Abstract
The fungal diversity in deep-sea environments has recently gained an increasing amount attention. Our knowledge and understanding of the true fungal diversity and the role it plays in deep-sea environments, however, is still limited. We investigated the fungal community structure in five sediments from a depth of ∼4000 m in the East India Ocean using a combination of targeted environmental sequencing and traditional cultivation. This approach resulted in the recovery of a total of 45 fungal operational taxonomic units (OTUs) and 20 culturable fungal phylotypes. This finding indicates that there is a great amount of fungal diversity in the deep-sea sediments collected in the East Indian Ocean. Three fungal OTUs and one culturable phylotype demonstrated high divergence (89%–97%) from the existing sequences in the GenBank. Moreover, 44.4% fungal OTUs and 30% culturable fungal phylotypes are new reports for deep-sea sediments. These results suggest that the deep-sea sediments from the East India Ocean can serve as habitats for new fungal communities compared with other deep-sea environments. In addition, different fungal community could be detected when using targeted environmental sequencing compared with traditional cultivation in this study, which suggests that a combination of targeted environmental sequencing and traditional cultivation will generate a more diverse fungal community in deep-sea environments than using either targeted environmental sequencing or traditional cultivation alone. This study is the first to report new insights into the fungal communities in deep-sea sediments from the East Indian Ocean, which increases our knowledge and understanding of the fungal diversity in deep-sea environments.
Introduction
Although once thought to be an uninhabitable milieu owing to its extreme conditions, the deep-sea is now recognized as a home to rich and largely microbial communities [1]. Whitman et al. [2] reported that deep-sea-derived microbial communities, mainly composed of bacteria and archaea, accounted for a total cellular carbon content of approximately 3×1017 g. Besides bacteria and archaea [3]–[5], fungi in deep-sea environments have been extensively studied [6]–[8]. The isolation of deep-sea fungi was first reported approximately 50 years ago from the Atlantic Ocean at a depth of 4450 m [9]. Recently, an increasing number of fungal species were found in several deep-sea environments, e.g. sediments from Gulf of Mexico [1] and Mariana Trench at 11500 m depth [10], calcareous sediments [11], the Chagos Trench at a depth of 5500 m [12] and the Central Indian Basin at about 5000 m depth [8]. Orsi et al. [13] revealed that the deep-sea sediments are vast habitats for fungal life where cell may live on geologic timescale and these active fungi have an overlooked role in organic carbon turnover, which provide a direct evidence for active fungal metabolism in the deep-sea environments. Despite recent advances, the distribution and diversity of fungal communities in deep-sea environments are still largely unknown. With the recent development of more advanced instruments designed for sampling and researching life at greater depths, there has been more interest in evaluating the diversity and ecological role of fungi from deep-sea environments [14]–[20].
Traditionally, fungal diversity studies on deep-sea environmental samples have been based on cultivation techniques [21]–[24]. Fungi from deep-sea environments do not necessarily require extreme culture conditions, and many studies on deep-sea fungi describe using cultivation methods under standard laboratory conditions [21], [22]. Currently, more than 120 fungal species have been isolated from deep-sea environments [6], [18], [21]–[24]. Targeted environmental sequencing analyses, however, have indicated that cultivable fungi are only a small fraction of the total number of fungi inhabiting deep-sea environments [7]. The molecular phylogenetic analysis of clone libraries constructed from environmental samples has become the gold-standard in fungal diversity research, and this technique is thought to be able to detect a wider range of fungi that more accurately represent the investigated environment [25]. To date, several unknown novel phylotypes including DSF-group with the phylum Ascomycota [17], [26], KML11 clade and Rozella in the new described Cryptomycota [26], [27] and BCGI clade [17], have been discovered by molecular phylogenetic analysis.
This technique of molecular phylogenetic analysis, however, can be easily biased at many steps of the process, such as PCR primer selection and the DNA extraction method used [20]. Previous studies have shown that the 18S rRNA technique is a valuable tool for assessing the global diversity of eukaryotes [28], [29]. This approach, however, is limited because identification is often restricted to the genus or family level [30]. Buchan et al. [31] reported that the internal transcribed spacer (ITS) regions in fungal rDNA exhibit a high degree of polymorphism between species and are thought to be highly conserved within species. ITS regions can provide better taxonomic resolution than 18S rDNA sequences [16].
To identify the maximum amount of fungal species in the five deep-sea sediments collected from the East Indian Ocean and to find new sequences for phylogenetic studies, environmental gene libraries were constructed after amplifying the sediment DNA using an ITS rRNA gene primer set. Furthermore, the culturable fungi in these sediments were also isolated and identified by amplifying and sequencing the ITS rRNA gene.
Materials and Methods
Ethics statement
All the five sampling locations in this study were included in high seas. Permits for sediment sampling were provided by Ministry of Foreign Affairs of the People’s Republic of China. No specific permissions were required for these locations and the field studies did not involve endangered or protected species.
Study site and sample collection
Five deep-sea sediment samples (A–E) were collected using a Remote Operated Vehicle (ROV) during the East Indian Ocean Open Cruise in March 2013 (Fig. S1 in File S1). The coordinates of Sample A–E were shown in Table 1. The collected sediment samples were mostly undisturbed and compact. The average length of the sediment cores collected from these locations was approximately 30 cm. Sub-cores of these samples were collected from a box corer using an alcohol-sterilized PVC cylinder that had a 5 cm inner diameter. Subsections of these samples were cut from the sediment sub-cores and immediately stored in sterile plastic bags to avoid any aerial contamination [18]. The bags were closed with rubber bands and transported to the laminar flow hood in the laboratory on board. A portion of sediment from the middle of each sub-sample that had not been in contact with the PVC cylinder wall was removed using an alcohol-flame-sterilized spatula and placed in a sterile vial for fungal isolation [21]. Fractions of these sediments were immediately stored at −20°C for direct DNA extraction after fungal isolation.
Table 1. The coordinates of Sample A–E.
| Samples | Latitude | Longitude | Depth (m) |
| A | −2°57′N | 95°19′E | 4810 |
| B | 00°00′N | 90°57′E | 4532 |
| C | 00°30′N | 82°03′E | 4530 |
| D | 7°57′N | 89°27′E | 4614 |
| E | 10°00′N | 84°33′E | 4571 |
Fungal isolation and identification
The cultivation and isolation methods for fungi from deep-sea environments do not differ fundamentally from the methods used for fungi from shallow marine environments. Physiological analyses have demonstrated that deep-sea-derived fungi are able to grow in deep-sea salinity and at low temperatures [32]. Three different methods were used for fungal isolation in this study, including the particle plating method [33], dilution plating method [21] and low temperature (10°C) incubation method. These fungal isolation methods are described in more detail in a recently published paper by Zhang et al. [18].
Fungal isolates were identified using a combination of morphology characteristics and the internal transcribed spacer (ITS) sequences. Total genomic DNA was extracted from all of the selected fungal strains using a method described by Lai et al. [16]. From the genomic DNA, nearly full-length ITS sequences were amplified by polymerase chain reaction with the primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) [34]. These fungal ITS gene sequencing and identification methods are described in more detail in a previously published paper by Toledo-Hernandez et al. [35].
DNA extraction, PCR amplification and clone library construction
Environmental DNA was isolated from two grams of sediment sample from each frozen subsection of the sediment cores using a soil DNA extraction kit (Omega Bio-Tek, Inc., Norcross, GA, USA) according to the manufacturer’s instructions and sterile techniques to avoid cross contamination. The DNA samples from the five sediments were amplified using the primers ITS1 and ITS4. The polymerase chain reaction mixture (20 µl) consisted of 2 µl 10× PCR buffer (500 mM KCl, 100 mM Tris-HCl, 15 mM 1% (w/v) MgCl2, and Triton X-100), 1.6 µl of 2.5 mM dNTP, 0.8 µl of each primer, 0.2 µl of 5 U Taq DNA polymerase (Takara Biotechnology Co., Ltd., Dalian, China), 13.6 µl of water, and 1.0 µl of template DNA (10–100 ng). PCR was conducted using an Eppendorf Mastercycler (Eppendorf German Co., Ltd., Hamburger, German) and the following program: denaturation at 95°C for 5 min, 25 cycles of 30 s at 95°C, 30 s at 55°C, and 90 s at 72°C, and a final extension at 72°C for 10 min. Reaction mixtures lacking template DNA were used as negative controls. Amplified products were gel-purified, ligated with pMD18-T easy vector (Takara Biotechnology Co., Ltd., Dalian, China) and transformed into Escherichia coli cells following the manufacturer’s instructions. Transformants were grown overnight at 37°C on Luria-Bertani agar containing 100 µg/ml ampicillin. The presence of insert was confirmed by PCR with M13 forward and reverse primers. One microliter of broth containing the clone was added to 25 µl of PCR reaction mixture. The PCR protocol included an initial hot start incubation (5 min at °C) followed by 34 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 1 min, and then a final extension at 72°C for 5 min [8]. Clones containing the positive insert were further processed for plasmid isolation and purification using the Millipore plasmid preparation Kit (Millipore, USA). Sequencing of the PCR products from the plasmids was conducted by Invitrogen (China). A total of five environmental gene libraries were constructed from the DNA samples from the five deep-sea sediments. Approximately 100 clones were screened from each library.
Phylogenetic analyses
All of the vector sequences from the sequenced fungal clones were analyzed using the rRNA Database Project CHECK_CHIMERA program to detect and eliminate the potential chimeric sequences. Pairwise alignment of the sequences was conducted using Clustal W in the MEGA software version 5.0. Conserved motifs were identified, and the sequences were trimmed manually. Clones were grouped into operational taxonomic units (OTUs) using a sequence similarity cut-off value of 98% and the Mothur software version 1.32.1 [36]. Rarefaction curves for the number of observed OTUs were calculated with fungal assemblage at each dataset. A representative sequence from each OTU was queried against an NCBI-GenBank BLASTN search.
Nucleotide sequence accession number
The ITS sequences for 20 culturable fungal isolate representatives and 45 uncultured fungal clone representatives obtained in this study were deposited in GenBank under accession numbers KJ173524–KJ17352 and KJ173554–KJ173590.
Results
Phylogeny of environmental fungal ITS-rDNA sequences
A total of 515 clones from five deep-sea sediment samples A–E (Fig. S1 in File S1) from the East Indian Ocean were sequenced. Of the resulting sequences, 445 sequences were found to be fungal, and a total of 45 operational taxonomic units (OTUs) (Table 2) were identified after clustering based on a 98% sequence identity criterion. The other 70 clones (∼13.6%) were eukaryotic or chimeric in nature and were excluded from this study. Rarefaction curves (Fig. S2 in File S1) were constructed for the ITS clone libraries from samples A–E. Rarefaction curves for three samples (B, C and D) demonstrated a plateau, which indicates that the number of sequences analyzed may sufficiently represent the fungal diversity in these samples. While the rarefaction curves of Sample A and E did not reach a plateau. It is likely that the fungal diversity of the two samples is higher than what was detected in this study.
Table 2. Phylogenetic affiliations of uncultured fungi obtained from deep-sea sediment samples A–E.
| OTU | Closest identified relative | The number of clones | ||||||
| no. | Taxon (Fungal phylum) | GenBank accession no. | Similarity % | A | B | C | D | E |
| OTU-01 | Phoma glomerata (Ascomycota) | EU273521 | 100 | 2 | 5 | |||
| OTU-02 | Cryptococcus curvatus (Basidiomycota) | KF472136 | 99 | 57 | ||||
| OTU-03 | Phoma herbarum (Ascomycota) | KC311476 | 99 | 1 | ||||
| OTU-04a | Rhizoscyphus ericae (Ascomycota) | JQ711893 | 98 | 1 | ||||
| OTU-05a | Sporobolomyces lactosus (Basidiomycota) | HQ914907 | 99 | 1 | ||||
| OTU-06a | Trichoderma asperellum (Ascomycota) | KC479819 | 100 | 1 | ||||
| OTU-07a | Trichosporon moniliiforme (Basidiomycota) | AF444415 | 100 | 27 | ||||
| OTU-08a | Uncultured fungus clone (Ascomycota) | GU211938 | 100 | 27 | ||||
| OTU-09a | Xeromyces bisporus (Ascomycota) | GU733338 | 99 | 9 | ||||
| OTU-10a | Uncultured ascomycete (Ascomycota) | EU046087 | 99 | 1 | ||||
| OTU-11 | Uncultured Geomyces (Ascomycota) | JQ346989 | 99 | 1 | ||||
| OTU-12 | Basidiomycete sp. (Basidiomycota) | EU871524 | 99 | 2 | ||||
| OTU-13 | Uncultured soil fungus (Basidiomycota) | DQ420877 | 100 | 14 | ||||
| OTU-14 | Cryptococcus fragicola (Basidiomycota) | AB035588 | 99 | 5 | ||||
| OTU-15 | Cryptococcus podzolicus (Basidiomycota) | FN428930 | 99 | 1 | ||||
| OTU-16a | Guehomyces pullulans (Basidiomycota) | AF444418 | 100 | 2 | ||||
| OTU-17 | Basidiomycete sp. (Basidiomycota) | EU871524 | 99 | 2 | ||||
| OTU-18 | Rhodotorula slooffiae (Basidiomycota) | AB566328 | 99 | 14 | ||||
| OTU-19 | Sterigmatomyces halophilus (Basidiomycota) | NR073302 | 100 | 37 | ||||
| OTU-20a | Uncultured compost fungus (Basidiomycota) | DQ365334 | 99 | 5 | ||||
| OTU-21 | Uncultured Mortierella (Zygomycota) | JF831505 | 100 | 1 | ||||
| OTU-22 | Uncultured Mortierella (Zygomycota) | JF831503 | 99 | 25 | ||||
| OTU-23a | Uncultured soil fungus (Ascomycota) | EU826926 | 99 | 2 | ||||
| OTU-24a | Alternaria alternata (Ascomycota) | GQ916545 | 99 | 39 | ||||
| OTU-25a | Cladosporium tenuissimum (Ascomycota) | AJ300331 | 100 | 3 | ||||
| OTU-26 | Alternaria sp. (Ascomycota) | KF888649 | 99 | 2 | ||||
| OTU-27 | Aspergillus penicillioides (Ascomycota) | AY373862 | 97 | 2 | ||||
| OTU-28 | Uncultured fungus clone (Ascomycota) | HQ143117 | 99 | 32 | ||||
| OTU-29 | Candida etchellsii (Ascomycota) | JQ653271 | 99 | 24 | ||||
| OTU-30 | Candida inconspicua (Ascomycota) | AB179767 | 99 | 6 | ||||
| OTU-31 | Candida sake (Ascomycota) | AJ549822 | 99 | 2 | ||||
| OTU-32 | Candida xylopsoci (Ascomycota) | FM178339 | 100 | 2 | ||||
| OTU-33a | Cladophialophora chaetospira (Ascomycota) | EU035404 | 93 | 2 | ||||
| OTU-34 | Cladosporium cladosporioides (Ascomycota) | GU932679 | 99 | 2 | ||||
| OTU-35a | Cladosporium sphaerospermum (Ascomycota) | GU017501 | 100 | 2 | ||||
| OTU-36a | Uncultured soil fungus (Basidiomycota) | DQ420860 | 89 | 2 | 1 | |||
| OTU-37a | Eurotium rubrum (Ascomycota) | AY373891 | 99 | 1 | ||||
| OTU-38a | Fusarium solani (Ascomycota) | JQ910159 | 100 | 5 | ||||
| OTU-39a | Galactomyces candidum (Ascomycota) | JN974290 | 100 | 22 | 19 | 11 | ||
| OTU-40a | Dipodascus australiensis (Ascomycota) | HQ115737 | 99 | 4 | ||||
| OTU-41 | Geomyces pannorum (Ascomycota) | DQ189228 | 99 | 1 | ||||
| OTU-42 | Hortaea werneckii (Ascomycota) | GQ334385 | 99 | 2 | ||||
| OTU-43a | Hypocrea virens (Ascomycota) | GU130297 | 100 | 1 | 12 | |||
| OTU-44 | Phoma sp. (Ascomycota) | HQ630999 | 98 | 1 | 1 | |||
| OTU-45 | Leptosphaeria sp. (Ascomycota) | AB752252 | 99 | 1 | ||||
| Total | 92 | 79 | 90 | 89 | 95 | |||
OTUs marked by a letter (a) are new reports for deep-sea environments. Bolded and italicized OTUs are affiliated with yeasts and filamentous fungi, respectively, and the remaining OTUs are affiliated with unidentified yeasts or filamentous fungi.
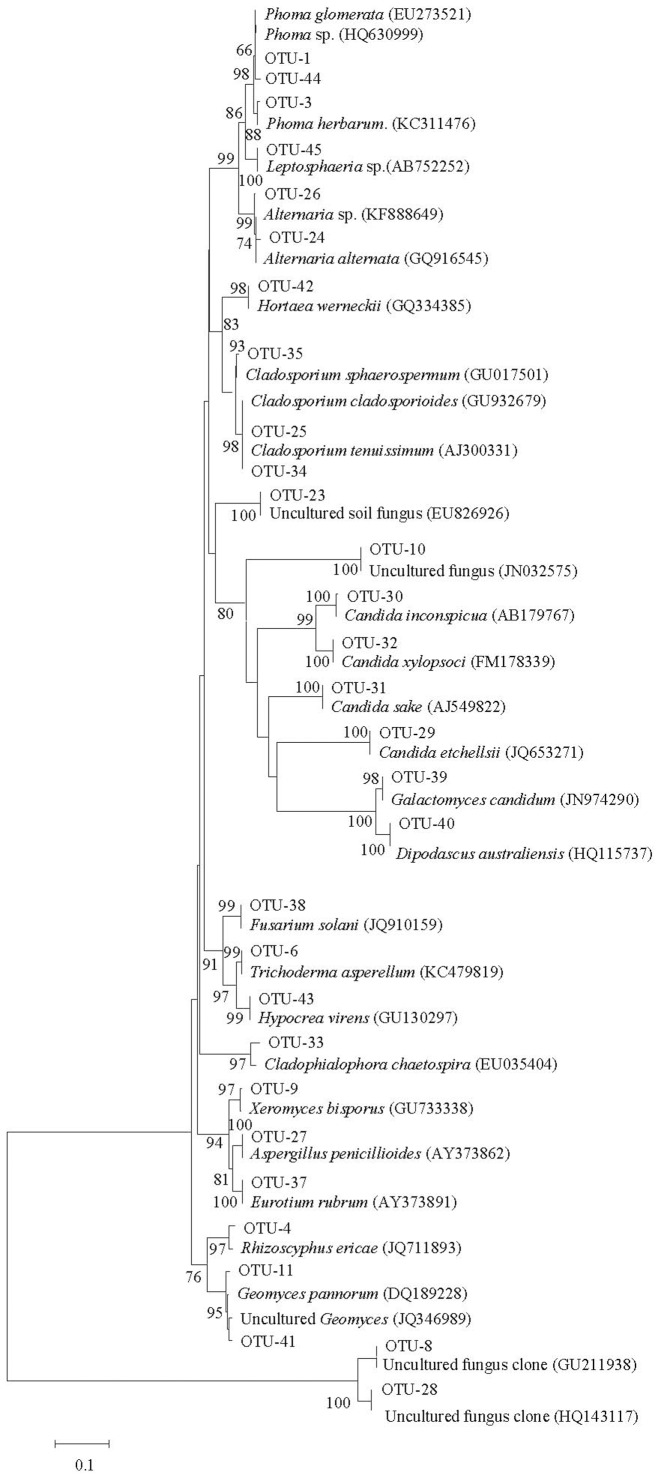
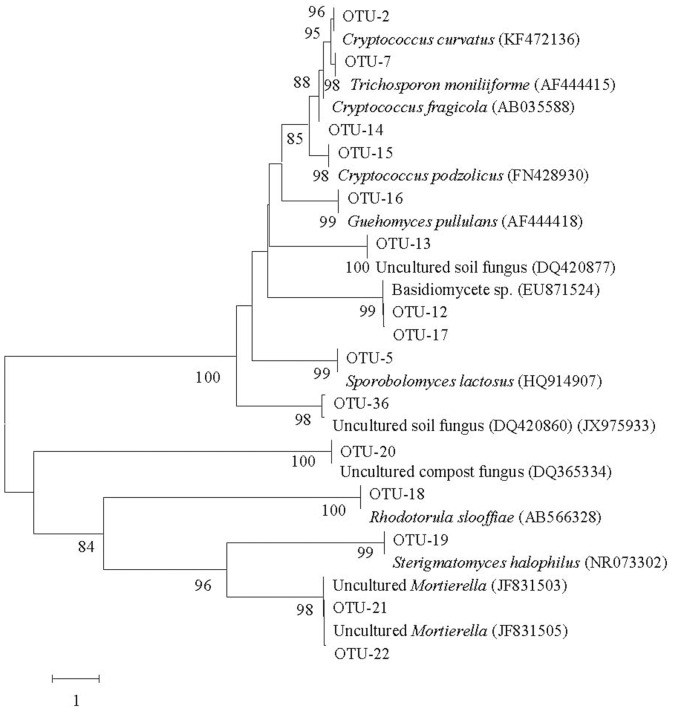
Most of the ITS sequences from the 45 OTUs demonstrated ≥98% similarity with sequences from their closest relative taxa in GenBank. Three new sequence types, however, only demonstrated 89%–97% similarity with the existing database. Therefore, these results from the phylogenetic analysis suggest that OTU-27, 33 and 36 are novel fungal taxa that are not closely related to previous identified fungal ITS sequences in public databases (Table 2). The composition indicates that a majority (419/445) of these amplified ITS sequences belong to the phyla Ascomycota (276 clones from 29 OTUs) and Basidomycota (143 clones from 24 OTUs) (Fig. 1 and 2). The other remaining sequences belong to the phylum Zygomycota (26 clones from OUT-21 and 22) (Fig. 2).
Figure 1. Neighbor-joining phylogenetic tree from analysis of ITS rDNA from 29 Ascomycota representative sequences in five libraries.
The numbers at the nodes are the percentages indicating the level of bootstrap support based on a neighbor-joining analysis of 1000 resampled data sets. Only values >50% are shown. The scale bar represents 0.1 substitutions per nucleotide position.
Figure 2. Neighbor-joining phylogenetic tree from analysis of ITS rDNA from 26 Basidomycota and Zygomycota representative sequences (OTU-21 and 22) in five libraries.
The numbers at the nodes are percentages indicating the level of bootstrap support based on a neighbor-joining analysis of 1000 resampled data sets. Only values >50% are shown. The scale bar represents 1 substitution per nucleotide position.
Furthermore, the phylogenetic analyses revealed that 240 clones from 17 OTUs were most closely related to cultivable yeast forms, including three species of genus Cryptococcus (63 clones), Galactomyces candidum (52 clones), Sterigmatomyces halophilus (37 clones), four species of genus Candida (34 clones), Trichosporon moniliiforme (27 clones), Rhodotorula slooffiae (14 clones), Basidiomycete sp. (4 clones), Dipodascus australiensis (2 clones), Guehomyces pullulans (2 clones), Hortaea werneckii (2 clones) and Sporobolomyces lactosus (one clone) (Fig. 1 and Table 2). Another 120 clones from 20 OTUs were closely related to filamentous fungi, including Aspergillus, Alternaria, Cladophialophora, Cladosporium, Eurotium, Fusarium, Geomyces, Hypocrea, Leptosphaeria, Mortierella, Phoma, Rhizoscyphus, Trichoderma and Xeromyces (Fig. 1 and Table 2). The remaining 85 clones from 8 OTUs were closely related to uncultured fungi from soil or plant ecological systems that have been deposited in the NCBI database [37]–[41].
Culturable fungal isolates and species richness
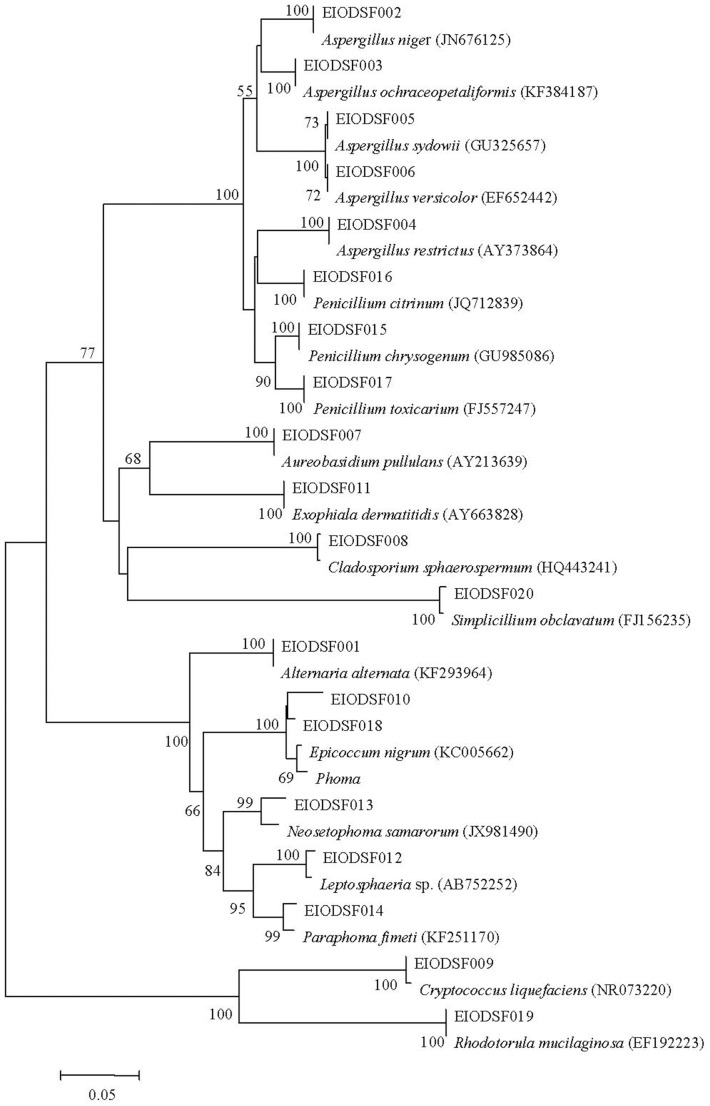
A total of 78 fungal isolates belonging to 20 phylotypes (Fig. 3) were recovered using traditional cultivation. The ITS sequencing results showed that most of these fungal sequences demonstrated >97% similarity with sequences from their closet relative species. The one isolate EIODSF013 (accession number KJ173536), however, only demonstrates 95% similarity with the existing sequence (accession number JX981490) in the NCBI database (Table 3). A multigene analysis combined with detailed morphological and ultra structural studies, however, are needed to determine the novelty of this isolate. Most of the 78 fungal isolates belonged to Ascomycota, including two yeast and 16 filamentous fungal species. Among these species, Aspergillus sp., Penicillium sp. and Simplicillium obclavatum were the most diverse and common, while Alternaria alternata, Aureobasidium pullulans, Cryptococcus liquefaciens, Exophiala dermatitidis, Epicoccum nigrum and Neosetophoma samarorum were the rarest fungal species with only single or double isolates. The remaining species occurred as several isolates.
Figure 3. Neighbor-joining phylogenetic tree from analysis of ITS sequences from fungi isolated from five deep-sea sediments from the East Indian Ocean.
The numbers at the nodes are the percentages indicating the level of bootstrap support based on a neighbor-joining analysis of 1000 resampled data sets. Only values >50% are shown. The scale bar represents 0.05 substitutions per nucleotide position.
Table 3. Phylogenetic affiliations of culturable fungi obtained from deep-sea sediment samples A–E.
| Isolates | Closest identified relative | The number of isolates | ||||||
| no. | Fungal genera or species (Phylum) | GenBank accession no. | Similarity % | A | B | C | D | E |
| EIODSF 001a | Alternaria alternata (Ascomycota) | KF293964 | 99% | 2 | ||||
| EIODSF 002 | Aspergillus niger (Ascomycota) | JN676125 | 99% | 4 | ||||
| EIODSF 003a | A. ochraceopetaliformis (Ascomycota) | KE384187 | 99% | 1 | 1 | 4 | 1 | |
| EIODSF 004 | A. restrictus (Ascomycota) | JX156352 | 99% | 2 | 1 | |||
| EIODSF 005 | A. sydowii (Ascomycota) | GU325657 | 99% | 2 | ||||
| EIODSF 006 | A. versicolor (Ascomycota) | EF652442 | 99% | 2 | 1 | 2 | ||
| EIODSF 007 | Aureobasidium pullulans (Ascomycota) | AY213639 | 99% | 2 | ||||
| EIODSF 008 | Cladosporium sphaerospermum (Ascomycota) | HQ443241 | 99% | 2 | 1 | 3 | ||
| EIODSF 009 | Cryptococcus liquefaciens (Basidiomycota) | AF145331 | 99% | 1 | ||||
| EIODSF 010 | Epicoccum nigrum (Ascomycota) | KC005662 | 98% | 1 | ||||
| EIODSF 011 | Exophiala dermatitidis (Ascomycota) | AY663828 | 99% | 1 | ||||
| EIODSF 012 | Leptosphaeria sp. (Ascomycota) | AB752252 | 99% | 6 | ||||
| EIODSF 013a | Neosetophoma samarorum (Ascomycota) | JX981490 | 95% | 1 | ||||
| EIODSF 014a | Paraphoma fimeti (Ascomycota) | KF251170 | 99% | 2 | 1 | |||
| EIODSF 015 | Penicillium chrysogenum (Ascomycota) | GU985086 | 99% | 2 | ||||
| EIODSF 016 | P. citrinum (Ascomycota) | JN624897 | 99% | 1 | 4 | 2 | ||
| EIODSF 017a | P. toxicarium (Ascomycota) | FJ557247 | 99% | 3 | ||||
| EIODSF 018 | Phoma sp. (Ascomycota) | EF120404 | 99% | 2 | 1 | 2 | ||
| EIODSF 019 | Rhodotorula mucilaginosa (Basidiomycota) | EF192223 | 99% | 4 | ||||
| EIODSF 020a | Simplicillium obclavatum (Ascomycota) | FJ156235 | 99% | 5 | 3 | 2 | 2 | 1 |
| Total | 27 | 7 | 10 | 14 | 20 | |||
Species marked by a letter (a) are new reports for deep-sea environments. Bolded and italicized isolates are included in yeasts and filamentous fungi, respectively.
Discussion
New insights into the fungal communities in deep-sea environments
The fungal diversity in deep-sea environments has recently gained an increasing amount of attention. Our knowledge and understanding of the true fungal diversity and the roles this diversity plays in deep-sea environments, however, is still limited. The aim of the present study was to obtain the maximum amount of fungal diversity from deep-sea sediments collected in the East Indian Ocean. A total of 45 fungal OTUs and 20 culturable fungal phylotypes were recovered in this study (Table 2 and 3), which revealed that there is a great amount of fungal diversity in the deep-sea sediments collected in the East Indian Ocean. Recently, many studies have shown that there is an increasing number of culturable fungal species and uncultured fungal clones in deep-sea sediments from the Central Indian Basin [8], [21], [42], South China Sea [16], [18] and Eastern Equatorial Pacific [43]. Few studies, however, have specifically focused on the fungal diversity in the deep-sea environments from the East Indian Ocean. This study is the first to report new insight into the fungal communities in deep-sea sediments from the East Indian Ocean using a combination of targeted environmental sequencing and cultivation. These findings increase our knowledge and understanding of the fungal diversity in deep-sea environments.
In this study, most of the clones (94.2%) detected by targeted environmental sequencing and all of the culturable fungal isolates recovered using traditional cultivation belonged to the phyla Ascomycota and Basidiomycota (Table 2 and 3). The remaining 5.8% clones (OTU-21 and 22) belonged to the phylum Zygomycota (Table 2). In addition to these three phyla, the phylum Chytridiomycota has also been detected in many deep-sea environments, e.g. Izu-Ogasawara Trench [6]. In this study, however, no putative Chytridiomycota sequences were detected in any of the five sediment samples collected from the East Indian Ocean. We do not believe that our methods were incapable of detecting these higher taxonomic groups because the same primer set has been shown to be able to detect a diverse range of these taxa from deep-sea environments [16], [44], [45]. Therefore, these results suggest that the fungal communities in the deep-sea sediments from the East Indian Ocean tend to be dominated by Ascomycota and Basidomycota, while other fungal taxonomic groups are rare or absent.
Notably, 44.4% (20 out of 45 OTUs, Table 2) fungal OTUs and 30% (6/20) culturable fungal phylotypes (Table 3) identified in this study are new reports for deep-sea sediments (Table 2 and 3). Some of these species demonstrated phylogenetic similarity to fungal species and genera known to be found in shallow-sea environments. For examples, A. alternata was found in tropic sea grass Enhalus acoroides [45]; E. rubrum, H. virens and Leptosphaeria sp. were found in marine mangroves [46]; A. ochraceopetaliformis and C. tenuissimum were found in marine corals and sponges [36], [47]; C. inconspicua may be found in crabs [48]. Other species were affiliated with culturable or uncultured fungi from soil or plant ecological systems. For examples, R. ericae was isolated from lodgepole pine in central British Columbia [49]; and D. australiensis was recovered from agricultural soil [50]; uncultured soil fungus (OTU-13) was detected in a genetically modified rice ecosystem [41]. These results may appear to be inconsistent, but previous studies have shown that while a majority of the fungal isolates from deep-sea environments were psychrotolerant, they grew more rapidly at 30°C than 5°C [42], [51]. Moreover, most of the fungi isolated from deep-sea environments are halotolerant and do not absolutely require seawater for growth [24], [32]. It is possible that there are no true indigenous fungi in deep-sea environments and that species from terrestrial environments have gradually adapted to deep-sea extreme conditions [6].
Yeast fungi detected by targeted environmental sequencing
Our targeted environmental sequencing results showed 53.9% fungal clones were closely related to yeast fungi. Among the yeast forms in this study, most of the clones represented phylotypes relevant to the genera Cryptococcus and Galactomyces. Some of Cryptococcus-related phylotypes are psychrotolerant and have been found in the majority of deep subseafloor samples from North Pond, Hydrate Ridge, Peru Margin and Eastern Equatorial Pacific [43], but Galactomyces-related phylotypes are rarely recovered from deep-sea environments. Four OTUs were closely related to Candida sp., which are considered to be associated with ecosystems in anaerobic environments [26].
In addition to the previously mentioned yeast phylotypes, Rhodotorula, Sterigmatomyces and Trichosporon yeasts were also found to be abundant in this study. Previous studies have shown that Rhodotorula spp. demonstrate remarkable ubiquity based on their presence in several different habitats, such as deep-sea sediments [52]. Phylotypes related to the genera Trichosporon and Sterigmatomyces are known pathogens or parasites of marine animals, which suggests that they may also be opportunistic pathogens or parasites of deep-sea animals [47], [53]. Furthermore, OUT-12 and 17 are related to unidentified Basidiomycetious yeasts (EU871524) and were detected in a water column from the Equatorial Indian Ocean (unpublished). The remaining yeast forms were singletons or doubletons, indicating the low abundance of these clones.
Filamentous fungi dominated the fungal community based on traditional cultivation
In this study, filamentous fungi dominated the fungal community using traditional cultivation. The five genera Aspergillus, Penicillium, Simplicillium, Cladosporium and Phoma were distributed in more than three sediments from the East Indian Ocean (Table 2). Members of the mycelia genera Aspergillus and Penicillium are known to be globally distributed fungal taxa. It seems doubtful that these fungal species are indigenous to deep-sea environments, and evidence of physiological adaption of these species to deep-sea environments had been reported by Raghukumar et al. [54]. Moreover, Aspergillus spp. were frequently detected in anaerobic marine sediments and were shown to play an important role in the denitrification process [20]. These findings suggest that these Aspergillus species may play a potentially versatile role for fungi in major ecological processes in the deep-sea environments. The genus Simplicillium was segregated from Verticillium and contains 10 species [55], [56]. These Simplicillium species occur in a broad range of ecological niches, such diseased plant tissue, soil, human nails, dog tissue and mushrooms [55], [57]. In this study, we report for the first time the presence of Simplicillium sp. in deep-sea sediments. Phoma species are known to be associated with not only land plants but also with marine plants. Previous studies have demonstrated that many terrestrial microorganisms could accumulate in deep-sea sediments [58], [59]. One terrestrial fungal genus, Cladosporium was isolated from three deep-sea sediments below 3000 m in this study, which indicates that sedimentation may be an important factor responsible for the accumulation of facultative marine fungi in deep-sea sediments.
In addition, only two isolates belonging to A. alternate were recovered using traditional cultivation, but this species was also detected by targeted environmental sequencing in this study. Therefore, A. alternata should be abundant in these deep-sea sediments from the East Indian Ocean. Previous studies have shown that the genera Alternaria was found in nearly every survey of free-living fungal communities associated with biological soil crusts [60], [61]. Few Alternaria sp., however, were detected in deep-sea environments.
Comparison of fungal community by targeted environmental sequencing and traditional cultivation
A distinct difference in the fungal community based on targeted environmental sequencing compared with traditional cultivation was that the Zygomycota spp. was not recovered by traditional cultivation but was detected by targeted environmental sequencing. Previous studies have also shown that Zygomycota spp. could be found in different deep-sea environments using targeted environmental sequencing [17], [24]. Currently, however, there are no reports of isolating Zygomycota species from deep-sea environment cultures. After a further comparison of the fungal phylotypes recovered using these two methods, it was found that the majority of the fungal phylotypes recovered using targeted environmental sequencing could not be recovered using a traditional cultivation method. This finding is consistent with the findings published by Le Calvez et al. [24], who reported that there are striking differences in the deep-sea fungal diversity results when using targeted environmental sequencing compared with traditional cultivation. These findings suggest that a combination of targeted environmental sequencing and traditional cultivation will generate a more accurate assessment of the fungal diversity in deep-sea environments compared with using targeted environmental sequencing or traditional cultivation alone. Furthermore, to obtain an even greater abundance of deep-sea fungi, it is necessary to combine the methods used in this study with other methods, such as the microscopic observation of samples appropriately staining, FISH, measuring ergosterol, metagenomic methods, and other new powerful tools expected to be developed in the future [62].
Supporting Information
Contains Fig. S1 Map of the East Indian Ocean, location and depth of the sampling site and Fig. S2 Rarefaction curves constructed for ITS clone libraries from each of the five sampling sites. ITS, internal transcribed spacer.
(DOCX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files, except that All the ITS sequences of 20 culturable fungal isolate representatives and 45 uncultured fungal clone representatives in the study are available from GenBank under accession numbers KJ173524–KJ17352 and KJ173554–KJ173590.
Funding Statement
The authors are grateful for the financial support provided by the National Basic Research Program of China (2010CB833803), National High Technology Research and Development Program of China (863 Program, 2012AA092104), Natural Science Foundation of China (41206139, 41376160), regional innovation demonstration project of Guangdong Province marine economic development (GD2012-D01-002), National Marine Public Welfare Research Project of China (201305017) and National Key Technologies R&D Program (2011BAE06B04-03). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Thaler AD, Dover CLV (2012) Vilgalys (2012) Ascomycete phylotypes recovered from a Gulf of Mexico methane seep are identical to an uncultured deep-sea fungal clade from the Pacific. Fungal Ecol 5: 270–273. [Google Scholar]
- 2. Whitman WB, Coleman DC, Wiebe WJ (1998) Prokaryotes: the unseen majority. Proc Acad Natl Sci USA 95: 6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. DeLong EF, Pace NR (2001) Environmental diversity of bacteria and archaea. Syst Biol 50: 470–478. [PubMed] [Google Scholar]
- 4. Sogin M, Morrison HG, Huber JA, Welch DM, Huse SM, et al. (2006) Microbial diversity in the deep sea and underexplored “rare biosphere”. Proc Natl Acad Sci USA103: 12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luna GM, Stumm K, Pusceddu A, Danovaro R (2009) Archaeal diversity in deep-sea sediments estimated by means of different terminal-restriction fragment length polymorphisms (T-RFLP) Protocols. Curr Microbiol 59: 356–361. [DOI] [PubMed] [Google Scholar]
- 6. Nagano Y, Nagahama T (2012) Fungal diversity in deep-sea extreme environments. Fungal Ecol 5: 463–471. [DOI] [PubMed] [Google Scholar]
- 7. Nagano Y, Nagahama T, Hatada Y, Nunoura T, Takami H, et al. (2010) Fungal diversity in deep-sea sediments-the presence of novel fungal groups. Fungal Ecol 3: 316–325. [Google Scholar]
- 8. Singh P, Raghukumar C, Verma P, Shouche Y (2012) Assessment of fungal diversity in deep-sea sediments by multiple primer approach. World J Microbiol Biotechnol 28: 659–667. [DOI] [PubMed] [Google Scholar]
- 9. Roth FJ, Orpurt PA, Ahearn DJ (1964) Occurrence and distribution of fungi in a subtropical marine environment. Can J Bot 42: 375–383. [Google Scholar]
- 10. Takami H, Inoue A, Fuji F, Horikoshi K (1997) Microbial flora in the deepest sea mud of the Mariana Trench. FEMS Microbiol Lett 152: 279–285. [DOI] [PubMed] [Google Scholar]
- 11. Raghukumar C, Raghukumar S (1998) Barotolerance of fungi isolated from deep-sea sediments of the Indian Ocean. Aquat Microb Ecol 15: 153–163. [Google Scholar]
- 12. Raghukumar C, Raghukumar S, Sheelu G, Gupta S, Nagendernath B, et al. (2004) Buried in time: culturable fungi in a deep-sea sediment core from the Chagos Trench, Indian Ocean. Deep Sea Res I 51: 1759–1768. [Google Scholar]
- 13. Orsi WD, Edgcomb VP, Christman GD, Biddle JF (2013) Gene expression in the deep biosphere. Nature 499: 205–210. [DOI] [PubMed] [Google Scholar]
- 14. Bhadury P, Bik H, Lambshead JD, Austen MC, Smerdon GR, et al. (2011) Molecular diversity of fungal phylotype co-amplified alongside nematodes from coastal and deep-sea marine environments. PloS One 6(10): e26445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takishita K, Tsuchiya M, Reimer JD, Maruyama T (2006) Molecular evidence demonstrating the basidiomycetous fungus Cryptococcus curvatus is the dominant microbial eukaryotic in sediment at the Kuroshima Knoll methane seep. Extremophiles 10: 165–169. [DOI] [PubMed] [Google Scholar]
- 16. Lai X, Cao L, Tan H, Fang S, Huang Y, et al. (2007) Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J 1: 756–762. [DOI] [PubMed] [Google Scholar]
- 17. Nagahama T, Takahashi E, Nagano Y, Abdel-Wahab MA, Miyazaki M (2011) Molecular evidence that deep-branching fungi are major fungal components in deep-sea methane cold seep sediments. Environ Microbiol 13: 2359–2370. [DOI] [PubMed] [Google Scholar]
- 18. Zhang XY, Zhang Y, Xu XY, Qi SH (2013) Diverse deep-sea fungi from the South China Sea and their antimicrobial activity. Curr Microbiol 67: 525–530. [DOI] [PubMed] [Google Scholar]
- 19. Zhang XY, Xu XY, Peng JX, Ma CF, Nong XH, et al. (2014) Antifouling potentials of eight deep-sea-derived fungi from the South China Sea. J Ind Microbiol Biotechnol 41: 741–748. [DOI] [PubMed] [Google Scholar]
- 20. Jebaraj CS, Raghukumar C, Behnke A, Stoeck T (2010) Fungal diversity in oxygen-depleted regions of the Arabian Sea revealed by targeted environmental sequencing combined with cultivation. FEMS Microbiol Ecol 71: 399–412. [DOI] [PubMed] [Google Scholar]
- 21. Damare S, Raghukumar C, Raghukumar S (2006) Fungi in deep-sea sediments of the central Indian basin. Deep-sea Res Part I 53: 14–27. [Google Scholar]
- 22. Burgaud G, Arzur D, Durand L, Cambon-Bonavita MA, Barbier G (2010) Marine culturable yeasts in deep-sea hydrothermal vents: species richness and association with fauna. FEMS Microbiol Ecol 73: 121–133. [DOI] [PubMed] [Google Scholar]
- 23. Gadanho M, Sampaio JP (2005) Occurrence and diversity of yeasts in the mid-Atlantic ridge hydrothermal fields near the Azores Archipelago. Microb Ecol 50: 408–417. [DOI] [PubMed] [Google Scholar]
- 24. Le Calvez T, Burgaud G, Mahe S, Barbier G, Vandenkoornhuyse P (2009) Fungal diversity in deep sea hydrothermal ecosystems. Appl Environ Microbiol 75: 6415–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pang KL, Mitchell JI (2005) Molecular approaches for assessing fungal diversity in marine substrata. Bot Mar 48: 332–347. [Google Scholar]
- 26. Bass D, Howe A, Brown N, Barton H, Demidova M, et al. (2007) Yeast forms dominate fungal diversity in the deep oceans. Proc Biol Sci 274: 3069–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lara E, Moreira D, Lopez-Garcia P (2010) The environmental clade LKM11 and Rozella Form the deepest branching cladeof fungi. Protist 161: 116–121. [DOI] [PubMed] [Google Scholar]
- 28. Stocek T, Taylor GT, Epstein S (2003) Novel eukaryotes from the permanently anoxic Cariaco Basin (Caribbean Sea). Appl Environ Microbiol 69: 5656–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stocek T, Epstein S (2003) Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygendepleted marine environments. Appl Environ Microbiol 69: 2657–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anderson IC, Cairney JWG (2004) Diversity and ecology of soil fungal communities: increased understanding through the application of molecular techniques. Environ Microbiol 6: 769–779. [DOI] [PubMed] [Google Scholar]
- 31. Buchan A, Newell SY, Moretam JIL, Moranm MA (2002) Analysis of internal transcribed spacer (ITS) regions of rRNA genes in fungal communities in a southeastern US salt marsh. Microb Ecol 43: 329–340. [DOI] [PubMed] [Google Scholar]
- 32. Burgaud G, Calvez TL, Arzur D, Vandenkoornhuyse P, Barbier G (2009) Diversity of culturable marine filamentous fungi from deep-sea hydrothermal vents. Environ Microbiol 11: 1588–1600. [DOI] [PubMed] [Google Scholar]
- 33. Bills GF, Polishook JD (1994) Abundance and diversity of microfungi in leaf litter of a lowland rain forest in Costa Rica. Mycologia 86: 187–198. [Google Scholar]
- 34.White TJ, Bruns TD, Lee SB, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. San Diego: Academic. 315–322. [Google Scholar]
- 35. Toledo-Hernandez C, Zuluaga-Montero A, Bones-Gonzalez A, Rodriguez JA, Sabat AM, et al. (2008) Fungi in healthy and diseased sea fan (Gorgonia ventalina): is Aspergillus sydowii always the pathogen? Coral Reefs 27: 707–714. [Google Scholar]
- 36. Singh P, Raghukumar C, Verma P, Shouche Y (2011) Fungal community analysis in the deepsea sediments of the Central Indian Basin by culture-independent approach. Microb Ecol 61: 507–517. [DOI] [PubMed] [Google Scholar]
- 37. Allison SD, McGuire KL, Treseder KK (2010) Resistance of microbial and soil properties to warming treatment seven years after boreal fire. Soil Biol Biochem 42: 1872–1878. [Google Scholar]
- 38. Urban A, Puschenreiter M, Strauss J, Gorfer M (2008) Diversity and structure of ectomycorrhizal and co-associated fungal communities in a serpentine soil. Mycorrhiza 18: 339–354. [DOI] [PubMed] [Google Scholar]
- 39. Waldrop MP, Zak DR, Blackwood CB, Curtis CC, Tilman D (2006) Resource availability controls fungal diversity across a plant diversity gradient. Ecol Lett 9: 1127–1135. [DOI] [PubMed] [Google Scholar]
- 40. Rao S, Hyde KD, Pointing SB (2013) Comparison of DNA and RNA, and cultivation approaches for the recovery of terrestrial and aquatic fungi from environmental samples. Curr Microbiol 66: 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee SH, Kim CG, Kang H (2011) Temporal dynamics of bacterial and fungal communities in a genetically modified (GM) rice ecosystem. Microb Ecol 61: 646–659. [DOI] [PubMed] [Google Scholar]
- 42. Singh P, Raghukumar C, Verma P, Shouche Y (2010) Phylogenetic diversity of culturable fungi from the deep-sea sediments of the Central Indian Basin and their growth characteristics. Fungal Divers 40: 89–102. [Google Scholar]
- 43. Orsi W, Biddle JF, Edgcomb V (2013) Deep sequencing of subseafloor eukaryotic rRNA reveals active fungi across marine subsurface provinces. PLoS One 8(2): e56335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Z, Nilsson RH, Lopez-Giraldez F, Zhuang WY, Dai YC, et al. (2011) Tasting soil fungal diversity with earth tongues: phylogenetic test of SATe alignments for environmental ITS data. PLoS One 6(4): e19039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sakayaroj J, Preefanon S, Supaphon O, Jones EBG, Phongpaichit S (2010) Phylogenetic diversity of endophyte assemblages associated with tropical seagrass Enhalus acoroides from Thailand. Fungal Divers 41: 1–19. [Google Scholar]
- 46. Liu T, Li ZL, Wang L, Tian L, Pei YH, et al. (2011) A new alkaloid from the marine-derived fungus Hypocrea virens . Nat Prod Res 25: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 47. Liu WC, Li CQ, Zhu P, Yang JL, Cheng KD (2010) Phylogenetic diversity of culturable fungi associated with two marine sponges: Haliclona simulans and Gelliodes carnosa, collected from the Hainan Island coastal waters of the South China Sea. Fungal Divers 42: 1–15. [DOI] [PubMed] [Google Scholar]
- 48. De Araujo FV, Soares CA, Hagler AN, Mendonca-Hagler LC (1995) Ascomycetous yeast communities of marine invertebrates in a southeast Brazilian mangrove ecosystem. Antonie van Leeuwenhoek 68: 91–99. [DOI] [PubMed] [Google Scholar]
- 49. Jones MD, Phillips LA, Treu R, Ward V, Berch SM (2012) Functional responses of ectomycorrhizal fungal communities to long-term fertilization of lodgepole pine (Pinus contorta Dougl. ex Loud. var. latifolia Engelm.) stands in central British Columbia. Appl Soil Ecol 60: 29–40. [Google Scholar]
- 50. Gorfer M, Blumhoff M, Klaubauf S, Urban A, Inselsbacher E, et al. (2011) Community profiling and gene expression of fungal assimilatory nitrate reductases in agricultural soil. ISME J 5: 1771–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Damare S, Raghukumar C (2008) Fungi and macroaggregation in deep-sea sediments. Microb Ecol 56: 168–177. [DOI] [PubMed] [Google Scholar]
- 52. Nagahama T, Hamamoto M, Nakase T, Takami H, Horikoshi K (2001) Distribution and identification of red yeasts in deep-sea environments around the northwest Pacific Ocean. Antonie van Leeuwenhoek 80: 101–110. [DOI] [PubMed] [Google Scholar]
- 53. Edgcomb VP, Beaudoin D, Gast R, Biddle JF, Teske A (2011) Marine subsurface eukaryotes: the fungal majority. Environ Microbiol 13: 172–183. [DOI] [PubMed] [Google Scholar]
- 54. Raghukumar C, Raghukumar S, Sheelu G, Gupta SM, Nagender NB, et al. (2004) Buried in time: culturable fungi in a deep-sea sediment core from the Chagos Trench, Indian Ocean. Deep-sea Res Part I 51: 1759–1768. [Google Scholar]
- 55. Zare R, Gams W (2001) A revision of Verticillium section Prostrata. IV. The genera Lecanicillium and Simplicillium gen. nov. Nova Hedwigia 73: 1–50. [Google Scholar]
- 56. Zhao D, Liu B, Li LY, Zhu XF, Wang YY, et al. (2013) Simplicillium chinense: a biological control agent against plant parasitic nematodes, Biocontrol Sci Technol. 23: 980–986. [Google Scholar]
- 57. Zare R, Gams W (2008) A revision of Verticillium fungicola species complex and its affinity with the genus Lecanicillium . Mycol Res 112: 811–824. [DOI] [PubMed] [Google Scholar]
- 58. Baross JA, Hanus FJ, Morita RY (1975) Survival of human enteric and other sewage microorganisms under simulated deep-sea conditions. Appl Microbiol 30: 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pivkin MV (2000) Filamentous fungi associated with holothurians from the sea of Japan, off the primorye coast of Russia. Biol Bull 198: 101–109. [DOI] [PubMed] [Google Scholar]
- 60. Bates ST, Garcia-Pichel F (2009) A culture-independent study of free-living fungi in biological soil crusts of the Colorado Plateau: their diversity and relative contribution tomicrobial biomass. Environ Microbiol 11: 56–67. [DOI] [PubMed] [Google Scholar]
- 61. Green LE, Porras-Alfaro A, Sinsabaugh RL (2008) Translocation of nitrogen and carbon integrates biotic crust and grass production in desert grassland. J Ecol 96: 1076–1085. [Google Scholar]
- 62.Nagahama T, Nagano Y (2012) Cultured and uncultured fungal diversity in deep-sea environments. In: Raghukumar C, editor. Biology of Marine Fungi. Germany: Springer. pp. 173–187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contains Fig. S1 Map of the East Indian Ocean, location and depth of the sampling site and Fig. S2 Rarefaction curves constructed for ITS clone libraries from each of the five sampling sites. ITS, internal transcribed spacer.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files, except that All the ITS sequences of 20 culturable fungal isolate representatives and 45 uncultured fungal clone representatives in the study are available from GenBank under accession numbers KJ173524–KJ17352 and KJ173554–KJ173590.