Summary
Background
To function in diverse cellular processes, the dynamic behavior of microtubules (MTs) must be differentially regulated within the cell. In budding yeast, the spindle position checkpoint (SPOC) inhibits mitotic exit in response to mispositioned spindles. To maintain SPOC-mediated anaphase arrest, astral MTs must maintain persistent interactions with and/or extend through the bud neck. However, the molecular mechanisms that ensure the stability of these interactions are not known.
Results
The presence of a MT extending through and/or interacting with the bud neck is maintained by spatial control of catastrophe and rescue, which extends MT lifetime >25-fold and controls the length of dynamic MTs within the bud compartment. Moreover, the single kinesin-8 motor, Kip3, alternately mediates both catastrophe and rescue of the bud MT. Kip3 accumulates in a length-dependent manner along the lattice of MTs within the bud. Yet, it induces catastrophe spatially near the bud tip. Rather, this accumulation of Kip3 facilitates its association with depolymerizing MT plus-ends, where Kip3 promotes rescue before MTs exit the bud. MT rescue within the bud requires the tail domain of Kip3, whereas the motor domain mediates catastrophe at the bud tip. In vitro, Kip3 exerts both stabilizing and destabilizing effects on reconstituted yeast MTs.
Conclusions
The kinesin-8 Kip3 is a multifunctional regulator that differentially stabilizes and destabilizes specific MTs. Control over MT catastrophe and rescue by Kip3 defines the length and lifetime of MTs within the bud compartment of cells with mispositioned spindles. This subcellular regulation of MT dynamics is critical to maintain mitotic arrest in response to mispositioned spindles.
Introduction
Microtubules (MTs) are essential cytoskeletal filaments comprised of polymerized tubulin that function in cellular processes such as chromosome segregation, cell migration and intracellular transport [1]. MTs display dynamic instability, the inherent property to stochastically transition from periods of growth to shortening, termed catastrophe, and vice versa, called rescue [2]. To function in diverse processes, dynamic instability must be regulated in space and time within a cell. Determining the mechanisms that spatially and temporally control MT dynamics is critical to understand how cells achieve complex MT-mediated functions.
One class of MT regulators are the plus-end tracking proteins, or +TIPs, which typically localize to the plus-end of polymerizing, but not depolymerizing MTs [3]. The +TIP kinesin-8 is a conserved class of MT motor proteins that possess plus-end directed motility and also regulate MT dynamics [4]. The kinesin-8 from budding yeast, Kip3, and from human, Kif18A, can accelerate MT depolymerization in vitro [5–7], and Kip3 elevates catastrophe frequency in the cell [5]. Furthermore, in vitro, Kip3 motility generates a concentration gradient along the lattice of stabilized MTs that increases toward the plus-end. As a result, Kip3 depolymerizes MTs in a length-dependent manner [6]. Loss of kinesin-8 leads to increased MT length in multiple species [8–12], and loss of the fission yeast kinesin-8 selectively decreases the catastrophe frequency of longer MTs [13]. However, whether Kip3, or kinesin-8 in general, destabilize MTs by a length-dependent mechanism in vivo has not been established.
In addition to destabilizing activities, kinesin-8 also display MT stabilizing properties. In budding yeast, Kip3 decreases MT depolymerization rate and increases rescue [5]. Fission yeast Klp5/6 enhances MT nucleation in vitro and in vivo [14]. Human Kif18A promotes pausing of dynamic MTs in vitro and dampens kinetochore oscillations in vivo [15, 16]. Thus kinesin-8 is a multifunctional protein that can stabilize and/or destabilize MTs. How these functions may be coordinated to regulate MT behavior in vivo is not clear.
The budding yeast, Saccharomyces cerevisiae, undergoes asymmetric cell division in which cytokinesis occurs at the junction between the mother and bud compartments, known as the bud neck. To achieve chromosome segregation, two mechanisms function to position the mitotic spindle across the neck prior to cytokinesis; reviewed in [17]. During early spindle formation the Kar9-dependent mechanism positions one spindle pole near the bud neck. During anaphase the Dynein-dependent mechanism pulls this pole across the neck into the bud. A surveillance mechanism named the spindle position checkpoint (SPOC) inhibits mitotic progression until anaphase spindles span the bud neck, when a signaling cascade called the mitotic exit network then triggers spindle disassembly and cytokinesis.
Bud neck-associated proteins play critical roles in the SPOC (reviewed in [18]), and perturbations of astral MT association with the bud neck inhibit SPOC function [19, 20]. When anaphase spindles are mispositioned within the mother, astral MTs typically pass through the neck into the bud [21, 22], and it was proposed that interaction between MTs and the bud neck is required for SPOC function [20]. Indeed, disruption of these MTs by laser severing causes aberrant mitotic exit when spindles are mispositioned [22]. Although the molecular nature of this interaction is unknown, astral MTs passing through the bud neck and/or extending into the bud compartment satisfy the requirement for the SPOC to delay mitotic exit. SPOC-mediated arrest can persist for hours [21], yet, the lifetime of yeast MTs is only ~3.5 minutes [23]. How these short-lived MTs can persistently contact or extend through the bud neck is not known.
We show that when anaphase spindles are mispositioned, the persistence of an astral MT extending through the bud neck is achieved by spatial control of MT dynamics. The kinesin-8, Kip3, increases the catastrophe frequency specifically at the bud tip, which limits MT length in the bud. Additionally, Kip3 elevates rescue frequency near the bud neck, which promotes rescue of the same MT before it depolymerizes out of the bud. By alternately stabilizing and destabilizing MTs, Kip3 increases their lifetime within the bud, which is required to prevent mitotic exit, and maintain genome stability when spindles are mispositioned.
Results
Microtubule lifetime is differentially regulated within cells with mispositioned spindles
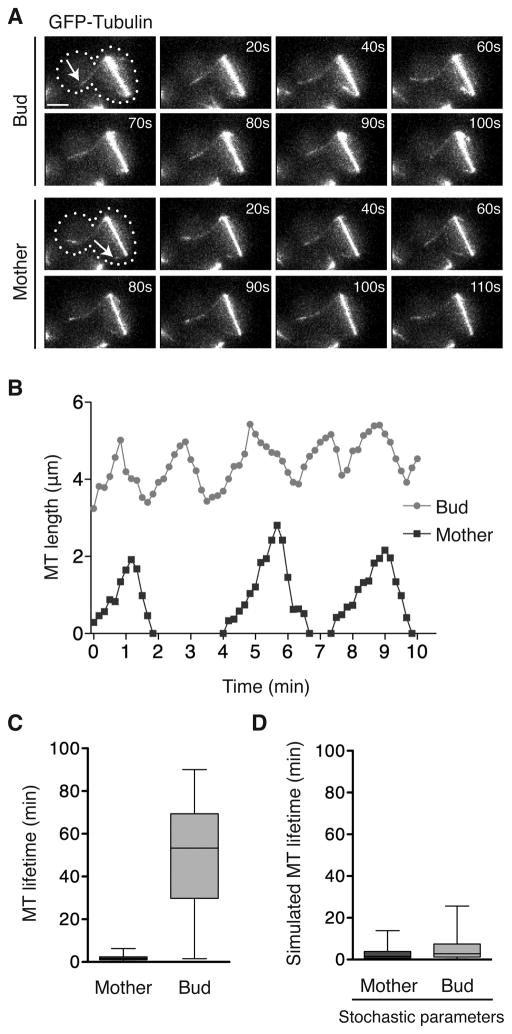
We sought to determine how persistent interaction between astral MTs and the bud neck is maintained in cells with mispositioned anaphase spindles. Cells lacking the motor protein Dynein (dyn1Δ) are deficient in positioning the spindle across the bud neck. To reproducibly observe mispositioned spindles, we monitored GFP-Tub1 labeled MTs in dyn1Δ cells (Figures 1A, 1B, and S1A–B). The mean astral MT lifetime within the mother is 2 minutes, comparable to lifetimes during other stages of the cell cycle. Strikingly, the mean lifetime of MTs that protrude into the bud compartment is 50 minutes (Figure 1C). This difference does not originate from a specific spindle pole since other MTs from the same pole that remain in the mother only persist for ~2 minutes. Similarly, MT lifetime is extended in the bud of other mutants that display mispositioned spindles. Mother MTs in num1Δ cells have a 2.5 minute average lifetime, while bud MTs last 47 minutes (Figure S1C). Correspondingly, in act5Δ cells the lifetime in the mother and bud is 2.5 and 42 minutes, respectively (Figure S1C). Thus, MT lifetime is differentially regulated in the mother and bud compartments of cells with mispositioned anaphase spindles. Furthermore, persistent interaction between astral MTs and the bud neck is achieved not by the repeated growth of new MTs, but through the regulation of specific MTs.
Figure 1. Microtubule lifetime is differentially regulated within cells with mispositioned spindles.
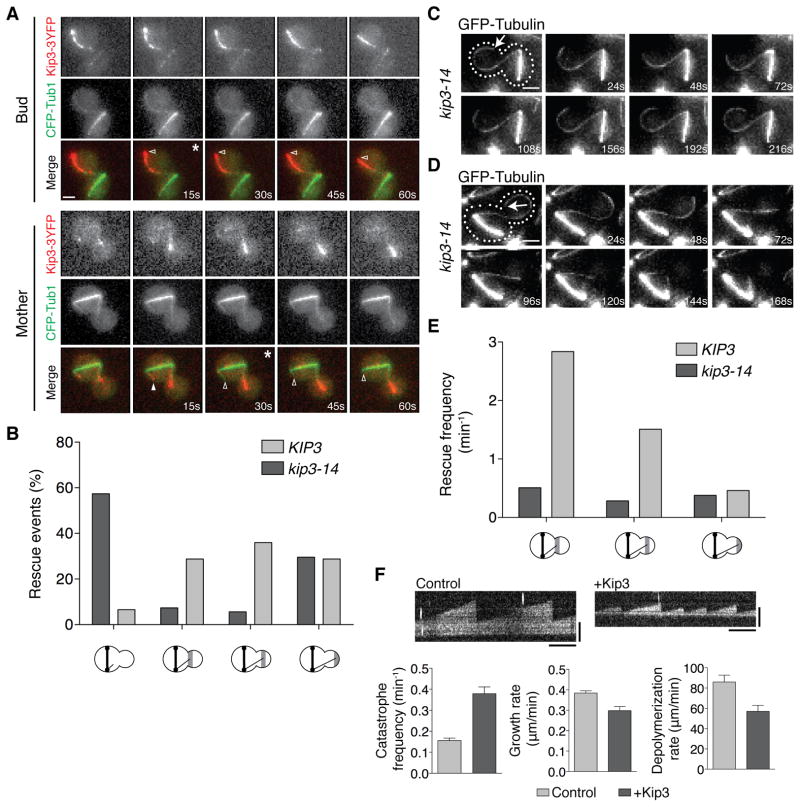
(A) Time-lapse images of MTs in the bud and mother MTs compartments of a dyn1Δ cell with mispositioned spindle (dotted line: cell shape, arrow: plus-end). Catastrophe occurs at 60 s for bud, and 80 s for mother MT. Scale bar is 2μm. See also Movie S1. (B) MT lifetime history plots from the cell in (A). (C) Boxplot of MT lifetimes. Boxes encompass 25th to 75th percentiles, and bars extend from minimum to maximum values. n > 39 for both. P < 0.0001. (D) Stochastic simulation of MT lifetime using the dynamic instability parameters measured in the mother or bud compartments (Table S1). n = 200 for both. See also Figure S1.
To understand how MT lifetime is extended in the bud, we monitored the dynamic behavior of individual MTs. Rather than pausing in the bud, MT plus-ends remain dynamic throughout the cell. When MTs undergo stochastic catastrophe and rescue in the absence of extensive pausing, their behavior can be described by four parameters of dynamic instability: the polymerization and depolymerization rates along with the catastrophe and rescue frequencies. Of these parameters, MTs in the bud exhibit a ~2-fold increase in mean rescue frequency, and a ~25% reduction in mean depolymerization rate relative to the mother (Table S1). To test whether these changes are sufficient to explain the extended MT lifetime observed in the bud, we performed stochastic simulations of individual MT dynamics. We first modeled MTs using the dynamic instability parameters measured in the mother (Table S1). In agreement with observed mother lifetimes, the average simulated lifetime of these MTs within the bud is 2.6 minutes (Figure 1D). We then modeled MTs using the dynamic instability parameters measured in the bud (Table S1). In contrast to the observed 50 minute lifetime, the simulated lifetime is only 5 minutes (Figure 1D). These simulations reveal that the differences in the mean values of dynamic instability parameters cannot account for the increased MT lifetime in the bud.
Spatial regulation of catastrophe and rescue keeps dynamic microtubules within the bud
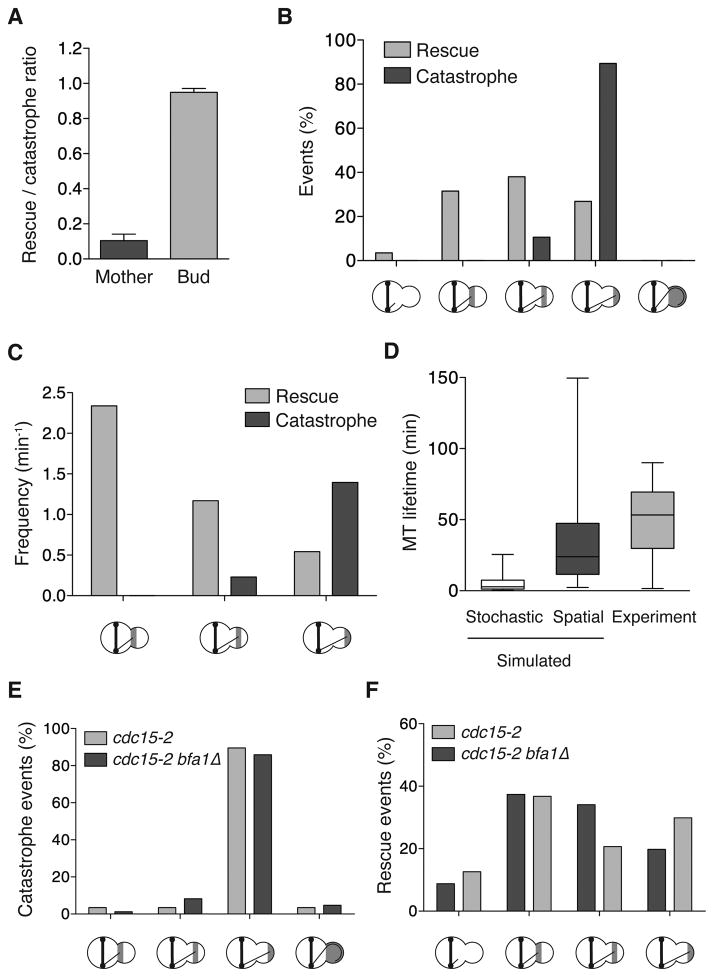
To determine how dynamic MTs are maintained in the bud, we measured the rescue/catastrophe ratio in each compartment. The majority of mother MTs that undergo catastrophe fail to rescue before depolymerizing completely, and the mean rescue/catastrophe ratio is ~0.1. Within the bud, almost all catastrophes are followed by rescue before the MT escapes the bud, and the ratio is elevated to > 0.9 (Figure 2A). As a result, bud MTs display ongoing cycles of polymerization and depolymerization, which underlies the dramatic increase in lifetime specifically within the bud (Figure 1B and S1A–B).
Figure 2. Spatial regulation of catastrophe and rescue extends microtubule lifetime in the bud and is independent of the SPOC.
(A) MT rescue to catastrophe ratio. Mean ± SEM. n > 41 for both. P < 0.0001. (B) Location of rescue and catastrophe in the bud. The X-axis demarks events within each third of the bud. The left category represents MTs that depolymerize out of the bud. The right represents MTs that overgrow the bud and curl around the bud tip prior to catastrophe. n > 169 for both. (C) Rescue and catastrophe frequencies, calculated from 119 and 99 minutes of growth and shortening time, respectively. (D) Simulated MT lifetime within the bud. The Stochastic category applies the mean bud-specific transition frequencies as in Figure 1D. The Spatial category applies the spatially resolved frequencies shown in (C) within each third of the bud. n = 200 for both. The Experiment category is the observed lifetime from Figure 1C. (E and F) Location of MT transitions in control (dyn1Δ cdc15-2) and dyn1Δ cdc15-2 bfa1Δ cells (37°C, 1 h). n > 85 for each category. See also Figure S2.
To determine whether MT behavior is subject to spatial regulation we monitored the position of catastrophe and rescue events. Within the bud ~90% of catastrophes occur in the vicinity of the bud tip which prevents MTs from overgrowing the bud (Figure 2B). Indeed, the catastrophe frequency is sharply elevated near the bud tip (Figure 2C). Conversely, rescue events are distributed throughout the bud (Figure 2B). Further analysis revealed that rescue frequency is also spatially regulated: it is lowest near the bud tip and significantly elevated near the neck (Figure 2C). Thus, MT dynamics are differentially controlled within the bud itself. Moreover, this subcellular regulation effectively tunes the length of these dynamic MTs to match the cellular compartment.
To determine if spatial variation in catastrophe and rescue frequencies is sufficient to explain the extended lifetime of bud MTs, we again simulated MTs using the spatially resolved transition frequencies shown in Figure 2C. Applying these spatially resolved values increases the average lifetime of simulated MTs from 5 to 33 minutes (Figure 2D), approaching the mean lifetime (50 min) measured in vivo. Refining our measurements of spatial variation in transition frequencies would likely reduce further the difference between simulated and measured MT lifetimes. Even so, these results reveal that the spatial regulation of catastrophes and rescues largely dictates the extended lifetime of the bud MTs.
When spindles are mispositioned, persistent MT interaction with the bud neck is required for the SPOC to delay mitotic exit [20, 22]. Thus the SPOC itself may function to retain MTs within the bud. To test this we inactivated the SPOC by deleting BFA1 [24]. These cells fail to delay mitotic exit when spindles are mispositioned. Thus, in order to analyze MT behavior, we also inactivated the mitotic exit network with the cdc15-2 mutation to prevent mitotic exit and cytokinesis. In either control (cdc15-2) or cdc15-2 bfa1Δ cells with mispositioned spindles, MT catastrophes are restricted to the bud tip (Figure 2E). Additionally, the spatial distribution of rescues is similar, and MTs rarely exit the bud (Figures 2F). Therefore, in mitotic exit network-inactivated cells, SPOC function is not required to spatially regulate MT transitions within the bud.
The Kar9 pathway links spindle orientation to cell polarity and is implicated in MT stability within the bud prior to anaphase [19, 25]. We hypothesized that Kar9 may mediate bud MT dynamics in anaphase cells with mispositioned spindles. Since kar9Δ is synthetic lethal with dyn1Δ, we combined kar9Δ with a temperature sensitive allele of Dynactin, act5ts. In both act5ts and act5ts kar9Δ cells, catastrophes and rescues are regulated within the bud as in dyn1Δ cells (Figures S2A–B). Thus, Kar9 does not mediate the MT transitions that extend lifetime within the bud.
Kip3 induces catastrophe spatially at the bud tip
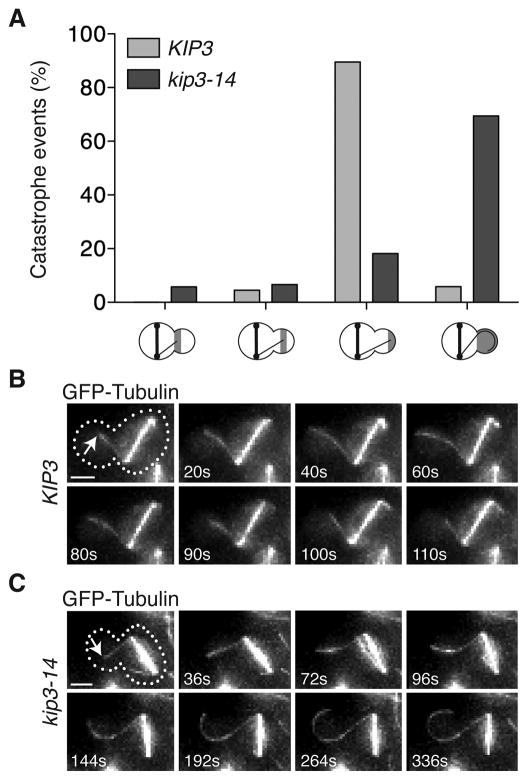
We sought to determine whether the kinesin-8, Kip3, mediates MT catastrophe in the bud. Since kip3Δ is synthetic lethal with dyn1Δ, we utilized the temperature sensitive kip3-14 allele [8]. At the restrictive temperature, dyn1Δ kip3-14 cells are inviable, Kip3-14-3YFP localization is lost on all MTs, and astral MT length is increased as in kip3Δ cells (Figures S3A–C). Thus, kip3-14 is a null allele at the restrictive temperature.
We next examined MT behavior in dyn1Δ kip3-14 cells with mispositioned spindles. In control cells ~90% of catastrophe events occur in the bud tip region at the restrictive temperature (Figures 3A and 3B). In dyn1Δ kip3-14 cells, however, MT catastrophe is severely compromised. Instead, MTs continue to grow and curl around the bud tip (Figures 3A and 3C). Kip3 is thus required to induce MT catastrophe specifically at the bud tip.
Figure 3. Kip3 induces catastrophe at the bud tip.
(A) Location of catastrophes in control (dyn1Δ KIP3) and dyn1Δ kip3-14 cells (37°C, 1 h). n > 153 for both. X-axis is labeled as in Figure 2B. (B) MT catastrophe at the bud tip in a dyn1Δ KIP3 cell (dotted line: cell shape, arrow: plus-end). Catastrophe occurs at 60 s. See also Movie S1. (C) MT overgrowing the bud tip in a dyn1Δ kip3-14 cell. MT reaches the bud tip at 96 s. See also Movie S2. Scale bars are 2μm. See also Figure S3.
Kip3 depolymerizes MTs in a length-dependent manner in vitro, and it was postulated that Kip3 might selectively induce catastrophe of longer MTs in vivo [6]. Thus, we reasoned that bud MTs might become long enough to accumulate a threshold amount of Kip3 that triggers catastrophe. However, MT length at the time of catastrophe ranges widely from 4–12μm, and when categorized according to length, the longer MTs always reach the bud tip before undergoing catastrophe (Figure S3D). Together these results suggest the MT-destabilizing activity of Kip3 may be spatially regulated in the bud.
Kip3 accumulates in a gradient along MTs in the bud compartment
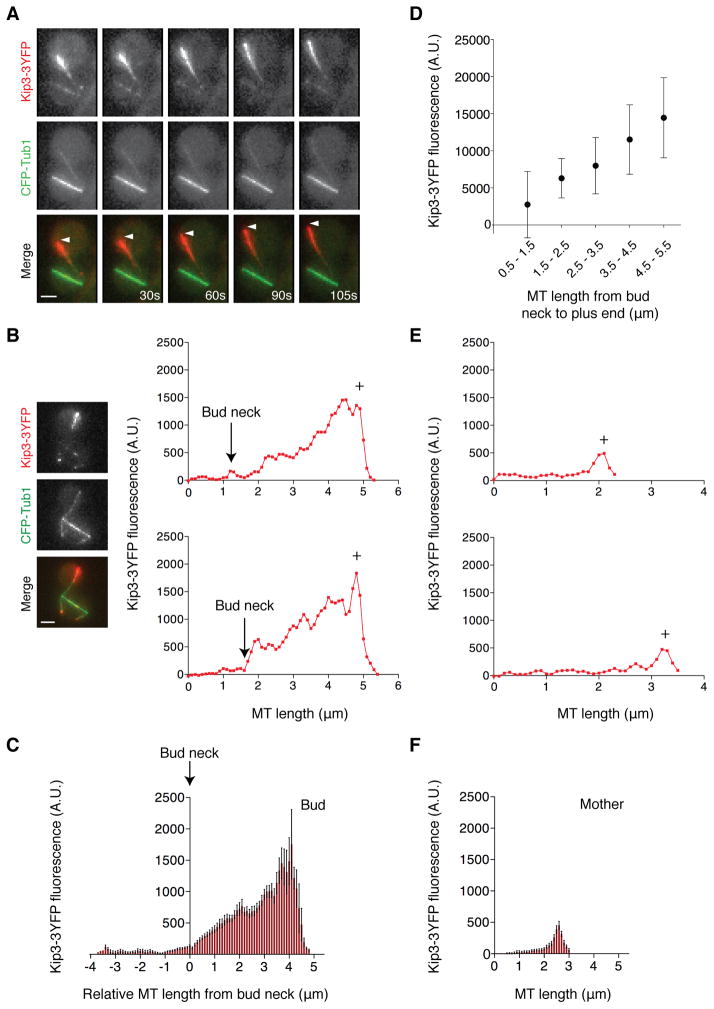
In vivo, Kip3-3YFP typically localizes as puncta on the plus-ends of growing MTs, and is absent during depolymerization [5]. Strikingly, we found that Kip3-3YFP coats a large region of the growing MT throughout the bud (Figure 4A). This localization is reminiscent of that observed in vitro, where Kip3 forms a concentration gradient along the MT lattice that increases toward the plus-end [6, 26]. Indeed, Kip3-3YFP concentrates progressively toward the plus-end of bud MTs (Figure 4B). The Kip3-3YFP gradient does not start at the MT minus-end, but rather increases inside the bud compartment (Figure 4B). When MTs are aligned with respect to where they cross the bud neck, Kip3-3YFP accumulates as a gradient only along the MT portion occupying the bud (Figure 4C). Moreover, this is accompanied by a length-dependent increase in Kip3-3YFP at MT plus-ends (Figures 4D and S4B). Similar Kip3-3YFP accumulation along bud MTs also occurs in both num1Δ and act5Δ cells (Figure S4D). By contrast, MTs contained entirely within the mother continue to display Kip3-3YFP puncta at their plus-ends (Figures 4E and 4F). Taken together, Kip3 localizes as a gradient along the MT lattice within the bud compartment of cells with mispositioned spindles.
Figure 4. Kip3 accumulates along the length of microtubules in the bud.
(A) Time-lapse images of MTs (CFP-Tub1, green) and Kip3-3YFP (red) in a cell with mispositioned spindle (arrowhead: growing MT). See also Movie S3. (B) Representative examples of Kip3-3YFP fluorescence along bud MTs. The spindle pole is at 0μm and the plus-end (+) is toward the right. Micrographs depict a representative cell. (C) Kip3-3YFP fluorescence along bud MTs aligned so that 0μm represents where each MT crosses the neck. Mean ± SEM. n = 46. (D) Kip3-3YFP fluorescence at the plus-end of growing bud MTs. Mean ± SD.n = 94. (E) Representative examples of Kip3-3YFP fluorescence along mother MTs. (F) Kip3-3YFP fluorescence along mother MTs aligned so their plus-ends overlap at the right. Mean ± SEM. n = 15 MTs with lengths 2–2.6μm. Fluorescence scale is identical in (B, C, E, and F). Scale bars are 2μm. See also Figure S4.
Kip3 associates with depolymerizing microtubules and is required for rescue within the bud
Kip3 generally reduces MT depolymerization rate and increases rescue frequency in cells [5, 27]. However, the physiological role for this stabilizing function has not been demonstrated. In the mother, the plus-end localization of Kip3 is lost during depolymerization (Figure 5A). However, the Kip3 accumulation along the lattice of bud MTs results in continued localization to the plus-end throughout depolymerization (Figure 5A). Thus, we hypothesized that Kip3 functions as a bud-specific MT stabilizer in these cells.
Figure 5. Kip3 associates with depolymerizing microtubule plus-ends and promotes rescue within the bud.
(A) Time-lapse images of MTs (CFP-Tub1, green) and Kip3-3YFP (red) in cells with a mispositioned spindle (open arrowhead: depolymerizing MT, closed arrowhead: growing MT, asterisk: catastrophe). Scale bar is 2μm. (B) Location of rescues in control (dyn1Δ KIP3) and dyn1Δ kip3-14 cells (37°C, 1 h). n > 153 for both. (C) Rescue while the MT remains in contact with the bud tip in dyn1Δ kip3-14 cells (dotted line: cell shape, arrow: plus-end). Rescue occurs at 72 s. See also Movie S4. (D) MT escaping the bud in dyn1Δ kip3-14 cells. Scale bars are 2μm. See also Movie S5. (E) Rescue frequencies in control (dyn1Δ KIP3) and dyn1Δ kip3-14 cells (37°C, 1 h), calculated from 75 and 68 minutes of total shortening time, respectively. (F, top) Kymographs of reconstituted yeast MTs in the absence (Control) and presence (+Kip3) of 10nM Kip3. Vertical bars are 3μm and horizontal bars are 200 s. (F, bottom) Dynamic instability parameters for yeast MTs ± 10nM Kip3. Catastrophe frequency ± SD (P < 0.0001). Mean growth and depolymerization ± SEM (P < 0.002 for both). For control and +Kip3, n = 169 and 139 for catastrophe, 31 and 18 for growth, and 33 and 37 for depolymerization, respectively. In (B) and (E) the X-axis is labeled as in Figure 2B. See also Figure S5.
To determine if Kip3 mediates MT stabilization in the bud, we observed its influence on the dynamic behavior of MTs. The growth rate is similar for mother and bud MTs even though bud MTs accumulate more Kip3 (Table S1). Also, the growth rate of bud MTs is similar in the presence or absence of Kip3, suggesting that Kip3 accumulation does not lead to progressive destabilization (Table S1). In fact, MTs in the bud depolymerize significantly more slowly than in the mother (Table S1). This effect is Kip3-dependent since bud MTs depolymerize 2-fold faster without Kip3 (Table S1). These data suggest that Kip3 accumulation along the bud MT may stabilize, rather than destabilize the MT.
We next inactivated Kip3-14 and monitored rescue in the bud. In control cells, rescue occurs throughout the bud, and few MTs exit the bud (Figure 5B). In contrast, rescue is severely compromised in kip3-14 cells (Figure 5B), which lack Kip3-14 localization along bud MTs (Figure S3E). In these cells, most MTs initially overgrow the bud and curl around the bud tip (Figures 3A and 3C). Following catastrophe, ~30% of these MTs rescue while still curled along the bud tip (Figure 5B and 5C). However, when MTs depolymerize away from the bud tip, they nearly always fail to rescue before exiting the bud (Figures 5B and 5D). Indeed, rescue frequency is severely reduced in the absence of Kip3 (Figure 5E). Thus Kip3 is required for the bud-specific MT rescue in cells with mispositioned spindles.
Kip3 promotes catastrophe and slows depolymerization of yeast microtubules in vitro
We tested whether Kip3 can directly mediate stabilization and destabilization of dynamic MTs in vitro. The effect of Kip3 on MT dynamics has been reported using mammalian tubulin [28, 29]. To move toward more physiological conditions we reconstituted dynamic MTs using purified budding yeast tubulin (Figure 5F and S5). Dynamic MTs were grown from stabilized seeds using 1.4μM tubulin. Addition of 10nM Kip3 results in a 2.5-fold increase in catastrophe frequency (Figure 5F). Additionally, Kip3 reduces MT growth rate ~25% (Figure 5F). These destabilizing effects on yeast MTs are similar to results obtained using mammalian MTs [28, 29]. In addition to destabilizing MTs in vitro, Kip3 also stabilizes them by decreasing the depolymerization rate 33% (Figure 5F). Yeast MTs typically do not display rescues in vitro [30, 31], and we did not observe rescues in the absence or presence of Kip3. Nonetheless, these data show using yeast MTs that the kinesin-8, Kip3, directly destabilizes MTs, yet also directly stabilizes them by dampening the depolymerization rate.
Microtubule rescue in the bud requires the carboxy-tail domain of Kip3
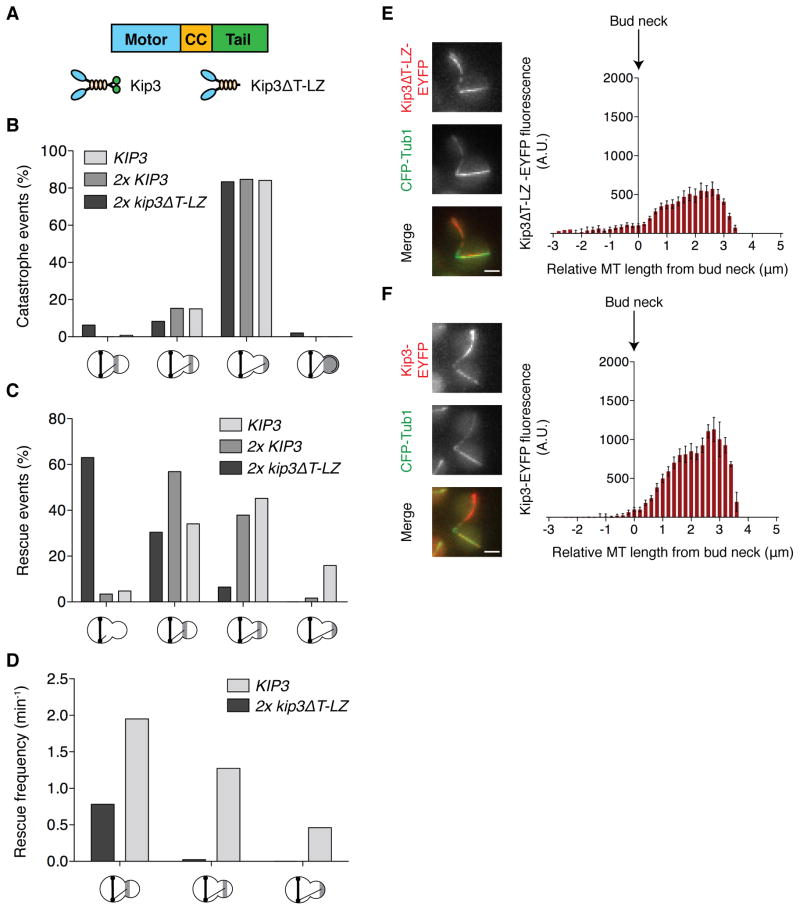
The C-terminal tail domain of Kip3 binds tubulin and MTs, and contributes to the general MT stabilizing effects observed for Kip3 [27]. Thus, we tested the role of the tail domain in MT rescue using the ‘tail-less’ Kip3, Kip3ΔT-LZ (Figure 6A) [27]. Due to reduced processivity and depolymerase activity, cells require two copies of the EYFP-tagged, kip3ΔT-LZ-EYFP, for complete localization and in vivo complementation [27]. Similarly, cells require two copies of non-tagged kip3ΔT-LZ to complement wild-type resistance to the MT destabilizing compound benomyl (Figure S6A). In 2x kip3ΔT-LZ cells, catastrophes are induced specifically at the bud tip, demonstrating that the Kip3 motor domain and coiled-coil region is sufficient for this function (Figure 6B). However, ~65% of depolymerizing MTs fail to rescue before exiting the bud (Figures 6C). Correspondingly, rescue frequency is severely reduced throughout the bud (Figure 6D). This decreased rescue is not simply a consequence of a potential increase in MT destabilization in 2x kip3ΔT-LZ cells because catastrophe and rescue remain properly regulated in 2x KIP3 cells (Figure 6B and 6C), which display further increased benomyl sensitivity (Figure S6A). Thus, bud-specific MT rescue requires the tail domain of Kip3.
Figure 6. The tail domain of Kip3 is required for microtubule rescue within the bud.
(A) Molecular organization of Kip3 and ‘tail-less’ Kip3ΔT-LZ. The leucine zipper (LZ) mediates Kip3ΔT-LZ dimerization. (B and C) Location of catastrophe and rescue. For KIP3, 2x KIP3 and 2x kip3ΔT-LZ cells (all dyn1Δ) n > 46 for each category. See also Movie S6. (D) Rescue frequencies in the bud, calculated from 79 and 166 minutes of MT shortening time for KIP3 and 2x kip3ΔT-LZ cells, respectively. (E) Kip3ΔT-LZ-EYFP fluorescence along bud MTs in 2x kip3ΔT-LZ cells expressing one non-tagged and one EYFP-tagged copy. MTs are aligned so 0μm is where each crosses the neck. Mean ±SEM. n = 25. (F) Kip3-EYFP fluorescence along bud MTs as in (E). n = 16. In (E) and (F) the fluorescence scale is identical and the micrographs depict a representative cell. Scale bars are 2μm. In (B–D) X-axes are labeled as in Figure 2B. See also Figure S6.
We next determined the localization of Kip3ΔT-LZ-EYFP on bud MTs. When dyn1Δ is combined with 2x kip3ΔT-LZ-EYFP cell viability is reduced and cells display abnormal morphologies. Thus, we used cells with one of the two kip3ΔT-LZ copies tagged with EYFP, in which morphology resembles control strains. In these cells Kip3ΔT-LZ-EYFP signal remains low within the mother, but rises steadily along the MT segment within the bud (Figure 6E). Consistent with 50% of the Kip3ΔT-LZ protein tagged by EYFP, the signal is correspondingly lower than in control cells containing 100% Kip3-EYFP (Figure 6F). Together, these data reveal that Kip3ΔT-LZ localizes properly, yet fails to induce rescue within the bud.
Spatial regulation of microtubule dynamics by Kip3 is required for SPOC-mediated mitotic arrest
Persistent interaction between astral MTs and the bud neck is needed to delay mitotic exit when spindles are mispositioned [22]. By extending MT lifetime in the bud, Kip3 can achieve this persistent interaction. Therefore, we predicted that 2x kip3ΔT-LZ cells, in which stabilization of bud MTs is compromised, would undergo aberrant mitotic exit with mispositioned spindles. The proportion of dyn1Δ cells with mispositioned spindles increases at 14°C, and these cells remain arrested in anaphase [32]. However, when this arrest fails the cells divide with improperly segregated nuclei. We found that 21% of KIP3 cells have mispositioned anaphase spindles, yet only 2% underwent aberrant mitotic exit (Figure 7A). By contrast, 39% of 2x kip3ΔT-LZ cells underwent aberrant mitotic exit (Figure 7A). Thus, the ability of Kip3 to promote rescue, and maintain dynamic MTs within the bud, is essential for maintaining mitotic arrest in cells with mispositioned spindles.
Figure 7. Kip3-mediated microtubule stabilization in the bud is required to prevent aberrant mitotic exit.
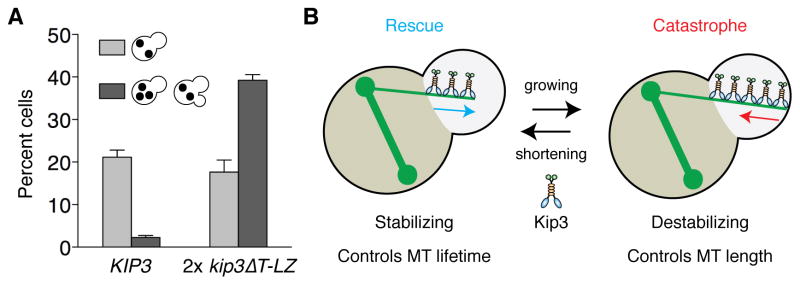
(A) Percent cells that are arrested (light) or underwent aberrant mitotic exit (dark) with a mispositioned spindle. Control (dyn1Δ KIP3) and dyn1Δ 2x kip3ΔT-LZ strains were grown at 14°C for 24 h, fixed, and stained with DAPI. Mean ± SD. n > 100 in each experiment. P < 0.001 for aberrant mitotic exit. (B) Model for how Kip3 activities spatially regulate MT dynamics to control astral MT length and lifetime. See discussion for further details.
Discussion
Here, we demonstrate that the kinesin-8, Kip3, functions as a MT stabilizer and destabilizer, and both functions are regulated spatially. Together these activities control the length and lifetime of specific MTs in cells with mispositioned spindles (Figure 7B). Kip3 sharply elevates catastrophe frequency at the bud tip, which limits MT length. Conversely, Kip3 increases rescue frequency near the bud neck to extend MT lifetime. The prolonged lifetime allows persistent MT interactions with the bud neck to delay mitotic exit, and maintain genome stability when spindles are mispositioned
In vitro Kip3 accumulates on MTs in a length-dependent manner, leading to length-dependent depolymerization [6]. In cells with mispositioned spindles Kip3 also accumulates in a length-dependent manner, yet induces catastrophe almost exclusively near the bud tip. Thus we hypothesize that the destabilizing activity of Kip3 may be spatially regulated at the bud tip. It was proposed that in fission yeast, the destabilizing activity of Klp5/6 is enhanced when MTs encounter the cell cortex and the resulting compressive forces slow MT growth [13, 14]. Kip3 could induce catastrophe by a similar mechanism. Another possibility is that other +TIPs block Kip3 access to the plus-end during MT growth. Such ‘end shielding’ could be relieved at the bud tip. One candidate is the +TIP Bim1, which together with Kip3 regulates MTs at the bud cortex in early mitosis [33]. Finally, the destabilizing activity of Kip3 may be enhanced, directly or indirectly, by factors at the bud tip.
Kip3 accumulates as a gradient along the MT lattice within the bud, which allows it to associate with the plus-end throughout depolymerization. We hypothesize that this accumulation may stabilize depolymerizing MTs. Length-dependent Kip3 accumulation is also apparent with regard to the total length of bud MTs (Figure S4C), and to what extent the increased length of these MTs may contribute to plus-end accumulation is not known. However, bud MTs with short or long segments in the mother display Kip3 gradients specifically within the bud (Figure S4A), suggesting bud-specific factors also influence Kip3 localization. For instance Bud6 influences Kip3 dynamic accumulation on astral MTs near the bud cortex prior to anaphase [33]. Interestingly, Bud6 is also implicated in delaying mitotic exit when spindles are mispositioned [19]. In order to investigate MT regulation in cells with mispositioned spindles we inhibited the Dynein pathway. Thus it remains unclear to what extent Dynein function may influence Kip3 activities or the dynamic behavior of MTs in the bud. Nonetheless our results reveal a physiological role for the MT stabilizing activity of Kip3.
Kip3 accumulation along bud MTs is inversely correlated with Kip3-mediated rescue frequency. This relationship could result from several potential, but not mutually exclusive mechanisms. Kip3 depolymerizes MTs cooperatively in vitro [26], and it was proposed that higher concentrations of Kip3 may cooperate to destabilize MTs, while at lower concentrations the stabilizing effect becomes prominent [27]. Rescue frequency within the bud is consistent with such a model. Another possibility is that the rescue promoting activity of Kip3 is locally enhanced. For example, in the nucleus Kip3 promotes rescue by transporting the MT polymerase Stu2 to the plus-end [34]. However, Kip3ΔT-LZ retains the motor domain sufficient for interaction with Stu2 [34], yet fails to induce rescue. Perhaps another factor increases the MT stabilizing potential of Kip3 near the bud neck.
Kip3 could promote rescue directly, or, indirectly through cooperation with other MT regulators. The tail domain can bind to MTs and tubulin [27]. Thus, the two tails in a Kip3 dimer may stabilize MTs by crosslinking adjacent protofilaments or by binding to structural elements unique to the depolymerizing plus-end. The tail could also mediate interactions with additional MT stabilizers. Another possibility is that Kip3 prevents MTs from leaving the bud strictly by slowing depolymerization. In this case the rescue frequency would not be predicted to change in the absence of Kip3. However, in kip3-14 cells, rescue frequency is significantly diminished, which would suggest that dampening depolymerization may increase the probability of rescue either directly, or in cooperation with additional factors. Although we did not observe Kip3-mediated rescue in vitro, reconstituted yeast MTs depolymerize ~30-fold faster than in vivo. Thus, rescue may require additional factors that further reduce the depolymerization rate and/or promote rescue. One candidate is the +TIP, Bim1, that decreases MT depolymerization rate in vitro [35]. In contrast to its mammalian homologue, EB1, Bim1 remains associated with depolymerizing plus-ends in vivo [36]. Thus, it will prove insightful to determine the effect of Kip3 in combination with other MT regulatory proteins on MT dynamics in vitro.
Spatial regulation of MT stability is a critical aspect of diverse cellular functions. We show that MT transition frequencies are spatially controlled by Kip3 to define the length and lifetime of specific MTs. Physiologically this regulation serves to prevent mitotic exit when spindles are mispositioned. In fission yeast, MT catastrophe at the cell poles [37], in part controlled by kinesin-8 [13, 14], is essential to properly position the nucleus [38]. During metaphase congression, kinetochore MTs undergo indefinite, yet spatially coordinated, cycles of catastrophe and rescue [39]. Kinesin-8 is required for metaphase congression in Drosophila S2 and human HeLa cells [40, 41]. Moreover, the human kinesin-8, Kif18A, accumulates as a gradient on kinetochore MTs [41], stabilizes dynamic MTs in vitro [15, 16], and has a nucleotide-dependent MT destabilizing activity [7, 42]. Thus, the spatial regulation of MTs by kinesin-8 may be a conserved mechanism to differentially control the length or lifetime of cellular MTs.
Experimental Procedures
Yeast strains
Yeast strains are derivatives of S288C background (Table S2). Kip3-14 has mutations L76P and L176P within the motor domain. Construction and verification of 2x kip3ΔT-LZ was designed using SnapGene software (GSL Biotech, LLC).
Cell imaging and analysis
Cells with mispositioned anaphase spindles had a straight spindle that spanned the diameter of the mother cell with neither pole in direct proximity to the bud neck. Cells that displayed aberrant morphologies such as grossly enlarged size, spindle breakdown, or hyper-elongated spindle in the mother were excluded. Images in figures represent maximum fluorescence intensity Z-projections.
Protein purification
Kip3 (6xHis-Kip3) was purified from baculovirus-infected insect cells essentially as described [5]. Yeast tubulin (β-tubulin-6xHis) was purified as described previously [43].
Supplementary Material
Highlights.
Kip3 controls microtubules (MTs) to delay mitotic exit due to mispositioned spindles
The kinesin-8 Kip3 both stabilizes and destabilizes MTs in the bud compartment
Kip3 accumulates in a MT length-dependent manner but induces catastrophe spatially
Kip3 extends MT lifetime by promoting rescue of depolymerizing MTs within the bud
Acknowledgments
We thank H. Amin, J. Briguglio, M. Glotzer, D. Kovar, E. Munro, and I. Sagot for helpful feedback on this manuscript. We thank S. Biggins, J. Cooper, A. Hoyt, J. Moore, D. Pellman, S. Rice, and X. Su for helpful reagents. Y.F. was the recipient of a National Institutes of Health predoctoral training fellowship (T32 GM07183). This work was supported by a National Institutes of Health grant (R01GM094313) to M.L.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nogales E. Structural insights into microtubule function. Annu Rev Biochem. 2000;69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- 2.Mitchison TJ, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 3.Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- 4.Su X, Ohi R, Pellman D. Move in for the kill: motile microtubule regulators. Trends Cell Biol. 2012;22:567–75. doi: 10.1016/j.tcb.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta ML, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- 6.Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- 7.Mayr MI, Hümmer S, Bormann J, Grüner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 8.Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West RR, Malmstrom T, Troxell CL, McIntosh JR. Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol Biol Cell. 2001;12:3919–3932. doi: 10.1091/mbc.12.12.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gandhi R, Bonaccorsi S, Wentworth D, Doxsey S, Gatti M, Pereira A. The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol Biol Cell. 2004;15:121–131. doi: 10.1091/mbc.E03-05-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rischitor P, Konzack S, Fischer R. The Kip3-like kinesin KipB moves along microtubules and determines spindle position during synchronized mitosis in Aspergillus nidulans hyphae. Eukaryotic Cell. 2004;3:632–45. doi: 10.1128/EC.3.3.632-645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stout JR, Yount AL, Powers JA, Leblanc C, Ems-McClung SC, Walczak CE. Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Mol Biol Cell. 2011;22:3070–3080. doi: 10.1091/mbc.E11-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tischer C, Brunner D, Dogterom M. Force- and kinesin-8-dependent effects in the spatial regulation of fission yeast microtubule dynamics. Mol Syst Biol. 2009;5:250. doi: 10.1038/msb.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erent M, Drummond DR, Cross RA. S. pombe kinesins-8 promote both nucleation and catastrophe of microtubules. PLoS ONE. 2012;7:e30738. doi: 10.1371/journal.pone.0030738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stumpff J, Du Y, English CA, Maliga Z, Wagenbach M, Asbury CL, Wordeman L, Ohi R. A Tethering Mechanism Controls the Processivity and Kinetochore-Microtubule Plus-End Enrichment of the Kinesin-8 Kif18A. Mol Cell. 2011;43:764–775. doi: 10.1016/j.molcel.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du Y, English CA, Ohi R. The Kinesin-8 Kif18A Dampens Microtubule Plus-End Dynamics. Curr Biol. 2010;20:374–80. doi: 10.1016/j.cub.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 17.Barral Y, Liakopoulos D. Role of spindle asymmetry in cellular dynamics. Int Rev Cell Mol Biol. 2009;278:149–213. doi: 10.1016/S1937-6448(09)78004-9. [DOI] [PubMed] [Google Scholar]
- 18.Merlini L, Piatti S. The mother-bud neck as a signaling platform for the coordination between spindle position and cytokinesis in budding yeast. Biol Chem. 2011;392:805–812. doi: 10.1515/BC.2011.090. [DOI] [PubMed] [Google Scholar]
- 19.Huisman SM, Bales OAM, Bertrand M, Smeets MFMA, Reed SI, Segal M. Differential contribution of Bud6p and Kar9p to microtubule capture and spindle orientation in S. cerevisiae. J Cell Biol. 2004;167:231–244. doi: 10.1083/jcb.200407167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adames NR, Oberle JR, Cooper JA. The surveillance mechanism of the spindle position checkpoint in yeast. J Cell Biol. 2001;153:159–168. doi: 10.1083/jcb.153.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bloecher A, Venturi GM, Tatchell K. Anaphase spindle position is monitored by the BUB2 checkpoint. Nat Cell Biol. 2000;2:556–558. doi: 10.1038/35019601. [DOI] [PubMed] [Google Scholar]
- 22.Moore JK, Magidson V, Khodjakov A, Cooper JA. The spindle position checkpoint requires positional feedback from cytoplasmic microtubules. Curr Biol. 2009;19:2026–2030. doi: 10.1016/j.cub.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maddox P, Chin E, Mallavarapu A, Yeh E, Salmon ED, Bloom K. Microtubule dynamics from mating through the first zygotic division in the budding yeast Saccharomyces cerevisiae. J Cell Biol. 1999;144:977–987. doi: 10.1083/jcb.144.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pereira G, Höfken T, Grindlay J, Manson C, Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- 25.Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 2003;112:561–574. doi: 10.1016/s0092-8674(03)00119-3. [DOI] [PubMed] [Google Scholar]
- 26.Varga V, Leduc C, Bormuth V, Diez S, Howard J. Kinesin-8 Motors Act Cooperatively to Mediate Length-Dependent Microtubule Depolymerization. Cell. 2009;138:1174–1183. doi: 10.1016/j.cell.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 27.Su X, Qiu W, Gupta ML, Jr, Pereira-Leal JB, Reck-Peterson SL, Pellman D. Mechanisms Underlying the Dual-Mode Regulation of Microtubule Dynamics by Kip3/Kinesin-8. Mol Cell. 2011;43:751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardner MK, Zanic M, Gell C, Bormuth V, Howard J. Depolymerizing Kinesins Kip3 and MCAK Shape Cellular Microtubule Architecture by Differential Control of Catastrophe. Cell. 2011;147:1092–1103. doi: 10.1016/j.cell.2011.10.037. [DOI] [PubMed] [Google Scholar]
- 29.Su X, Arellano-Santoyo H, Portran D, Gaillard J, Vantard M, Thery M, Pellman D. Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat Cell Biol. 2013;15:948–957. doi: 10.1038/ncb2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta ML, Bode CJ, Thrower DA, Pearson CG, Suprenant KA, Bloom KS, Himes RH. beta-Tubulin C354 mutations that severely decrease microtubule dynamics do not prevent nuclear migration in yeast. Mol Biol Cell. 2002;13:2919–2932. doi: 10.1091/mbc.E02-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bode CJ, Gupta ML, Suprenant KA, Himes RH. The two alpha-tubulin isotypes in budding yeast have opposing effects on microtubule dynamics in vitro. EMBO Rep. 2003;4:94–99. doi: 10.1038/sj.embor.embor716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li YY, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.ten Hoopen R, Cepeda-García C, Fernández-Arruti R, Juanes MA, Delgehyr N, Segal M. Mechanism for astral microtubule capture by cortical Bud6p priming spindle polarity in S. cerevisiae. Curr Biol. 2012;22:1075–1083. doi: 10.1016/j.cub.2012.04.059. [DOI] [PubMed] [Google Scholar]
- 34.Gandhi SR, Gierliński M, Mino A, Tanaka K, Kitamura E, Clayton L, Tanaka TU. Kinetochore-Dependent Microtubule Rescue Ensures Their Efficient and Sustained Interactions in Early Mitosis. Dev Cell. 2011;21:920–933. doi: 10.1016/j.devcel.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blake-Hodek KA, Cassimeris L, Huffaker TC. Regulation of Microtubule Dynamics by Bim1 and Bik1, the Budding Yeast Members of the EB1 and CLIP-170 Families of Plus-End Tracking Proteins. Mol Biol Cell. 2010;21:2013–23. doi: 10.1091/mbc.E10-02-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolyniak MJ, Blake-Hodek K, Kosco K, Hwang E, You L, Huffaker TC. The regulation of microtubule dynamics in Saccharomyces cerevisiae by three interacting plus-end tracking proteins. Mol Biol Cell. 2006;17:2789–2798. doi: 10.1091/mbc.E05-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brunner D, Nurse P. CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell. 2000;102:695–704. doi: 10.1016/s0092-8674(00)00091-x. [DOI] [PubMed] [Google Scholar]
- 38.Tran PT, Marsh L, Doye V, Inoue S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inoue S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savoian MS, Glover DM. Drosophila Klp67A binds prophase kinetochores to subsequently regulate congression and spindle length. J Cell Sci. 2010;123:767–776. doi: 10.1242/jcs.055905. [DOI] [PubMed] [Google Scholar]
- 41.Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peters C, Brejc K, Belmont L, Bodey AJ, Lee Y, Yu M, Guo J, Sakowicz R, Hartman J, Moores CA. Insight into the molecular mechanism of the multitasking kinesin-8 motor. EMBO J. 29:3437–3447. doi: 10.1038/emboj.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta ML, Bode CJ, Georg GI, Himes RH. Understanding tubulin-Taxol interactions: mutations that impart Taxol binding to yeast tubulin. Proc Natl Acad Sci USA. 2003;100:6394–6397. doi: 10.1073/pnas.1131967100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.