Abstract
The ability to withstand mitochondrial damage is especially critical for cells such as neurons that survive long-term. We report that cytochrome c (cyt c), a key trigger of apoptosis that is released upon mitochondrial permeabilization, is targeted for proteasome-mediated degradation in postmitotic neurons but not in normal proliferating cells. Importantly, an unbiased siRNA screen identifiedp53 associated Parkin-like cytoplasmic protein (PARC/CUL9) as an E3 ligase that targets cyt c for degradation. PARC/CUL9 levels were markedly elevated with neuronal differentiation and over expression of PARC/CUL9 was sufficient to promote cyt c degradation. Conversely, PARC/CUL9 deficiency made neurons more vulnerable to mitochondrial damage, compromising their ability to survive long-term. Degradation of cyt c by an identical mechanism was also seen in brain tumor cells, highlighting this as an important strategy engaged by neurons and cancer cells to ensure optimal long-term survival.
Keywords: apoptosis, cytochrome c, PARC, CUL9, ubiquitin, Apaf-1, neurons, cancer, mitochondria, proteasome, Parkin
Introduction
Mitochondrial permeabilization and cyt c release are key events that trigger caspase activation and apoptosis in mammalian cells (1). Indeed, mitochondrial release of cyt c has been considered the point of commitment to death for most mitotic cells (2, 3). However, postmitotic cells such as neurons can restrict apoptosis even after cyt c release. Strictly regulating apoptosis after the point of cyt c release is particularly important for neurons so that they can recover from any mitochondrial damage and survive long-term (4). Neurons have been shown to strictly inhibit caspases by the X-linked Inhibitor of Apoptosis Protein (XIAP) and by the maintenance of a highly reduced cellular environment that prevents cyt c oxidation and restricts its pro-apoptotic activity (5, 6). Importantly, sympathetic neurons have the ability to recover from mitochondrial permeabilization if caspase activation is restricted (7, 8). However, the exact fate of cytosolic cyt c in these situations when mitochondria are permeabilized but the cells survive remains unknown.
Several factors have been identified to regulate cyt c-mediated caspase activation. For example, physiological levels of potassium (9), nucleotides (10), and tRNA (11) can inhibit apoptosome formation. Additionally, various proteins including PHAPI, TUCAN, and Aven can modulate apoptosome-mediated caspase activation (12). Oxidation (13) and nitrosylation (14) have also been reported to increase the proapoptotic activity of cyt c.
Apoptosis inhibition is also a fundamental hallmark of cancer cells (15). Indeed, a strict regulation of apoptosis post-cyt c release appears to be adapted by cancer cells for their survival (3, 4, 12). For example, like neurons, many cancer cells are resistant to cytosolic cyt c (6). In addition, both neurons and cancer cells utilize glucose extensively and engage the pentose phosphate pathway to generate a highly reducing cellular environment that limits the ability of cyt c to activate caspases and induce apoptosis (6). These results have brought into focus the possibility that the multiple mechanisms evolved by neurons to restrict apoptosis may be similar to those adapted by mitotic cells during cancer progression.
Here, we describe a novel mechanism in which mitochondrially-released cyt c is targeted for rapid degradation in postmitotic neurons and cancer cells when apoptosis is restricted. Importantly, we discovered that cytosolic cyt c is targeted for degradation by PARC/CUL9, an E3 ligase closely related to Parkin. These results highlight cyt c degradation as an important survival mechanism engaged by both neurons and cancer cells and identify a novel function of PARC/CUL9 in maintaining cell survival after mitochondrial damage.
Results
Cytosolic cyt c is targeted for proteasome-mediated degradation in neurons
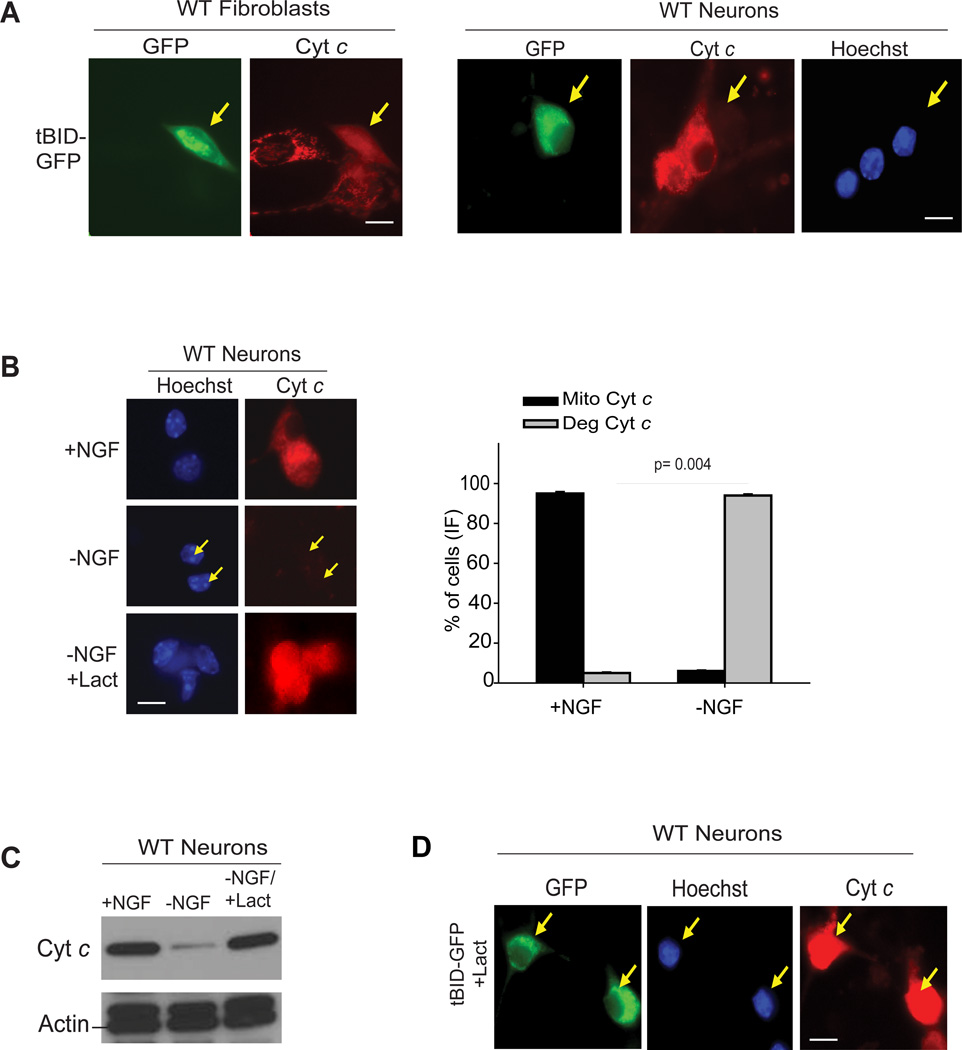
To examine the status of cytoplasmic cyt c that is released from the mitochondria but unable to engage apoptosis, we injected neurons and fibroblasts with tBID, which induces mitochondrial permeabilization, in the presence of a caspase inhibitor. As expected, after release from mitochondria, cyt c accumulated in the cytosol of fibroblasts. In neurons, however, cyt c released from the mitochondria was targeted for degradation (Fig. 1A). Degradation of cytosolic cyt c was also observed in neurons when cyt c release was induced by physiological stimuli, such as nerve growth factor (NGF) deprivation (Figure 1B, Supplementary Fig. S1A). This degradation was proteasome dependent as addition of the proteasome inhibitors lactacystin or bortezomib completely prevented the degradation of cyt c and resulted in its accumulation in the cytosol (Fig. 1B-D and Supplementary Fig. S1B). The degradation of cyt c is likely an important mechanism for neuronal survival as it would allow neurons to withstand mitochondrial damage and decrease any risk of apoptosis caused by the accidental release of cyt c.
Figure 1.
(A) Fibroblasts and sympathetic neurons were injected with tBid-GFP (+zVAD-FMK, 50 µM) (arrows). After 12-24 hrs, cells were immunostained for cyt c. (B) Neurons were either maintained in NGF (+NGF), deprived of NGF for 48h without lactacystin (−NGF), or with lactacystin (10 µM)(−NGF+Lact), in the presence of QVD-fmk (25 µM) and the status of cyt c was assessed by immunocytochemistry. Quantification of (B) shown as percentage of cells with mitochondrial (Mito) or degraded (Deg) cyt c. Data are presented as mean ± SEM of three experiments. (C) Neurons were treated as shown and the status of cyt c was assessed by Western blots. Data are representative of three experiments. (D) Neurons were injected with tBid-GFP (arrows) in the presence of lactacystin (10 µM) and the status of cyt c was assessed by immunocytochemistry. Scale bars, 50µm. All images are representative of three or four independent experiments.
Low Apaf-1 levels are a key determining factor for cyt c degradation
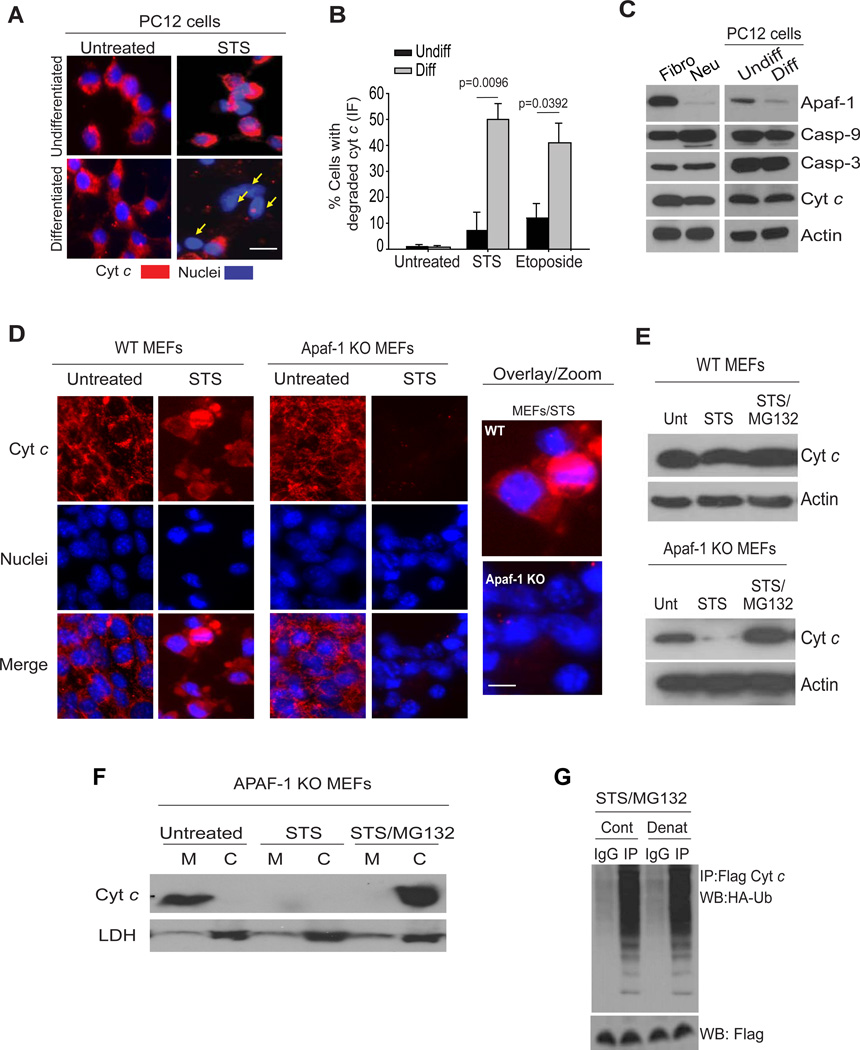
We next examined whether the degradation of cyt c was also seen in other postmitotic cells. Indeed, we found that cardiomyocytes and myotubes also rapidly degrade cyt c after its release from mitochondria (Supplementary Fig. S1C). To examine whether cellular differentiation into a postmitotic state engages the pathway of cyt c degradation, we used the rat pheochromocytoma PC12 cells, which can be maintained either in a mitotic undifferentiated state or can be differentiated into neuronal-like cells in response to the addition of NGF (16, 17). Undifferentiated or neuronally-differentiated PC12 cells were treated with staurosporine to induce the release of cyt c from mitochondria and its status was assessed by immunofluorescence. In contrast to the accumulation of cyt c in the cytosol seen in undifferentiated PC12 cells, cytosolic cyt c was markedly degraded in the differentiated PC12 cells (Fig. 2A and 2B). Degradation was not stimuli-specific as other apoptotic stimuli, such as DNA damage also induced degradation of cytosolic cyt c in differentiated but not mitotic PC12 cells (Fig. 2B).
Figure 2.
(A) Undifferentiated or differentiated PC12 cells were treated with STS (1µM) in the presence of QVD-fmk (25 µM) for 18 hours followed by immunofluorescence for cyt c. (B) Quantification of cells that have released and degraded cyt c in the cytosol (Etoposide: 10 µM for 18 hours). Data are presented as mean ± SEM of three experiments. (C) Comparison of apoptotic proteins between fibroblasts (Fibro) and neurons (Neu) and between undifferentiated (Undiff) and differentiated (Diff) PC12 cells. Data are representative of three experiments. (D) MEFs isolated from either wild-type (WT) or Apaf-1 deficient (Apaf-1 KO) mice were left untreated or treated with staurosporine (STS, 1 µM) for 24 hrs and the status of cyt c was examined by immunocytochemistry (D) or Western blot analysis (E). Wild-type MEFs were treated with QVD-OPH (25 µM) to prevent caspase activation. MG132 (10 µM) was added for 24 hrs. (F) Apaf-1 KO MEFs were treated with STS or STS plus MG132. Subcellular fractions (M, mitochondrial; C, cytosolic) were analyzed by Western blots for cyt c or the cytosolic marker lactate dehydrogenase (LDH). (G) Apaf-1 knockout MEFs were transfected with Flag-cyt c and HA-Ub-wt constructs. Cells were treated with staurosporine (STS; 1 µM) and MG132 (10 µM) for 16 hours. A subset of samples (Denat) were first heated in the presence of 1% SDS to disassociate protein-protein interactions before being subjected to immunoprecipitation. Immunoprecipitates with Flag antibody (cyt c) were probed for HA-Ub by Western blot. Scale bars, 50µm. All Western blot images are representative of three independent experiments.
To gain insight into the mechanism by which neuronal differentiation engages the pathway for cyt c degradation, we focused on the key components of the apoptosome: mitochondrial-released cyt c binds to Apaf-1 to promote caspase-9 and caspase-3 activation, which results in cell death (1). We found that levels of Apaf-1 were selectively reduced in neurons and differentiated PC12 cells while caspase-9 and caspase-3 levels remained unchanged (Fig.2C)(17);Apaf-1 levels are also reduced in postmitotic cardiomyocytes and myotubes (18, 19). Low Apaf-1 levels could leave cytosolic cyt c available as a target for degradation. Therefore, we asked whether deletion of Apaf-1 could confer the ability to degrade cyt c in mitotic fibroblasts (MEFs), which otherwise accumulate cytosolic cyt c. As expected, staurosporine-induced release of mitochondrial cyt c in wild-type fibroblasts resulted in its accumulation in the cytosol. In striking contrast, Apaf-1-deficient fibroblasts treated with staurosporine readily degraded cytosolic cyt c; this degradation was mediated via the ubiquitin proteasome pathway as it was inhibited by the proteasome inhibitor MG132 (Fig. 2D, 2E, and Supplementary Fig. S2 A, S2B). Sub cellular fractionation of these cells also showed the degradation of mitochondrially-released cyt c after staurosporine treatment and its accumulation in the cytosol in the presence of the proteasome inhibitor MG132 (Fig. 2F). These results reveal that the mechanism targeting cytosolic cyt c for degradation is not unique to postmitotic cells and show that mitochondrial-released cyt c that is unable to bind Apaf-1 is targeted for degradation by the proteasome.
We then examined the ubiquitination of cyt c using Apaf-1 deficient fibroblasts expressing HA-tagged-Ubiquitin (Ub) and Flag-cyt c. Robust ubiquitination of mitochondrially-released cyt c was detected in cells treated with staurosporine in the presence of proteasome inhibitors (Fig.2G and Supplementary Fig. S2C). To rule out a possibility that the higher molecular-weight bands were from cyt c-associated proteins rather than cyt c itself, the protein mixture was first heated in the presence of 1% SDS to disassociate protein-protein interactions before being subjected to immunoprecipitation. Our results show that these high molecular-weight bands corresponded to ubiquitinated cyt c as they were detected even under denaturing conditions (Fig.2G).
Neuroblastoma and glioblastoma cells degrade cytosolic cyt c
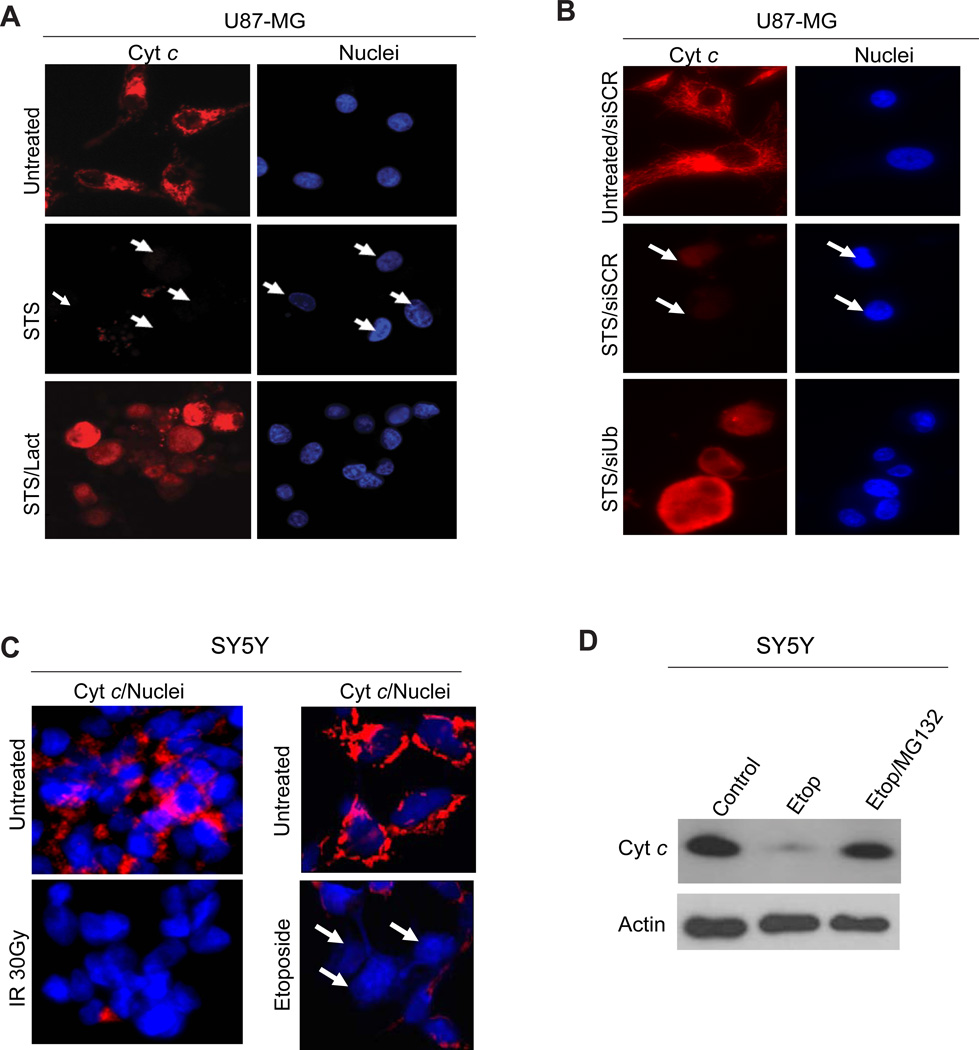
Our previous studies have shown that the apoptotic brake sengaged by neurons to ensure their long-term survival have been adapted by many cancer cells to evade apoptosis (4, 6). Inactivating cyt c could allow cancer cells to survive apoptotic stresses that result in mitochondrial damage. To investigate this possibility we used neuroblastoma and glioblastoma cell lines and subjected them to various apoptosis stressors and examined the status of cytosolic cyt c in the absence or presence of proteasome inhibitors. Indeed, we found that cyt c released from mitochondria is targeted for degradation via the ubiquitin-proteasome pathway in glioblastoma (U87-MG) and neuroblastoma (SH-SY5Y) cell lines (Fig.3A-C). Degradation of cytosolic cyt c in these cancer cells was seen in response to multiple apoptotic stimuli including staurosporine, DNA damage by etoposide, and gamma irradiation. Importantly, proteasome inhibition with lactacystin or MG132 or knockdown of ubiquitin blocked the degradation of cytosolic cyt c (Fig.3A-D).
Figure 3.
(A) U87-MG cells were treated with staurosporine (STS, 1 µM) in the presence or absence of the proteasome inhibitor lactacystin (plus the caspase inhibitor QVD-OPH (25 µM)) for 24 hours. Cells were immunostained for cyt c(red). Hoechst was used to label the nuclei. Representative image shows that cyt c degradation is blocked by addition of lactacystin (Lact). (B) U87-MG cells were transfected with siRNA to ubiquitin (siUb) or scrambled siRNA (siSCR) as control. 72 hours later, cells were treated with staurosporine (STS) for 24 hours in the presence of caspase inhibitors. Cells were then immunostained using cyt c antibody (red). Representative images show that ubiquitin is required for cyt c degradation. (C) SH-SY5Y cells were irradiated with 30 Gyor treated with 10 µM etoposide (Etop)and cultured for 24 hrs in the presence of the caspase inhibitor QVD-OPH. The status of cyt c (red) was assessed by immunostaining. Hoechst was used to label the nuclei. All images are representative of three independent experiments.
Cyt c degradation is mediated by the E3 ligase PARC/CUL9
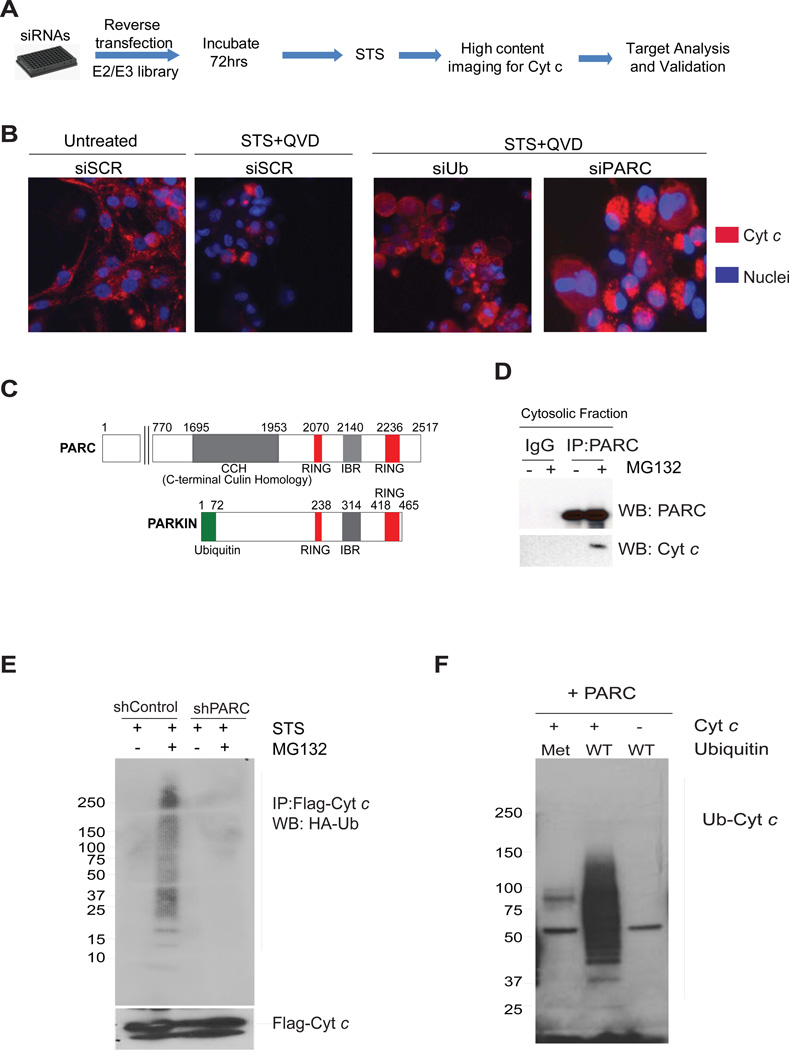
To identify the specific E3 ligase that ubiquitinates cytosolic cyt c, we took an unbiased approach and screened a siRNA library of human E3 ligases (Supplementary Table 1). U87-MG cells were reverse transfected with siRNAs and after a period of three days to allow for the silencing of target genes, cells were treated with staurosporine (in the presence of a caspase inhibitor) to induce the release of cyt c. Transfections were performed in 96-well plates with each well having pools of 4 siRNAs targeting each gene. A siRNA against ubiquitin, which blocks cyt c degradation, and a scrambled siRNA were included as positive and negative controls in each plate in this screen. After 24 hours of staurosporine treatment to allow for the release of cyt c from mitochondria, cyt c was stained by immunofluorescence. The status of cytosolic cyt c (degraded or accumulated) in each of the wells was then scored by two independent investigators to identify potential E3 ligases that, when down regulated, caused the cytosolic accumulation of mitochondrially-released cyt c (Fig. 4A). Following the primary screen and secondary validation (using three independent, non-pooled siRNAs), we identified PARC/CUL9 (p53 associated Parkin-like cytoplasmic protein)as the only E3 ligase that targeted cyt c for degradation in this assay (Fig.4B).
Figure 4.
(A) Flow diagram of the siRNA screen used for the identification of E3 ligases that target cyt c for degradation. (B) U87-MG cells were transfected with control scrambled (siSCR), ubiquitin (siUb)or PARC (siPARC) siRNAs and treated with staurosporine (1 µM) in the presence of QVD-fmk (25 µM) (see methods). The status of cyt c was assessed by immunostaining. (C) Schematic of PARC and Parkin proteins showing homologous regions. (D) Immunoprecipitation using cytosolic fractions demonstrates cyt c and PARC binding in U87-MG cells treated with staurosporine (STS) and MG132. (E) Ubiquitination assay using U87-MG control and shPARC knockdown cells transfected with Flag-cyt c and HA-ubiquitin, and then treated with staurosporine. Immunoprecipitates with Flag antibody (cyt c) were probed for HA-Ub by Western blot. (F) In vitro ubiquitination assay using immunoprecipitated PARC, recombinant cyt c and recombinant ubiquitin, either wild type (WT) or mutant. Recombinant mutant of ubiquitin (Met: Methylated ubiquitin-unable to form poly ubiquitin chains) was used as negative control. Data are representative of three experiments.
PARC, also known as Cullin-9 (Cul9), belongs to the Cull in family of E3 ligases (20, 21). Interestingly, PARC/CUL9 has high homology with the C-terminus of Parkin (21), an E3 ligase mutated in Parkinson’s disease (Fig. 4C)(22, 23). While Parkin is known to target damaged mitochondria for degradation through mitophagy (24), the substrates of PARC/CUL9had remained unknown. We examined whether cytosolic cyt c interacted with PARC/CUL9in cells upon mitochondrial permeabilization. U87-MG cells were treated with staurosporine in the presence of proteasome inhibitor, and PARC/CUL9 was then immunoprecipitated from cytosolic fractions of these cells and probed for cyt c binding. Our results show that cyt c, and PARC/CUL9 form a complex upon mitochondrial permeabilization in these cells (Fig.4D).
To determine whether PARC/CUL9 was responsible for cyt c ubiquitination, we stably knocked down PARC/CUL9 in U87-MG cells and assessed cyt c ubiquitination. Ubiquitination of cyt c was markedly reduced in cells that were stably knocked-down for PARC/CUL9 (Fig.4E). To confirm these results, we performed an in vitro ubiquitination assay using immunoprecipitated PARC/CUL9 and recombinant cyt c and ubiquitin. These in vitro ubiquitination experiments showed cyt c poly ubiquitination, which could be inhibited by the addition of methylated ubiquitin (Fig.4F).
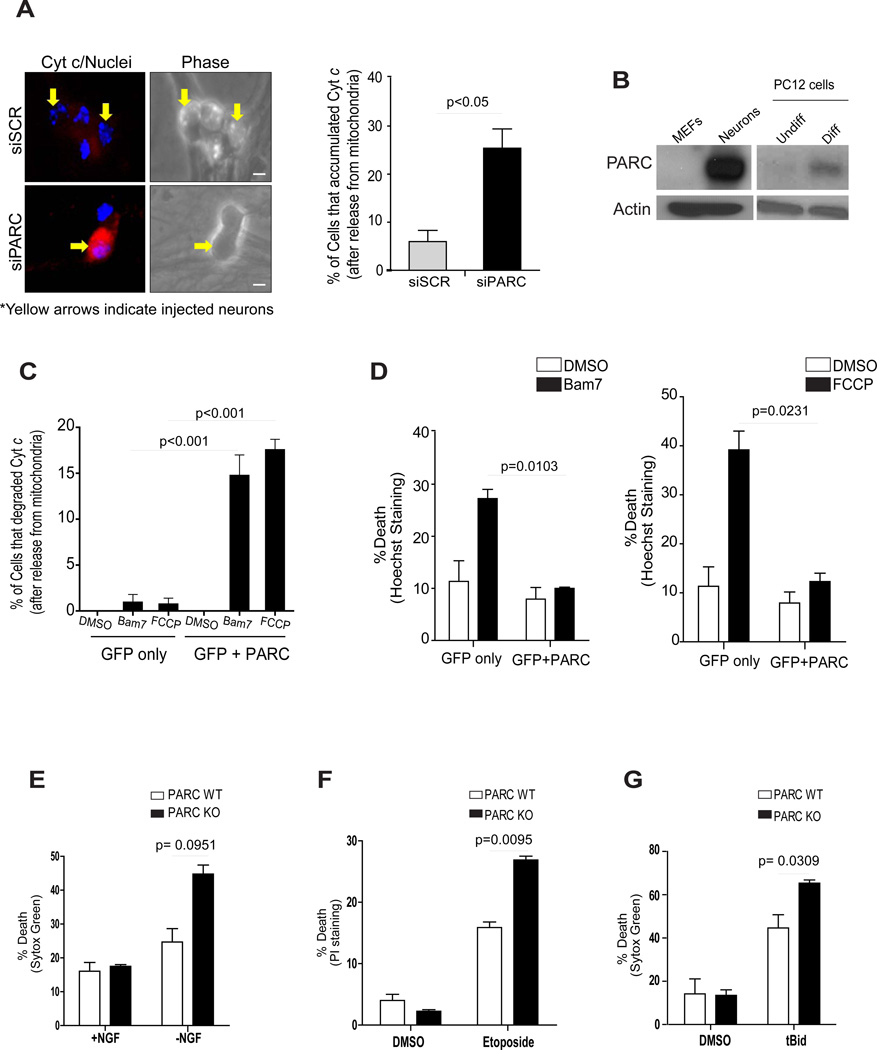
We next examined whether PARC/CUL9 also mediated the degradation of cytosolic cyt c in the multiple cell types where cyt c degradation was observed. In U87-MG cells, knockdown of PARC/CUL9 resulted in an increased cytosolic accumulation of cyt c in response to staurosporine (Supplementary Fig. S3A). Importantly, knockdown of PARC/CUL9 in Apaf-1 knockout MEFs also resulted in the accumulation of mitochondrial-released cyt c (Supplementary Fig. S3B). Importantly, to determine whether cyt c degradation in neurons was also mediated by PARC/CUL9, we microinjected neurons with scrambled siRNA or siRNA to PARC/CUL9, and then deprived of NGF to induce the release of cyt c. While neurons injected with control siRNA showed robust degradation of mitochondrial-released cyt c, knockdown of PARC/CUL9 caused increased accumulation of cyt c in the cytosol (Fig.5A).
Figure 5.
(A) One day after micro-injection with siSCR or siPARC, sympathetic neurons were deprived of NGF for 24 hours in the presence of QVD-fmk (25 µM) and status of cyt c was determined by immunofluorescence and quantified. Error bars represent ± SD for triplicate experiments. (B) Western blot analysis comparing the levels of PARC between MEFs and sympathetic neurons as well as between undifferentiated (Undiff) and differentiated (Diff) PC12 cells. (C, D) HeLa cells were transfected with pCDNA3 empty vector or pCDNA3-HA-PARC and pEGFP. Three days later cells were treated with Bam7 or FCCP for 10 hours and the status of cyt c was determined by immunostaining (C) and cell death was quantified by nuclear morphology using Hoechst staining (D). (E) PARC knockout and wild-type neurons were deprived of NGF for 10 hours or treated with 10 µM Etoposide for 96 hours (F). Death was measured by the vital dyes Sytox Green or Propidium Iodide. Error bars represent ± SD for triplicate experiments. (G) PARC knockout and wild-type neurons were deprived of NGF in the presence of cycloheximide for 48 hours and then microinjected with tBid (8 mM). Death was measured by Sytox Green. Error bars represent ± SD for triplicate experiments.
PARC/CUL9 promotes cell survival after mitochondrial damage
We examined whether PARC/CUL9 was expressed at higher levels in cells that degraded cytosolic cyt c. PARC/CUL9 levels were strikingly elevated in neurons and differentiated PC12 cells (that degrade cyt c) as compared to fibroblasts and undifferentiated PC12 cells (that do not degrade cyt c) (Fig.5B). Interestingly, Apaf-1 levels in the U87-MG and SH-SY5Y cell lines that degrade cyt c were lower than in normal fibroblasts where as PARC/CUL9 levels were comparable (Supplementary Fig. S4A). Thus, while basal PARC/CUL9 expression in the context of low Apaf-1 levels appears to be sufficient for cytosolic cyt c degradation, neuronal differentiation is accompanied not only by a decrease in Apaf-1 but also by a marked increase in PARC/CUL9.
Next, we examined whether over expression of PARC/CUL9 could promote survival in cells that were exposed to mitochondrial damage. We focused on cells that lacked the capability of degrading mitochondrially-released cyt c. HeLa cells and wild-type MEFs were treated with the Bax activator compound Bam7 (25) or the mitochondrial uncoupler FCCP to induce mitochondrial damage and release of cyt c. We found that PARC/CUL9 over expression enabled these cells to degrade cytosolic cyt c (Fig.5C). Importantly, PARC/CUL9 over expression also conferred significant protection in response to mitochondrial damage in these cells (Fig.5D, Supplementary Fig. S4B). Similar protection was also seen with PARC/CUL9 over expression in undifferentiated PC12 cells (Supplementary Fig. S4C).
Conversely, we examined whether PARC/CUL9 deficiency made neurons more vulnerable to mitochondrial damage. We obtained sympathetic neurons from PARC/CUL9-deficient mice (26) and treated them with various insults that induced mitochondrial damage. Our results show that PARC/CUL9-deficient neurons were more sensitive to mitochondrial damage induced by NGF deprivation, etoposide or tBid injection when compared to wild-type neurons (Fig.5E-G).
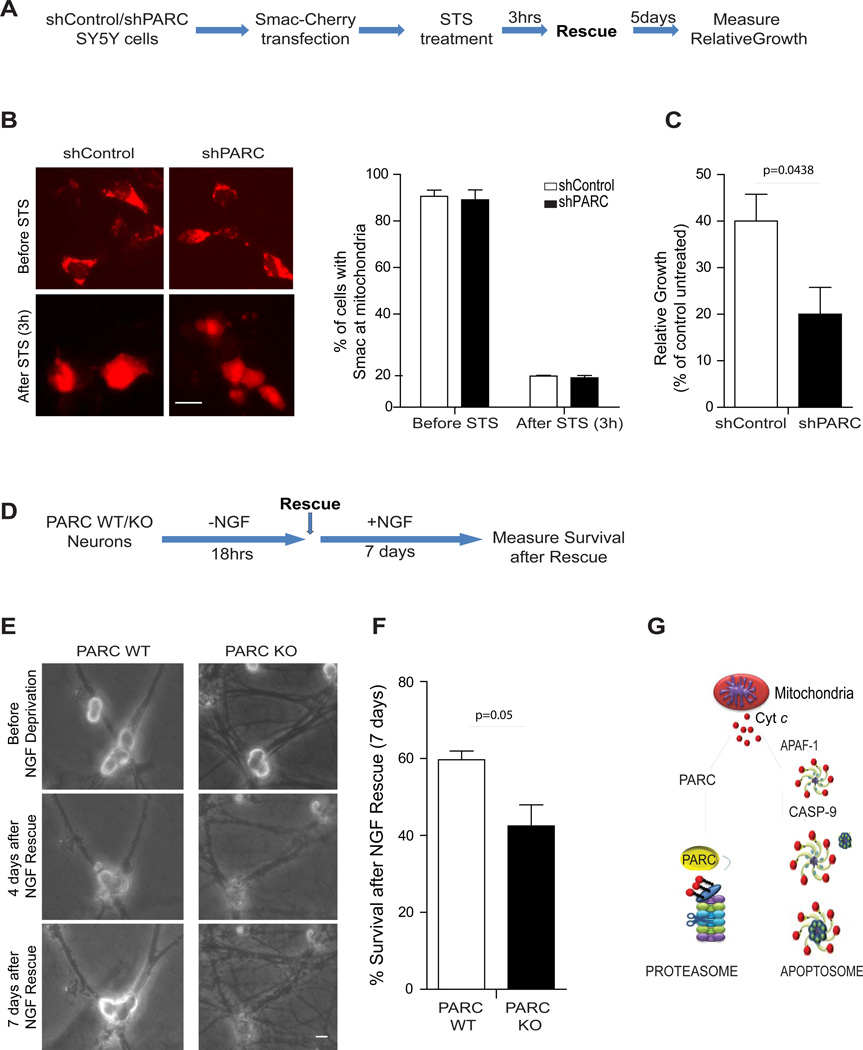
To critically determine whether cells expressing PARC/CUL9 are better equipped to recover after mitochondrial damage, we examined long-term survival in both cancer cells and neurons. We focused first on SH-SY5Y cells and determined the time point after apoptotic treatment at which most cells had undergone mitochondrial outer membrane permeabilization (MOMP) using the Smac (1–60)-mCherry construct (27) (Fig.6A). We found that most cells had released Smac to the cytosol (an indicator of MOMP) after 3 hours of staurosporine treatment (500 nM) (Fig.6B). Importantly, down regulation or over expression of PARC/CUL9 had no effect on the release of Smac from mitochondria, indicating that the kinetics of MOMP were not affected by PARC/CUL9 (Fig. 6B and Supplementary Fig. S4D) Control or PARC/CUL9-deficient SH-SY5Y cells were treated with staurosporine for 3 hours, washed extensively and assessed for survival 5 days later. Our results show that PARC/CUL9-expressing cells had a significant growth advantage compared to those that were knocked down for PARC/CUL9(Fig.6C).
Figure 6.
(A) Flow diagram of the assessment of long-term survival of wild type and PARC-knockdown SY5Y cells. (B) Images of shControl and shPARC SY5Y cells transfected with Smac-cherry and treated with STS for 3hrs. The percentage of cells that have released mitochondrial Smac and undergone mitochondrial outer membrane permeabilization (MOMP) was quantified. Error bars represent ± SD for triplicate experiments (Right panel). (C) After 3 hours of STS treatment cells were washed several times and complete media was re-added to the STS-treated cultures and 5 days later, relative growth was determined by fixating and staining the colonies with methylene blue (as described in material and methods). (D) Flow diagram of the NGF deprivation and rescue (with NGF readdition) assay used to determine the long-term survival of wild type and PARC-deficient sympathetic neurons. (E) Images of the exact same field of neurons taken before treatment, 4 days after recovery and 7 days after recovery. (F) Quantification of the percent survival after 18 hrs of NGF deprivation and 7 days of NGF rescue. Error bars represent ± SD for triplicate experiments. All images are representative of three independent experiments. (G) Proposed model for cyt c regulation after its release from mitochondria. Scale bars, 50µm
To examine the importance of PARC/CUL9 on long-term survival after mitochondrial damage in neurons, we isolated sympathetic neurons from wild-type and PARC/CUL9 knockout mice. Neurons were deprived of NGF for 18 hours, a period of time that was previously shown to induce MOMP in these cells (8). Neurons were then rescued by addition of NGF-containing media and left in culture for 7 additional days (Fig.6D). After 7 days of NGF re-addition, the rescued neurons were clearly identifiable with large and phase-bright cell bodies, whereas the non-rescued neurons became atrophic and appeared degenerated (Fig. 6E). Thus, after 7 days of NGF rescue, it was possible to unequivocally distinguish between those cells that were rescued by NGF re-addition and those cells that had already committed to die and were not able to recover with NGF re-addition. Our results with this NGF rescue assay show that PARC/CUL9-deficient neurons were less capable of recovery after mitochondrial damage (Fig. 6F).
Together, these results show that neurons and cancer cells can respond to mitochondrial damage by degrading the mitochondrially-released cyt c and limiting the risk of apoptosis. Our results also identify PARC/CUL9 as an E3 ligase that targets cyt c degradation and show that PARC/CUL9 can modulate cell survival and allow recovery from situations of mitochondrial damage.
Discussion
Despite the central role of cyt c in activating apoptosis, the mechanism by which cyt c itself is regulated in cells is poorly understood. In fact, the mitochondrial release of cyt c has been considered to be the point-of-no-return in the apoptotic cascade because of the mitochondrial damage incurred as a consequence of mitochondrial permeabilization occurring prior to cyt c release in cells (3). Thus, a cell could die of mitochondrial dysfunction and energetic failure even if caspase activation is blocked. However, sympathetic neurons have been shown to maintain survival and recover even after mitochondrial permeabilization and cyt c release (7, 8). Our results here show that neurons that have released cyt c but are unable to activate caspases engage a mechanism to rapidly degrade cyt c and eliminate any further risk of activating the apoptotic machinery.
A question that emerged was whether the ability to degrade cyt c was particular to postmitotic cells such as neurons or whether the degradation machinery was present in all cells but selectively engaged in postmitotic cells. Our results show that a key feature common to multiple postmitotic cells where cyt c degradation was observed was that these cells had low levels of Apaf-1. As Apaf-1 is the main protein cyt c binds to upon its release from mitochondria, low Apaf-1 levels could leave an excess of cyt c available for degradation. Also, since the interaction between Apaf-1 and cyt c is thought to be transient (28), all cytosolic cyt c could become available for degradation if apoptosis is unable to proceed. Our results show that Apaf-1 deficiency alone is able to confer the ability of degrading cyt c to mitotic cells such as fibroblasts that otherwise do not degrade cytosolic cyt c. These results are significant because they imply that the machinery that targets cytosolic cyt c for degradation is not unique to postmitotic cells. Indeed, Apaf-1 deficient neural precursor cells have also been shown to degrade cyt c (29, 30). Together, these results point to Apaf-1 as a key factor that determines whether cyt c remains stable in the cytosol or is targeted for degradation once it is released from the mitochondria.
Using an unbiased siRNA screen, we identified PARC/CUL9 as an E3 ligase that targets cyt c, once released from the mitochondria, for degradation. While PARC/CUL9 has been identified as an E3 ligase due the presence of RING and Cull in domains (21, 26, 31), our results identify cyt c as its first known substrate. As cyt c degradation occurs only after its release from mitochondria, any previous proteomic approach to identify the PARC/CUL9 substrates would have failed to detect cyt c as it would have remained in the mitochondria under normal conditions. Our data show that PARC/Cul9 could ubiquitinate cyt c in vitro and its knockdown increased the accumulation of cytosolic cyt c in cells (Fig. 4, 5). However, a fraction of cells still degraded cyt c even in the absence of PARC/Cul9, indicating that an alternative E3 ligase could also target cyt c for degradation.
The C-terminus of PARC/CUL9 is highly similar to Parkin, a known E3 ligase with mutations that are associated with Parkinson’s disease (23). Parkin is well known to be recruited to damaged mitochondria and promote its degradation (24). The high degree of structural similarity between PARC/CUL9 and Parkin (Fig.5C) suggests that like Parkin, PARC/CUL9 may also be important for promoting neuronal survival after mitochondria injury. Consistent with this model, our results show thatPARC/CUL9-deficient neurons were more vulnerable to multiple situations of mitochondrial damage. PARC/CUL9 over expression was also sufficient to degrade cytosolic cyt c and promote survival in mitotic cells exposed to mitochondrial damage. Importantly, we also examined the ability of PARC/CUL9 to regulate neuronal recovery after mitochondrial damage by conducting a long-term survival assay. Indeed, our results show that PARC/CUL9-deficient neurons were less capable of recovery after mitochondrial damage and rescue (Fig.6). While PARC/CUL9 can reduce the risk of apoptosis by targeting mitochondrially-released cyt c for degradation and promoting neuronal recovery, it is important to point out that sympathetic neurons with significant dysfunctional mitochondria cannot be maintained alive indefinitely. These neurons are remarkably capable of maintaining mitochondrial membrane potential despite the loss of cyt c for a period in part by maintaining ATP generation via glycolys is (8, 32, 33). Our results reiterate the notion that sympathetic neurons are not irreversibly committed to die upon mitochondrial permeabilization and that PARC/CUL9 is important for optimal neuronal rescue during this time period.
As in neurons, the ability to evade apoptosis is one of the hallmarks of cancer cells (15). In previous reports, we identified the strict inhibition of caspases by XIAP and the reducing cellular environment as two mechanisms by which apoptosis is selectively restricted in neurons and cancer cells (4, 6). Neurons and cancer cells have also been shown to recover from mitochondrial permeabilization if apoptosis is restricted (7, 8, 27). Here we find that cytosolic cyt c is degraded in cancer cells just as observed in neurons, indicating that these cells engage redundant mechanisms to ensure survival in situations of mitochondrial damage. Degradation of cytosolic cyt c could also be important even if apoptosis is inhibited as continued accumulation of cytosolic cyt c over a long period of time could be detrimental to cells. For example, while the binding of cyt c to Apaf-1 via the WD40 domain has been best studied (34–36), it is conceivable that cyt c may also interact with other WD40 domain-containing proteins with negative consequences. The mechanism by which cyt c is degraded in both neurons and cancer cells involves proteasomal targeting of cyt c by the same E3 ligase PARC/CUL9. An important implication of these results is that even though neurons and cancer cells are markedly different cell types, they are strikingly similar with regards to apoptosis restriction and that the survival mechanisms evolved by neurons may be co-opted by cancer cells to evade apoptosis.
Materials and Methods
Sympathetic neuronal cultures
Primary sympathetic neurons were dissected from the superior cervical ganglia of postnatal day 0–1 mice (P0–P1). Cells were plated on collagen coated dishes at a density of 60 000 cells per well for Western or RT-PCR analysis, or 10 000 cells per well for microinjection, survival counts or immunofluorescence experiments. Sympathetic neurons were grown for 4–5 days in NGF-containing media before treating them with experimental conditions. For treatments, etoposide was used at a concentration of 20 uM. For NGF deprivation, cultures were rinsed three times with medium lacking NGF, followed by the addition of goat anti-NGF neutralizing antibody to this media in the presence of caspase inhibitors (QVD-OPH or z-VAD-fmk). ICR outbreed mice (Harlan) were used for isolation of neurons in all experiments.
For the assessment of cell viability, sympathetic neurons were grown in NGF-containing medium (AM50) for 4–5 d, and then either maintained in AM50 or treated with various conditions. For NGF deprivation, cultures were rinsed twice with medium lacking NGF (AM0: AM50 medium without NGF), followed by addition of AM0 containing goat anti–NGF. For rescue experiments in which NGF was re-added to NGF-deprived cultures, cultures were rinsed three times and incubated in the NGF-containing medium (AM50) for seven additional days. After 7 days of NGF readdition, the rescued neurons were clearly identifiable with large and phase-bright cell bodies, whereas the non-rescued neurons became atrophic and appeared degenerated.
XIAP or PARC/CUL9 deficient neurons were isolated from XIAP or PARC/CUL9 knockout C57BL/6 mice, respectively; neurons from wild-type littermate controls were used in those experiments. For isolation of Apaf-1 deficient knockout fibroblasts, Apaf-1 deficient C57BL/6mice were used; wild-type littermates were used as controls in these experiments. Apaf-1deficient mice were generated by Joachim Herz (UT Southwestern) and were kindly provided by the laboratory of Dr. Susan Ackerman (Jackson Laboratories).
siRNA transfections and screen
To identify the specific E3 ligase that targets cytosolic cyt c for degradation, we screened a siRNA library of human E3 ligases from Dharmacon (Ref Seq database v5.0-8.0). All transfections were performed with 50 nmol/L final concentration of siGENOME SMART pools (4 siRNAs per gene). Each well had pools of 4 siRNAs that targeted individual genes and 6 wells per gene were plated (for 3 untreated and 3 treated conditions) on each 96 well plate. Each 96-well plate was analyzed in triplicate for a total of 9 independent untreated control and 9 independent treated conditions. Each plate also had a scrambled siRNA as a negative control and siRNA to ubiquitin as a positive control (again, 6 wells per plate in triplicates). U87-MG cells were reverse transfected with the siRNAs and after a period of three days to allow for the silencing of target genes, cells were either left untreated or treated with staurosporine (for 24 hrs) to induce the release of cyt c. The status of cyt c was then assessed by immunofluorescence to identify potential E3 ligases that, when down regulated, caused the cytosolic accumulation of mitochondrial-released cyt c. A siRNA against ubiquitin, which blocks cyt c degradation (in the presence of caspase inhibitors) and causes cell death (in the absence of caspase inhibitors), was included as a positive control in the screen. Reverse transfections were performed by mixing appropriate Dharmafect (Thermo Fisher) with siRNA pools and incubating for 20 minutes followed by delivery of trypsin-derived single-cell suspensions. Following the primary screen a secondary validation was performed using three independent (not pooled) siRNAs (from Sigma) using the same assay to determine the status of cytosolic cyt c.
Immunofluorescence analysis
The status of cyt c (whether intact in the mitochondria or released) was examined by immunostaining with cyt c antibodies. Briefly, sympathetic neurons were cultured for 4 days after which they were either left untreated or deprived of NGF for various time points. Neurons and other cell lines were fixed in 4% paraformaldehyde and incubated overnight in anti-cyt c primary antibody (556432, BD Biosciences) followed by a 2 h incubation with anti-mouse Cy3 secondary antibody (Jackson Labs). Nuclei were stained with Hoechst 33258 (Molecular Probes).
Microinjection and quantitation of cell survival
The method of microinjecting sympathetic neurons has been described previously (6, 37). After injections, the number of viable cells injected was determined by counting rhodamine-positive cells that had intact phase-bright bodies. The nucleus was injected with 200 ng/ml of the plasmid DNA along with 50 ng/ml of EGFP-expressing DNA (Clonetech laboratories Inc.) in microinjection buffer. After 24 h to allow for expression, GFP-expressing cells were identified by fluorescence microscopy. The survival of these double-injected cells was assessed by morphological criteria (38). For injection of siRNA, neurons were dissected from postnatal day 2 mice, cultured for 3 days and microinjected with siRNA to PARC/CUL9 (siPARC) or scrambled control (siSCR) in the presence of rhodamine.
For experiments in which neuronal survival was assessed in response to mitochondrial damage, neurons were deprived of NGF in the presence of cycloheximide for 48 hours (to relieve the XIAP inhibition of caspases) (5, 39) before microinjection with tBid peptide (8 mM) for 10 hours.
Image acquisition and processing
Images were acquired by an ORCA-ER digital B/W CCD camera (Hamamatsu) mounted on a DMIRE2 inverted fluorescence microscope (Leica) using Metamorph version 7.6 software (Molecular Devices). For most quantification experiments three-hundred cells were analyzed in triplicate samples. The data presented in the figures show the percentage of apoptosis ± SEM of three independent experiments. In the case of the siRNA screen, a high content imaging scope was used to take 18 images of each of the wells of a 96 well plate format, using a 20X /0.8 NA objective. The High Content Imaging instrument was the Array Scan VTI HCS Reader (Thermo Scientific). Confocal images were taken on an Olympus FV1000 confocal microscope.
Ubiquitination Assays
Cells were transfected with 2 ug of Flag-Cyt c and HA-ubiquitin constructs. Twenty-four hours later, cells were treated with staurosporine in the presence or absence of MG132 for 16 hours. Cells were then lysed in RIPA buffer containing protease inhibitors and NEM. For denaturing conditions, cells were lysed in 1% SDS and boiled for 10 minutes. Samples were then diluted with 0.5% NP40 and subjected to immunoprecipitation. One milligram of total protein was incubated with EZ-view Flag beads overnight. After stringent washes, 30 ul of Laemmli sample buffer containing 2% (v/v) β-mercaptoethanol was added to samples. Samples were then boiled for 5 min and Western blot analysis was performed using an HRP-conjugated-anti-HA antibody (Roche).
For the in vitro ubiquitination assays, PARC/CUL9 was immunoprecipitated from U87-MG cells, washed 5 times with high salt buffer (containing 500 mM NaCl) and followed by 5 washes with PBS. The in vitro reaction was then set up as follows: Reaction volumes of 40 ul containing 3.5 mM Mg ATP (Boston Biochem), 3.5 mM of Energy Regeneration solution (Boston Biochem), 40 nM human ubiquitin activating enzyme, 150 nM HsUbc7, 500 nM recombinant cyt c, were incubated for 2 h at 37°C in the presence of 10 uM unlabeled ubiquitin (wild type or mutant). Reactions were quenched with 15 ul of Laemmli sample buffer containing 2% (v/v) B-mercaptoethanol and boiled for 5 min. Ubiquitination was detected by Western blot.
Long-term survival in SY5Y control and shPARC/CUL9 cells
Relative cell numbers after treatment were determined by fixing and staining the colonies with methylene blue (Sigma-Aldrich) dissolved in 50% methanol. Color extraction was performed using 0.5 M hydrochloric acid, and the staining (which is proportional to cell number) was quantitated by measuring absorbance at 595 nM. All experiments were conducted three times in duplicate.
Statistical Analysis
p values for the differences in means were calculated with an unpaired, two-tailed Student’s t test.
Supplementary Material
Acknowledgements
We thank Angelique Whitehurst, Gary Johnson, Cam Patterson, Doug Cyr, Holly McDonough, Pamela Lockyer, Matthew Smith, and Melissa Srougi for technical support. We also thank all the members of the Deshmukh and Gershon labs for critical review of this manuscript. The Bax inhibitor Bam7 was kindly provided by Dr. Loren Walensky (Harvard Medical School) and the Smac(1–60) mCherry construct was kindly provided by Dr. Douglas R. Green (St. Jude Children’s hospital). This work was supported by NIH grants GM078366 (to MD), AHA and ABTA Fellowships (to VG). Confocal imaging was supported by NINDS Center Grant P30 NS04892.
Footnotes
Author contributions: V.G., V.S., J.S., A.J.K., A.E., Y.H., A.C., M.D. planned and conducted the experiments. B.G. and N.S. contributed to the siRNA screen. X.P. and Y. X. provided a key reagent and technical expertise. V.G. and M.D. wrote the manuscript.
Competing interest: the authors disclose no competing interests.
Data and materials availability: Data and materials are available upon request.
Supplemental Materials
Fig. S1. Degradation of mitochondrially-released cyt c in neurons, cardiomyocytes and myotubes.
Fig. S2. Cyt c ubiquitination and degradation in Apaf-1-deficient cells.
Fig. S3. Inhibition of cyt c degradation in PARC/CUL9-deficient cells.
Fig. S4. PARC/CUL9 over expression inhibits cell death post-MOMP.
Table 1. List of genes included in the siRNA library of human E3 ligases from Dharmacon.
Supplemental materials and methods
References
- 1.Wang X. Genes Dev. 2001;15:2922. [PubMed] [Google Scholar]
- 2.Green DR, Kroemer G. Science. 2004;305:626. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 3.Tait SW, Green DR. Nat. Rev. Mol. Cell Biol. 2010;11:621. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 4.Wright KM, Deshmukh M. Cell Cycle. 2006;5:1616. doi: 10.4161/cc.5.15.3129. [DOI] [PubMed] [Google Scholar]
- 5.Potts PR, Singh S, Knezek M, Thompson CB, Deshmukh M. J. Cell Biol. 2003;163:789. doi: 10.1083/jcb.200307130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaughn AE, Deshmukh M. Nat. Cell Biol. 2008;10:1477. doi: 10.1038/ncb1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinou I, et al. J. Cell Biol. 1999;144:883. doi: 10.1083/jcb.144.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshmukh M, Kuida K, Johnson EM., Jr J. Cell Biol. 2000;150:131. doi: 10.1083/jcb.150.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cain K, Langlais C, Sun XM, Brown DG, Cohen GM. J. Biol. Chem. 2001;276:41985. doi: 10.1074/jbc.M107419200. [DOI] [PubMed] [Google Scholar]
- 10.Chandra D, et al. Cell. 2006;125:1333. doi: 10.1016/j.cell.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 11.Mei Y, et al. Mol. Cell. 2010;37:668. doi: 10.1016/j.molcel.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schafer ZT, Kornbluth S. Dev. Cell. 2006;10:549. doi: 10.1016/j.devcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 13.Brown GC, Borutaite V. Biochim. Biophys. Acta. 2008;1777:877. doi: 10.1016/j.bbabio.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 14.Schonhoff CM, Gaston B, Mannick JB. J. Biol. Chem. 2003;278:18265. doi: 10.1074/jbc.M212459200. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan D, Weinberg Robert A. Cell. 2011;144:646. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Greene LA. J. Cell Biol. 1978;78:747. doi: 10.1083/jcb.78.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright KM, Linhoff MW, Potts PR, Deshmukh M. J. Cell Biol. 2004;167:303. doi: 10.1083/jcb.200406073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith MI, Huang YY, Deshmukh M. PLoS ONE. 2009;4:e5097. doi: 10.1371/journal.pone.0005097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potts MB, Vaughn AE, McDonough H, Patterson C, Deshmukh M. J. Cell Biol. 2005;171:925. doi: 10.1083/jcb.200504082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merlet J, Burger J, Gomes JE, Pintard L. Cell. Mol. Life Sci. 2009;66:1924. doi: 10.1007/s00018-009-8712-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikolaev AY, Li M, Puskas N, Qin J, Gu W. Cell. 2003;112:29. doi: 10.1016/s0092-8674(02)01255-2. [DOI] [PubMed] [Google Scholar]
- 22.Vaughan JR, Davis MB, Wood NW. Ann. Human Genet. 2001;65:111. doi: 10.1017/S0003480001008557. [DOI] [PubMed] [Google Scholar]
- 23.Dawson TM, Ko HS, Dawson VL. Neuron. 2010;66:646. doi: 10.1016/j.neuron.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Youle RJ, Narendra DP. Nat. Rev. Mol. Cell. Biol. 2011;12:9. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gavathiotis E, Reyna DE, Bellairs JA, Leshchiner ES, Walensky LD. Nat. Chem. Biol. 2012;8:639. doi: 10.1038/nchembio.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pei XH, et al. Cancer Res. 2011;71:2969. doi: 10.1158/0008-5472.CAN-10-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tait SW, et al. Dev. Cell. 2010;18:802. doi: 10.1016/j.devcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Twiddy D, et al. J. Biol. Chem. 2004;279:19665. doi: 10.1074/jbc.M311388200. [DOI] [PubMed] [Google Scholar]
- 29.Cozzolino M, et al. Cell Death Differ. 2004;11:1179. doi: 10.1038/sj.cdd.4401476. [DOI] [PubMed] [Google Scholar]
- 30.Ferraro E, et al. Mol. Biol. Cell. 2008;19:3576. doi: 10.1091/mbc.E07-09-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarikas A, Hartmann T, Pan Z-Q. Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang LK, Schmidt RE, Johnson EM., Jr J. Cell Biol. 2003;162:245. doi: 10.1083/jcb.200302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang LK, Johnson EM., Jr J. Cell Biol. 2002;157:771. doi: 10.1083/jcb.200112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srinivasula SM, Ahmad M, T F-A, Alnemri ES. Mol. Cell. 1998;1:949. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 35.Hu Y, Ding L, Spencer DM, Nunez G. J. Biol Chem. 1998;273:33489. doi: 10.1074/jbc.273.50.33489. [DOI] [PubMed] [Google Scholar]
- 36.Acehan D, et al. Mol. Cell. 2002;9:423. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.