl2 strains identified the presence of DCL2/AGL2-derived transposable element-specific siRNAs (data not shown), indicating targeting of TE RNA by the antiviral defense RNA-silencing pathway. However, no significant increase in TE transcript accumulation was observed in RNA-seq analysis of the rdr knockout strains, including the triple knockout strain (Table S1). In this regard, Nolan et al. [11] reported that silencing of the introduced LINE1-like transposon Tad in N. crassa required one of the two Dicer genes and Argonaute gene qde-2, but surprisingly, not the RdRP gene qde1. The mechanism leading to the generation of TE-specific siRNAs in C. parasitica requires additional investigation.
The determination that C. parasitica rdr genes do not play a significant role in the RNA-silencing antiviral defense response, the primary aim of the study, is important information with bearing on future investigations. Disruption of individual and combinations of rdr genes also resulted in no observable changes in growth, colony morphology, asexual sporulation or virulence on chestnut. Similar results were observed for individual argonaute disruption mutants and individual and double dicer gene-disruption mutants in the absence of virus infection. Thus, the RNA-silencing components in C. parasitica also appear not to contribute to developmental, metabolic or morphological functions during vegetative growth, as has been reported for dcl2 and rdr3 disruption mutants of Trichoderma atroviride [56]. Reports of a role for RNA-silencing pathways in stress responses have been mixed. The N. crassa gdq-1 rdr and dicer gene mutant strains were reported to be more sensitive to histidine and DNA-damaging agents, such as hydroxyurea [31], while C. neoformans RNA-silencing mutants showed no alterations in stress responses [12]. The C. parasitica individual and combined rdr mutants and Δdcl2 failed to show any altered response to histidine or hydroxyurea (Table 2) and appear not to be involved in stress responses. However, the extensive characterization reported here does not eliminate possible roles for the C. parasitica rdrs in co-suppression or meiotic silencing, a determination that will require the development of additional experimental capabilities with this fungus.
Supporting Information
Disruption of C. parasitica rdr1 in EP155. The 4957 bp fragment (850 bp left arm + rdr1 gene 3763 bp + 344 bp right arm) is replaced by pro-trpC-hph cassette (2172 bp). The primer pair R1f (310 bp upstream of the disruption fragment) and R1r (727 bp downstream of the disruption fragment) was designed to confirm the disruption yielding a 6.0 kb fragment from the wild-type strain and a 3.2 kb fragment from Δrdr1.
(TIF)
Disruption of C. parasitica rdr2 . Δrdr2 was generated by the replacement of the 1.5 kb fragment in the rdr2 gene with a 3-kb hph cassette. The disruption confirmation primer R2f/R2r pair amplified the intact rdr2 gene (∼4.1 kb) in the wild-type strain and a 5.7 kb fragment in Δrdr2.
(TIF)
Disruption of C. parasitica rdr4 gene. A fragment including 37 bp upstream of rdr4 gene start codon and 3836 bp of the rdr4 gene containing two alternative spliced introns (showing in dashed lines in the wild type strain) was replaced with the loxP-flanked hph cassette (1.4 kb). Primer pair R4f/R4r yielded a 2.4 kb PCR product for Δrdr4 versus a 4.8 kb fragment for the wild-type strain.
(TIF)
RNA-seq results for identified transposon element sequences for strains Δrdr1/2/3 and Δ rdr4 .
(XLSX)
Acknowledgments
We thank Jennifer van Renterghem for technical assistance, Dr. Chris Schardl, University of Kentucky, for the plasmids pKAES173 and pKAES175 and Dr. William Reid, University of Maryland, for assistance in submission of the RNA-seq data to the NCBI sequence read archive.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. The raw RNA-seq data have now been deposited in the NCBI’s sequence read archive (SRA) under BioProject PRJNA258639. All other data are contained within the paper and supporting information.
Funding Statement
This work was supported by the University of Maryland and by Public Health Service grants GM555981 and AI076219 (to DLN). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dang YK, Yang QY, Xue ZH, Liu Y (2011) RNA Interference in Fungi: Pathways, Functions, and Applications. Eukaryotic Cell 10: 1148–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chang S-S, Zhang Z, Liu Y (2012) RNA interference pathways in fungi: mechanisms and functions. Ann. Rev. Microbiol. 66: 305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cortesi P, McCulloch CE, Song H, Lin H, Milgroom MG (2001) Genetic control of horizontal virus transmission in the chestnut blight fungus, Cryphonectria parasitica . Genetics 159: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Biella S, Smith ML, Aist JR, Cortesi P, Milgroom MG (2002) Programmed cell death correlates with virus transmission in a filamentous fungus. Proc. Biol. Soc. 269: 2269–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi GH, Dawe AL, Churbanov A, Smith ML, Milgroom MG, et al. (2012) Molecular characterization of vegetative incompatibility genes that restrict hypovirus transmission in the chestnut blight fungus Cryphonectria parasitica . Genetics 190: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang D-X, Spiering MJ, Dawe AL, Nuss DL (2014) Vegetative incompatibility loci with dedicated roles in allorecognition restrict mycovirus transmission in Chestnut Blight fungus. Genetics 197: 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kinsey JA (1990) Tad, a LINE-like transposable element of Neurospora can transpose between nuclei in heterokaryons. Genetics 126: 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kempken F, Kuck U (1998) Evidence for circular transposition derivatives from the fungal hAT-transposon Restless. Current Genet. 34: 200–203. [DOI] [PubMed] [Google Scholar]
- 9. Kinsey JA, Garrett-Engele PW, Cambareri EB, Selker EU (1994) The Neurospora transposon Tad is sensitive to repeat-induced point mutation (RIP). Genetics 138: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou Y, Cambareri EB, Kinsey JA (2001) DNA methylation inhibits expression and transposition of the Neurospora Tad retrotransposon. Genetics 265: 748–754. [DOI] [PubMed] [Google Scholar]
- 11. Nolan T, Braccini L, Azzalin G, De Toni A, Macino G, et al. (2005) The post-transcriptional gene silencing machinery functions independently of DNA methylation to repress a LINE-like retrotransposon in Neurospora crassa . Nucleic Acids Research 33: 1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janbon G, Maeng S, Yang D-H, Ko Y-J, Jung K-W, Moyrand F, et al. (2010) Characterization of the role of RNA silencing components in Cryptococcus neoformans . Fungal Genetics and Biology 47: 1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nicolas FE, Moxon S, de Haro JP, Calo S, Grigoriev IV, et al. (2010) Endogenous short RNAs generated by Dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides . Nucleic Acids Research 38: 5535–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang X-B, Wu Q, Ito T, Cillo F, Li W-X, et al. (2010) RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nunes CC, Gowda M, Sailsbery J, Xue M, Chen F, et al. (2011) Diverse and tissue-enriched small RNAs in the plant pathogenic fungus Magnaporthe oryzae . BMC Genomics 12: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Segers GC, Zhang X, Deng F, Sun Q, Nuss DL (2007) Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. USA 104: 12902–12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hammond TM, Andrewski MD, Roossinck MJ, Keller NP (2008) Aspergillus mycoviruses are targets and suppressors of RNA silencing. Eukaryotic Cell 7: 350–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ding SW (2010) RNA based antiviral immunity. Nat. Rev. Immunol. 10: 632–644. [DOI] [PubMed] [Google Scholar]
- 19. Hammond SM (2005) Dicing and Slicing: The core machinery of the RNA interference pathway. FEBS Lett. 579: 5822–5829. [DOI] [PubMed] [Google Scholar]
- 20. Sun Q, Choi GH, Nuss DL (2009) A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc. Natl. Acad. Sci. USA 106: 17927–17932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang X, Nuss DL (2008) A host dicer is required for defective viral RNA production and recombinant virus vector RNA instability for a positive sense RNA virus. Proc. Natl. Acad. Sci. USA 105: 16749–16754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC (2000) An RNA dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, et al. (2010) Arabidopsis RNA-dependent RNA polymerase and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell tpc.109.073056. [DOI] [PMC free article] [PubMed]
- 24. Felix M-A, Ashe A, Piffaretti J, Wu G, Nuez I, et al. (2011) Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to Nodaviruses. PLoS Biol. 9, e1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu R, Yigit E, Li WX, Ding S-W (2009) An RIG-I-like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabitis elegans . PLoS Pathogen 5, e100028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schott DH, Cureton DK, Whelan SP, Hunter CP (2005) An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 102: 18420–18424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cogoni C, Macino G (1999) Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399: 166–169. [DOI] [PubMed] [Google Scholar]
- 28. Calo S, Nicolas FE, Vila A, Torres-Martinez S, Ruiz-Vazquez RM (2012) Two distinct RNA-dependent RNA polymerases are required for initiation and amplification of RNA silencing in the basal fungus Mucor circinelloides . Molecular Microbiology 83: 379–394. [DOI] [PubMed] [Google Scholar]
- 29. Shiu PK, Raju NB, Zickler D, Metzenberg RL (2001) Meiotic silencing by unpaired DNA. Cell 107: 905–916. [DOI] [PubMed] [Google Scholar]
- 30. Shiu PK, Metzenberg RL (2002) Meiotic silencing by unpaired DNA properties, regulation and suppression. Genetics 161: 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee H-C, Chang S-S, Choudhary S, Aalto AP, Maiti M, et al. (2009) qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459: 274–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang X, Hsueh Y-P, Li W, Floyd A, Skalsky R, et al. (2011) Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes and Development 24: 2566–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lan X, Yao Z, Zhou Y, Shang J, Lin H, et al. (2008) Deletion of the cpku80 gene in the chestnut blight fungus, Cryphonectria parasitica, enhances gene disruption efficiency. Curr. Genet. 53: 59–66. [DOI] [PubMed] [Google Scholar]
- 34. Chen B, Choi GH, Nuss DL (1994) Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 264: 1762–1764. [DOI] [PubMed] [Google Scholar]
- 35. Hillman BI, Shapira R, Nuss DL (1990) Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 80: 950–956. [Google Scholar]
- 36. Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2– a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Churchill ACL, Ciufetti LM, Hansen DR, Van Etten, HD, Van Alfen, NK (1990) Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 17: 25–31. [Google Scholar]
- 38. Kuhn R, Torres RM (2002) Cre/loxP recombination system and gene targeting. Methods Mol. Biol. 180: 175–204. [DOI] [PubMed] [Google Scholar]
- 39. Zhang D-X, Lu H-L, Liao X, St. Leger RJ, Nuss DL (2013) Simple and efficient recycling of fungal selectable marker genes with the Cre-LoxP recombination system via anastomosis. Fungal Genet. Biol. 61: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Florea S, Andreeva K, Machado C, Mirabito PM, Schardl CL (2009) Elimination of marker genes from transformed filamentous fungi by unselected transient transfection with a Cre-expressing plasmid. Fungal Genet. Biol. 46: 721–730. [DOI] [PubMed] [Google Scholar]
- 41. Segers GC, van Wezel R, Zhang X, Hong Y, Nuss DL (2006) Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryotic Cell 5: 896–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Spiering MJ, Faulkner JR, Zhang D-X, Machado C, Grossman RB, et al. (2008) Role of the LolP cytochrome P450 monooxygenase in loline alkaloid biosynthesis. Fungal Genetics and Biol. 45: 1307–1314. [DOI] [PubMed] [Google Scholar]
- 43. Zhang D-X, Stromberg AJ, Spiering MJ, Schardl CL (2009) Coregulated expression of loline alkaloid-biosynthesis genes in Neotyphodium uncinatum cultures. Fungal Genet. Biol. 46: 517–530. [DOI] [PubMed] [Google Scholar]
- 44. Suzuki N, Geletka LM, Nuss DL (2000) Essential and dispensable virus-encoded replication elements revealed by efforts to develop hypoviruses as gene expression vectors. J. Virol. 74: 7568–7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang X, Segers G, Sun Q, Deng F, Nuss DL (2008) Characterization of hypovirus-derived small RNAs generated in the chestnut blight fungus by an inducible DCL-2 dependent pathway. J. Virol. 82: 2613–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Catalanotto C, Azzalin G, Macino G, Cogoni C (2002) Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora . Genes & Development 16: 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Salgado PS, Koivunen MRL, Makeyev EV, Bamford DH, Stuart DI, et al. (2006) The structure of an RNAi polymerase links RNA silencing and transcription. PLoS Biology 4: 2274–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wassenegger M, Krczal G (2006) Nomenclature and functions of RNA-directed RNA polymerases. Trends in Plant Science 11: 142–151. [DOI] [PubMed] [Google Scholar]
- 49. Cerutti H, Casas-Mollano JA (2006) On the origin and functions of RNA-mediated silencing: from protists to man. Current Genetics 50: 81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nuss DL (2011) Mycoviruses, RNA silencing and viral RNA recombination. Adv. Vir. Res. 80: 25–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hammond TM, Keller NP (2005) RNA silencing in Aspergillus nidulans is independent of RNA-dependent RNA polymerase. Genetics 169: 607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nakayashiki H, Kadotani N, Mayama S (2006) Evolution and diversification of RNA silencing proteins in fungi. J. Mol. Evol. 63: 127–135. [DOI] [PubMed] [Google Scholar]
- 53. Choudhary S, Lee H-C, Maiti M, He Q, Cheng P, et al. (2007) A double-stranded-RNA response program important for RNA interference efficiency. Mol. Cell. Biol. 27: 3995–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chicas A, Cogoni C, Macino G (2004) RNAi-dependent and RNAi-independnet mechanisms contribute to silencing of RIPed sequences in Neurospora crassa. Nuc. Acids Res. 32: 4237–4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Murata T, Kadotani N, Yamaguchi M, Tosa Y, Mayama S, et al. (2007) SiRNA-dependent and-independent post-transcriptional cosuppression of the LTR-retrotransposon MAGGY in the phytopathogenic fungus Magnaporthe oryzae. Nuc. Acids Res. 35: 5987–5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Carreras-Villasenor N, Esquivel-Naranjo EU, Villalobos-Escobedo JM, Abreu-Goodger C Herrera-Estrella A (2013) The RNAi machinery regulates growth and development in the filamentous fungus Trichoderma atroviride . Molecular Microbiology 89: 96–112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Disruption of C. parasitica rdr1 in EP155. The 4957 bp fragment (850 bp left arm + rdr1 gene 3763 bp + 344 bp right arm) is replaced by pro-trpC-hph cassette (2172 bp). The primer pair R1f (310 bp upstream of the disruption fragment) and R1r (727 bp downstream of the disruption fragment) was designed to confirm the disruption yielding a 6.0 kb fragment from the wild-type strain and a 3.2 kb fragment from Δrdr1.
(TIF)
Disruption of C. parasitica rdr2 . Δrdr2 was generated by the replacement of the 1.5 kb fragment in the rdr2 gene with a 3-kb hph cassette. The disruption confirmation primer R2f/R2r pair amplified the intact rdr2 gene (∼4.1 kb) in the wild-type strain and a 5.7 kb fragment in Δrdr2.
(TIF)
Disruption of C. parasitica rdr4 gene. A fragment including 37 bp upstream of rdr4 gene start codon and 3836 bp of the rdr4 gene containing two alternative spliced introns (showing in dashed lines in the wild type strain) was replaced with the loxP-flanked hph cassette (1.4 kb). Primer pair R4f/R4r yielded a 2.4 kb PCR product for Δrdr4 versus a 4.8 kb fragment for the wild-type strain.
(TIF)
RNA-seq results for identified transposon element sequences for strains Δrdr1/2/3 and Δ rdr4 .
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. The raw RNA-seq data have now been deposited in the NCBI’s sequence read archive (SRA) under BioProject PRJNA258639. All other data are contained within the paper and supporting information.