Abstract
Cancer immunotherapy generally offers limited clinical benefit without coordinated strategies to mitigate the immunosuppressive nature of the tumor microenvironment. Critical drivers of immune escape in the tumor microenvironment include tumor-associated macrophages (TAM) and myeloid-derived suppressor cells (MDSC), which not only mediate immune suppression but also promote metastatic dissemination and impart resistance to cytotoxic therapies. Thus, strategies to ablate the effects of these myeloid cell populations may offer great therapeutic potential. In this report, we demonstrate in a mouse model of pancreatic ductal adenocarcinoma (PDAC) that inhibiting signaling by the myeloid growth factor receptor CSF1R can functionally reprogram macrophage responses that enhance antigen presentation and productive anti-tumor T cell responses. Investigations of this response revealed that CSF1R blockade also upregulated T cell checkpoint molecules, including PDL1 and CTLA4, thereby restraining beneficial therapeutic effects. We found that PD1 and CTLA4 antagonists showed limited efficacy as single agents to restrain PDAC growth, but that that combining these agents with CSF1R blockade potently elicited tumor regressions, even in larger established tumors. Taken together, our findings provide a rationale to reprogram immunosuppressive myeloid cell populations in the tumor microenvironment under conditions that can significantly empower the therapeutic effects of checkpoint-based immunotherapeutics.
Keywords: macrophage, chemoresistance, pancreatic cancer, immune checkpoint, innate immunity, cytokines
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal human malignancies. Current therapies are ineffective at treating late stage disease. The few durable responses to therapy seen in PDAC patients are often associated with significant cytotoxic lymphocyte (CTL) infiltration into tumor tissue, suggesting that effective immunotherapy would hold promise to improve patient outcome (1, 2). However, attempts to use immunotherapeutics as single agents have achieved only limited clinical success (3, 4). While multiple factors can contribute to the resistance of PDAC to immunotherapies, one dominant player is the presence of a suppressive immune microenvironment. Critical drivers of this immunosuppressive microenvironment include tumor-associated macrophages (TAMs), monocytic myeloid-derived suppressor cells (Mo-MDSCs), and granulocytic MDSCs (G-MDSCs). These leukocytes can also promote tumor cell proliferation, confer resistance to cytotoxic stress, and facilitate metastatic dissemination (5, 6). Therefore, high numbers of tumor-infiltrating myeloid cells often correlate with early local or metastatic relapse, leading to poor survival in pancreatic cancer patients (7–9). Therapeutics that can reprogram these myeloid responses might overcome immunosuppression to enhance responses to immunotherapy. Previous work by our group and others demonstrated that combining cytotoxic chemotherapy with the blockade of colony-stimulating factor 1 receptor (CSF1R), which is prominently expressed by monocytes, Mo-MDSCs, and macrophages, results in improved anti-tumor T cell responses (10–12). These data suggest that CSF1R blockade could be effective at alleviating local tumor-induced immune suppression and bolstering the response to immunotherapy.
In this report, we investigate the mechanisms by which inhibition CSF1R signaling alleviates immune suppression. We demonstrate that CSF1/CSF1R blockade not only decreases the number of TAMs, but also reprograms remaining TAMs to support antigen presentation and bolster T cell activation within the tumor microenvironment. This in-turn leads to reduced immune suppression and elevated interferon responses, which restrain tumor progression. However, in response to reduced immune suppression programmed death 1 ligand 1 (PDL1) is up-regulated on tumor cells and cytotoxic T lymphocyte antigen 4 (CTLA4) on T cells. These checkpoint molecules limit the potential of CSF1R inhibition to stimulate anti-tumor immunity. While both programmed cell death protein 1 (PD1) and CTLA4 antagonists demonstrate limited ability to restrain PDAC growth in this mouse model, similar to reported efficacy as single agents in PDAC patients (3, 4). However, CSF1R blockade overcomes these limitations to achieve regression in even well-established tumors. These data suggest that reprogramming myeloid cell responses via CSF1/CSF1R blockade could improve the efficacy of checkpoint-based immunotherapeutics.
Methods
Pancreatic cancer tissue microarray cohort and analysis
Tissue microarray (TMA) studies were conducted on surgically resected PDAC specimens from 60 patients diagnosed in the Department of Pathology at Washington University. Patients underwent pancreaticoduodenectomy followed by adjuvant chemotherapy. Fifty-nine of the sixty patients did not receive neoadjuvant therapy. To assemble TMAs, clearly defined areas of tumor tissue were demarcated and two biopsies (1.0-mm diameter) were taken from each donor block. The Washington University School of Medicine ethics committee approved this study. Fully automated image acquisition was performed using an Aperio ScanScope XT Slide Scanner system with a 20× objective (Aperio Technologies) to capture whole-slide digital images. Fluorescent staining analysis was performed using MetaMorph software.
Immunohistochemistry(IHC)
Tissues were fixed in 10% formalin, embedded in paraffin, and dehydrated in 70% ethanol. Five-μm-thick sections were deparaffinized in xylene, rehydrated in graded ethanol, and subjected to antigen retrieval by steam heating in Citra™ antigen retrieval solution (BioGenex). CSF1 was stained with clone 2D10 at 1:100 (Thermo) and detected using indirect immunofluorescence.
Cell lines and constructs
KC cells were derived from PDAC tumor tissue obtained from p48-CRE/LSL-KRas/p53flox/flox mice (backcrossed C57/B6, n=6 by speed congenic) by our laboratory. Kras-INK (KI) cells were obtained from Dr. Hanahan’s laboratory (17, 18). All cell lines were negative for MAP and mycoplasma. Subsets of these cells were labeled with a polycistronic click beetle red luciferase-mCherry reporters.
Orthotopic model and preclinical animal cohorts
Syngeneic orthotopic PDAC tumors were established by surgical implantation, as previously described (19). Briefly, we injected 200,000 cells in 50 μl Matrigel (BD-Biosciences) into each mouse’s pancreas. Cohorts of mice were randomized into different treatment groups by either bioluminescence imaging on day 12 or gross palpation of the pancreas. Mice were treated with 50 mg/kg Gemcitabine (GEM; Hospira) by intravenous (i.v.) injection into the right retro-orbital sinus every 4–5 days. Preclinical studies were conducted with 10–15 10-week-old female mice per group. Tumor burden was measured by establishing gross wet weight of the pancreas/tumor and comparing it to that of five parallel mice sacrificed at the beginning of treatment. All studies involving animals were approved by the Washington University School of Medicine Institutional Animal Studies Committee.
CSF1R inhibitors, CSF1 neutralizing antibodies, and checkpoint antagonists
CSF1 neutralizing antibody (clone 5A1, BioXCell) was administered via intraperitoneal (i.p.) injection every 4–5 days, with the 1st injection containing 1 mg and subsequent injections 0.5 mg. CSF1R inhibitors (CSF1Ri) were provided by Plexxikon Inc. PLX3397 is a selective bispecific inhibitor for c-Fms and the c-Kit receptor tyrosine kinases (13–15). GW2580 has been described in detail previously (16). Both GW2580 and PLX3397 were administered at 800 mg/kg in chow. CTLA4 and PD1 antagonists (clones UC10-4F10 and RMP1-14, BioXCell) were given every 4–5 days at 250 and 200 μg/dose, respectively.
Flow cytometry analysis
Single-cell suspensions were prepared from dissected pancreatic tumors by manual mincing using a scalpel, followed by enzymatic digestion with 3.0 mg/ml collagenase A (Roche) and DNase I (Sigma) for 30 min at 37°C with constant stirring. Digestion mixtures were quenched by 10% fetal bovine serum (FBS), and filtered through 40-μm nylon strainers (Fisher Scientific). Cells were incubated for 10 min at 4°C with rat anti-mouse CD16/CD32 mAb (eBiosciences) at 1:200 dilution. Cells were washed twice in PBS/BSA and incubated for 20 min with 100 μl of fluorophore-conjugated anti-mouse antibodies (CD3e (145-2C11), CD4 (6K1.5), CD8a (53–6.7), CD11b (M1/70), CD11c (N418), CD19 (MB19-1), Ly6C (HK1.4), CD45 (30-F11), CD115 (AFS98), F4/80 (BM8), MHCII (M5/114.15.2), FoxP3 (FJK-16s), CD44 (IM7), CD69 (H1.2F3), PD1 (J43), PDL1 (MIH5), PDL2 (122), CTLA4 (UC10-4B9), IgG2α/κ (eBR2a), (all from eBioscience) and/or Ly6G (1A8, BioLegend), and CD206 (MR5D3, AbDSerotec) using the manufacturers’ recommended concentrations. Data acquisition was performed on the LSR-II system (BD Biosciences), and FlowJo software version 9.2 (Tree Star) was used for analysis.
Additional details in Supplemental Data
Results
CSF1 is overexpressed by human PDAC cells
Previously, we reported that inhibition of CSF1/CSF1R signaling could improve the efficacy of chemotherapy in murine PDAC models by enhancing chemotherapy-induced anti-tumor immunity (11). However, the mechanisms by which inhibition of CSF1/CSF1R signaling regulates anti-tumor immunity are not well understood. To determine the cellular sources of CSF1 and CSF1R in human pancreatic cancer patients, we analyzed TMAs constructed from 77 cases of invasive PDAC and 4 samples of normal pancreatic tissue. IHC staining showed that CSF1 is frequently, but not exclusively, expressed by malignant PDAC cells (Figure 1A). In addition, tumors frequently had elevated expression of CSF1 compared to normal tissue. PDAC cells in 70% of tumor specimens exhibited moderate to high levels of CSF1 expression (Figure 1A–C). By contrast, CSF1R was frequently detected in the tumor stroma, while only ~10% of the tumors examined had CSF1R expression in the epithelial compartment (Figure 1A and D). These observations are consistent with other reports (20, 21) and suggest that PDAC tumor cells frequently produce high levels of CSF1.
Figure 1. PDAC tumors overexpress CSF1.
A–B) Immunohistochemical analysis of CSF1 expression in normal pancreas and PDAC tissue. Representative immunofluorescent images are shown. C–D) Stratification of patient PDAC samples based on expression levels of CSF1 and CSF1R (n=4 normal and 77 PDAC).
Inhibition of CSF1R signaling reprograms the tumor microenvironment
In order to understand the impact of CSF1R signaling on the tumor microenvironment, we compared the gene expression profile of PDAC tumor tissue following treatment with either CSF1R inhibitors (CSF1Ri) or vehicle. Towards this end, we orthotopically implanted KI PDAC tumor cells into syngeneic mice. This cell line produces high levels of CSF1 but does not express CSF1R (11). Starting on day 14 post-implantation, we treated mice with either vehicle or the CSF1R tyrosine kinase inhibitor, PLX3397. Additional details on PLX3397 can be found in the Methods section and published elsewhere (13, 14, 16, 22). Eight days of CSF1Ri treatment resulted in a significant reduction in the number of tumor-infiltrating CD11b+Ly6G−Ly6CLoF4/80HiMHCII+ macrophages and CD11b+Ly6G−Ly6CHi monocytes/ Mo-MDSCs, but not CD11b+Ly6G+Ly6C+MHCIILow G-MDSCs (Figure 2A, Figure S1). Microarray analyses of whole tumor tissue mRNA expression revealed 204 downregulated and 158 upregulated genes following CSF1Ri treatment (Figure 2B, Table S1). As expected, expression of genes indicative of macrophage infiltration, including Cd68, Mrc1, Msr1, and Csf1r, were decreased in CSF1Ri-treated tumors (Figure 2D). The list of downregulated genes was enriched for molecules involved in “inflammatory responses, chemotaxis, myeloid leukocyte-mediated immunity, and proteolysis,” consistent with the decreased number of infiltrating macrophages (Figure 2C–D). The list of upregulated genes was enriched for molecules involved in “antigen presentation, allograft rejection, interferon responses, and TH1 immunity” (Figure 2C). This is consistent with the idea that CSF1R blockade can overcome immune suppression. Corresponding to these altered pathways, genes indicative of cytotoxic T lymphocyte (CTL) responses (Ifng, Cd3e, Cd8a, and Prf1), T cell recruitment (Cxcl10, Ccl3, and Ccl4), and interferon responses (e.g. Ifng, Stat1, Irf1, and Irf9) were upregulated (Figure 2E). Array results were also validated by quantitative real-time PCR (qRT-PCR) on a second set of samples (Figure 2F). To determine the impact of these alterations, we applied these gene lists to existing gene expression datasets from PDAC patients (23) and found that the core elements of the downregulated gene list were indicative of poor clinical outcomes (Figure 2G). Taken together, these results suggest that: (1) inhibition of CSF1R signaling in the stromal compartment decreases myeloid responses and reprograms the tumor microenvironment to support T cell-mediated anti-tumor immunity and (2) these changes could improve patient outcomes.
Figure 2. CSF1/CSF1R blockade reprograms the tumor immune microenvironment.
A) Leukocyte infiltration in KI tumors from mice treated with vehicle or CSF1Ri (PLX3397) for 8 days. The frequency of CD11b+CD3/19−Ly6G−Ly6CLoF4/80HiMHCII+ macrophages, CD11b+Ly6G−Ly6CHi Mo-MDSC, and CD11b+Ly6GHiLy6C+MHCIIlow/− G-MDSC subsets is depicted as the mean percentage over total live cells.
B) Cluster analysis of differential gene expression (Table S1) in vehicle- and CSF1Ri-treated tumors.
C) Table of biologic processes enriched in “upregulated” or “downregulated” genes (DAVID analysis).
D–E) Selected gene sets are displayed with associated biological activities.
F) qRT-PCR analysis of orthotopic KI tumor tissue following treatment with vehicle or CSF1Ri for 8 days. Graph depicts mean fold change compared to vehicle.
G) Kaplan Meier analysis of patient cohorts stratified by expression level of genes down-regulated from the analysis in (B).
In all panels n=4–6 mice/group and * denotes p<0.05 (Mann-Whitney U-test), unless specified.
CSF1/CSF1R signal blockade selectively kills CD206Hi TAMs
To determine how inhibition of CSF1/CSF1R signaling impacts myeloid responses, we treated tumor-bearing mice with CSF1 neutralizing antibodies for 6, 12, 24, or 48 hours or 8 days and analyzed tumor-infiltrating myeloid cell composition and cell death at these time points. Within the first 6 hours of αCSF1 treatment, total TAM numbers began to decrease. By 8 days, TAM numbers had decreased by ~60% (Figure 3B). TAMs are a heterogeneous population of macrophages with diverse biological activities (24–28). While classical activation of macrophages can restrain cancer development, alternative activation often plays a pro-tumorigenic role (29, 30). Distinct surface markers have been used to distinguish between classically and alternatively activated macrophages. Murine PDAC tumors contain a distinct subset of CD206Hi TAMs (Figures 3A, S1), and their counterparts in human pancreatic cancer have been associated with poor clinical outcomes (7). Quantification of CD206Hi and CD206Low TAM subsets revealed that αCSF1 treatment for 8 days led to a >90% depletion of CD206Hi TAMs, while CD206Low TAMs decreased by only ~45% (Figure 3C–D). Similar results were seen following CSF1Ri treatment (Figure 3G). The loss of CD206Hi TAMs could result from either preferential killing of this TAM subset or altered CD206 expression. To distinguish between these possibilities, we analyzed the kinetics of macrophage cell death. We found that in PDAC tumors, CD206Hi TAMs experienced significantly higher levels of cell death following αCSF1 treatment than CD206Low TAMs (Figure 3C–E). These data suggest that CD206Hi TAMs are more sensitive to the CSF1R signal blockade. Consistent with this differential sensitivity, we found that CD206Hi TAMs express higher levels of CSF1R (Figure 3F). In addition, while total Mo-MDSCs (CD11b+/Ly6G−/Ly6C+) did not demonstrate decreased infiltration until after 8 days of αCSF1 treatment, CD206Hi Mo-MDSCs were markedly reduced as early as 12 hours after CSF1 neutralization (Figure S2A). By contrast, the number of CD206Low Mo-MDSCs, CD11b+/Ly6G+/Ly6C−/MHCII+ mature granulocytes, and CD11b+/Ly6G+/Ly6C+ G-MDSCs remained unaffected until after 8 days of CSF1/CSF1R blockade (Figure S2B). Taken together, these data suggest that the blockade of CSF1/CSF1R signaling preferentially, but not exclusively, depletes CD206Hi TAMs and CD206Hi Mo-MDSCs in pancreatic tumors.
Figure 3. CSF1/CSF1R signaling blockade reprograms TAM response.
A) Representative flow cytometry plots with gating strategy to identify mature granulocytes, G-MDSCs, Mo-MDSCs, and TAM subsets.
B–D) Frequency of total, CD206Hi and CD206Low TAMs in orthotopic KI tumors treated with αCSF1 for 6 hours–8 days. Mean percentage of macrophages over total cells is depicted.
C) Representative analysis of MHCII and CD206 expression in TAMs following 8-day treatment with vehicle or αCSF1.
E) Analysis of dead (live/dead blue dye+) CD206Hi and CD206Low TAMs in PDAC tumors from (B).
F) CSF1R expression by MFI in CD206Hi and CD206Low TAMs in vehicle-treated mice from (B).
G) CD206 expression by MFI and CD206Hi TAM number following 8 days of αCSF1 treatment.
H) qRTPCR analysis on CD11b+Ly6G/C−F4/80+MHCII+ TAMs sorted from KI tumors following 8-day treatment with vehicle or αCSF1.
I) MHCII expression by MFI in TAMs from (H).
All graphs depict means values or normalized fold change +/−SEM, n=4–6 mice/group and * denotes p<0.05 by unpaired t-test or Mann-Whitney U-test.
CSF1/CSF1R signaling blockade reprograms TAMs
Despite extensive loss of macrophages and Mo-MDSCs, 40–50% of TAMs remain after αCSF1 or CSF1Ri treatment. To determine whether CSF1 blockade reprograms the remaining macrophages to support anti-tumor activities, we FACS sorted TAMs from 8-day vehicle or αCSF1-treated mice bearing established KI tumors and compared their gene expression profiles. TAMs from αCSF1-treated tumors displayed reduced expression of immunosuppressive molecules, including Pdcd1lg2, Il10, Arg1, Tgfb1and Ccl22. By contrast, anti-tumor immunity genes, such as Il12a, Ifna, Ifnb1, Ifng, Cxcl10, and Nos2, were upregulated (Figure 3H). We also observed markedly increased surface expression of MHCII after CSF1 or CSF1R inhibition (Figure 3I). Taken together, these data suggest that the CSF1/CSF1R blockade reprograms remaining TAMs to support anti-tumor interferon responses and T cell activities.
CSF1/CSF1R signal blockade alters the function of TAMs and dendritic cells
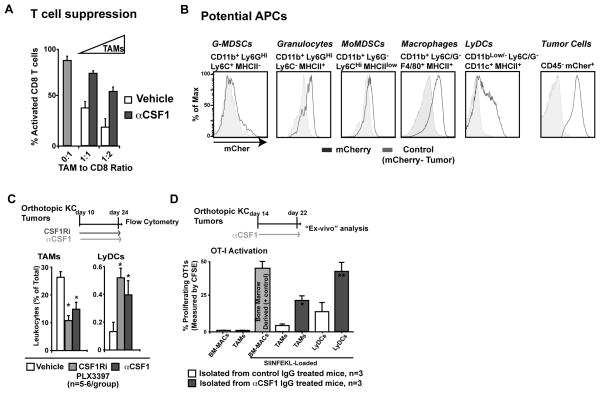
Based on the observed differences in cytokine profiles among TAMs, we predicted that CSF1/CSF1R blockade might also alter the ability of macrophages to suppress T cell functions. To address this hypothesis, we assessed the immunosuppressive activity and antigen presentation capacity of macrophages in PDAC tumors from mice following CSF1 blockade. Consistent with the reduced expression of immunosuppressive factors (Figure 3H), we found that fluorescence-activated cell sorted TAMs from 8-day αCSF1-treated mice had significantly reduced ability to block CD8+ T cell activation in ex vivo assays (Figure 4A). These data suggest that the TAMs that remain after CSF1 blockade have reduced immunosuppressive activity.
Figure 4. CSF1/CSF1R signaling blockade enhances TAM support for CTL responses.
A) Analysis of T cell suppression by TAMs from vehicle- or αCSF1-treated mice. TAMs were isolated by FACS and assayed for their ability to suppress splenic CD8+ T cell proliferation following anti-CD3/CD28 stimulation. The mean number of proliferation cycles is depicted after 70 hours. Representative data from two replicate experiments (n=3 mice/group).
B) Flow cytometry analysis of tumor-derived mCherry fluorescence in tumor-infiltrating leukocytes. Representative plots from 5 mice are depicted.
C) Frequency of CD11b+/Ly6G−/Ly6CLo/F4/80Hi/MHCII+ TAMs and CD11bLow/−/Ly6GC−/CD19−/CD11c+/MHCII+ Lymphoid DCs in orthotopic KI tumors after 8 days of αCSF1 or CSF1Ri treatment.
D) TAMs and LyDCs were isolated by FACS from mice in (C), loaded with SIINFEKL peptide, co-cultured with splenic OT1 cells for 18 hours. OT1 proliferation was measured by CFSE dilution. Results reflect two triplicate experiments using 3 mice/group.
All graphs depict mean values +/− SEM. * denotes p<0.05 by unpaired t-test.
We also analyzed how CSF1 blockade might impact the number and function of antigen presenting cells (APCs) in the tumor microenvironment. To identify potential APCs in PDAC tumors, we orthotopically implanted mCherry-labeled KI tumor cells. This model allowed us to identify potential APCs by their uptake of tumor antigens, based on their mCherry fluorescence (Figure 4B, (31)). We were able to detect tumor-derived mCherry signal in granulocytes, monocytes, TAMs, and dendritic cells (DCs) (Figure 4B). The highest levels of mCherry uptake were observed in TAMs and a subset of CD11blow/−/Ly6G/C−/CD19−/CD11c+/MHCII+ cells, presumably lymphoid-like DCs (LyDCs). CSF1/CSF1R blockade did not affect mCherry uptake. Interestingly, unlike in TAMs, CSF1/CSF1R blockade significantly increased the number of tumor-infiltrating LyDCs and their surface expression of MHCII (Figure 4C, and Figure S2C-E). Because of the high level of tumor antigen uptake by TAMs and LyDCs, we tested the ability of these two cell types to present antigen to naïve CD8+ T cells and stimulate their proliferation. We isolated TAMs and LyDCs from orthotopic KC tumors obtained from mice treated with either vehicle or αCSF1 for 8 days. These leukocytes were then loaded with SIINFEKL peptide and assessed for their ability to activate OT1 T cells. While macrophages and LyDCs isolated from vehicle-treated tumors had very limited ability to activate T cells, αCSF1 treatment significantly enhanced the capacity of these two cell types to induce CD8+ T cell proliferation (Figure 4D). Taken together, these data suggest that CSF1 blockade alleviates immunosuppressive activities and enhances APC potential in both TAMs and tumor-infiltrating LyDCs.
CSF1/CSF1R blockade modestly increases anti-tumor T cell activity
To further understand how the blockade of CSF1/CSF1R signaling might reprogram the tumor microenvironment to regulate tumor progression, we assessed alterations in tumor-infiltrating T lymphocytes and tumor growth following CSF1 or CSF1R blockade in established murine PDAC tumors. Mice bearing established (12 days, ~1cm) orthotropic KI or PAN02 tumors were treated with αCSF1 IgGs or CSF1Ri. Tumor progression was modestly reduced by αCSF1 or CSF1Ri treatment as a single agent (Figure 5A–C). This reduction in tumor growth correlated with increases in CD3+CD8+ CTLs and CD3+CD4+ effectors T cells, decreases in CD4+ Foxp3+ T regulatory cells (TRegs), and significantly improved effector-to-TReg ratios (Figure 5D–E). While the majority of tumor-infiltrating CD8+ CTLs had a CD69+, CD44+, and CD62L− activated phenotype, CSF1R blockade led to a modest increase in both the number of CD69+ CD8+ T cells (65% to 76%) and the level of CD44 expression (Figure 5F). The observed increase in T cell numbers and enhancement of activation status correspond to our results from gene expression profiling in Figure 2.
Figure 5. CSF1/CSF1R blockade bolsters T cell responses.
A–C) Mice bearing established orthotopic KI or PAN02 tumors were treated with vehicle, CSF1Ri, or αCSF1. Tumor burden is displayed as mean tumor weight (n=10–15 mice/group), normalized to five mice sacrificed at the start of treatment (“RX Start”).
D–E) Analysis of tumor-infiltrating CD3+CD8+CTLs, CD3+CD4+Foxp3− effector T cells, and CD4+Foxp3+ Treg from mice in (A–B) is depicted as mean percentage over total live cells (n=6 mice/group). The mean effector (CTL + CD4+ effector)-to- TReg ratio is also depicted.
F) CD69, CD44, CTLA4, and PD1 expression in CD3+CD8+CTLs from mice in (A) is depicted as both MFI and percentage of positive cells. Representative plots are depicted.
* denotes p<0.05 by Mann-Whitney and n=5–6 in all panels.
CSF1/CSF1R signal blockade alters T cell checkpoint signaling
Although the CSF1/CSF1R blockade enhanced T cell infiltration, we hypothesized that anti-tumor immunity might be limited via the engagement of T cell checkpoints. We found that approximately 70% of activated CTLs had a high level of PD1 expression, which was unaffected by CSF1R blockade. By contrast, CTLA4 expression on CD8+ CTLs was significantly upregulated by CSF1R inhibition (Figure 5F). Along these lines, our array analysis (Figure 2) showed that Cd274 (PDL1) was significantly upregulated following CSF1R blockade. We verified these results using qRT-PCR, and found that both Cd274 and Ctla4, but not Pdcd1lg2 (PDL2), are upregulated in tumor tissues following CSF1 or CSF1R blockade (Figure 6A–B). These data suggest that while CSF1 blockade reprograms the tumor microenvironment to enhance effector T cell infiltration, engagement of T cell checkpoints is also enhanced.
Figure 6. CSF1/CSF1R signaling blockade elevates PDL1 expression in tumor cells.
A–B) qRT-PCR analysis of KI tumors following 8-day treatment with vehicle, CSF1Ri or αCSF1.
C) PDL1 and PDL2 expression in denoted tumor-infiltrating myeloid cells from orthotopic KI tumors treated with vehicle or CSF1Ri. Representative FACS plots and MFI are depicted.
D) Mean percentage of PDL1+ and PDL2+ TAMs and monocytes.
E) Mean percentage of PDL1+ PDAC cells in orthotopic KI tumors from mice treated with vehicle, CSF1Ri, or αCSF1. PDAC cells were identified as CD45− mCherry+.
F–G) PD1 expression in tumor-infiltrating myeloid cells following vehicle or CSF1Ri treatment. Representative expression plots, MFI and positive cells percentage data are depicted.
All graphs depict means values +/−SEM, n=3–7 mice/group. * denotes p<0.05 by unpaired t-test.
To determine the cellular sources of these molecules, we analyzed PDL1, PDL2, and PD1 expression on tumor cells and tumor-infiltrating myeloid cells from vehicle- or CSF1Ri-treated mice. We found that TAMs expressed high levels of PD1, PDL1, and PDL2, but consistent with a decreased immunosuppressive capacity, tumor-infiltrating macrophages from CSF1Ri-treated mice had markedly decreased PDL2 and PD1 expression (Figure 6C, 6F). CSF1Ri treatment also decreased the total number of PD1- and PDL2-positive TAMs (Figure 6D). Similar effects were also seen with αCSF1 treatment (data not shown). Neither Mo-MDSCs nor G-MDSCs expressed significant levels of PDL2. While CSF1R blockade did not alter PD1 or PDL1 expression in G-MDSCs, PDL1 expression was modestly elevated in Mo-MDSCs following CSF1Ri treatment.
Expression of PDL1, PD1, and PDL2 has been reported on human PDAC tumor cells, potentially allowing them to evade immune surveillance by suppressing T cell function. To determine if CSF1R blockade affects the expression of these molecules on PDAC cells, we used mCherry-expressing KI or KC cells to identify tumor cells in vivo. We found that both KI and KC cells express PDL1 at modest levels in vivo, but neither cell line expresses PDL2 or PD1 (Figure 6C, 6F, and not shown). However, following CSF1 or CSF1R blockade, the number of PDL1+ tumor cells and overall expression level of PDL1 was markedly upregulated on PDAC tumor cells (Figure 6C, 6E). These observations correspond with the increased mRNA levels of Cd274 identified by array analysis and qRT-PCR validation (Figure 2, 6A). Taken together, these results suggest that while CSF1/CSF1R blockade reprograms macrophage responses to bolster CTL responses, this reprogramming also leads to upregulation of PDL1 on tumor cells and CTLA4 on T cells. These checkpoints will likely limit the efficacy of observed anti-tumor immune responses.
CSF1R blockade enhances responses to checkpoint immunotherapy
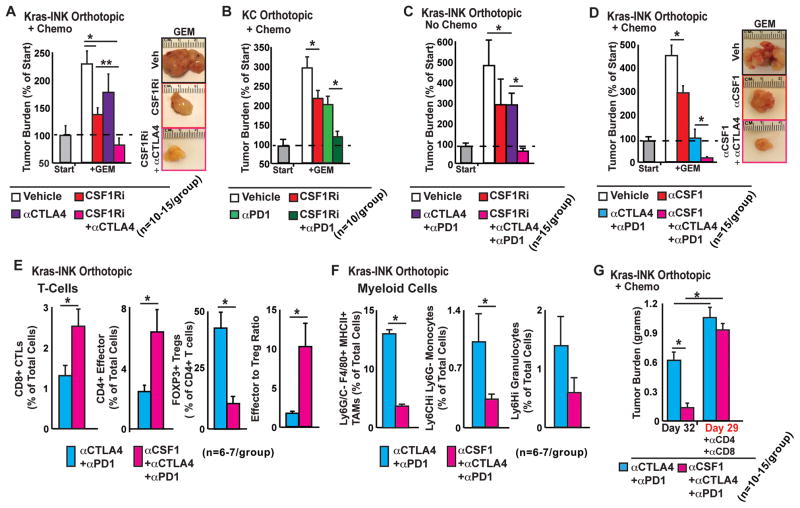
Based on the above data, we hypothesized that CSF1 or CSF1R blockade could enhance PDAC responses to PD1- and/or CTLA4-antagonist based immunotherapy. To assess this hypothesis, we treated mice bearing established KI tumors with αPD1 or αCTLA4 with or without CSF1Ri in combination with gemcitabine (GEM). PD1 and CTLA4 antagonists in combination with GEM had only limited efficacy at blunting the progression of established tumors (Figure 7A–B). By contrast, the combination of CSF1R blockade with either PD1 or CTLA4 antagonists reduced tumor progression by more than 90%. Since combined PD1 and CTLA4 antagonist therapy is being tested clinically for the treatment of both melanoma and PDAC, we also tried this combined therapeutic approach. In the absence of chemotherapy, even combined αPD1/αCTLA4 treatment only limited tumor progression by ~50%. However, the addition of CSF1R blockade to αPD1/αCTLA4 treatment completely blocked tumor progression and even regressed established tumors by 15% (Figure 7C). When CSF1 blockade was combined with αPD1/αCTLA4 and GEM treatment, we observed complete tumor regression in 30% of animals and an average tumor regression of ~85% (Figure 7D). Similar results were seen in orthotopic KC tumors, and when the less potent CSF1R inhibitor, GW2850, was used (Figure 7B, S3A–B). Analysis of T cell responses following combined therapy with αCSF1 and αPD1/αCTLA4 antagonists demonstrated increased CD8+ CTL and CD4+ effector T cell infiltration and decreased CD4+ Foxp3+ TReg numbers (Figure 7E). In addition, the number of TAMs, Mo-MDSCs, and G-MDSCs decreased following this combined therapeutic regimen (Figure 7F).
Figure 7. CSF1/CSF1R signaling blockade enhances T cell checkpoint immunotherapy.
A–D) Mice bearing orthotopic KI or KC tumors were treated with vehicle, CSF1Ri, or αCSF1, +/− GEM +/− αPD1, and +/−αCTLA4. The tumor burden is displayed as mean tumor weight (n=10–15 mice/group), normalized to five mice sacrificed at the start of treatment (“Start”).
E) Frequency of tumor-infiltrating CD3+CD8+CTLs, CD3+CD4+Foxp3− T effectors, and Foxp3+ CD4+ TRegs from mice in (D) is depicted as mean percentage of total live cells (n=6 mice/group). Mean effector (CTL + CD4+ effector) to TReg ratio is depicted.
F) Flow cytometric analysis of tumor-infiltrating CD11b+Ly6C/G−F4/80+MHCII+ TAMs, CD11b+Ly6C+Ly6G− Mo-MDSCs, and CD11b+ Ly6C+Ly6G+MHCII− G-MDSCs from mice in (D) is depicted as mean percentage of total cells (n=6 mice/group).
G) Mice bearing orthotopic KI tumors were treated with GEM, αPD1, αCTLA4, vehicle or αCSF1, +/− αCD4 and αCD8. The tumor burden is displayed as mean tumor weight (n=10–15 mice/group).
All graphs depict mean values +/− SEM and * denotes p<0.05 by unpaired t-test and/or Mann-Whitney U-test.
To determine if alterations in tumor burden in CSF1Ri treatment mice were due to increased T cells responses we conducted CD4 and CD8 T depletion studies and found that CSF1R blockade no longer improved checkpoint-based therapy (Figure 7G). Taken together, these results suggest that CSF1/CSF1R blockade improve checkpoint immunotherapy by enhancing CD4+ and CD8+ T cell activities.
Discussion
In this report, we show that blockade of CSF1/CSF1R signaling in pancreatic tumors depletes CD206Hi TAMs and reprograms remaining macrophages to support anti-tumor immunity. The blockade alone modestly enhances anti-tumor interferon responses, promotes CTL infiltration, and slows tumor progression. However, the therapeutic effect is limited by the induction of T cell checkpoint molecules, including PDL1 on tumor cells and CTLA4 on T cells. Addition of the CSF1/CSF1R blockade markedly improved the efficacy of αPD1 and αCTLA4 checkpoint immunotherapy and led to the regression of even well-established PDAC tumors. These data suggest that CSF1/CSF1R signaling may be an effective therapeutic target to reprogram the immunosuppressive microenvironment of human PDAC tumors and enhance the efficacy of immunotherapy.
Recent data from several groups suggest that inhibition of CSF1R signaling alters the immunologic responses of tumor-infiltrating macrophages in several cancer types (10–12, 32–34). Mok et al. targeted CSF1R signaling using the compound PLX3397 in a murine melanoma model; PLX3397 treatment depleted >80% of TAMs, leaving behind a small population of MHCIIHi macrophages (10). These effects led to increased efficacy of adoptively transferred T cell based therapies. These data agree with our report here. In addition, recent work by Pyonteck et al. has shown that blockade of CSF1R signaling, using the small molecule inhibitor BLZ945, significantly blunts murine glioma tumor growth by reprogramming macrophage responses (32). In contrast to pancreas, melanoma and breast models, macrophage numbers in these murine glioma studies were not reduced. Instead, TAM survival was sustained by tumor-derived factors. However, in glioma, CSF1R blockade impairs the tumor-promoting functions of TAMs and regresses established tumors. Taken together, these results suggest that CSF1/CSF1R signaling can regulate both the number and the function of TAMs, but these activities may be highly dependent on tumor-type/tissue-specific factors.
One possible mechanism by which CSF1Ri reprograms the remaining TAMs is that CSF1R signaling may promote tumor-promoting macrophage phenotypes, while its blockade polarizes TAMs into the anti-tumor phenotype. In a study by Fleetwood et al., macrophages cultured in CSF1 or CSF2 demonstrated different cytokine profiles and transcription activity(35). For example, in response to lipopolysaccharide, CSF2-derived macrophages preferentially produce IL-6, IL-12, and TNFα, while CSF1-derived macrophages produce IL-10 and CCL-2, but not IL-12. These data suggest that the exact cytokine milieu differentially program macrophages to play diverse roles. Intriguingly PDAC tumors can also produce high levels of CSF2 (36, 37), which could reprogram TAMs toward DC-like phenotypes when unopposed by CSF1R signaling.
Alternative to TAMs being reprogrammed by CSF1Ri, another possible mechanism is that CSF1R signaling blockade selects for a subset of tumor-restraining macrophages that are insensitive to the CSF signal kills-off a subset of TAMs that have a pro-tumor phenotype. In many physiological and pathological settings, including cancers, macrophages are composed of heterogeneous subsets of populations with distinct functions (24). These subsets may depend on different factors for their survival, proliferation, and effector functions. Selection pressure due to CSF1 signal blockade may have enriched for subsets of anti-tumor macrophages in PDAC tissue that are less dependent on CSF1 signaling for their survival. Our analysis of cell death in CD206HiMHCIILow vs. CD206LoMHCIIHi TAM sensitivity to αCSF1 IgG supports this hypothesis (Figure 3B). While both CD206Hi and CD206Low TAM populations had detectable cell death upon CSF1 neutralization, the CD206Hi populations were preferentially depleted. The CD206Hi TAM subset had significantly higher CSF1R expression levels, suggesting that this population may be more dependent on the CSF1 signal. Taken together, the heterogeneity of macrophages within the tumor tissue suggests that subsets of TAMs can be targeted to modulate the tumor microenvironment and enhance tumor elimination.
CD206 is expressed in many subsets of myeloid cells other than macrophages, including immature dendritic cells and monocytes (38). Whether CD206 expression is correlated to differential activation status in these cell types is not known. Interestingly, Tie2+ monocytes almost uniformly express CD206 (39). It remains to be seen whether the loss of CD206Hi tumor-infiltrating monocytes upon αCSF1 treatment (Figure S2A) involves the Tie2+ monocytes and/or affects tumor vasculature.
Although CSF1/CSF1R blockade enhances the anti-tumor activity of myeloid cells and T cell responses, its efficacy can be blunted by upregulation of immune checkpoint molecules, especially PDL1. While tumor intrinsic pathways have been reported to drive PDL1 expression in tumor cells (4), multiple lines of evidence suggest that PDL1 expression by epithelial tumors is an adaptive response to interferon signaling from tumor stroma. Several groups have reported that IFNγ and IFNα directly lead to the upregulation of PDL1 (40–43). Consistent with these studies, in vitro treatment with recombinant IFNγ markedly upregulated PDL1 expression in our PDAC cell lines (not shown). Given the elevated expression of interferons and interferon response genes in CSF1Ri-treated PDAC tumor tissue, we reason that CSF1Ri-mediated interferon production might drive the upregulation of PDL1 in PDAC cells, an inherent limitation of this therapy.
Even though T cell checkpoint inhibitors alone have achieved impressive clinical benefits in some other cancers, particularly melanoma (44, 45), their application in pancreatic cancer as single agents has had limited efficacy (3). This is potentially due to the immunosuppressive microenvironment of PDAC tissue, which could be alleviated by therapeutic strategies that reprogram dominant myeloid responses to allow for effective checkpoint therapy.
Supplementary Material
Acknowledgments
DGD acknowledges generous support from a Lustgarten Innovation Award, an Edward Mallinckrodt Jr. Award, The Cancer Research Foundation, and a Siteman Cancer Center Career Development Award and R01 CA177670-01. DCL acknowledge the Siteman Cancer Frontier Fund and NCI R01 CA168863-01. MAM acknowledges funding from the Siteman Cancer Center Cancer Biology Pathway. AWG acknowledges Washington University Clinical and Translational Grant KL2TR000450. TMN acknowledges NCI grant T32 CA 009621. This work was supported by the Genome Technology Access Center which is partially supported by NCI Cancer Center Support Grant #P30 CA91842 and ICTS/CTSA Grant# UL1RR024992.
Abbreviations
- CCL
C-C motif ligand
- CSF1
colony-stimulating factor 1
- CSF2
colony-stimulating factor 2
- CSF1R
colony-stimulating factor 1 receptor
- CTL
cytotoxic T lymphocyte
- DC
dendritic cells
- GEM
gemcitabine
- G-MDSC
granulocytic myeloid-derived suppressor cell
- IL
interleukin
- MDSC
myeloid derived suppressor cell
- M-MDSC
monocytic myeloid-derived suppressor cell
- PDAC
pancreatic ductal adenocarcinoma
- TAM
tumor-associated macrophage
- TReg
regulatory T cell
References
- 1.Gunturu KS, Rossi GR, Saif MW. Immunotherapy updates in pancreatic cancer: are we there yet? Therapeutic advances in medical oncology. 2013;5:81–9. doi: 10.1177/1758834012462463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lutz E, Yeo CJ, Lillemoe KD, Biedrzycki B, Kobrin B, Herman J, et al. A lethally irradiated allogeneic granulocyte-macrophage colony stimulating factor-secreting tumor vaccine for pancreatic adenocarcinoma. A Phase II trial of safety, efficacy, and immune activation. Ann Surg. 2011;253:328–35. doi: 10.1097/SLA.0b013e3181fd271c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Royal RE, Levy C, Turner K, Mathur A, Hughes M, Kammula US, et al. Phase 2 trial of single agent Ipilimumab (anti-CTLA-4) for locally advanced or metastatic pancreatic adenocarcinoma. Journal of immunotherapy. 2010;33:828–33. doi: 10.1097/CJI.0b013e3181eec14c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le DT, Lutz E, Uram JN, Sugar EA, Onners B, Solt S, et al. Evaluation of ipilimumab in combination with allogeneic pancreatic tumor cells transfected with a GM-CSF gene in previously treated pancreatic cancer. J Immunother. 2013;36:382–9. doi: 10.1097/CJI.0b013e31829fb7a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coussens LM, Pollard JW. Leukocytes in Mammary Development and Cancer. Cold Spring Harb Perspect Biol. 2010 doi: 10.1101/cshperspect.a003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollard JW. Trophic macrophages in development and disease. Nat Rev Immunol. 2009;9:259–70. doi: 10.1038/nri2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ino Y, Yamazaki-Itoh R, Shimada K, Iwasaki M, Kosuge T, Kanai Y, et al. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br J Cancer. 2013;108:914–23. doi: 10.1038/bjc.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balaz P, Friess H, Kondo Y, Zhu Z, Zimmermann A, Buchler MW. Human macrophage metalloelastase worsens the prognosis of pancreatic cancer. Ann Surg. 2002;235:519–27. doi: 10.1097/00000658-200204000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurahara H, Shinchi H, Mataki Y, Maemura K, Noma H, Kubo F, et al. Significance of M2-polarized tumor-associated macrophage in pancreatic cancer. The Journal of surgical research. 2011;167:e211–9. doi: 10.1016/j.jss.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 10.Mok S, Koya RC, Tsui C, Xu J, Robert L, Wu L, et al. Inhibition of CSF-1 Receptor Improves the Antitumor Efficacy of Adoptive Cell Transfer Immunotherapy. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-13-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchem JB, Brennan DJ, Knolhoff BL, Belt BA, Zhu Y, Sanford DE, et al. Targeting Tumor-Infiltrating Macrophages Decreases Tumor-Initiating Cells, Relieves Immunosuppression, and Improves Chemotherapeutic Responses. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-12-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1:54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008;105:3041–6. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plexxikon, Inc, assignee. Molecular Scaffolds for Kinase Ligand Development. United States: 2005. [Google Scholar]
- 15.DeNardo DG, Brennan D, Rexhapaj E, Ruffel B, Shiao S, Gallagher WM, et al. Leukocyte complexity in breast cancer predicts overall survival and functionally regulates response to chemotherapy. Cancer Discovery. 2011;1:54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conway JG, McDonald B, Parham J, Keith B, Rusnak DW, Shaw E, et al. Inhibition of colony-stimulating-factor-1 signaling in vivo with the orally bioavailable cFMS kinase inhibitor GW2580. Proc Natl Acad Sci U S A. 2005;102:16078–83. doi: 10.1073/pnas.0502000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nature medicine. 2011;17:500–3. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy LD, Sahraei M, Subramani DB, Besmer D, Nath S, Tinder TL, et al. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene. 2011;30:1449–59. doi: 10.1038/onc.2010.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim MP, Evans DB, Wang H, Abbruzzese JL, Fleming JB, Gallick GE. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nature protocols. 2009;4:1670–80. doi: 10.1038/nprot.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pyonteck SM, Gadea BB, Wang HW, Gocheva V, Hunter KE, Tang LH, et al. Deficiency of the macrophage growth factor CSF-1 disrupts pancreatic neuroendocrine tumor development. Oncogene. 2011 doi: 10.1038/onc.2011.337. [DOI] [PubMed] [Google Scholar]
- 21.Jiao X, Sherman BT, Huang da W, Stephens R, Baseler MW, Lane HC, et al. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–6. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schubert C, Schalk-Hihi C, Struble GT, Ma HC, Petrounia IP, Brandt B, et al. Crystal structure of the tyrosine kinase domain of colony-stimulating factor-1 receptor (cFMS) in complex with two inhibitors. The Journal of biological chemistry. 2007;282:4094–101. doi: 10.1074/jbc.M608183200. [DOI] [PubMed] [Google Scholar]
- 23.Stratford JK, Bentrem DJ, Anderson JM, Fan C, Volmar KA, Marron JS, et al. A six-gene signature predicts survival of patients with localized pancreatic ductal adenocarcinoma. PLoS Med. 2010;7:e1000307. doi: 10.1371/journal.pmed.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Movahedi K, Laoui D, Gysemans C, Baeten M, Stange G, Van den Bossche J, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer research. 2010;70:5728–39. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 25.Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. 2010;22:231–7. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–61. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 27.Mantovani A. From phagocyte diversity and activation to probiotics: back to Metchnikoff. Eur J Immunol. 2008;38:3269–73. doi: 10.1002/eji.200838918. [DOI] [PubMed] [Google Scholar]
- 28.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–69. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol. 2009;27:451–83. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 31.Engelhardt JJ, Boldajipour B, Beemiller P, Pandurangi P, Sorensen C, Werb Z, et al. Marginating dendritic cells of the tumor microenvironment cross-present tumor antigens and stably engage tumor-specific T cells. Cancer cell. 2012;21:402–17. doi: 10.1016/j.ccr.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19:1264–72. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Priceman SJ, Sung JL, Shaposhnik Z, Burton JB, Torres-Collado AX, Moughon DL, et al. Targeting distinct tumor-infiltrating myeloid cells by inhibiting CSF-1 receptor: combating tumor evasion of antiangiogenic therapy. Blood. 2010;115:1461–71. doi: 10.1182/blood-2009-08-237412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strachan DC, Ruffell B, Oei Y, Bissell MJ, Coussens LM, Pryer N, et al. CSF1R inhibition delays cervical and mammary tumor growth in murine models by attenuating the turnover of tumor-associated macrophages and enhancing infiltration by CD8 T cells. Oncoimmunology. 2013;2:e26968. doi: 10.4161/onci.26968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleetwood AJ, Lawrence T, Hamilton JA, Cook AD. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J Immunol. 2007;178:5245–52. doi: 10.4049/jimmunol.178.8.5245. [DOI] [PubMed] [Google Scholar]
- 36.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. Oncogenic Kras-Induced GM-CSF Production Promotes the Development of Pancreatic Neoplasia. Cancer cell. 2012;21:836–47. doi: 10.1016/j.ccr.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bayne LJ, Beatty GL, Jhala N, Clark CE, Rhim AD, Stanger BZ, et al. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer cell. 2012;21:822–35. doi: 10.1016/j.ccr.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Dyken SJ, Locksley RM. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annual review of immunology. 2013;31:317–43. doi: 10.1146/annurev-immunol-032712-095906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pucci F, Venneri MA, Biziato D, Nonis A, Moi D, Sica A, et al. A distinguishing gene signature shared by tumor-infiltrating Tie2-expressing monocytes, blood “resident” monocytes, and embryonic macrophages suggests common functions and developmental relationships. Blood. 2009;114:901–14. doi: 10.1182/blood-2009-01-200931. [DOI] [PubMed] [Google Scholar]
- 40.Chen J, Feng Y, Lu L, Wang H, Dai L, Li Y, et al. Interferon-gamma-induced PD-L1 surface expression on human oral squamous carcinoma via PKD2 signal pathway. Immunobiology. 2012;217:385–93. doi: 10.1016/j.imbio.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 41.Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Science translational medicine. 2013;5:200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terawaki S, Chikuma S, Shibayama S, Hayashi T, Yoshida T, Okazaki T, et al. IFN-alpha directly promotes programmed cell death-1 transcription and limits the duration of T cell-mediated immunity. J Immunol. 2011;186:2772–9. doi: 10.4049/jimmunol.1003208. [DOI] [PubMed] [Google Scholar]
- 43.Rowe JH, Ertelt JM, Way SS. Innate IFN-gamma is essential for programmed death ligand-1-mediated T cell stimulation following Listeria monocytogenes infection. J Immunol. 2012;189:876–84. doi: 10.4049/jimmunol.1103227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369:134–44. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–33. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.