Abstract
The influence of diet and genetics was investigated in a healthy white person who had distinctly low methylprednisolone clearance. Pharmacokinetic and pharmacodynamic parameter values were similar on 2 occasions during the consumption of a low-carbohydrate diet and a Weight Watchers diet, indicating that the decreased clearance was unlikely attributable to a change in diet composition. Although the subject was found to be homozygous for CYP3A5*3, genetic findings were not significant for a number of other CYP3A4 and CYP3A5 allelic variants. Because of the high prevalence of CYP3A5*3/*3 in whites and because 5 of 7 white control subjects are also homozygous for CYP3A5*3, this genotype cannot fully explain the reduced metabolism of the drug. Other genetic or contributing factors might have been involved. New polymerase chain reaction–based genotyping methods for functionally defective CYP3A5*6, *8, *9, and *10 alleles were developed in this study. These assays will be useful for CYP3A5 genotype analysis in future clinical studies.
Keywords: Methylprednisolone clearance, steroid pharmacodynamics, low-carbohydrate diet, CYP3A5*3 allele, genotyping method
Corticosteroids are widely used for a variety of diseases because of their anti-inflammatory and immunosuppressive properties. Most steroids are metabolized predominantly in the liver by cytochrome P450 3A (CYP3A) isoenzymes, which are important enzymes that catalyze the metabolism of endogenous steroids, procarcinogens, and approximately 50% of all drugs. CYP3A enzymes are known to be affected by numerous enzyme inducers and inhibitors,1 and a number of induction and inhibitory drug interactions have been documented for corticosteroids.2-5 Presumably, the steroid-drug interactions are attributable to the alternations in CYP3A-mediated corticosteroid metabolism.
The overall activity of CYP3A is contributed by 4 members of the CYP3A subfamily: CYP3A4, CYP3A5, CYP3A7, and CYP3A43. Among all, CYP3A4 and CYP3A5 are the 2 major CYP3A isoenzymes expressed in human adult livers, and CYP3A7 is the major fetal isoform.6 Whereas CYP3A4 expression varies as much as 40-fold among liver and small intestinal tissues,7 significant amounts of CYP3A5 protein are found only in the presence of the wild-type CYP3A5*1 allele.8,9 Because CYP3A4 and CYP3A5 have similar structures and overlapping substrate specificities,10,11 it is difficult to segregate the relative contribution of these 2 enzymes to CYP3A-mediated drug metabolism and to clearly distinguish their role in the genotype-phenotype relationship.
The molecular basis for alternations in CYP3A activity has been investigated recently. Numerous single nucleotide polymorphisms (SNPs) have been identified in the 5′-flanking and coding regions of CYP3A4 and CYP3A5 genes;12 some of them have been shown to be associated with altered activity in vitro.8,13-18 However, none of these SNPs have been shown consistently to contribute significantly to variations in drug clearance in vivo. One of the most often studied alleles, CYP3A5*3, which represents an A to G transversion at nucleotide 6986 that creates a cryptic splice site resulting in the generation of a premature stop codon and a truncated CYP3A5 protein,8 has been shown to be associated with low expression of CYP3A5 protein.8,9 This allele exhibits diversified ethnic differences. Allelic frequency of CYP3A5*3 was found to be 0.93 for whites, 0.32 for African Americans, 0.63 for Hispanics, 0.73 for East Asians, and 0.60 for South Asians,19 contributing to the expression of CYP3A5 protein in 33% and 60% of livers from whites and African Americans, respectively.8
In this report, we describe a case in which a 27-year-old healthy white man exhibited reduced methylprednisolone clearances causing prolonged corticosteroid pharmacodynamics while consuming a low-carbohydrate diet and a Weight Watchers diet. Newly developed genetic tests and previously reported methods for CYP3A4/5 SNPs were applied to help understand genotype-phenotype association in this person.
CASE REPORT
In 2003, we conducted a study at Mercer University that sought to determine the pharmacokinetics and pharmacodynamics of methylprednisolone in healthy subjects with different histamine N-methyltransferase (HNMT) C314T genotypes.20 Ten subjects participated and completed the study. We found that 1 subject had distinctly low methylprednisolone clearance, which approximated a 43% decrease compared to that of other subjects. Moreover, the suppressive effects of methylprednisolone on cortisol and whole blood histamine concentrations were prolonged.
Further investigations revealed that this subject had been consuming a low-carbohydrate, high-protein, high-fat diet for about 2.5 months before his participation in the pharmacokinetic study in March. His initial weight was 109 kg, and it declined to 84 kg about 5 months after the start of the low-carbohydrate diet. After several months of the low-carbohydrate diet consumption, the subject developed mild hypertension and stopped the diet. He then switched to consuming a Weight Watchers diet, in which approximately 25% to 30% of calories were derived from fat and 40% to 50% from carbohydrate.
We speculated that the decreased metabolism of methylprednisolone in this subject could be attributed to the consumption of the low-carbohydrate diet, presumably because of an inhibition of CYP3A activity. Meanwhile, we also suspected that genetic variations in CYP3A4 and CYP3A5 might explain the reduced clearance. To confirm and determine the cause of decreased methylprednisolone clearance and prolonged cortisol and whole blood histamine suppression in this subject, we repeated the pharmacokinetic study at the National Institutes of Health (NIH) in August 2004, when the subject was no longer following the low-carbohydrate diet and was consuming the Weight Watchers diet. We also performed genetic analysis to determine the presence of various SNPs in the CYP3A4 and CYP3A5 genes that have been shown to alter CYP3A activity. Six days before this study, the subject was started on ibuprofen and empiric clindamycin therapy for his root canal. With permission from his oral surgeon, the antibiotic was stopped 2 days before the initiation of the study because clindamycin is minimally metabolized by CYP3A.
MATERIALS AND METHODS
Two separate pharmacokinetic studies were performed at the Center for Clinical Research, Mercer University Southern School of Pharmacy, Atlanta, Georgia, and the NIH, Bethesda, Maryland. Both studies were approved by the corresponding institutional review board. Written informed consent was obtained from the study participant. Consent for additional DNA analysis was also obtained from 7 other subjects who participated in the first pharmacokinetic study at Mercer University. Before each pharmacokinetic study, the health status of the study subject was assessed and ascertained by medical and drug history, physical examination, urinalysis, complete blood chemistry (liver and renal function tests), and hematologic (complete blood count) profile. Screening electrocardiogram was also performed in the first study.
Study Procedures
Each pharmacokinetic study consisted of a baseline (24 hours, no drug) and a methylprednisolone phase (32 or 52 hours, with drug). The subject was fasted for 10 hours before the initiation of both phases and for 2 hours after receiving the methylprednisolone during the drug phase.
For the first pharmacokinetic study, the subject underwent the baseline phase 2 weeks before the methylprednisolone phase. During the baseline phase, no drug was given, and a 9-mL blood sample was collected for plasma cortisol and whole blood histamine concentrations every 2 hours for 24 hours. During the methylprednisolone phase, the subject was given an intravenous bolus dose of 0.6 mg/kg (ideal body weight) methylprednisolone sodium succinate (Solu-Medrol; Upjohn, Kalamazoo, Mich) over 2 minutes. Ten milliliters of blood was drawn at 0, 0.25, 0.5, 1, 1.5, 2, 3, and 4 hours, and then every 2 hours until 24 hours, 28, and 32 hours for the determination of plasma methylprednisolone and cortisol and whole blood histamine concentrations.
For the second study, the methylprednisolone phase was performed immediately after the baseline phase, in which 8 mL of blood was sampled for each time point as described above. After the collection of the last blood sample for the baseline phase, methylprednisolone 0.6 mg/kg (ideal body weight) was administered intravenously to the subject. Eight milliliters blood was then obtained at 0.25, 0.5, 1, 1.5, 2, 3, and 4 hours, and then every 2 hours until 24 hours, and every 4 hours until 52 hours postdose. A 20-mL EDTA blood sample was also collected during this second study for genetic analysis.
Nutritional Assessment
Retrospective dietary information for the low-carbohydrate diet was obtained from the subject by a diet questionnaire and an interview with a registered dietician. For the Weight Watchers diet, specifics about a typical daily intake were obtained by a dietician, and the composition of the diet was analyzed using the software Food Processor (ESHA Research, Salem, Ore).
Analytical Assays
Plasma cortisol and methylprednisolone concentrations were determined simultaneously by a normal phase high-performance liquid chromatography method as previously reported.21 Whole blood histamine concentrations were determined by a commercial enzyme immunoassay kit, according to the manufacturer’s instructions (Immunotech, Marseille, France)
Pharmacokinetic and Pharmacodynamic Analysis
Methylprednisolone pharmacokinetics and pharmacodynamics were analyzed as previously described.20 In brief, methylprednisolone plasma concentrations were fitted with one compartment model. Cortisol and whole blood histamine suppression were assessed by indirect response models, with baseline cortisol analyzed by Fourier analysis.
Determination of Variant CYP3A4 and CYP3A5 Alleles
CYP3A5*3, *6, *8, *9, and *10, and CYP3A4*17 were determined by newly developed polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) tests; methods for CYP3A5*3 and CYP3A4*17 have been reported earlier.16 Whereas PCR-based methods for CYP3A*6 have been described previously by others,22 different primer sequences and restriction enzyme were used in our assay. To design primers for the specific amplification, genomic DNA sequences of 4 human CYP3As (CYP3A4, CYP3A5, CYP3A7, and CYP3A43) were obtained from GenBank (Accession number NG_000004.1), and corresponding regions for each allele were aligned using Vector NTI 8.0. DNA was amplified in a 25-uL reaction mixture containing 40 ng genomic DNA, 0.4 μM primers, 0.4 mM of each dNTP, and 2.5 units AmpliTaq Gold (PerkinElmer Life Analytical Sciences, Boston, Mass). Thermal cycling conditions included 38 cycles at 94°C for 30 seconds, 56°C for 20 seconds, and 72°C for 30 seconds. An initial denaturation step of 95°C for 10 minutes and a final extension step of 72°C for 5 minutes were included. PCR products were purified by QIAquick PCR purification kit (QIAGEN, Valencia, Calif), and specific amplification of the target gene was confirmed by direct sequencing. Five uL of the purified products was directly digested with the restriction enzyme, followed by separation on 3% agarose gel. Table I summarizes the primer sequences and restriction enzymes used and the resultant fragment lengths for each specific allele. Genomic DNAs containing known allele were obtained from previous studies and used as controls to assure specific amplification and complete digestion.15,17
Table I.
Primer Sequences and Restriction Enzymes for the Newly Developed Polymerase Chain Reaction–Restriction Fragment Length Polymorphism (PCR-RFLP) Methods
| Fragment Size (bp)
|
||||||
|---|---|---|---|---|---|---|
| Allele | Primers (5′→3′) | Enzyme | wt/wt | wt/vt | vt/vt | Reference |
| CYP3A4*17 | FP:CTGGACATGTGGGTTTCCTGT | BpmI | 290 | 290, 153, 137 | 153, 137 | 16 |
| RP:AGCAGTTATTTTTAAGAGAGAAAGATAAAT | ||||||
| CYP3A5*3 | FP:CTTTAAAGAGCTCTTTTGTCTcTCA | BseMII | 197 | 197, 162, 35 | 162, 35 | 16 |
| RP:GAAGCCAGACTTTGATCATTATG | ||||||
| CYP3A5*6 | FP:CACAAGACCCCTTTGTGGAGAGCACTcA | BseMII | 128, 38 | 166, 128, 38 | 166 | — |
| RP:CTTTTAAGTGGATGAATTATACGATAT | ||||||
| CYP3A5*8 | FP:CCTGAGTAACTCACCAGCCCTCTG | BsrGI | 204 | 204, 120, 84 | 120, 84 | — |
| RP:ACCATAAGAAGCAAAAGAGGAAGCTCAAGC | ||||||
| CYP3A5*9 | FP:GATTTCATCTAAGCTGTGATGTTG | BtsI | 142, 77, 24 | 142, 101, 77, 24 | 142, 101 | — |
| RP:GGTCATCCCCTCACCTTATTGGGCAAcACTG | ||||||
| CYP3A5*10 | FP:GACCCAGAAACTGCATTGGCATGAGaT | BglII | 151 | 151, 130, 21 | 130, 21 | — |
| RP:CCTTCAGTTAAAAAAATTCTTAATAAAACATAC | ||||||
Mismatched nucleotides with the CYP3A5 sequence are underlined with the lower case. FP, forward primer; RP, reverse primer; wt, wild type; vt, variant.
The existence of defective CYP3A4/5 SNP for CYP3A4*2, *5, *6, *8, *11, *12, *13, and *16, and CYP3A5*7, was examined by direct DNA sequencing after PCR amplification, which was carried out in a 25-μL reaction mixture containing 40 ng genomic DNA, 0.4 μM primers, 0.4 mM of each dNTP, and 2.5 units AmpliTaq Gold. Thermal cycling conditions and primers were the same as reported in the literature.13,14,23 PCR products were purified by QIAquick PCR purification kit before sequencing. Each exon was sequenced in both directions using ABI Prism 3100 Genetic Analyzer (PerkinElmer Cetus, Foster City, Calif) and screened for specific mutations. Using the primers as mentioned above, CYP3A4 exons 5, 7, 9, and 11 were fully sequenced.
For comparison, 7 other white subjects who participated in the first study were used as controls, and the presence of the CYP3A5*3 allele was also determined.
RESULTS
The subject tolerated the study procedures well in both studies except that he experienced drowsiness in the morning after methylprednisolone administration in the first study. Otherwise, he had no other complaints and did not experience any significant adverse reactions.
During the first study period with the low-carbohydrate diet, the subject limited carbohydrate intake to between 20 and 25 g/d, which approximated 5% of total calories. The amount of protein and fat consumed daily could not be estimated because the subject could not recall this information. He ran 2 times per week and was moderately active at work as a physical therapist assistant. During the consumption of the Weight Watchers diet, the total calories for a typical daily intake were calculated to be approximately 2000 calories, with 200 g carbohydrate, 106 g protein, 80 g fat, and 16.5 g dietary fiber. This intake was represented as 41% carbohydrate, 22% protein, and 37% fat of total calories. The subject participated in light-to-moderate activity that burned on average around 250 to 300 calories 2 times per week.
Figure 1 compares methylprednisolone concentration versus time curves for the study participant during the consumption of the low-carbohydrate diet and the Weight Watchers diet with the mean profiles for control subjects who are heterozygous and homozygous for the CYP3A5*3 allele. The decrease in methylprednisolone disposition and increase in area under the concentration versus time curve from time zero to infinity (AUC) for the study subject are clearly demonstrated in Figure 1.
Figure 1.
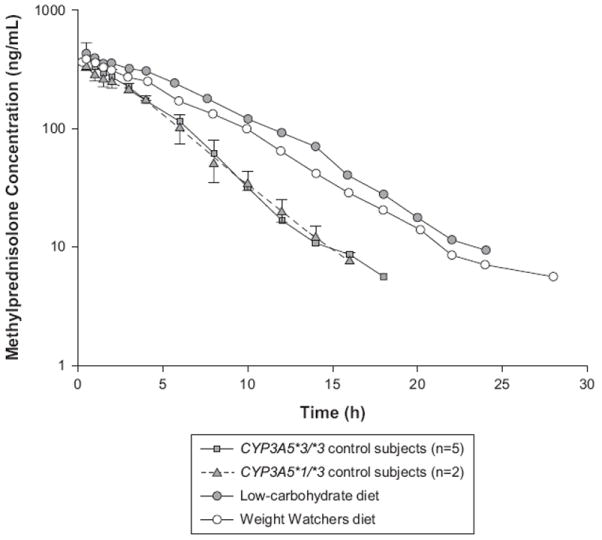
Plasma methylprednisolone concentration versus time curves for the study subject during the consumption of a low-carbohydrate diet and a Weight Watchers diet and the mean profiles for the control subjects who are heterozygous and homozygous for the CYP3A5*3 allele.
Table II summarizes the pharmacokinetic and pharmacodynamic parameter values for the subject during the consumption of the low-carbohydrate diet and the Weight Watchers diet. Parameter estimates obtained from the control subjects were also included. Methylprednisolone pharmacokinetic parameter values were similar between the 2 diets, with a clearance of 13.6 L/h and 16.5 L/h for the low-carbohydrate and the Weight Watchers diet, respectively. These clearances were 43% and 31% lower than the median methylprednisolone clearance obtained from the control subjects. Likewise, half-life and the AUC were around 1.8-fold and 1.6-fold higher for this subject than the median values for the control subjects. Apparently, no differences were observed in the volume of distribution between the 2 diets.
Table II.
Summary of Pharmacokinetic and Pharmacodynamic Parameters for the Study Subject During the Consumption of a Low-Carbohydrate Diet and a Weight Watchers Diet and for the Control Subjects
| Parameters | Low-Carbohydrate Diet | Weight Watchers Diet | Control Subjects |
|---|---|---|---|
| Methylprednisolone pharmacokinetics | |||
| CL, L/h | 13.6 | 16.5 | 23.9 (21.7-26.3)a |
| Vd, L | 90.8 | 101 | 97.7 (78.2-108)a |
| t½, h | 4.64 | 4.25 | 2.62 (2.300-3.230)a |
| AUC, ng·h/mL | 3231 | 2655 | 1679 (1491-1950)a |
| Cortisol suppression | |||
| Kout,h−1 | 0.318 | 0.255 | 0.347 (0.225-0.354)b |
| IC50, ng/mL | 1.00 | 2.53 | 1.43 (1.37-2.32)b |
| Whole blood histamine suppression | |||
| kH, h−1 | 10.1 | 11.7 | 16.1 (6.00-27.4)a |
| , h−1 | 0.309 | 0.374 | 0.319 (0.272-0.352)a |
| IC50, ng/mL | 2.75 | 1.85 | 9.99 (5.20- 22.3)a |
Data are presented as median (range). CL, clearance; Vd, volume of distribution; t½, half-life; AUC, area under the concentration versus time curve from time zero to infinity; kout, first-order elimination rate constant of cortisol; kH, overall disappearance of histamine from the central blood compartment; , zero-order rate of return of histamine to the blood compartment; IC50, concentration of methylprednisolone producing 50% suppression of cortisol circadian secretion or causing 50% inhibition of the return rate of histamine into the blood compartment.
n = 7. See text for explanation for the number of subjects.
n = 3. See text for explanation for the number of subjects.
The suppressive effects of methylprednisolone on cortisol secretion and whole blood histamine concentrations for a typical control subject and this study participant are illustrated in Figures 2 and 3. For a typical control subject, cortisol concentrations rapidly declined in a first-order fashion to reach a nadir at about 10 hours. The levels remained suppressed for a prolonged period of time until 20 to 22 hours, followed by a rapid return toward baseline at about 24 to 28 hours. Similarly, whole blood histamine concentrations decreased to a minimum at 8 to 12 hours and then slowly increased and returned to the baseline level at 24 to 32 hours. For the study participant, cortisol secretion was suppressed up to 48 hours when cortisol concentrations started to return toward baseline at 52 hours, whereas whole blood histamine concentrations were suppressed until 22 hours and returned back to baseline level at 44 to 48 hours.
Figure 2.
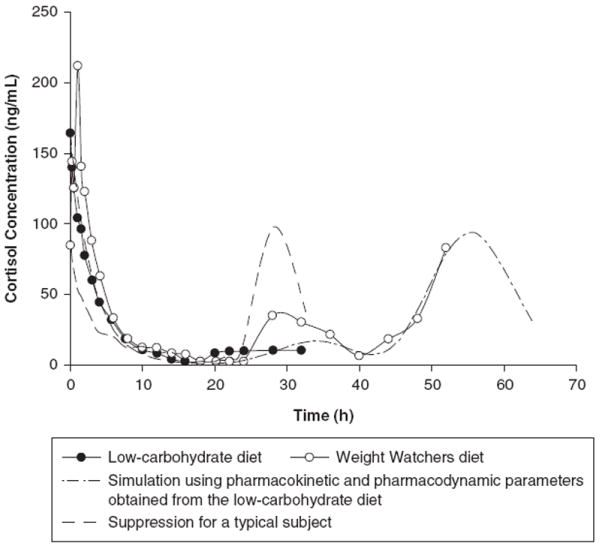
Suppression of cortisol secretion for a typical control subject and the study participant during the consumption of a low-carbohydrate diet and a Weight Watchers diet.
Figure 3.
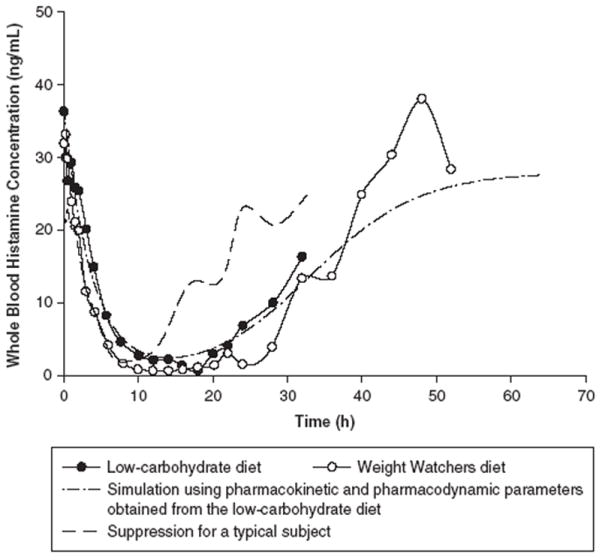
Suppression of whole blood histamine for a typical control subject and the study participant while he was consuming a low-carbohydrate diet and a Weight Watchers diet.
Because subjects with HNMT C314T polymorphism tended to be less sensitive to the suppressive effects of methylprednisolone on cortisol secretion and because this subject has a wild-type HNMT genotype,20 pharmacodynamic parameter values for cortisol suppression were compared only to 3 control subjects who have the same HNMT genotype. Whereas most of the pharmacodynamic parameters were not apparently different, the concentration causing 50% inhibition (IC50) for whole blood histamine suppression was lower for the study participant than that for the control subjects. Otherwise, these parameter values were found to be similar during the consumption of the low-carbohydrate diet and the Weight Watchers diet for the study participant.
Genetic analysis showed that the study subject is homozygous for the variant CYP3A5*3 allele, whereas he is negative for CYP3A4*2, *5, *6, *8, *11, *12, *13, *16 and *17, and CYP3A5*6, *7, *8, *9, and *10. Table III summarizes the characteristics of all variant alleles for CYP3A4 and CYP3A5 and lists the results for the alleles that were tested in this study. Among the 7 control white subjects, 2 were found to be heterozygous and 5 were homozygous for the CYP3A5*3 allele.
Table III.
Characteristics of All Variant Alleles for CYP3A4 and CYP3A5 and the Results for the Alleles That Were Tested in This Study
| Allele | Results | Single Nucleotide Polymorphism | Amino Acid Change | Phenotypic Change | Frequency (%) | Reference |
|---|---|---|---|---|---|---|
| CYP3A4 | ||||||
| *2 | wt/wt | 15713T>C | Ser222Pro | ↓ activity | 2.7 W; 0 B; 0 C | 13 |
| *3 | ND | 23172T>C | Met445Thr | ↔ activity | 0.47 W, 0 B, 1.6 C | 13,14 |
| *4 | ND | 15820C>G | Ile118Val | 1.5 C | 58 | |
| *5 | wt/wt | 15820C>G | Pro218Arg | 1.0 C | 58 | |
| *6 | wt/wt | 17776insA | Frame shift, early stop condon | 0.5 C | 58 | |
| *7 | ND | 6004G>A | Gly56Asp | ↔ activity | 1.41 W | 14 |
| *8 | wt/wt | 13908G>A | Arg130Gln | No detectable protein | 0.33 W | 14 |
| *9 | ND | 14292G>A | Val170Ile | ↔ activity | 0.24 W | 14 |
| *10 | ND | 14304G>C | Asp174His | ↔ activity | 0.24 W | 14 |
| *11 | wt/wt | 21867C>T | The363Met | ↓lower protein, ↔ activity | 0.34 W | 14 |
| *12 | wt/wt | 21896C>T | Leu373Phe | ↓activity | 0.34 W | 14 |
| *13 | wt/wt | 22026C>T | Pro416Leu | No detectable protein | 0.34W | 14 |
| *14 | ND | 44T>C | Leu15Pro | 0 W, 0 B | 59 | |
| *15 | ND | 14269 G>A | Arg162Gln | 0 W; 2.0 B | 59 | |
| *16 | wt/wt | 15603C>G | Thr185Ser | 0 B, 1.4 J, 5.0 M | 59,62 | |
| *17 | wt/wt | 15615T>C | Phe189Ser | ↓ activity | 2.0 W | 15 |
| *18 | ND | 20070T>C | Leu293Pro | ↑activity | 2.0 A | 15 |
| *19 | ND | 23237C>T; 20230G>A | Pro467Ser | ↔ activity | 2.0 A | 15 |
| *20 | ND | 25889insA | Frameshift | No activity | Westlind-Johnsson et al., in press CPT | |
| CYP3A5 | ||||||
| *2 | ND | 27289C>A | Thr398Asn | 0 B; 0 C | 23,63,64 | |
| *3 | vt/vt | 6986A>G | Splicing defect, premature stop condon, truncated protein | Severely ↓ mRNA expression and activity | 95.0 W; 27.0 B; 73.0 C; 76.0 J; 70.0 K | 8,18,23 |
| *4 | ND | 14665A>G | Gln200Arg | 1.0 C | 64 | |
| *5 | ND | 12952T>C | Splicing defect | 1.0 C, 0.3 J | 18,64 | |
| *6 | wt/wt | 14690G>A | Splicing defect resulting in frameshift, truncated protein | None or severely ↓activity | 13.0 B | 8,23 |
| *7 | wt/wt | 27131-32insT | Frame shift | 10.0 B | 23 | |
| *8 | wt/wt | 3699C>T | Arg28Cys | ↓ activity | 0 W, 4.0 B, 0 A | 17 |
| *9 | wt/wt | 19386G>A | Ala337Thr | ↓ activity | 0 W, 0 B, 2.0 A | 17 |
| *10 | wt/wt | 6986A>G; 29753T>C; 31611C>T | Splicing defect; Phe446Ser | ↓activity | 2.0 W, 0 B, 0 A | 17 |
ND, not determined; wt, wild type; vt, variant; W, white; B, black; C, Chinese; J, Japanese; M, Mexican, A, Asian; K, Korean.
DISCUSSION
Drug metabolism is an important determinant that contributes significantly to variations in the pharmacokinetics of drugs. It can be affected by a number of factors that include age, sex, disease states, environment determinants, and genetic characteristics,24 leading to large interindividual variability in pharmacokinetic parameters such as clearance and AUC for some drugs. In this case study, we attempted to determine whether 2 of these factors, diet and genetics, were associated with low methylprednisolone clearance for a healthy white subject.
Initially, we expected that the decreased clearance of the study subject was probably related to diet instead of genetics, because methylprednisolone pharmacokinetics is very sensitive to enzyme inhibition or induction, attributable to the presence of the methyl group at the 6α position. Results from the repeated study showed that pharmacokinetic parameters were very similar between the low-carbohydrate diet and the Weight Watchers diet, indicating that the low clearance was unlikely attributed to the consumption of the low-carbohydrate diet, implying further that CYP3A activity was not significantly inhibited. Although previous studies have shown that a low-carbohydrate, high-protein diet significantly increased the clearance of theophylline, propranolol, antipyrine, and aminopyrine,25-29 these drugs are metabolized by multiple CYP enzymes, and the distinct effects of the diet on various specific enzymes including CYP3A could not be inferred. Methylprednisolone pharmacodynamics appears to be similar between the low-carbohydrate diet and the Weight Watchers diet because all pharmacodynamic parameters except one were not different during the consumption of the 2 diets.
Slight differences in pharmacokinetic parameter values were observed between the low-carbohydrate and the Weight Watchers diet; these small variations were likely associated with the weight gain of the subject from 88.6 to 95.2 kg in the second diet period. A previous study demonstrated a decreased disposition of methylprednisolone in obesity; absolute clearance was approximately 40% lower in obese subjects who were at least 35% over ideal body weight (ie, total body weight/ideal body weight [TBW/IBW] >1.35) than those within 10% of IBW.30 Our subject was overweight but was not overly obese during the first study (TBW/IBW, 1.21), and methylprednisolone clearance was slightly increased during the second study when his TBW/IBW was 1.30. Furthermore, mean clearance ranged from 21.5 to 25 L/h for 17 other healthy males whose weight was within 120% IBW in 3 previous studies.3,31,32 Taken together, it does not appear that the low clearance in our subject was related to his increased body weight.
Half-life is an important determinant of corticosteroid pharmacodynamics because the duration of action is determined by how long plasma concentrations are maintained above the IC50 for various responses.2 The mean increased half-life for the study subject was 4.4 hours, which was about 1.5-fold larger than the values obtained from the control subjects and other reports.2-4,31-33 This extended half-life prolonged the suppressive effects of methylprednisolone on cortisol secretion and whole blood histamine concentrations. Cortisol and whole blood histamine remained suppressed for a total of approximately 30 and 20 hours, respectively, versus a 10- to 14-hour suppression for a typical control subject. It is noteworthy that the second study was lengthened to confirm the prolonged suppression by capturing additional data for the returns of cortisol and whole blood histamine. The duration of responses was estimated by stimulating the suppression using pharmacokinetic and pharmacodynamic parameter values obtained from the first study. It is observed that the actual effect curves for the Weight Watchers diet were almost superimposed on the simulated curves, suggesting that the concentration versus time curves were well characterized by pharmacokinetic-pharmacodynamic modeling, and the pharmacological responses of cortisol and whole blood histamine to methylprednisolone can be precisely estimated and predicated.
Genetic analysis revealed that the study subject is homozygous for CYP3A5*3, but he does not have CYP3A5*6, *7, *8, *9, *10 alleles. These results are not unexpected, as CYP3A5*3/*3 was found in more than 80% of whites,19 and the rest of the variant alleles have not been identified or were rarely found in whites.8,17,19,23 Although CYP3A5*3/*3 was associated with low CYP3A5 protein expression8,9 causing minimal contribution of CYP3A5 to the total CYP3A protein,9 it does not appear that this genotype was the sole reason for the low methylprednisolone clearance in this subject, as CYP3A5*3/*3 is highly prevalent in whites and 5 of 7 control subjects are also homozygous for the CYP3A5*3 allele.
Previous studies reported conflicting results on the association of CYP3A5 genotype with the pharmacokinetics of the probe drugs midazolam and nifedipine.34-39 Whereas midazolam clearance was higher or tended to be higher in CYP3A5*3/*3 than in CYP3A5*1/*3 cancer patients,34,35 no differences were found in midazolam and nifedipine clearance between different CYP3A5 genotype groups.36-39 Likewise, positive association of CYP3A5 genotype with pharmacokinetic parameter values was found for the immunosuppressants tacrolimus and sirolimus40-44 but not cyclosporine.40,45 Of note, these immunosuppressants are also substrates for the effux pump p-glycoprotein (P-gp) in addition to the metabolizing enzymes CYP3A4 and CYP3A5. Therefore, genetic variations in the multidrug resistance (MDR)-1 gene could also possibly affect their concentrations and pharmacokinetics, although it has not been consistently demonstrated to be the case.45-50 Methylprednisolone has been shown to be a substrate for P-gp in vitro and its absorption was restricted in rat small intestine.51-53 However, it seems unlikely that MDR-1 polymorphisms had significant influence on the hepatic metabolism of methylprednisolone in our subject, because P-gp plays a more significant role in intestinal than hepatic metabolism54 and because methylprednisolone was given intravenously to this subject.
It is suspected that the decreased clearance in the study subject might be related more closely with variations in CYP3A4 than those of CYP3A5. It has been demonstrated that for livers and intestines with homozygous CYP3A5*3/*3, CYP3A4 was the predominate protein in these tissues.9,55 Moreover, 6β-hydroxylation of testosterone in vitro was reduced significantly for CYP3A5 when compared to CYP3A4.11,56,57 The contribution of intrinsic CYP3A5 activity to the overall metabolism of methylprednisolone may be limited as well. Our result showing similar methylprednisolone clearance between control subjects who are heterozygous and homozygous for the CYP3A5*3 allele seems to be supportive of this notation (Figure 1); the presence of the CYP3A5*1 allele resulting in functional CYP3A5 protein did not significantly increase methylprednisolone clearance in CYP3A5*3 heterozygotes.
Our genetic analysis ruled out the presence of 9 CYP3A4 allelic variants including CYP3A4*2, *5, *6, *8, *11, *12, *13, *16, and *17, for which in vitro phenotypic changes have been reported (Table III). This finding is not surprising because allelic frequency for these variants was low and some of these alleles were found only in Chinese, Japanese, or Mexican persons.13,14,58,59 These results indicate that the reduced methylprednisolone clearance was not associated with most of the currently known CYP3A4 variants and are consistent with previous findings showing no significant correlation between CYP3A4 genetic variants and low midazolam hydroxylation activity or high dextromethorphan/3-methoxymorphinan ratio in a small number of liver samples and healthy volunteers.59,60 Recently, a new CYP3A4*20 allele with no in vitro activity has been reported.12 It is possible that the decreased methylprednisolone metabolism in our study subject might be related to this allele or other as yet unidentified polymorphisms in CYP3A4.
The current study was performed based on the premise that methylprednisolone is metabolized primarily by CYP3A enzymes. However, other hydroxyl metabolites and glucuronide conjugate have been identified from urine in patients receiving high-dose pulse methylprednisolone therapy.61 Thus, reduced clearance in our study participant could also be attributed to variations in these other metabolic pathways. In addition, our study was limited by the lack of CYP3A phenotyping with a validated probe drug such as midazolam to demonstrate a true reduction in CYP3A activity in our subject.
CONCLUSION
The low methylprednisolone clearance in the study subject does not appear to be associated with a change in diet composition and the CYP3A5*3/*3 genotype. The involvement of other genetic or contributing factors cannot be excluded. New genotyping methods were developed to detect CYP3A5*6, *8, *9, and *10 alleles that carry functionally defective CYP3A5 SNP.
Acknowledgments
This work was supported in part by the American Foundation for Pharmaceutical Education (ACPE) and the Burroughs Wellcome Fund through the American Association of Colleges of Pharmacy New Investigators Program for Pharmacy Faculty (AACP-NIP) Award and in part by Grant No. 24211 from the National Institute of General Medical Sciences, National Institutes of Health.
References
- 1. [December 27, 2005];Cytochrome P450 Drug-Interaction Table. 2005 Nov 23; Available at: http://medicine.iupui.edu/flockhart/table.htm.
- 2.Jusko WJ, Ludwig EA. Corticosteroids. In: Evans WE, Schentag JJ, Jusko WJ, editors. Applied Pharmacokinetics: Principles of Therapeutic Drug Monitoring. Vancouver, Wash: Applied Therapeutics, Inc; 1992. pp. 27-1–27-34. [Google Scholar]
- 3.Booker BM, Magee MH, Blum RA, Lates CD, Jusko WJ. Pharmacokinetic and pharmacodynamic interactions between diltiazem and methylprednisolone in healthy volunteers. Clin Pharmacol Ther. 2002;72:370–382. doi: 10.1067/mcp.2002.127944. [DOI] [PubMed] [Google Scholar]
- 4.Slayter KL, Ludwig EA, Lew KH, Middleton E, Jr, Ferry JJ, Jusko WJ. Oral contraceptive effects on methylprednisolone pharmacokinetics and pharmacodynamics. Clin Pharmacol Ther. 1996;59:312–321. doi: 10.1016/S0009-9236(96)80009-9. [DOI] [PubMed] [Google Scholar]
- 5.McCrea JB, Majumdar AK, Goldberg MR, et al. Effects of the neurokinin1 receptor antagonist aprepitant on the pharmacokinetics of dexamethasone and methylprednisolone. Clin Pharmacol Ther. 2003;74:17–24. doi: 10.1016/S0009-9236(03)00066-3. [DOI] [PubMed] [Google Scholar]
- 6.Schuetz JD, Beach DL, Guzelian PS. Selective expression of cytochrome P450 CYP3A mRNAs in embryonic and adult human liver. Pharmacogenetics. 1994;4:11–20. doi: 10.1097/00008571-199402000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54:1271–1294. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- 8.Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 9.Lin YS, Dowling AL, Quigley SD, et al. Co-regulation of CYP3A4 and CYP3A5 and contribution to hepatic and intestinal midazolam metabolism. Mol Pharmacol. 2002;62:162–172. doi: 10.1124/mol.62.1.162. [DOI] [PubMed] [Google Scholar]
- 10.Thummel KE, Wilkinson GR. In vitro and in vivo drug interactions involving human CYP3A. Annu Rev Pharmacol Toxicol. 1998;38:389–430. doi: 10.1146/annurev.pharmtox.38.1.389. [DOI] [PubMed] [Google Scholar]
- 11.Williams JA, Ring BJ, Cantrell VE, et al. Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab Dispos. 2002;30:883–891. doi: 10.1124/dmd.30.8.883. [DOI] [PubMed] [Google Scholar]
- 12. [December 27, 2005];CYP3A4 allele nomenclature. 2005 Aug 30; Available at: http://www.imm.ki.se/CYPalleles/cyp3a4.htm.
- 13.Sata F, Sapone A, Elizondo G, et al. CYP3A4 allelic variants with amino acid substitutions in exons 7 and 12: evidence for an allelic variant with altered catalytic activity. Clin Pharmacol Ther. 2000;67:48–56. doi: 10.1067/mcp.2000.104391. [DOI] [PubMed] [Google Scholar]
- 14.Eiselt R, Domanski TL, Zibat A, et al. Identification and functional characterization of eight CYP3A4 protein variants. Pharmacogenetics. 2001;11:447–458. doi: 10.1097/00008571-200107000-00008. [DOI] [PubMed] [Google Scholar]
- 15.Dai D, Tang J, Rose R, et al. Identification of variants of CYP3A4 and characterization of their abilities to metabolize testosterone and chlorpyrifos. J Pharmacol Exp Ther. 2001;299:825–831. [PubMed] [Google Scholar]
- 16.Lee SJ, Bell DA, Coulter SJ, Ghanayem B, Goldstein JA. Recombinant CYP3A4*17 is defective in metabolizing the hypertensive drug nifedipine, and the CYP3A4*17 allele may occur on the same chromosome as CYP3A5*3, representing a new putative defective CYP3A haplotype. J Pharmacol Exp Ther. 2005;313:302–309. doi: 10.1124/jpet.104.078758. [DOI] [PubMed] [Google Scholar]
- 17.Lee SJ, Usmani KA, Chanas B, et al. Genetic findings and functional studies of human CYP3A5 single nucleotide polymorphisms in different ethnic groups. Pharmacogenetics. 2003;13:461–472. doi: 10.1097/00008571-200308000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Saeki M, Saito Y, Nakamura T, et al. Single nucleotide polymorphisms and haplotype frequencies of CYP3A5 in a Japanese population. Hum Mutat. 2003;21:653. doi: 10.1002/humu.9147. [DOI] [PubMed] [Google Scholar]
- 19.Xie HG, Wood AJ, Kim RB, Stein CM, Wilkinson GR. Genetic variability in CYP3A5 and its possible consequences. Pharmacogenomics. 2004;5:243–272. doi: 10.1517/phgs.5.3.243.29833. [DOI] [PubMed] [Google Scholar]
- 20.Hon YY, Jusko WJ, Spratlin VE, Jann MW. Altered methylprednisolone pharmacodynamics in healthy subjects with histamine n-methyltransferase C314T genetic polymorphism. J Clin Pharmacol. doi: 10.1177/0091270006286434. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebling WF, Szefler SJ, Jusko WJ. Analysis of cortisol, methylprednisolone, and methylprednisolone hemisuccinate: absence of effects of troleandomycin on ester hydrolysis. J Chromatogr. 1984;305:271–280. [PubMed] [Google Scholar]
- 22.Lee SJ, Goldstein JA. Functionally defective or altered CYP3A4 and CYP3A5 single nucleotide polymorphisms and their detection with genotyping tests. Pharmacogenomics. 2005;6:357–371. doi: 10.1517/14622416.6.4.357. [DOI] [PubMed] [Google Scholar]
- 23.Hustert E, Haberl M, Burk O, et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics. 2001;11:773–779. doi: 10.1097/00008571-200112000-00005. [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson GR. Pharmacokinetics: the dynamics of drug absorption, distribution, and elimination. In: Hardman JG, Limbird LE, Gilman AG, editors. Goodman and Gilman’s The Pharmacological Basis of Therapeutics. New York, New York: McGraw-Hill; 2001. pp. 3–29. [Google Scholar]
- 25.Alvares AP, Anderson KE, Conney AH, Kappas A. Interactions between nutritional factors and drug biotransformations in man. Proc Natl Acad Sci U S A. 1976;73:2501–2504. doi: 10.1073/pnas.73.7.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kappas A, Anderson KE, Conney AH, Alvares AP. Influence of dietary protein and carbohydrate on antipyrine and theophylline metabolism in man. Clin Pharmacol Ther. 1976;20:643–653. doi: 10.1002/cpt1976206643. [DOI] [PubMed] [Google Scholar]
- 27.Anderson KE, Conney AH, Kappas A. Nutrition and oxidative drug metabolism in man: relative influence of dietary lipids, carbohydrate, and protein. Clin Pharmacol Ther. 1979;26:493–501. doi: 10.1002/cpt1979264493. [DOI] [PubMed] [Google Scholar]
- 28.Fagan TC, Walle T, Oexmann MJ, et al. Increased clearance of propranolol and theophylline by high-protein compared with high-carbohydrate diet. Clin Pharmacol Ther. 1987;41:402–406. doi: 10.1038/clpt.1987.48. [DOI] [PubMed] [Google Scholar]
- 29.Juan D, Worwag EM, Schoeller DA, Kotake AN, Hughes RL, Frederiksen MC. Effects of dietary protein on theophylline pharmacokinetics and caffeine and aminopyrine breath tests. Clin Pharmacol Ther. 1986;40:187–194. doi: 10.1038/clpt.1986.162. [DOI] [PubMed] [Google Scholar]
- 30.Dunn TE, Ludwig EA, Slaughter RL, Camara DS, Jusko WJ. Pharmacokinetics and pharmacodynamics of methylprednisolone in obesity. Clin Pharmacol Ther. 1991;49:536–549. doi: 10.1038/clpt.1991.64. [DOI] [PubMed] [Google Scholar]
- 31.Kong AN, Ludwig EA, Slaughter RL, et al. Pharmacokinetics and pharmacodynamic modeling of direct suppression effects of methylprednisolone on serum cortisol and blood histamine in human subjects. Clin Pharmacol Ther. 1989;46:616–628. doi: 10.1038/clpt.1989.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lew KH, Ludwig EA, Milad MA, et al. Gender-based effects on methylprednisolone pharmacokinetics and pharmacodynamics. Clin Pharmacol Ther. 1993;54:402–414. doi: 10.1038/clpt.1993.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiss WG, Slaughter RL, Ludwig EA, Middleton E, Jr, Jusko WJ. Steroid dose sparing: pharmacodynamic responses to single versus divided doses of methylprednisolone in man. J Allergy Clin Immunol. 1990;85:1058–1066. doi: 10.1016/0091-6749(90)90051-5. [DOI] [PubMed] [Google Scholar]
- 34.Wong M, Balleine RL, Collins M, Liddle C, Clarke CL, Gurney H. CYP3A5 genotype and midazolam clearance in Australian patients receiving chemotherapy. Clin Pharmacol Ther. 2004;75:529–538. doi: 10.1016/j.clpt.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Goh BC, Lee SC, Wang LZ, et al. Explaining interindividual variability of docetaxel pharmacokinetics and pharmacodynamics in Asians through phenotyping and genotyping strategies. J Clin Oncol. 2002;20:3683–3690. doi: 10.1200/JCO.2002.01.025. [DOI] [PubMed] [Google Scholar]
- 36.Shih PS, Huang JD. Pharmacokinetics of midazolam and 1’-hydroxymidazolam in Chinese with different CYP3A5 genotypes. Drug Metab Dispos. 2002;30:1491–1496. doi: 10.1124/dmd.30.12.1491. [DOI] [PubMed] [Google Scholar]
- 37.Floyd MD, Gervasini G, Masica AL, et al. Genotype-phenotype associations for common CYP3A4 and CYP3A5 variants in the basal and induced metabolism of midazolam in European- and African-American men and women. Pharmacogenetics. 2003;13:595–606. doi: 10.1097/00008571-200310000-00003. [DOI] [PubMed] [Google Scholar]
- 38.Yu KS, Cho JY, Jang IJ, et al. Effect of the CYP3A5 genotype on the pharmacokinetics of intravenous midazolam during inhibited and induced metabolic states. Clin Pharmacol Ther. 2004;76:104–112. doi: 10.1016/j.clpt.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Fukuda T, Onishi S, Fukuen S, et al. CYP3A5 genotype did not impact on nifedipine disposition in healthy volunteers. Pharmacogenomics J. 2004;4:34–39. doi: 10.1038/sj.tpj.6500218. [DOI] [PubMed] [Google Scholar]
- 40.Hesselink DA, van Schaik RH, van der Heiden IP, et al. Genetic polymorphisms of the CYP3A4, CYP3A5, and MDR-1 genes and pharmacokinetics of the calcineurin inhibitors cyclosporine and tacrolimus. Clin Pharmacol Ther. 2003;74:245–254. doi: 10.1016/S0009-9236(03)00168-1. [DOI] [PubMed] [Google Scholar]
- 41.Zheng H, Webber S, Zeevi A, et al. Tacrolimus dosing in pediatric heart transplant patients is related to CYP3A5 and MDR1 gene polymorphisms. Am J Transplant. 2003;3:477–483. doi: 10.1034/j.1600-6143.2003.00077.x. [DOI] [PubMed] [Google Scholar]
- 42.Goto M, Masuda S, Kiuchi T, et al. CYP3A5*1-carrying graft liver reduces the concentration/oral dose ratio of tacrolimus in recipients of living-donor liver transplantation. Pharmacogenetics. 2004;14:471–478. doi: 10.1097/01.fpc.0000114747.08559.49. [DOI] [PubMed] [Google Scholar]
- 43.Anglicheau D, Le Corre D, Lechaton S, et al. Consequences of genetic polymorphisms for sirolimus requirements after renal transplant in patients on primary sirolimus therapy. Am J Transplant. 2005;5:595–603. doi: 10.1111/j.1600-6143.2005.00745.x. [DOI] [PubMed] [Google Scholar]
- 44.Thervet E, Anglicheau D, King B, et al. Impact of cytochrome p450 3A5 genetic polymorphism on tacrolimus doses and concentration-to-dose ratio in renal transplant recipients. Transplantation. 2003;76:1233–1235. doi: 10.1097/01.TP.0000090753.99170.89. [DOI] [PubMed] [Google Scholar]
- 45.Anglicheau D, Thervet E, Etienne I, et al. CYP3A5 and MDR1 genetic polymorphisms and cyclosporine pharmacokinetics after renal transplantation. Clin Pharmacol Ther. 2004;75:422–433. doi: 10.1016/j.clpt.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 46.Anglicheau D, Verstuyft C, Laurent-Puig P, et al. Association of the multidrug resistance-1 gene single-nucleotide polymorphisms with the tacrolimus dose requirements in renal transplant recipients. J Am Soc Nephrol. 2003;14:1889–1896. doi: 10.1097/01.asn.0000073901.94759.36. [DOI] [PubMed] [Google Scholar]
- 47.Macphee IA, Fredericks S, Tai T, et al. Tacrolimus pharmacogenetics: polymorphisms associated with expression of cytochrome p4503A5 and P-glycoprotein correlate with dose requirement. Transplantation. 2002;74:1486–1489. doi: 10.1097/00007890-200212150-00002. [DOI] [PubMed] [Google Scholar]
- 48.von Ahsen N, Richter M, Grupp C, Ringe B, Oellerich M, Armstrong VW. No influence of the MDR-1 C3435T polymorphism or a CYP3A4 promoter polymorphism (CYP3A4-V allele) on dose-adjusted cyclosporin A trough concentrations or rejection incidence in stable renal transplant recipients. Clin Chem. 2001;47:1048–1052. [PubMed] [Google Scholar]
- 49.Chowbay B, Cumaraswamy S, Cheung YB, Zhou Q, Lee EJ. Genetic polymorphisms in MDR1 and CYP3A4 genes in Asians and the influence of MDR1 haplotypes on cyclosporin disposition in heart transplant recipients. Pharmacogenetics. 2003;13:89–95. doi: 10.1097/00008571-200302000-00005. [DOI] [PubMed] [Google Scholar]
- 50.Mai I, Stormer E, Goldammer M, et al. MDR1 haplotypes do not affect the steady-state pharmacokinetics of cyclosporine in renal transplant patients. J Clin Pharmacol. 2003;43:1101–1107. doi: 10.1177/0091270003257222. [DOI] [PubMed] [Google Scholar]
- 51.Saitoh H, Hatakeyama M, Eguchi O, Oda M, Takada M. Involvement of intestinal P-glycoprotein in the restricted absorption of methylprednisolone from rat small intestine. J Pharm Sci. 1998;87:73–75. doi: 10.1021/js970163u. [DOI] [PubMed] [Google Scholar]
- 52.Oka A, Oda M, Saitoh H, Nakayama A, Takada M, Aungst BJ. Secretory transport of methylprednisolone possibly mediated by P-glycoprotein in Caco-2 cells. Biol Pharm Bull. 2002;25:393–396. doi: 10.1248/bpb.25.393. [DOI] [PubMed] [Google Scholar]
- 53.Yates CR, Chang C, Kearbey JD, et al. Structural determinants of P-glycoprotein-mediated transport of glucocorticoids. Pharm Res. 2003;20:1794–1803. doi: 10.1023/b:pham.0000003377.39548.f6. [DOI] [PubMed] [Google Scholar]
- 54.Lin JH. Drug-drug interaction mediated by inhibition and induction of P-glycoprotein. Adv Drug Deliv Rev. 2003;55:53–81. doi: 10.1016/s0169-409x(02)00171-0. [DOI] [PubMed] [Google Scholar]
- 55.Westlind-Johnsson A, Malmebo S, Johansson A, et al. Comparative analysis of CYP3A expression in human liver suggests only a minor role for CYP3A5 in drug metabolism. Drug Metab Dispos. 2003;31:755–761. doi: 10.1124/dmd.31.6.755. [DOI] [PubMed] [Google Scholar]
- 56.Patki KC, Von Moltke LL, Greenblatt DJ. In vitro metabolism of midazolam, triazolam, nifedipine, and testosterone by human liver microsomes and recombinant cytochromes p450: role of cyp3a4 and cyp3a5. Drug Metab Dispos. 2003;31:938–944. doi: 10.1124/dmd.31.7.938. [DOI] [PubMed] [Google Scholar]
- 57.Huang W, Lin YS, McConn DJ, et al. Evidence of significant contribution from CYP3A5 to hepatic drug metabolism. Drug Metab Dispos. 2004;32:1434–1445. doi: 10.1124/dmd.104.001313. [DOI] [PubMed] [Google Scholar]
- 58.Hsieh KP, Lin YY, Cheng CL, et al. Novel mutations of CYP3A4 in Chinese. Drug Metab Dispos. 2001;29:268–273. [PubMed] [Google Scholar]
- 59.Lamba JK, Lin YS, Thummel K, et al. Common allelic variants of cytochrome P4503A4 and their prevalence in different populations. Pharmacogenetics. 2002;12:121–132. doi: 10.1097/00008571-200203000-00006. [DOI] [PubMed] [Google Scholar]
- 60.Garcia-Martin E, Martinez C, Pizarro RM, et al. CYP3A4 variant alleles in white individuals with low CYP3A4 enzyme activity. Clin Pharmacol Ther. 2002;71:196–204. doi: 10.1067/mcp.2002.121371. [DOI] [PubMed] [Google Scholar]
- 61.Vree TB, Lagerwerf AJ, Verwey-van Wissen CP, Jongen PJ. High-performance liquid chromatography analysis, preliminary pharmacokinetics, metabolism and renal excretion of methylprednisolone with its C6 and C20 hydroxy metabolites in multiple sclerosis patients receiving high-dose pulse therapy. J Chromatogr B Biomed Sci Appl. 1999;732:337–348. doi: 10.1016/s0378-4347(99)00292-3. [DOI] [PubMed] [Google Scholar]
- 62.Fukushima-Uesaka H, Saito Y, Watanabe H, et al. Haplotypes of CYP3A4 and their close linkage with CYP3A5 haplotypes in a Japanese population. Hum Mutat. 2004;23:100. doi: 10.1002/humu.9210. [DOI] [PubMed] [Google Scholar]
- 63.Jounaidi Y, Hyrailles V, Gervot L, Maurel P. Detection of CYP3A5 allelic variant: a candidate for the polymorphic expression of the protein? Biochem Biophys Res Commun. 1996;221:466–470. doi: 10.1006/bbrc.1996.0618. [DOI] [PubMed] [Google Scholar]
- 64.Chou FC, Tzeng SJ, Huang JD. Genetic polymorphism of cytochrome P450 3A5 in Chinese. Drug Metab Dispos. 2001;29:1205–1209. [PubMed] [Google Scholar]


