Abstract
Background
CXCL12 is a small chemotactic cytokine belonging to the CXC chemokine family expressed in various organs. It contributes to the migration, invasion and angiogenesis of cancer cells. Recently, the CXCL12 G801A polymorphism was shown to be associated with an increased risk of various kinds of cancers, but the results were too inconsistent to be conclusive.
Methods
To solve the problem of inadequate statistical power and conflicting results, a meta-analysis of published case-control studies was performed, including 4,435 cancer cases and 6,898 controls. Odds ratios (ORs) and their 95% confidence intervals (CIs) were used to determine the strength of association between CXCL12 G801A polymorphism and cancer risk.
Results
A significant association between CXCL12 G801A polymorphism and cancer risk was found under all genetic models. Further, subgroup analysis stratified by ethnicity suggested a significant association between CXCL12 G801A polymorphism and cancer risk in the Asian subgroup under all genetic models. However, in the Caucasian subgroup, a significant association was only found under an additive genetic model and a dominant genetic model. The analysis stratified by cancer type found that CXCL12 G801A polymorphism may increase the risk of breast cancer, lung cancer, and “other” cancers. Based on subgroup stratified by source of controls, a significant association was observed in hospital-based studies under all genetic models.
Conclusions
The CXCL12 G801A polymorphism is associated with an increased risk of cancer based on current published data. In the future, large-scale well-designed studies with more information are needed to better estimate possible gene-gene or gene-environment interactions.
Introduction
Chemokines are small glycoproteins that contribute to the regulation of various biological processes [1]. CXCL12, also known as stromal cell-derived factor 1(SDF-1), is a small chemotactic cytokine belonging to the CXC chemokine family that is constitutively expressed in various organs [2]. It contributes to the regulation of leukocyte trafficking and many essential biological processes, including cardiac and neuronal development, stem cell motility, neovascularization, and tumorigenesis [3]–[7].
CXCL12 binds primarily to the CXCR4 receptor, resulting in a CXCL12/CXCR4 receptor-ligand system involving a one-on-one interaction [8], [9]. CXCR4 may play a vital role in the metastatic processes of many types of cancers, including colorectal, breast and oral squamous cell carcinoma [10]–[12]. Further research has emphasized the key role of CXCR4 in tumor cell malignancy; the activation of CXCR4 by CXCL12 has been shown to induce the migration, invasion and angiogenesis of tumor cells [13], [14].
CXCL12 is located on chromosome 10q11.1 and has a G→A mutation at position 801 in the 3′-untranslated region in its β transcriptional splice variant [15], [16]. The CXCL12 G801A polymorphism may be essential to increasing the production of a CXCL12 protein that has been shown to be associated with an increased risk of various kinds of cancers, such as breast cancer, lung cancer and lymphoma [17]–[19]. Recently, numerous studies have shown that the CXCL12 G801A polymorphism occurs in different types of cancers, but the results have been too inconsistent to be conclusive. In addition, the sample size of each study is relatively small; thus, their statistical power is too low to detect associations between the CXCL12 G801A polymorphism and cancer risk. Meta-analysis is a powerful method for resolving inconsistent findings from a relatively large number of subjects. To solve the problem of inadequate statistical power and conflicting results, we performed this meta-analysis of published case-control studies.
Materials and Methods
Literature Search
Two investigators independently searched for eligible studies of the associations between CXCL12 G801A polymorphism and cancer risk. Studies published through March 2014 were identified through a computerized search of PubMed without language limitation. The key words used in this search were as follows: (CXCL12, SDF-1 or rs1801157) and (cancer, tumor, carcinoma or neoplasm) and polymorphism. The references of all identified publications were also searched for additional studies. Studies included in this meta-analysis had to meet the following inclusion criteria: (a) used a case-control study design, (b) evaluated CXCL12 G801A polymorphism and cancer risk, (c) reported detailed genotype frequencies of cases and controls or these could be calculated from the text of the manuscript, and (d) the control subjects were in agreement with the Hardy-Weinberg equilibrium (HWE).
Data Extraction
Two investigators extracted the data independently, and disagreements were settled by discussion. The following data were extracted from the eligible studies: the first author's name, year of publication, country of origin, ethnicity, the source of controls, and numbers of genotyped cases and controls. If the data was not available, study authors were contacted to request missing data.
Statistical Analysis
ORs and their 95% CIs were used to determine the strength of association between the CXCL12 G801A polymorphism and cancer risk. The significance of the pooled OR was determined using the Z test, and P<0.05 was considered statistically significant. Additive (A vs. G), dominant (GA+AA vs. GG), and recessive (AA vs. GG+GA) genetic models were investigated. Subgroup analysis was performed by ethnicity, cancer type (if one cancer type contained less than two studies, it was defined as “other”), and source of controls, either hospital or population controls. HWE was tested using the chi-square test among controls, and P<0.05 was considered a significant departure from HWE. If the P value for heterogeneity was >0.05 and I 2<50%, indicating an absence of heterogeneity between studies, the fixed-effects model (the Mantel-Haenszel method) was used. In contrast, if either the P value for heterogeneity was ≤0.05 or I 2 was ≥50%, indicating heterogeneity among the studies, the more appropriate random-effects model (the DerSimonian and Laird method) was used. Sensitivity analyses were performed to assess the stability of the results. Funnel plots and Egger's linear regression test were used to diagnose potential publication bias, and P<0.05 was used to indicate possible publication bias. All analyses were performed using Stata software. P values were based on two-sided tests.
Results
Characteristics of Eligible Studies
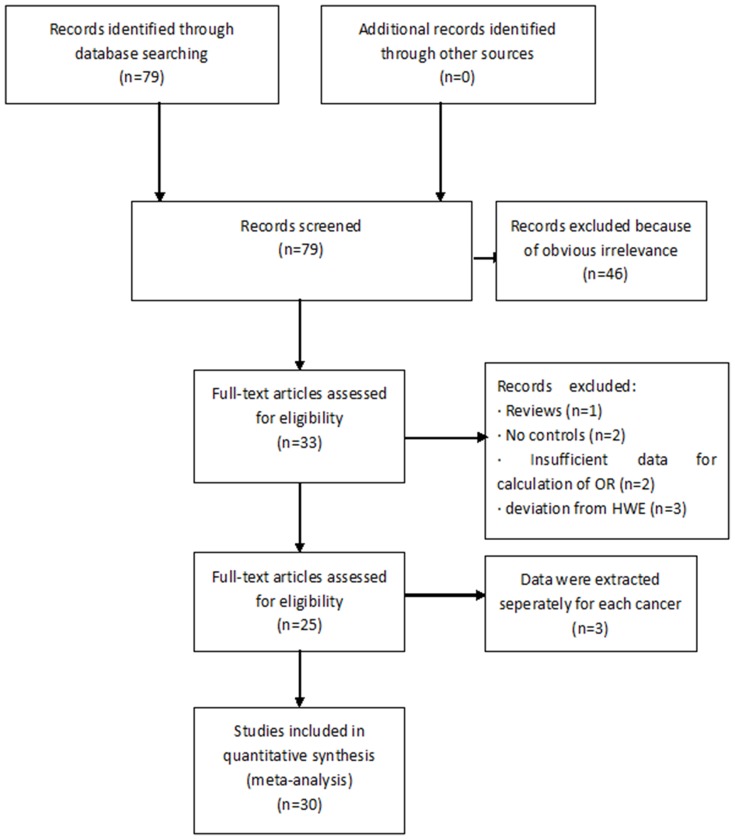
Our meta-analysis was performed according to guidelines of the “Preferred Reporting Items for Systematic Reviews and Meta-Analyses” (PRISMA) statement (Checklist S1) and “Meta-analysis on Genetic Association Studies” statement (Checklist S2). Figure 1 graphically illustrates the study flow chart. The literature search yielded 79 potentially relevant articles. After screening the titles and abstracts, 46 articles were excluded because of obvious irrelevance. In addition, after reading the full text of the 33 remaining articles, 8 articles were excluded for the following reasons: article was a review (n = 1), articles lacked controls (n = 2), articles had insufficient data (n = 2), and articles deviated from HWE (n = 3). Articles that reported data for different types of cancers were treated as independent studies. Thus, 25 articles [17]–[41] (30 independent case-control studies) met the inclusion criteria; they included 4,435 cancer cases and 6,898 controls. Data collected from the included studies are summarized in Table 1.
Figure 1. Flow chart of study selection in the meta-analysis.
Table 1. Characteristics of eligible studies included in the meta-analysis.
| author | year | cancer | country | ethnicity | control source | HWE | Cases | controls | ||||
| GG | GA | AA | GG | GA | AA | |||||||
| Zafiropoulos [17] | 2004 | breast cancer | Greece | Caucasian | HB | 0.764 | 98 | 136 | 30 | 101 | 92 | 19 |
| 2004 | bladder cancer | Greece | Caucasian | HB | 0.124 | 31 | 32 | 5 | 67 | 71 | 10 | |
| 2004 | skin cancer | Greece | Caucasian | HB | 0.262 | 64 | 38 | 8 | 169 | 164 | 30 | |
| Razmkhah [18] | 2005 | lung cancer | Iran | Asian | HB | 0.504 | 25 | 38 | 9 | 145 | 97 | 20 |
| Razmkhah [20] | 2005 | breast cancer | Iran | Asian | HB | 0.682 | 105 | 139 | 34 | 101 | 67 | 13 |
| Hidalgo-Pascual [21] | 2007 | colorectal cancer | Spain | Caucasian | PB | 0.77 | 212 | 128 | 9 | 319 | 172 | 25 |
| Hirata [22] | 2007 | prostate cancer | Japan | Asian | HB | 0.651 | 72 | 78 | 17 | 91 | 63 | 13 |
| Dimberg [23] | 2007 | colorectal cancer | Sweden | Caucasian | HB | 0.117 | 84 | 62 | 5 | 81 | 56 | 4 |
| de Oliveira [24] | 2007 | CML | Brazil | Caucasian | HB | 0.628 | 10 | 11 | 4 | 39 | 18 | 3 |
| Khademi [25] | 2008 | head and neck cancer | Iran | Asian | HB | 0.504 | 64 | 84 | 8 | 145 | 97 | 20 |
| Vairaktaris [26] | 2008 | oral cancer | Mixed | Caucasian | PB | 0.448 | 104 | 51 | 4 | 55 | 41 | 5 |
| Lin [27] | 2009 | breast cancer | China | Asian | HB | 0.621 | 106 | 98 | 16 | 175 | 136 | 23 |
| Vazquez-Lavista [28] | 2009 | bladder cancer | Mexico | Mixed | PB | 0.822 | 29 | 15 | 3 | 83 | 39 | 4 |
| de Oliveira [19] | 2009 | breast cancer | Brazil | Caucasian | HB | 0.939 | 59 | 41 | 3 | 61 | 32 | 4 |
| 2009 | NHL | Brazil | Caucasian | HB | 0.356 | 36 | 33 | 1 | 59 | 26 | 5 | |
| 2009 | HL | Brazil | Caucasian | HB | 0.356 | 22 | 10 | 4 | 59 | 26 | 5 | |
| Kruszyna [29] | 2010 | breast cancer | Poland | Caucasian | PB | 0.686 | 123 | 61 | 9 | 136 | 58 | 5 |
| Kruszyna [30] | 2010 | laryngeal cancer | Poland | Caucasian | PB | 0.114 | 69 | 46 | 3 | 181 | 67 | 2 |
| Lee [31] | 2011 | NSCLC | China | Asian | HB | 0.379 | 99 | 112 | 36 | 171 | 136 | 21 |
| de Oliveira [32] | 2011 | breast cancer | Brazil | Caucasian | HB | 0.758 | 32 | 21 | 2 | 37 | 15 | 2 |
| Cacina [33] | 2012 | endometrial cancer | Turkey | Asian | HB | 0.061 | 49 | 52 | 12 | 69 | 64 | 6 |
| Tee [34] | 2012 | cervical cancer | Taiwan | Asian | HB | 0.697 | 37 | 29 | 10 | 164 | 140 | 33 |
| Kucukgergin [35] | 2012 | bladder cancer | Turkey | Asian | HB | 0.35 | 58 | 58 | 26 | 94 | 80 | 23 |
| Liarmakopoulos [36] | 2013 | gastric cancer | Greece | Caucasian | HB | 0.116 | 39 | 43 | 6 | 205 | 229 | 46 |
| Perim [37] | 2013 | ALL | Brazil | Caucasian | PB | 0.72 | 33 | 18 | 3 | 46 | 11 | 1 |
| Razmkhah [38] | 2013 | gastric cancer | Iran | Asian | HB | 0.504 | 66 | 48 | 10 | 145 | 97 | 20 |
| 2013 | colorectal cancer | Iran | Asian | HB | 0.504 | 62 | 39 | 8 | 145 | 97 | 20 | |
| Shi [39] | 2013 | colorectal cancer | Taiwan | Asian | PB | 0.1 | 141 | 113 | 4 | 248 | 52 | 0 |
| Kontogianni [40] | 2013 | breast cancer | Greece | Caucasian | HB | 0.585 | 114 | 118 | 29 | 247 | 198 | 35 |
| Cai [41] | 2013 | renal cell cancer | China | Asian | HB | 0.127 | 150 | 111 | 61 | 237 | 136 | 29 |
CML: chronic myeloid leukemia; NHL: non-hodgkin lymphoma; HL: hodgkin lymphoma; NSCLC: non small cell lung cancer; ALL:acute lymphocytic leukemia; HB: hospital-based; PB:population-based.
Results of the Meta-analysis
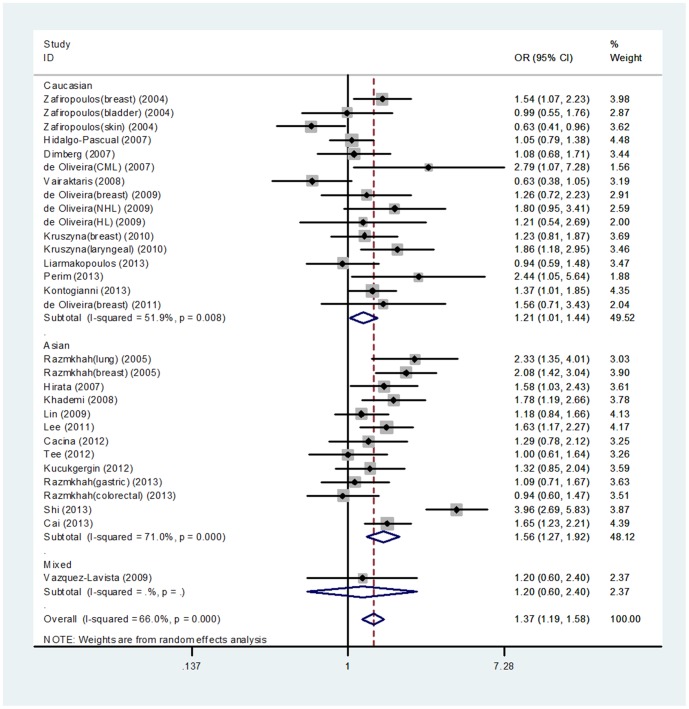
A significant association between CXCL12 G801A polymorphism and cancer risk was found under an additive genetic model (OR = 1.30, 95% CI = 1.16–1.45), a dominant genetic model (OR = 1.37, 95%CI = 1.19–1.58), and a recessive genetic model (OR = 1.38, 95% CI = 1.13–1.69). Subgroup analysis stratified by ethnicity also suggested a significant association between CXCL12 G801A polymorphism and cancer risk in the Asian subgroup under an additive genetic model (OR = 1.45, 95% CI = 1.23–1.70), a dominant genetic model (OR = 1.56, 95%CI = 1.27–1.92) (Figure 2), and a recessive genetic model (OR = 1.71, 95% CI = 1.41–2.07). In the Caucasian subgroup, a significant association was found under an additive genetic model (OR = 1.16, 95%CI = 1.00–1.34) and a dominant genetic model (OR = 1.21, 95%CI = 1.01–1.44).
Figure 2. Forest plot of CXCL12 G801A polymorphism and cancer risk under a dominant genetic model (GA+AA vs. GG) stratified by ethnicity.
Furthermore, in the analysis by stratified cancer type, a significantly increased risk was found in breast cancer and lung cancer under all genetic models. In addition, under the additive and dominant genetic models, a significantly increased risk was found in “other” cancers. However, no significant association with this polymorphism was observed in bladder, colorectal and gastric cancers. Base on subgroup analysis by source of controls (hospital or population controls), a significant association was observed in hospital-based studies under all genetic models (Table 2).
Table 2. Pooled ORs and 95% CIs of the association between CXCL12 G801A polymorphism and cancer risk.
| A vs G | GA/AA vs GG | AA vs GA/GG | |||||||
| OR(95%CI) | I 2(%) | P-value | OR(95%CI) | I 2(%) | P-value | OR(95%CI) | I 2(%) | P-value | |
| overall | 1.30(1.16–1.45) | 67.3 | 0.000 | 1.37(1.19–1.58) | 66.0 | 0.000 | 1.38(1.13–1.69) | 33.3 | 0.041 |
| ethnicity | |||||||||
| Asian | 1.45(1.23–1.70) | 70.8 | 0.000 | 1.56(1.27–1.92) | 71.0 | 0.000 | 1.71(1.41–2.07) | 40.8 | 0.062 |
| Caucasian | 1.16(1.00–1.34) | 52.9 | 0.007 | 1.21(1.01–1.44) | 51.9 | 0.008 | 1.11(0.87–1.41) | 12.0 | 0.316 |
| Cancer type | |||||||||
| Breast cancer | 1.32(1.17–1.48) | 0.0 | 0.537 | 1.43(1.23–1.66) | 0.0 | 0.436 | 1.41(1.06–1.87) | 0.0 | 0.843 |
| Bladder cancer | 1.22(0.97–1.55) | 0.0 | 0.577 | 1.19(0.87–1.62) | 0.0 | 0.731 | 1.58(0.96–2.61) | 0.0 | 0.744 |
| Lung cancer | 1.65(1.34–2.04) | 0.0 | 0.610 | 1.80(1.36–2.39) | 17.6 | 0.271 | 2.24(1.41–3.57) | 0.0 | 0.475 |
| colorectal cancer | 1.33(0.76–2.34) | 91.6 | 0.000 | 1.43(0.73–2.80) | 91.7 | 0.000 | 0.86(0.53–1.41) | 36.6 | 0.192 |
| Gastric cancer | 0.98(0.77–1.25) | 0.0 | 0.505 | 1.02(0.74–1.39) | 0.0 | 0.637 | 0.87(0.48–1.55) | 0.0 | 0.476 |
| Others | 1.31(1.06–1.61) | 68.8 | 0.000 | 1.36(1.05–1.75) | 66.7 | 0.001 | 1.47(0.96–2.26) | 51.3 | 0.020 |
| Source of controls | |||||||||
| HB | 1.27(1.15–1.41) | 49.1 | 0.004 | 1.34(1.18–1.52) | 45.0 | 0.011 | 1.49(1.28–1.75) | 28.1 | 0.105 |
| PB | 1.42(0.94–2.14) | 86.8 | 0.000 | 1.49(0.93–2.40) | 86.6 | 0.000 | 1.09(0.69–1.73) | 46.1 | 0.085 |
Sensitivity Analysis
A single study was excluded each time to evaluate the effect of an individual study on the combined ORs and 95% CIs. The omission of any single study did not significantly change the pooled effects of the additive, dominant and recessive genetic models; these findings confirmed that the meta-analysis results were statistically robust and that our results were reliable and stable (data not shown).
Publication Bias
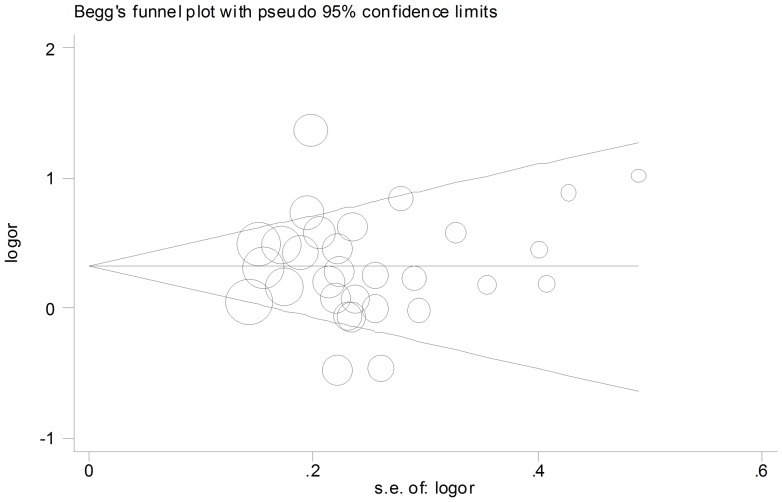
Begg's funnel plot and Egger's test were performed to assess the publication bias of this set of publications. The shape of the funnel plot did not show obvious publication bias (Figure 3). Similarly, Egger's test revealed no evidence of publication bias (P = 0.996 for the additive genetic model; P = 0.953 for the dominant genetic model; and P = 0.342 for the recessive genetic model).
Figure 3. Funnel plot for studies of the association of CXCL12 G801A polymorphism and cancer risk under a dominant genetic model (GA+AA vs. GG).
Discussion
CXCL12 is primarily produced by stromal cells and is important for the growth, angiogenesis and metastasis of tumor cells [42], [43]. The CXCL12 G801A polymorphism may be essential to increasing the production of CXCL12 protein. Furthermore, overexpression of CXCL12 is associated with the development and metastasis of many kinds of cancers. The CXCL12 G801A polymorphism has been investigated in various types of cancers. However, the results of previous studies conflicted about the association between CXCL12 G801A polymorphism and cancer risk. In order to resolve this controversy, the present meta-analysis, which included 4,435 cases and 6,898 controls from 30 case-control studies, explored the association between CXCL12 G801A polymorphism and cancer risk. Our results indicated that CXCL12 G801A polymorphism was associated with an increased risk of cancers.
Additionally, our study contributes the results of subgroup analyses stratified by ethnicity, cancer type and source of controls. Our results indicated that the CXCL12 G801A polymorphism was associated with an increased risk of cancers, especially for breast and lung cancer. However, no significant association was observed for bladder, colorectal and gastric cancers. This is maybe because cancers types differ by carcinogenic mechanisms and environmental exposures and have disparate responses to CXCL12 G801A genotypes. In addition, gene-gene and gene-environment interactions may influence the association between CXCL12 G801A polymorphism and susceptibility to specific cancers [44]–[49]. Furthermore, for some cancer types defined as “other”, only a few studies were published; therefore, it was difficult to detect small, but meaningful associations. Consequently, large-scale and detailed studies are needed to examine these relationships.
In the subgroup analysis by ethnicity, the CXCL12 G801A polymorphism was found to confer an increased cancer risk among Asians under all the genetic models, whereas in the Caucasian subgroup, a significant association was only observed under an additive genetic model and a dominant genetic model. The mechanism that explains this ethnic difference is unknown, but differences in genetic backgrounds and life-styles may contribute to different genetic characteristics and susceptibility to specific cancers. In the present meta-analysis, we failed to find significant relationships between CXCL12 G801A polymorphism and cancer risk in ethnic groups besides Asian and Caucasian. Therefore, more studies in other ethnic groups may be necessary for further progress in this area.
In the subgroup analysis stratified by the source of controls, significant associations were observed in hospital-based studies but not in population-based studies. However, most of the included studies were hospital-based because hospital controls are more readily available. Therefore, the findings in this subgroup should be interpreted with caution. Additional population-based studies are needed to better evaluate this association.
We identified previous genome-wide studies relevant to our research, such as those conducted in breast cancer and lung cancer [50]–[55]. However, these studies were not included in our analysis because their raw data was not available. No significant association between CXCL12 G801A polymorphism and cancer risk was observed in those studies, which conflicts with our results. Possible reasons for this inconsistency are that genome-wide association studies are limited by their relatively small samples and can't contain all kinds of populations.
Two meta-analyses similar to that presented herein were performed by Gong et al. [56] in 2012 and MA et al. [57] in 2012, who also investigated the influence of CXCL12 G801A polymorphism on susceptibility to cancers, with similar conclusions. There were two main differences between these two studies and our study. First, the study of Gong et al. included two articles that deviated from HWE, which were excluded from our study. Second, the literature searches of the two prior meta-analyses were conducted before October 2011 and May 2011, respectively. Since then, several additional studies of the CXCL12 G801A polymorphism and cancer risk were published. Therefore, the sample was larger and the statistical power was greater in our meta-analysis.
We conducted the largest and most comprehensive quantitative meta-analysis of the relationship between CXCL12 G801A polymorphisms and cancer risk. Nevertheless, we recognize some limitations of this meta-analysis. First, our meta-analysis was based primarily on unadjusted ORs with 95% CIs because potential correlative factors, such as environmental factors and other lifestyle habits, were not available. Second, the meta-analysis was limited by the relatively small number of available studies, which limited our ability to perform subgroup analysis for every type of cancer. Third, our analysis was limited to Asian and Caucasian ethnicities, and it is uncertain whether these results are generalizeable to other populations. In addition, cancer is a multi-factorial disease that results from complex interactions between many genetic and environmental factors. Therefore, a single gene or single environmental factor is unlikely to explain cancer susceptibility.
In conclusion, this meta-analysis suggested that CXCL12 G801A polymorphism was associated with an increased risk of cancer based on current published data. In the future, large-scale well-designed studies with more information about potential correlative factors are needed to better estimate possible gene-gene or gene-environment interactions.
Supporting Information
PRISMA 2009 Checklist.
(DOC)
Meta-analysis on Genetic Association Studies Checklist.
(DOC)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no funding or support to report.
References
- 1. Locati M, Murphy PM (1999) Chemokines and chemokine receptors: biology and clinical relevance in inflammation and AIDS. Annu Rev Med 50: 425–440. [DOI] [PubMed] [Google Scholar]
- 2. Muller A, Homey B, Soto H, Ge N, Catron D, et al. (2001) Involvement of chemokine receptors in breast cancer metastasis. Nature 410: 50–56. [DOI] [PubMed] [Google Scholar]
- 3. Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, et al. (1998) Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci USA 95: 9448–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barbieri F, Bajetto A, Porcile C, Pattarozzi A, Massa A, et al. (2006) CXC receptor and chemokine expression in human meningioma: SDF1/CXCR4 signaling activates ERK1/2 and stimulates meningioma cell proliferation. Ann NY Acad Sci 1090: 332–343. [DOI] [PubMed] [Google Scholar]
- 5. Hattori K, Heissig B, Tashiro K, Honjo T, Tateno M, et al. (2001) Plasma elevation of stromal cell derived factor-1 induces mobilization of mature and immature hematopoietic progenitor and stem cells. Blood 97: 3354–3360. [DOI] [PubMed] [Google Scholar]
- 6. Lane WJ, Dias S, Hattori K, Heissig B, Choy M, et al. (2000) Stromalderived factor 1-induced megakaryocyte migration and platelet production is dependent on matrix metalloproteinases. Blood 96: 4152–4159. [PubMed] [Google Scholar]
- 7. Petit I, Jin D, Rafii S (2007) The SDF-1-CXCR4 signaling pathway: a molecular hub modulating neo-angiogenesis. Trends Immunol 28: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bleul CC, Farzan M, Choe H, Parolin C, Clark-Lewis I, et al. (1996) The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature 382: 829–833. [DOI] [PubMed] [Google Scholar]
- 9. Tachibana K, Hirota S, Iizasa H, Yoshida H, Kawabata K, et al. (1998) The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature 393: 591–594. [DOI] [PubMed] [Google Scholar]
- 10. Schimanski CC, Schwald S, Simiantonaki N, Jayasinghe C, Gönner U, et al. (2005) Effect of chemokine receptors CXCR4 and CCR7 on the metastatic behavior of human colorectal cancer. Clin Cancer Res 11: 1743–1750. [DOI] [PubMed] [Google Scholar]
- 11. Kato M, Kitayama J, Kazama S, Nagawa H (2003) Expression pattern of CXC chemokine receptor-4 is correlated with lymph node metastasis in human invasive ductal carcinoma. Breast Cancer Res 5: R144–R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uchida D, Begum NM, Almofti A, Nakashiro K, Kawamata H, et al. (2003) Possible role of stromal-cell-derived factor-1/CXCR4 signaling on lymph node metastasis of oral squamous cell carcinoma. Exp Cell Res 290: 289–302. [DOI] [PubMed] [Google Scholar]
- 13. Chen Y, Stamatoyannopoulos G, Song CZ (2003) Down-regulation of CXCR4 by inducible small interfering RNA inhibits breast cancer cell invasion in vitro. Cancer Res 63: 4801–4804. [PubMed] [Google Scholar]
- 14. Mori T, Doi R, Koizumi M, Toyoda E, Ito D, et al. (2004) CXCR4 antagonist inhibits stromal cell-derived factor 1-induced migration and invasion of human pancreatic cancer. Mol Cancer Ther 3: 29–37. [PubMed] [Google Scholar]
- 15. Shirozu M, Nakano T, Inazawa J, Tashiro K, Tada H, et al. (1995) Structure and chromosomal localization of the human stromal cell-derived factor 1 (SDF-1) gene. Genomics 28: 495–500. [DOI] [PubMed] [Google Scholar]
- 16. Watanabe MA, de Oliveira Cavassin GG, Orellana MD, Milanezi CM, Voltarelli JC, et al. (2003) SDF-1 gene polymorphisms and syncytia induction in Brazilian HIV-1 infected individuals. Microb Pathog 35: 31–34. [DOI] [PubMed] [Google Scholar]
- 17. Zafiropoulos A, Crikas N, Passam AM, Spandidos DA (2004) Significant involvement of CCR2-64I and CXCL12-3a in the development of sporadic breast cancer. J Med Genet 41: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Razmkhah M, Doroudchi M, Ghayumi SMA, Erfani N, Ghaderi A (2005) Stromal cell-derived factor-1 (SDF-1) gene and susceptibility of Iranian patients with lung cancer. Lung Cancer 49: 311–315. [DOI] [PubMed] [Google Scholar]
- 19. de Oliveira KB, Oda JM, Voltarelli JC, Nasser TF, Ono MA, et al. (2009) CXCL12 rs1801157 Polymorphism in Patients with Breast Cancer, Hodgkin's Lymphoma, and Non-Hodgkin's Lymphoma. J Clin Lab Anal 23: 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Razmkhah M, Talei AR, Doroudchi M, Khalili-Azad T, Ghaderi A (2005) Stromal cell-derived factor-1 (SDF-1) alleles and susceptibility to breast carcinoma. Cancer Lett 225: 261–266. [DOI] [PubMed] [Google Scholar]
- 21. Hidalgo-Pascual M, Galan JJ, Chaves-Conde M, Ramírez-Armengol JA, Moreno C, et al. (2007) Analysis of CXCL12 3′UTR G>A polymorphism in colorectal cancer. Oncol Rep 18: 1583–1587. [DOI] [PubMed] [Google Scholar]
- 22. Hirata H, Hinoda Y, Kikuno N, Kawamoto K, Dahiya AV, et al. (2007) CXCL12 G801A polymorphism is a risk factor for sporadic prostate cancer susceptibility. Clin Cancer Res 13: 5056–5062. [DOI] [PubMed] [Google Scholar]
- 23. Dimberg J, Hugander A, Löfgren S, Wågsäter D, et al. (2007) Polymorphism and circulating levels of the chemokine CXCL12 in colorectal cancer patients. Int J Mol Med 19: 11–15. [PubMed] [Google Scholar]
- 24. de Oliveira CE, Cavassin GG, Perim Ade L, Nasser TF, de Oliveira KB, et al. (2007) Stromal cell-derived factor-1 chemokine gene variant in blood donors and chronic myelogenous leukemia patients. J Clin Lab Anal 21: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Khademi B, Razmkhah M, Erfani N, Gharagozloo M, Ghaderi A, et al. (2008) SDF-1 and CCR5 genes polymorphism in patients with head and neck cancer. Pathol Oncol Res 14: 45–50. [DOI] [PubMed] [Google Scholar]
- 26. Vairaktaris E, Vylliotis A, Spyridonodou S, Derka S, Vassiliou S, et al. (2008) A DNA polymorphism of stromal-derived factor-1 is associated with advanced stages of oral cancer. Anticancer Res 28: 271–275. [PubMed] [Google Scholar]
- 27. Lin GT, Tseng HF, Yang CH, Hou MF, Chuang LY, et al. (2009) Combinational polymorphisms of seven CXCL12-related genes are protective against breast cancer in Taiwan. OMICS 13: 165–172. [DOI] [PubMed] [Google Scholar]
- 28. Vázquez-Lavista LG, Lima G, Gabilondo F, Llorente L (2009) Genetic association of monocyte chemoattractant protein 1 (MCP-1) -2518 polymorphism in Mexican patients with transitional cell carcinoma of the bladder. Urology 74: 414–418. [DOI] [PubMed] [Google Scholar]
- 29. Kruszyna Ł, Lianeri M, Rubis B, Knuła H, Rybczyńska M, et al. (2010) CXCL12-3′ G801A polymorphism is not a risk factor for breast cancer. DNA Cell Biol 29: 423–427. [DOI] [PubMed] [Google Scholar]
- 30. Kruszyna L, Lianeri M, Rydzanicz M, Szyfter K, Jagodziński PP (2010) SDF1-3′A gene polymorphism is associated with laryngeal cancer. Pathol Oncol Res 16: 223–227. [DOI] [PubMed] [Google Scholar]
- 31. Lee YL, Kuo WH, Lin CW, Chen W, Cheng WE, et al. (2011) Association of genetic polymorphisms of CXCL12/SDF1 gene and its receptor,CXCR4, to the susceptibility and prognosis of non-small cell lung cancer. Lung Cancer 73: 147–152. [DOI] [PubMed] [Google Scholar]
- 32. de Oliveira KB, Guembarovski RL, Oda JM, Mantovani MS, Carrera CM, et al. (2011) CXCL12 rs1801157 polymorphism and expression in peripheral blood from breast cancer patients. Cytokine 55: 260–265. [DOI] [PubMed] [Google Scholar]
- 33. Cacina C, Bulgurcuoglu-Kuran S, Iyibozkurt AC, Yaylim-Eraltan I, Cakmakoglu B (2012) Genetic variants of SDF-1 and CXCR4 genes in endometrial Carcinoma. Mol Biol Rep 39: 1225–1229. [DOI] [PubMed] [Google Scholar]
- 34. Tee YT, Yang SF, Wang PH, Tsai HT, Lin LY, et al. (2012) G801A polymorphism of human stromal cell-derived factor 1 gene raises no susceptibility to neoplastic lesions of uterine cervix. Int J Gynecol Cancer 22: 1297–1302. [DOI] [PubMed] [Google Scholar]
- 35. Kucukgergin C, Isman FK, Dasdemir S, Cakmakoglu B, Sanli O, et al. (2012) The role of chemokine and chemokine receptor gene variants on the susceptibility and clinicopathological characteristics of bladder cancer. Gene 511: 7–11. [DOI] [PubMed] [Google Scholar]
- 36. Liarmakopoulos E, Theodoropoulos G, Vaiopoulou A, Rizos S, Aravantinos G, et al. (2013) Effects of stromal cell-derived factor-1 and survivin gene polymorphisms on gastric cancer risk. Mol Med Rep 7: 887–892. [DOI] [PubMed] [Google Scholar]
- 37. de Lourdes Perim A, Guembarovski RL, Oda JM, Lopes LF, Ariza CB, et al. (2013) CXCL12 and TP53 genetic polymorphisms as markers of susceptibility in a Brazilian children population with acute lymphoblastic leukemia (ALL). . Mol Biol Rep 40: 4591–4596. [DOI] [PubMed] [Google Scholar]
- 38. Razmkhah M, Ghaderi A (2013) SDF-1alpha G801A polymorphism in Southern Iranian patients with colorectal and gastric cancers. Indian J Gastroenterol 32: 28–31. [DOI] [PubMed] [Google Scholar]
- 39. Shi MD, Chen JH, Sung HT, Lee JS, Tsai LY, et al. (2013) CXCL12-G801A polymorphism modulates risk of colorectal cancer in Taiwan. Arch Med Sci 9: 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kontogianni P, Zambirinis CP, Theodoropoulos G, Gazouli M, Michalopoulos NV, et al. (2013) The impact of the stromal cell-derived factor-1-30A and E-selectin S128R polymorphisms on breast cancer. Mol Biol Rep 40: 43–50. [DOI] [PubMed] [Google Scholar]
- 41. Cai C, Wang LH, Dong Q, Wu ZJ, Li MY, et al. (2013) Association of CXCL12 and CXCR4 gene polymorphisms with the susceptibility and prognosis of renal cell carcinoma. Tissue Antigens 82: 165–170. [DOI] [PubMed] [Google Scholar]
- 42. Nagasawa T, Kikutani H, Kishimoto T (1994) Molecular cloning and structure of a pre-B-cell growth-stimulating factor. Proc Natl Acad Sci USA 91: 2305–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA (1996) A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1). J Exp Med 184: 1101–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singh V, Jaiswal PK, Kapoor R, Kapoor R, Mittal RD (2014) Impact of chemokines CCR5Δ32, CXCL12G801A, and CXCR2C1208T on bladder cancer susceptibility in north Indian population. Tumour Biol 35: 4765–4772. [DOI] [PubMed] [Google Scholar]
- 45. de Oliveira KB, Guembarovski RL, Guembarovski AM, da Silva do Amaral Herrera AC, Sobrinho WJ, et al. (2013) CXCL12, CXCR4 and IFNγ genes expression: implications for proinflammatory microenvironment of breastcancer. Clin Exp Med 13: 211–219. [DOI] [PubMed] [Google Scholar]
- 46. Sei S, Boler AM, Nguyen GT, Stewart SK, Yang QE, et al. (2001) Protective effect of CCR5 delta 32 heterozygosity is restricted by SDF-1 genotype in children with HIV-1 infection. AIDS 15: 1343–1352. [DOI] [PubMed] [Google Scholar]
- 47. Kerdivel G, Le Guevel R, Habauzit D, Brion F, Ait-Aissa S, et al. (2013) Estrogenic potency of benzophenone UV filters in breast cancer cells: proliferative and transcriptional activity substantiated by docking analysis. PLoS One 8: e60567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jin F, Brockmeier U, Otterbach F, Metzen E (2012) New insight into the SDF-1/CXCR4 axis in a breast carcinoma model: hypoxia-induced endothelial SDF-1 and tumor cell CXCR4 are required for tumor cell intravasation. Mol Cancer Res 10: 1021–1031. [DOI] [PubMed] [Google Scholar]
- 49. Beheshti A, Sachs RK, Peluso M, Rietman E, Hahnfeldt P, et al. (2013) Age and space irradiation modulate tumor progression: implications for carcinogenesis risk. Radiat Res 179: 208–220. [DOI] [PubMed] [Google Scholar]
- 50. Lan Q, Hsiung CA, Matsuo K, Hong YC, Seow A, et al. (2012) Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet 44: 1330–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shiraishi K, Kunitoh H, Daigo Y, Takahashi A, Goto K, et al. (2012) A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet 44: 900–903. [DOI] [PubMed] [Google Scholar]
- 52. Lee D, Lee GK, Yoon KA, Lee JS (2013) Pathway-Based Analysis Using Genome-wide Association Data from a Korean Non-Small Cell Lung Cancer Study. PLoS One 8: e65396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li N, Zhou P, Zheng J, Deng J, Wu H, et al. (2014) A polymorphism rs12325489C>T in the lincRNA-ENST00000515084 exon was found to modulate breast cancerrisk via GWAS-based association analyses. PLoS One 9: e98251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Véron A, Blein S, Cox DG (2014) Genome-wide association studies and the clinic: a focus on breast cancer. Biomark Med 8: 287–296. [DOI] [PubMed] [Google Scholar]
- 55. Zhang Y, Long J, Lu W, Shu XO, Cai Q, et al. (2014) Rare Coding Variants and Breast Cancer Risk: Evaluation of Susceptibility Loci Identified in Genome-Wide Association Studies. Cancer Epidemiol Biomarkers Prev 23: 622–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gong H, Tan M, Wang Y, Shen B, Liu Z, et al. (2012) The CXCL12 G801A polymorphism and cancer risk: Evidence from 17 Case-control studies. Gene 509: 228–231. [DOI] [PubMed] [Google Scholar]
- 57. Ma XY, Jin Y, Sun HM, Yu L, Bai J, et al. (2012) CXCL12 G801A polymorphism contributes to cancer susceptibility: a meta-analysis. Cell Mol Biol (Noisy-le-grand) 58 Suppl: OL1702–1708. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA 2009 Checklist.
(DOC)
Meta-analysis on Genetic Association Studies Checklist.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.