Abstract
Cell wall O-glycoproteins and N-glycoproteins are two types of glycomolecules whose glycans are structurally complex. They are both assembled and modified within the endomembrane system, i.e., the endoplasmic reticulum (ER) and the Golgi apparatus, before their transport to their final locations within or outside the cell. In contrast to extensins (EXTs), the O-glycan chains of arabinogalactan proteins (AGPs) are highly heterogeneous consisting mostly of (i) a short oligo-arabinoside chain of three to four residues, and (ii) a larger β-1,3-linked galactan backbone with β-1,6-linked side chains containing galactose, arabinose and, often, fucose, rhamnose, or glucuronic acid. The fine structure of arabinogalactan chains varies between, and within plant species, and is important for the functional activities of the glycoproteins. With regards to N-glycans, ER-synthesizing events are highly conserved in all eukaryotes studied so far since they are essential for efficient protein folding. In contrast, evolutionary adaptation of N-glycan processing in the Golgi apparatus has given rise to a variety of organism-specific complex structures. Therefore, plant complex-type N-glycans contain specific glyco-epitopes such as core β,2-xylose, core α1,3-fucose residues, and Lewisa substitutions on the terminal position of the antenna. Like O-glycans, N-glycans of proteins are essential for their stability and function. Mutants affected in the glycan metabolic pathways have provided valuable information on the role of N-/O-glycoproteins in the control of growth, morphogenesis and adaptation to biotic and abiotic stresses. With regards to O-glycoproteins, only EXTs and AGPs are considered herein. The biosynthesis of these glycoproteins and functional aspects are presented and discussed in this review.
Keywords: arabinogalactan protein, cell wall, endoplasmic reticulum, extensin, glycan, glycosyltransferase, Golgi apparatus, plants
INTRODUCTION
Plants synthesize glycoconjugates that are structurally diverse and complex reflecting the diversity of plant physiological functions. The glycomolecules are usually assembled and modified within the plant endomembrane system, including the endoplasmic reticulum (ER), the Golgi apparatus and secretory vesicles responsible for their transport to different cell compartments/organelles including the cell wall. Their synthesis involves a number of steps, beginning with the formation of activated nucleotide sugars such as NDP-sugars or NMP-sugars (Bar-Peled and O’Neill, 2011). After their synthesis in the cytosol, the nucleotide sugars are then actively transported into the ER and Golgi stacks where they serve as donor substrates during glycan synthesis. Glycosyltransferases (GTs) transfer specific sugars from activated nucleotide sugars to a specific glycan acceptor leading to the extension of the glycomolecule involved. This occurs through a stepwise and sequential process which involves a number of different GTs of the secretory system. It is worth noting that 1.8% of Arabidopsis thaliana’s genome currently encode GT genes representing more than 462 GTs in total (Ulvskov et al., 2013).
Among the different plant organelles, the plant cell wall is a polysaccharide-rich extracellular compartment (Albersheim et al., 2011). In addition to polysaccharides, the plant cell wall also contains a significant percentage (∼10–15%) of N- and O-glycosylated proteins that are relatively less studied with regards to their biosynthesis and function. Both the N- and the O-glycosylation of proteins has a significant impact on both their structural properties and biological activities (Varki, 1993). Glycosylation and glycan processing are major post-translational modifications (PTMs) that cell wall proteins undergo inside the cell, and are considered important for their proper function. Indeed, in general, glycans are involved in the control of protein folding, cellular targeting and mobility, as well as signaling for regulation of plant growth, defense and different interactions with the surrounding environment (Varki and Lowe, 2009; Larkin and Imperiali, 2011; Cannesan et al., 2012; Nguema-Ona et al., 2013; Chen et al., 2014).
The N-/O- glycosylation of cell wall proteins is critical for plant development and responses to stress. Understanding and controlling O- and N-glycosylation of secreted proteins is also important in plant biotechnological applications.
N-GLYCOSYLATED PROTEINS: SYNTHESIS AND FUNCTION
The N-glycosylation of proteins starts in the ER. ER-synthesizing events for N-glycans are highly conserved in all eukaryotes studied so far since they are instrumental for efficient protein folding (Aebi, 2013). The N-glycosylation pathway starts by the transfer en bloc of a lipid linked preassembled precursor (Glc3Man9GlcNAc2) by the oligosaccharyltransferase (OST) onto the N-glycosylation sites (Asn-X-Ser/Thr and/or Asn-X-Cys) of the nascent proteins (Burda and Aebi, 1999; Gil et al., 2009; Zielinska et al., 2010; Matsui et al., 2011). The α-glucosidases I and II then remove two glucose residues from the N-glycan resulting in the presence of only one terminal glucose on the glycoprotein. This allows its entry into the ER control quality cycle (Aebi, 2013). Once the glycoprotein is correctly folded, the last glucose residue is removed by the α-glucosidase II prior to its transport into the Golgi apparatus where further modifications occur including removal of mannose residues and sequential addition of specific sugars through the action of GTs resulting in the formation of complex-type N-glycans. In plants, many genes encoding for Golgi GTs have already been identified (Table 1). These include, for example, N-acetylglucosaminyltransferase I (GnT I; Bakker et al., 1999; Strasser et al., 1999a; Wenderoth and von Schaewen, 2000), N-acetylglucosaminyltransferase II (GnT II; Strasser et al., 1999b), core α-1,3-fucosyltransferase (α1,3-FuT; Leiter et al., 1999; Wilson et al., 2001a), β-1,2-xylosyltransferase (β1,2-XylT; Strasser et al., 2000; Pagny et al., 2003), Lewis-type α-1,4-fucosyltransferase (α1,4-FuT Bakker et al., 2001; Wilson et al., 2001b; Léonard et al., 2002), and β-1,3-galactosyltransferase (β1,3-GalT; Strasser et al., 2007). In contrast to the ER steps, evolutionary adaptation of N-glycan processing in the Golgi apparatus has given rise to a variety of organism-specific complex structures (Varki, 2011). Therefore, more complex plant N-glycans consist of specific glyco-epitopes such as core β-1, 2-xylose, core α-1,3-fucose residues, and Lewisa substitutions on the terminal position of the antenna (Figure 1A; Lerouge et al., 1998; Bardor et al., 2003, 2011; Strasser et al., 2004).When abnormally processed, N-glycosylated proteins cause major developmental disorders and are usually associated to diseases in mammals (Ioffe and Stanley, 1994; Metzler et al., 1994; Lowe and Marth, 2003; Hennet, 2012). In plants, abnormal N-glycosylated proteins rarely present developmental disorders under normal growth conditions (von Schaewen et al., 1993; Strasser et al., 2004). However, the cellulose-deficient Arabidopsis mutant rsw3 which is defective in the catalytic subunit of the α-glucosidase II presents radially swollen roots and a deficiency in cellulose content (Burn et al., 2002). Moreover, under stress conditions (e.g., salt), modified phenotypes such as abnormal plant growth (Strasser et al., 2007) or altered root growth in the Arabidopsis cgl mutants (Kang et al., 2008; von Schaewen et al., 2008), have been observed. Indeed, in these studies, reduced root growth and abnormal root morphology were observed for Arabidopsis plants cultivated on media containing high NaCl concentration. In contrast to Arabidopsis, a severe phenotype with arrested seedling development and premature death before reaching the reproductive stage has been reported recently for rice gntI mutant (Fanata et al., 2013). Such plants also present defects in cell wall composition, especially reduced cell wall thickness, and decreased in cellulose content as well as reduced sensitivity to cytokinin. Plant complex-type N-glycans are ascribed to many biological functions in relation with plant development that have been recently reviewed by Strasser (2014; this issue). These include effects on plant innate immunity, tolerance to abiotic stress and root development. Therefore these functional aspects will not be further described in this review.
Table 1.
Known enzymes involved in plant N- glycans and O- cell wall glycan biosynthesis.
| AGP glycan biosynthetic enzymes | CAZy family | Protein name | Origin | Reference |
|---|---|---|---|---|
| Hydroxyproline O-galactosyltransferase | GT31 | AtGALT2 | Arabidopsis thaliana | Basu et al. (2013) |
| β-1,3-galactosyltransferase | GT31 | At1g77810 | Arabidopsis thaliana | Qu et al. (2008) |
| β-1,6-galactosyltransferase | GT31 | AtGalT31A | Arabidopsis thaliana | Geshi et al. (2013) |
| – | GT29 | AtGalT29A | Arabidopsis thaliana | Dilokpimol et al. (2014) |
| Arabinofuranosyltransferase | GT77 | RAY1 | Arabidopsis thaliana | Gille et al. (2013) |
| β-glucuronosyltransferase | GT14 | AtGlcAT14A | Arabidopsis thaliana | Knoch et al. (2013) |
| α-1,2-fucosyltransferase | GT37 | AtFUT4 | Arabidopsis thaliana | Wu et al. (2010) |
| – | GT37 | AtFUT6 | Arabidopsis thaliana | Wu et al. (2010) |
| Extensin glycan biosynthetic enzymes | CAZy family | Protein name | Origin | Reference |
| Serine O-galactosyltransferase | unknown | SGT1 | Chlamydomonas reinhardtii; Arabidopsis thaliana | Saito et al. (2014) |
| Arabinosyltransferase | GT77 | RRA3 | Arabidopsis thaliana | Velasquez et al. (2011) |
| – | GT77 | XEG113 | Arabidopsis thaliana | Gille et al. (2009) |
| N-glycan biosynthetic enzymes | CAZy family | Protein name | Origin | Reference |
| Oligosaccharyltransferase | OST | Arabidopsis thaliana; Oryza sativa | Farid et al. (2013), Qin et al. (2013) | |
| α-glucosidase I | GCS I | Arabidopsis thaliana | Boisson et al. (2001) | |
| α-glucosidase II | GCS II | Solanum tuberosum | Taylor et al. (2000) | |
| α-mannosidase I | MNS 1-3 | Arabidopsis thaliana | Liebminger et al. (2009) | |
| N-acetylglucosaminyltransferase I | GT13 | GnT I | Arabidopsis thaliana; Nicotiana tabacum; Solanum tuberosum | Bakker et al. (1999), Strasser et al. (1999a), Wenderoth and von Schaewen (2000) |
| α-mannosidase II | GM II | Arabidopsis thaliana | Strasser et al. (2006) | |
| N-acetylglucosaminyltransferase II | GT16 | GnT II | Arabidopsis thaliana | Strasser et al. (1999b) |
| α-1,3 fucosyltransferase | GT10 | α-1,3-FuT | Vigna radiata; Arabidopsis thaliana; Medicago sativa | Leiter et al. (1999), Wilson et al. (2001a), Sourrouille et al. (2008) |
| β-1,2-xylosyltransferase | GT61 | β-1,2-XylT | Arabidopsis thaliana | Strasser et al. (2000), Pagny et al. (2003), Bencúr et al. (2005) |
| β-1,3-galactosyltransferase | GT31 | β-1,3-GalT | Arabidopsis thaliana | Strasser et al. (2007) |
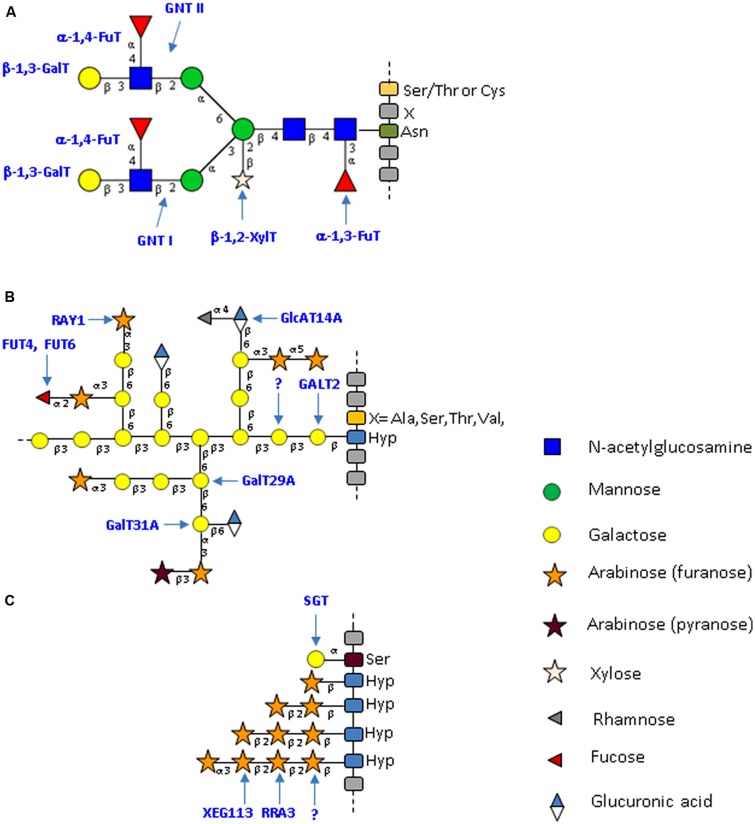
FIGURE 1.
Typical structure of plant N- and O-glycans from cell wall proteins. (A) Specific complex-type N-glycans attached to plant glycoproteins. This N-glycan results from the action of a plant-specific repertoire of glycosyltransferases that lead to the formation of a glycan bearing plant-specific glyco-epitopes such as a core β-1,2-xylose; a core α-1,3-fucose and a Lewisa antennae (Lerouge et al., 1998; Wilson et al., 2001b; Bardor et al., 2003, 2011). The N-glycan structures presented here are drawn according to the symbolic nomenclature adopted by the Consortium for Functional Glycomics (Varki et al., 2009). (B) Schematic representation of O-glycans (type II arabinogalactan) attached to AGPs. These glycans predominantly consist of arabinose and galactose. Minor sugars, such as glucuronic acid, fucose or rhamnose, are also present. The O-glycans are attached to non-contiguous Hyp residues. The model presented is modified from Tan et al. (2010) and Tryfona et al. (2012). (C) Schematic representation of O-glycans attached to plant EXT. These glycans consist of short chains of arabinose and on single galactose residues. The O-glycans are attached to contiguous Hyp residues. The model presented is modified from Saito et al. (2014). Yellow circle: galactose; green circle: mannose; blue square: N-acetylglucosamine; star: white xylose and red triangle: fucose; gray triangle: rhamnose; orange star: arabinose (furanose); purple star: arabinose (pyranose); blue/white diamond: glucuronic acid.
O-GLYCOSYLATED CELL WALL PROTEINS, ARABINOGALACTAN PROTEINS, AND EXTENSINS
Plant O-glycosylated cell wall proteins belong to the superfamily of hydroxyproline-rich glycoproteins (HRGPs). This superfamily of plant cell wall proteins which account for nearly 10% of the dry weight of the wall, is characterized by a high proline (Pro) content. Furthermore many of these Pro residues become hydroxylated (hydroxyproline, Hyp) during synthesis and consequently become glycosylated in various ways. Pro residues are distributed at different sites within the sequence and these patterns have suggested different classifications of HRGP members into different groups. EXTs and arabinogalactan proteins (AGPs) are two O-glycosylated HRGP subfamilies which have gained much attention (Kieliszewski and Shpak, 2001; Showalter, 2001; Schultz et al., 2002; Showalter et al., 2010; Kieliszewski et al., 2011; Lamport et al., 2011; Nguema-Ona et al., 2012, 2013; Tan et al., 2012; Velasquez et al., 2012). The nature of sugars being incorporated and the level of glycosylation vary between these two families, but also within the members of these subfamilies. For example, Kieliszewski et al. (2011) have shown that occurrence of contigs of 3–5 Hyp, preceded by a serine residue (Ser-Hyp4) led to the synthesis of a short arabinoside of 3–5 residues. Serine residue in the Ser-Hyp4 contig is often O-glycosylated with a single galactose (Velasquez et al., 2012; Saito et al., 2014). This action is performed by serine-O-galactosyltransferases (Ser-O-Gal-T), specific to plants (Saito et al., 2014). Non-contiguous Hyp residues rather lead to the synthesis of a large arabino-galactosylated glyco-epitope on the protein (Kieliszewski and Lamport, 1994; Shpak et al., 1999; Kieliszewski et al., 2011).
Arabinogalactan proteins and EXTs have been studied for decades, and shown to fulfill many functions related to development, and responses to biotic and abiotic stresses in plants (Hall and Cannon, 2002; Motose et al., 2004; Lee et al., 2005; Nguema-Ona et al., 2007, 2013; Seifert and Roberts, 2007; Cannon et al., 2008; Ellis et al., 2010; Lamport et al., 2011; Velasquez et al., 2011; Cannesan et al., 2012; Moore et al., 2014a,b). These studies have emphasized the importance of their O-glycan structures. Indeed, AGPs and EXTs are decorated with complex to simple carbohydrate-chains (#x1-1000doc) that are required for functionality of these glycomolecules. Until recently, the enzymes, as well as the molecular mechanisms controlling the synthesis of HRGP O-glycans, were poorly understood. A recent effort in the identification of the genes involved in the biosynthesis of HRGP O-glycans has considerably improved our understanding of the molecular events controlling the addition of sugars on these Hyp-rich proteins. The aim of this section is to bring together recent advances in the biosynthesis of HRGP O-glycans, with a focus on AGPs and EXTs. Structural and biological functions are also discussed.
AGPs AND EXTs: THE SYNTHESIS OF O-GLYCANS
PRO HYDROXYLATION OF HRGPs
Pro hydroxylation of plant cell wall HRGPs occurs predominantly on Hyp that are formed in the secretory pathway through the action of proline hydroxylases (P4Hs). In Arabidopsis, 13 P4Hs have been identified (Hieta and Myllyharju, 2002; Vlad et al., 2007; Velasquez et al., 2011). P4Hs are membrane-anchored enzymes (Yuasa et al., 2005). It is likely that Pro hydroxylation begins in the ER and continues in the Golgi apparatus. Detailed investigations of substrate affinity of two Arabidopsis P4-Hs, AtP4H1 and AtP4H2, showed that both AtP4H1 and AtP4H2 hydroxylate AGP-like and EXT-like synthetic peptides (Hieta and Myllyharju, 2002; Tiainen et al., 2005). However, the substrate specificity of the enzymes towards the two classes of synthetic peptides differed. Additional data showed that AtP4H2 poorly hydroxylated animal collagen, but did not hydroxylate animal hypoxia-inducible transcription factor (HIF); while AtP4H1 hydroxylates both animal Hyp-containing proteins collagen and HIF (Hieta and Myllyharju, 2002). Similarly, Velasquez et al. (2011) showed that some root hair-specific P4Hs are able to hydroxylate EXTs, and displayed almost no activity toward AGP-like peptides. Root hair morphology of Arabidopsis p4h mutants was dramatically altered. Complementation of these mutants with wild type genes restored the phenotype. In addition to being substrate-specific, Velasquez et al. (2011) also showed that some P4Hs were also cell type-specific: P4H2 and P4H5 being confined to trichoblast cells, while P4H13 being present in both trichoblast and atrichoblast cells. Recently, it has also been shown that different tomato P4Hs played a role in plant growth, and exhibited substrate- and tissue-specific activities (Fragkostefanakis et al., 2014). The authors have shown that silencing individual P4Hs result on an increased expansion of root and leaf cells in tomato. This increase correlated with a reduction in the amount of AGPs and possibly EXTs. Plants are therefore likely to regulate the secretion of various classes of HRGPs at different stages of development and/or responses to stress to perform specific functions in a given cell type or organ. After their synthesis and secretion, HRGPs may be modified and/or re-arranged in the cell wall, but this aspect has received little attention so far, particularly in the case of AGPs.
GLYCOSYLTRANSFERASES INVOLVED IN AGP O-GLYCAN BIOSYNTHESIS
Liang et al. (2010) has suggested that ∼15 GTs are involved in AGP glycan biosynthesis. Initiation of the biosynthesis requires the action of specific AGP O-Hyp Gal-T, able to initiate the galactosylation of hydroxylated residues on AGP backbone. Recently, an Arabidopsis Gal-T (AtGalT2), belonging to CAZy GT family 31 and containing a pfam 01762 domain encoding a Gal-T catalytic domain, able to add one galactosyl residue to Hyp residues of synthetic AGP-like peptides, has been identified (Basu et al., 2013). The authors showed that AtGalT2 was able to add one galactose residue to synthetic AGP-like peptide, and not to synthetic EXT-like peptides. AtGalT2 was also harboring a GAL-LECTIN binding domain pfam 00337. This domain, previously identified as a N-acetylgalactosaminyl GT (CAZy GT family 27), was involved in catalyzing the first steps of the glycosylation of mammalian mucins (Hassan et al., 2000; Wandall et al., 2007). AtGAlT2 was found to be located in the ER and in the Golgi apparatus, a pattern similar to the one displayed by P4Hs (Yuasa et al., 2005; Velasquez et al., 2011). It is possible that these two enzymes co-operate in plants to hydroxylate Pro residues and add the first galactosyl residue of the newly synthesized β-1,3- galactan chain. In addition to AtGalT2, Qu et al. (2008), using a combination of bioinformatic approaches, identified several additional Gal-Ts belonging to the GT family 31, and showed their putative involvement in the elongation of the β-1,3- galactan backbone of AGPs. For instance, the protein encoded by the gene At1g77810 was demonstrated to exhibit a specific β-1,3-Gal-T activity. An additional Gal-T activity that adds the second galactose to the Gal-Hyp nascent chains has also been partially characterized (Liang et al., 2010).
In addition to β-1,3-Gal-T, AGP glycan synthesis also requires the action of different other β-1,6-Gal-T, α-1,3- and α-1,5-arabinosyltransferase (Ara-T), β-glucuronosyltransferase (GlcA-T), and α-1,2-fucosyltransferase (FuT; Wu et al., 2010). Recently, two Arabidopsis Gal-Ts showing a β-1,6-Gal-T activity have been identified: AtGalT31A, a β-1,6-Gal-T which is classified into the CAZy GT family 31, is required for the addition of Gal residues to existing β-1,6- galactan chains (Geshi et al., 2013) while AtGalT29A (CAZy GT family 29) is required for the addition of galactose residues to β-1,3- and β-1,6- galactan chains (Dilokpimol et al., 2014). Both AtGalT31A and AtGalT29A are type II transmembrane proteins located in the Golgi apparatus. Traces of ER-localization previously observed with P4Hs and AtGalT2 were not found, suggesting that addition of β-1,6-galactose residues to the side chains of AGPs occurs later during the transit of nascent HRGPs into Golgi stacks. Using subcellular co-localization approaches, FRET acceptor photo-bleaching techniques as well as immuno-precipitation techniques, the authors showed that (i) AtGalt31A and AtGal29A were organized into heterodimer complexes, and (ii) this heterodimer had an enhanced enzymatic activity than the homodimer AtGalT31A/AtGalT31A, or AtGalT29A/AtGalT29A. AGP arabinogalactan chains are also modified with glucuronic acid (GlcA) residues. Knoch et al. (2013) have identified an Arabidopsis transferase belonging to the CAZy GT family 14, named AtGlcAT14A, exhibiting an AGP-specific GlcA-T activity, able to transfer GlcA residues both onto β-1,3- and β-1,6-galactan chains (see also Zhou et al., 2009; Ye et al., 2011). Interestingly, AtGlcAT14A was localized to the Golgi apparatus. AtGlcAT14A is co-expressed with AtGalT31A and co-localize in the Golgi apparatus. However, the FRET photo-bleaching acceptor technique showed that both enzymes did not physically interact. These findings suggest that all the enzymes involved in AGP glycan synthesis, although probably co-regulated, are not necessarily part of a unique multi-protein complex. Arabidopsis AGP glycans were also shown to contain fucose residues (Tryfona et al., 2012). Two Arabidopsis FuT AtFUT4 and AtFUT6, belonging to the CAZy GT family 37, were shown to specifically add fucose residues to tobacco arabinogalactosylated AGP glycan chains (Wu et al., 2010). Interestingly, de-arabinosylation of tobacco AGP glycans (using arabinofuranosidase) prevented the addition of fucose residue to the glycan, suggesting that arabinosylation was required for further addition of fucose by AtFUT4 and AtFUT6, supporting the arguments for sequential synthesis of AGP glycans along the Golgi cisternae. Biochemical data showed that both FuTs fucosylate AGP glycan in a different manner, most likely on different arabinose residues (Wu et al., 2010). Finally, arabinose, along with galactose, is the more abundant sugar found in AGP glycans. Recently, Gille et al. (2013) have identified an AGP-altered mutant of Arabidopsis named reduced arabinose yariv1 (ray1-1). Monosaccharide composition of a root AGP fraction precipitated with β-glucosyl Yariv, showed a significant decrease in arabinose content in the ray1-1 mutant, as compared to the wild type. In addition, the ray1-1 mutant showed a reduction in the length of its primary roots. RAY1-1 gene was found to encode for a CAZy GT family 77 Ara-T, localized in the Golgi apparatus (Gille et al., 2013). It is however, unknown if RAY1 is able to add arabinosyl residues to short oligo-arabinosides also found on AGPs.
EXT O-GLYCAN BIOSYNTHESIS
Extensin O-glycans consists of short arabinoside chains with single galactose residues, linked respectively to Hyp residues, and serine residues of the Ser-Hyp4 motifs. In contrast to the length and the molecular weight of arabinogalactan chains found in AGPs; EXT arabinoside chains are limited to 4–5 arabinosyl residues, predominantly β-1,2-linked. The number of enzymes required for their biosynthesis is also reduced to Ara-T initiating and elongating the arabinoside chains, and to the Ser-O-Gal-T, adding the single galactose residue to serine (Velasquez et al., 2012; Saito et al., 2014). Ser-O-Gal-T are type I transmembrane proteins, located in the ER and possibly in the cis-Golgi cisternae (Saito et al., 2014), and prior hydroxylation of Pro residues is required for galactosylation of serine residues on EXTs. It is unknown if this initial galactosylation is required for further EXT arabinosylation.
While the enzyme adding the first galactose residue to AGPs is now identified, the enzyme transferring the first arabinosyl-residue to O-Hyp EXT (and maybe on Ser-Hyp3 domains of certain AGPs; Qi et al., 1991), remains unidentified. However, Ara-Ts adding the second, the third, and then the fourth arabinose residue to O-Hyp EXTs have been identified. Indeed, Arabidopsis RRA1-3, XEG113, and ExAD were shown (or proposed for ExAD; Velasquez et al., 2012) to transfer respectively, the second, the third and the fourth arabinose residue β-1,2-linked to EXT (Egelund et al., 2007; Gille et al., 2009; Velasquez et al., 2011, 2012). XEG113 belongs to the CAZy GT family 77 and xeg113 mutants exhibited abnormally elongated hypocotyls under stress conditions. XEG113 was found to be associated with Golgi membranes (Gille et al., 2009). RRA3 is also a type II transmembrane protein, member of the CAZy GT family 77, localized in the Golgi apparatus, and shown to reduce root hair growth. Rra3 Arabidopsis mutants exhibited impaired root hairs. Using a base-mediated hydrolysis of the peptide backbone, followed by mass spectrometry analyses, the authors elegantly showed that XEG113 was responsible for the addition of the second arabinose residue to an elongating β-1,2- arabinan chain, while RRA3 was responsible for the addition of the third residue. RRA3, XEG113, P4H2, and P4H5 were co-expressed, also indicating that EXT glycan synthesis must be tightly regulated within the endomembrane system. Moreover, different cell wall related proteins including AtRSH1 (a classical EXT HRGP), AtLRX1 (a hybrid EXT HRGP), AtPRP1 (a proline-rich protein), and several peroxidase genes were also co-expressed with Ara-T and P4Hs, in Arabidopsis root hairs (Velasquez et al., 2011).
Together, these studies suggest that AGP and EXT glycan synthesis is initiated in the ER and continues in the Golgi apparatus, similarly to the N-glycosylation pathway. P4Hs and AtGalT2 may co-operate during hydroxylation and galactosylation of AGP in the ER and in the Golgi apparatus. Elongation and ramification of the AGP glycans would probably take place in different Golgi subcompartments before their export to the cell surface. But specific compartmentalization of the enzymes involved in HRGPs synthesis within specific Golgi cisternae is not yet established and requires further investigations. Such an arrangement has already been described for enzymes involved in the N-glycosylation of secreted proteins (Saint-Jore-Dupas et al., 2006) and for the synthesis of the hemicellulosic polysaccharide xyloglucan (Chevalier et al., 2010; Driouich et al., 2012).
AGPs AND EXTs: ROLE IN MORPHOLOGY AND DEVELOPMENT
Cell wall components are organized into networks of polysaccharides and glycoproteins which, apart from operating individually, are strongly interconnected (Carpita and Gibeaut, 1993; Burton et al., 2010; Albersheim et al., 2011). Indeed, it is widely acknowledged that cellulose microfibrils and hemicellulose constitute a primary network of polysaccharides, embedded into a second network made of pectic polysaccharides. Less often referred as such, O-Hyp cell wall proteins constitute the third network of the wall component. Bridges between these three networks do also exist and structural alteration occurring on a single cell wall component often affects overall cell wall architecture and integrity. Thus, structurally altered AGPs/EXTs weaken cell wall architecture (both covalently and non-covalently), and affect biological processes controlled by the cell wall compartment.
Indeed, most of the Arabidopsis mutants defective in one or more enzymes described above presented various developmental and morphological alterations. van Hengel and Roberts (2002) showed that the lack of fucose residue in the Arabidopsis mur1 mutant caused their roots to be shortened. This growth defect was due to structural modification of root AGPs (van Hengel and Roberts, 2002), as well as a result of altered rhamnogalacturonan-II synthesis, since the disorder was partially rescued by exogenous application of boric acid (O’Neill et al., 2001). Liang et al. (2013) showed that a deficiency in the genes AtFUT4 and AtFUT6 caused a reduction of root growth under saline stress conditions (see also Tryfona et al., 2014). The lack of fucose residue was proposed to affect intramolecular interactions between AGPs and other wall components. Similarly reduced galactosylation of AGPs in reb1-1 mutant of Arabidopsis caused strong swelling of trichoblast cells as well as reduced root growth (Andème-Onzighi et al., 2002; Nguema-Ona et al., 2006). The Arabidopsis mutant atglcat14a, deficient in an AGP-specific GlcA GT, showed an abnormal increase in root and hypocotyl length, when compared to the wild type (Knoch et al., 2013). Biochemical analysis in this mutant showed an alteration in the AGP composition and associated glycosidic linkages as compared to the wild type, suggesting that biochemical phenotype indirectly impacts cell elongation via an overall change in cell wall architecture and integrity. The mutation in AtGalT31A caused the arrest of the embryo development at the globular stage, while complementation of the mutant with AtGALT31A restored the wild type phenotype, thus linking the requirement of correctly glycosylated AGPs with the progression of embryogenesis beyond the globular stage (Geshi et al., 2013). However, a study of atgalt2 deficient Arabidopsis mutants showed that allelic mutant lines contained less Gal-T activity when compared to the wild type without displaying any significant alteration of the phenotype. Basu et al. (2013) suggested that other Hyp-O Gal-Ts may compensate for the loss of AtGalT2, and that examination of these mutants under non-physiological conditions, or the production of multigene mutants within this gene family may reveal novel phenotypes. Recently, an unusual AGP (named APAP1) was found to be covalently linked to pectin rhamnogalacturonan-I and to arabinoxylans (Tan et al., 2013). Absence of APAP1 in the corresponding mutant led to an increased extractability of pectins and xylans, thus suggesting the alteration of its overall wall architecture. Apap1 mutants exhibited a significant increase in the height inflorescence stem, although the overall morphology was comparable to that of the wild type.
Arabidopsis EXT-deficient or EXT-altered mutants also presented various developmental and morphological alterations, due to an alteration in their overall wall architecture. Velasquez et al. (2011) showed that disrupting the Pro hydroxylation and/or improper O-glycosylation impacted EXT ability to form covalent intra and inter-molecular network in the wall. Indeed, secondary helix conformation found in EXTs, required for normal catalysis of the di-isodityrosine bondages by wall peroxidases (Held et al., 2004), was altered in P4H-deficient and Ara-T-defective Arabidopsis mutants. The authors concluded that the absence, or the alteration of their Hyp-O-arabinosides, destabilized the EXT helical secondary structure, altering their ability to interact in the wall with other cell wall components, thus altering their structural function in muro. Interestingly, unlike in plants, the hydroxylated Pro residues of animal proteins are not glycosylated. Pro hydroxylation itself is sufficient for the conformational stability of animal Hyp-rich proteins such as collagen. This PTM is generally sufficient for proper functioning of such proteins. Indeed, Hyp stabilizes their triple helical structure at body temperature (Kivirikko and Pihlajaniemi, 1998; Myllyharju, 2003), and to date, no animal Hyp-containing proteins have been found to be glycosylated. O-glycosylation of Hyp is a rather plant-specific PTM, required for the proper functioning of plant Hyp-containing proteins including HRGPs. While the enzymes hydroxylating Pro residues on AGPs and EXTs are similar to mammalian systems, the ones that initiate and elongate the glycan chains of these HRGPs are unique to plants.
Furthermore, the study of the Arabidopsis rsh mutant, deficient in an EXT (RSH/EXT3) has also shown the importance of EXTs for normal plant cell wall architecture and function in development. RSH/EXT3 is an Arabidopsis EXT which was shown to play a key role during cytokinesis, by controlling cell plate formation (Hall and Cannon, 2002; Cannon et al., 2008). RSH/EXT3 positively charged was proposed to interact with negatively charged pectins to create a template for newly synthesized cell walls.
AGPs AND EXTs: ROLE IN BIOTIC STRESS
In addition to their role in morphology and growth, AGPs and EXTs were shown to play key roles in plant responses to biotic stress. Esquerré-Tugayé (1979) and Esquerré-Tugayé et al. (1979) have shown that plants respond to fungal infection by an increased secretion of HRGPs. Both AGPs (reviewed in Nguema-Ona et al., 2013) and EXTs were later on shown to play various roles in this response to pathogens. More specifically root apices and exudates were found to be enriched in AGPs (Figure 2), their chemical composition being different depending on both root tissues and plant species (Dolan et al., 1995; Durand et al., 2009; Cannesan et al., 2012). AGPs have long been suspected to be involved in root-microorganisms interaction including symbiotic associations (Scheres et al., 1990; Balestrini et al., 1996; Berry et al., 2002). For instance, alteration of AGP synthesis or secretion was shown to inhibit Rhizobium sp. YAS34 attachment to the root surface of Arabidopsis thaliana (Vicré et al., 2005; Figure 2). Xie et al. (2012) further demonstrated that AGPs from pea root exudates promote polar orientation and adhesion of Rhizobium leguminosarum. However, AGP functioning in root defense remained speculative until recently, as demonstrated by the study of Cannesan et al. (2012) on pea roots. The authors have shown that AGPs isolated from root cap (RC) and border cells are strong attractants of zoospores of the pathogenic oomycete Aphanomyces euteiches in vitro (Cannesan et al., 2012). The AGPs also inhibited in vitro cyst germination and the subsequent mycelium growth and propagation. These findings highlight the important contribution of AGPs in Aphanomyces euteiches root infection and show for the first time that AGPs are involved in controlling root-pathogenic oomycete interaction (see also Nguema-Ona et al., 2013).
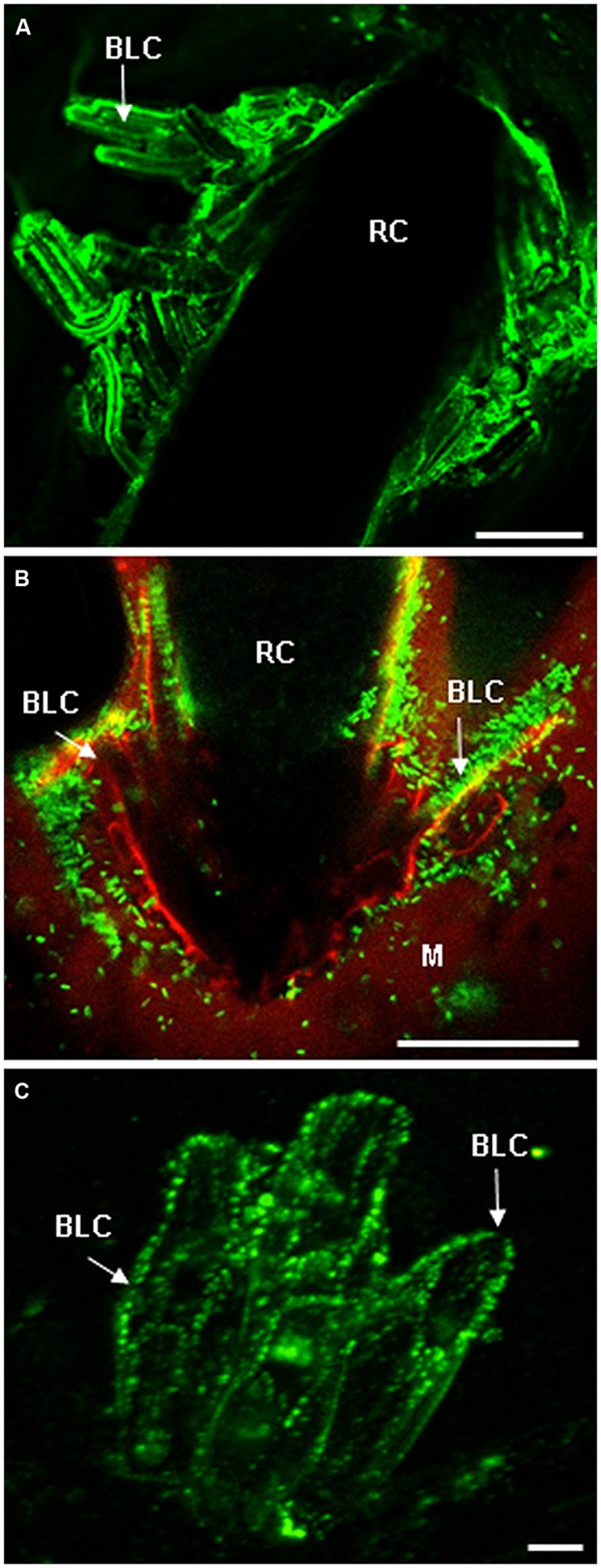
FIGURE 2.
Root cap (RC) and border cells are both enriched in AGP and EXT epitopes. (A) Immunostaining of AGP epitopes at the surface of RC and border-like cells of Brassica napus with the mAb JIM8 (from Cannesan et al., 2012 with permission). Root border-like cells are produced and released from the RC. (B) Micrographs showing the association between root border-like cells from Arabidopsis thaliana and Rhizobium sp. YAS34-GFP. The GFP-expressing bacteria appear green at the root surface (from Vicré et al., 2005 with permission). This association is AGP-dependant as demonstrated in Vicré et al. (2005). (C) Fluorescent micrographs of root border-like cells from flax (Linum usitatissimum) immunostained with the monoclonal antibody LM1 specific for EXT epitopes (from Plancot et al., 2013 with permission). Bars = 20 μm (A), 50 μm (B), and 8 μm (C). BLCs, border-like cells; M, mucilage; RC, root cap.
Extensins have also been shown to play a significant role in plant defense and protection against bioagressors. Immunolocalization studies using the mAbs JIM 20 and JIM 11 revealed the abundant presence of EXT epitopes in cell walls of the resistant wax gourd cultivar to Fusarium oxysporum as compared to susceptible cultivar (Xie et al., 2011). In addition, elicitation with fusaric acid or infection with F. oxysporum caused important decrease of the immunofluorescence in both resistant and susceptible cultivars. Also, elicitation of grapevine callus cultures resulted in both the insolubilization of a specific 89.9 kD EXT and the induction of the catalytic activity of an EXT peroxidase (Jackson et al., 2001). Furthermore, EXTs have been shown to accumulate in response to the pathogenic oomycete Sclerospora graminicola in resistant pearl millet cultivar (Deepak et al., 2007). The high content of EXTs was tightly correlated with an increase in the levels of isodityrosine and H2O2 suggesting cell wall strengthening in the resistant cultivar presumably to limit Sclerospora graminicola penetration and tissue infection.
More recently, the implication of EXTs as part of the innate immune response of root border-like cells (BLCs) of Arabidopsis thaliana and Linum usitatissimum has been investigated by Plancot et al. (2013). Root border cells from plants such as pea, soybean, or cotton are highly specialized in root protection and production of various anti-microbial compounds (Hawes et al., 2000, 2003). Although such a function still needs to be clearly established for BLCs, a class of border cells that is relatively less studied. Recent work suggests a role for these BLCs in root defense (Driouich et al., 2013). Plancot et al. (2013) have also demonstrated that, in response to elicitors (e.g., flagellin 22), a significant increase in the production of H2O2 was detected in root BLCs together with a strong activation of genes involved in EXT biosynthesis and cross-linking. This is consistent with the finding of Velasquez et al. (2011) which showed that EXTs biosynthesis genes were co-expressed with peroxidase genes. Interestingly, treatment with elicitors also caused modifications in the distribution of EXT epitopes within cell walls of root BLCs (Figure 2). The effect of elicitation on the pattern of labeling with the mAb LM1 was shown to depend on both the nature of elicitors and plant species. Elicitation with flagellin 22 almost abolished immunostaining of LM1-recognized epitopes reflecting reorganization of the EXT network within the cell wall due to extensive cross-linking. Such an oxidative cross-linking of EXTs may result in a reinforced glyco-network that enhances physical properties of the cell wall in both Arabidopsis thaliana and L. usitatissimum (Plancot et al., 2013). This reinforcement of the cell wall would in turn limit/prevent penetration and progression of pathogens within root tissues.
Together these findings strongly suggest that AGPs and EXTs are key components of root protection, and more specifically of root border cells. However, further investigations where root border cells are directly challenged with specific pathogens are needed to provide a biological context for these observations. So far, the immune response in roots remains poorly understood and appears to be highly complex and cell-type specific (Millet et al., 2010; Cannesan et al., 2012; Balmer and Mauch-Mani, 2013). To our knowledge, the only study that clearly demonstrated the relationship between the production of EXT and plant resistance to pathogens was performed in leaf tissues (Wei and Shirsat, 2006). In this study, over-expression of the EXT1 gene in leaves of Arabidopsis thaliana clearly limits the spreading of the pathogenic bacteria Pseudomonas syringae DC3000 within the tissues. Subsequently, the infection symptoms are significantly reduced. It is clear that the implication of EXT and AGP populations in root protection is far from being fully understood and more studies are needed to elucidate the role of individual HRGPs/or their glycans in resistance to biotic stress.
CONCLUSION AND OUTLOOK
Like N-glycoproteins, cell wall O-glycoproteins, AGPs and EXTs, are synthesized, assembled and modified within the secretory system. Their glycans, although structurally different and diverse, play a major role in their stability, activity and function. Both types of glycoproteins were shown to be involved in the control of many biological activities and physiological processes in various plant species. However, the specific role of each glycan type and the associated oligosaccharides in biological processes is not known. One of the important challenges for the future is to elucidate the contribution of each of these glycans (and associated sugars) in regulating cell growth, development and adaptation of plants to environmental stresses, either biotic or abiotic. Even more challenging is the search for potential relationships between a given glycan/oligosaccharide structure and a given function in a given tissue. For instance, how specific O-glycan structures regulate morphology, growth or biotic interactions of certain root cell types with microbes is a major issue that deserves further attention.
Recently, a number of the carbohydrate active enzymes involved in N- and O-glycan metabolism have been identified and have advanced our understanding of the biosynthetic machineries of these glycoproteins. How these enzymes are spatially organized and assembled within different compartments of the endomembrane system (i.e., specifically within Golgi subcompartments and Golgi-derived secretory vesicles) and how these are regulated during development is not fully understood and remains an exciting research opportunity for the future.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Part of the work described in this review was supported by the University of Rouen, and the GRR-Végétal-Agronomie-Sol-Innovation of Haute Normandie, Le Fonds Européen de Développement Regional (FEDER), and the ANR. We are grateful to our lab members for their active implication in various “Plant Glycobiology” projects and to Dr. C. Santaella (CNRS Cadarache-Université de Marseille) for her stimulating discussions during the course of our root-microbe interaction studies. The authors are thankful to Dr. J. P. Moore (University of Stellenbosch-Republic of South Africa) for careful reading of the manuscript.
REFERENCES
- Aebi M. (2013). N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833 2430–2437 10.1016/j.bbamcr.2013.04.001 [DOI] [PubMed] [Google Scholar]
- Albersheim P., Darvill A., Roberts K., Sederoff R., Staehelin A. (2011). Plant Cell Walls: from Chemistry to Biology. New York: Garland Science; 365–407 [Google Scholar]
- Andème-Onzighi C., Sivaguru M., Judy-March J., Baskin T. I., Driouich A. (2002). The reb1-1 mutation of Arabidopsis alters the morphology of trichoblasts, the expression of arabinogalactan-proteins and the organization of cortical microtubules. Planta 215 949–958 10.1007/s00425-002-0836-z [DOI] [PubMed] [Google Scholar]
- Bakker H., Lommen A., Jordi W., Stiekema W., Bosch D. (1999). An Arabidopsis thaliana cDNA complements the N-acetylglucosaminyltransferase I deficiency of CHO Lec1 cells. Biochem. Biophys. Res. Commun. 261 829–832 10.1006/bbrc.1999.1117 [DOI] [PubMed] [Google Scholar]
- Bakker H., Schijlen E., de Vries T., Schiphorst W. E., Jordi W., Lommen A., et al. (2001). Plant members of the α-1-3/4-fucosyltransferase gene family encode an α-1-4-fucosyltransferase, potentially involved in Lewis(a) biosynthesis, and two core α-1-3-fucosyltransferases. FEBS Lett. 507 307–312 10.1016/S0014-5793(01)02999-4 [DOI] [PubMed] [Google Scholar]
- Balestrini R., Hahn M. G., Faccio A., Mendgen K., Bonfante P. (1996). Differential localization of carbohydrate epitopes in plant cell walls in the presence and absence of arbuscular mycorrhizal fungi. Plant Physiol. 111 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer D., Mauch-Mani B. (2013). More beneath the surface? Root versus shoot antifungal plant defenses. Front. Plant Sci. 4:256 10.3389/fpls.2013.00256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardor M., Crémata J., Lerouge P. (2011). Glycan engineering in transgenic plants. Annu. Plant Rev. 41 409–424 10.1002/9781444391015.ch17 [DOI] [Google Scholar]
- Bardor M., Faveeuw C., Fitchette A., Gilbert D., Galas L., Trottein F., et al. (2003). Immunoreactivity in mammals of two typical plant glycoepitopes, core α-(1,3)-fucose and core xylose. Glycobiology 13 427–434 10.1093/glycob/cwg024 [DOI] [PubMed] [Google Scholar]
- Bar-Peled M., O’Neill A. (2011). Plant nucleotide sugar formation, interconversion and salvage by sugar recycling. Annu. Rev. Plant Biol. 62 127–155 10.1146/annurev-arplant-042110-103918 [DOI] [PubMed] [Google Scholar]
- Basu D., Liang Y., Liu X., Himmeldirk K., Faik A., Kieliszewski M. J., et al. (2013). Functional identification of a hydroxyproline-O-galactosyltransferase specific for arabinogalactan protein biosynthesis in Arabidopsis. J. Biol. Chem. 288 10132–10143 10.1074/jbc.M112.432609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencúr P., Steinkellner H., Svoboda B., Mucha J., Strasser R., Kolarich D., et al. (2005). Arabidopsis thaliana β-1,2-xylosyltransferase: an unusual glycosyltransferase with the potential to act at multiple stages of the plant N-glycosylation pathway. Biochem. J. 388 515–525 10.1042/BJ20042091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry A. M., Rasmussen U., Bateman K., Huss-Danell K., Lindwall S., Bergman B. (2002). Arabinogalactan proteins are expressed at the symbiotic interface in root nodules of Alnus spp. New Phytol. 155 469–479 10.1046/j.1469-8137.2002.00466.x [DOI] [PubMed] [Google Scholar]
- Boisson M., Gomord V., Audran C., Berger N., Dubreucq B., Granier F., et al. (2001). Arabidopsis glucosidase I mutants reveal a critical role of N-glycan trimming in seed development. EMBO J. 20 1010–1019 10.1093/emboj/20.5.1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P., Aebi M. (1999). The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426 239–257 10.1016/S0304-4165(98)00127-5 [DOI] [PubMed] [Google Scholar]
- Burn J. E., Hurley U. A., Birch R. J., Arioli T., Cork A., Williamson R. E. (2002). The cellulose-deficient Arabidopsis mutant rsw3 is defective in a gene encoding a putative glucosidase II, an enzyme processing N-glycans during ER quality control. Plant J. 32 949–960 10.1046/j.1365-313X.2002.01483.x [DOI] [PubMed] [Google Scholar]
- Burton R. A., Gidley M. J., Fincher G. B. (2010). Heterogeneity in the chemistry, structure and function of plant cell walls. Nat. Chem. Biol. 6 724–732 10.1038/nchembio.439 [DOI] [PubMed] [Google Scholar]
- Cannesan M. A., Durand C., Burel C., Gangneux C., Lerouge P., Ishii T., et al. (2012). Effect of arabinogalactan proteins from the root caps of pea and Brassica napus on Aphanomyces euteiches zoospore chemotaxis and germination. Plant Physiol. 159 1658–1670 10.1104/pp.112.198507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon M. C., Terneus K., Hall Q., Tan L., Wang Y., Wegenhart B. L., et al. (2008). Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. U.S.A. 105 2226–2231 10.1073/pnas.0711980105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita N. C., Gibeaut D. M. (1993). Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3 1–30 10.1111/j.1365-313X.1993.tb00007.x [DOI] [PubMed] [Google Scholar]
- Chen X. L., Shi T., Yang J., Shi W., Gao X., Chen D., et al. (2014). N-glycosylation of effector proteins by an α-1,3-mannosyltransferase is required for the rice blast fungus to evade host innate immunity. Plant Cell 26 1360–1376 10.1105/tpc.114.123588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier L., Bernard S., Ramdani Y., Lamour R., Bardor M., Lerouge P., et al. (2010). Subcompartment localization of the side chain xyloglucan-synthesizing enzymes within Golgi stacks of tobacco suspension-cultured cells. Plant J. 64 977–989 10.1111/j.1365-313X.2010.04388.x [DOI] [PubMed] [Google Scholar]
- Deepak S., Shailasree S., Kini R. K., Hause B., Shetty S. H., Mithöfer A. (2007). Role of hydroxyproline-rich glycoproteins in resistance of pearl millet against downy mildew pathogen Sclerospora graminicola. Planta 226 323–333 10.1007/s00425-007-0484-4 [DOI] [PubMed] [Google Scholar]
- Dilokpimol A., Poulsen C. P., Vereb G., Kaneko S., Schulz A., Geshi N. (2014). Galactosyltransferases from Arabidopsis thaliana in the biosynthesis of type II arabinogalactan: molecular interaction enhances enzyme activity. BMC Plant Biol. 14:90 10.1186/1471-2229-14-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L., Linstead P., Roberts K. (1995). An AGP epitope distinguishes a central metaxylem initial from other vascular initials in the Arabidopsis root. Protoplasma 189 149–155 10.1007/BF01280168 [DOI] [Google Scholar]
- Driouich A., Follet-Gueye M.-L., Bernard S., Kousar S., Chevalier L., Vicré-Gibouin M., et al. (2012). Golgi-mediated synthesis and secretion of matrix polysaccharides of the primary cell wall of higher plants. Front. Plant Sci. 3:79 10.3389/fpls.2012.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driouich A., Follet-Gueye M.-L., Vicré-Gibouin M., Hawes M. (2013). Root border cells and secretions as critical elements in plant host defense. Curr. Opin. Plant Biol. 16 489–495 10.1016/j.pbi.2013.06.010 [DOI] [PubMed] [Google Scholar]
- Durand C., Vicré-Gibouin M., Follet-Gueye M.-L., Duponchel L., Moreau M., Lerouge P., et al. (2009). The organization pattern of root border-like cells of Arabidopsis is dependent on cell wall homogalacturonan. Plant Physiol. 150 1411–1421 10.1104/pp.109.136382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelund J., Obel N., Ulvskov P., Geshi N., Pauly M., Bacic A., et al. (2007). Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol. Biol. 64 439–451 10.1007/s11103-007-9162-y [DOI] [PubMed] [Google Scholar]
- Ellis M., Egelund J., Schultz C. J., Bacic A. (2010). Arabinogalactan protein: key regulators at the cell surface. Plant Physiol. 153 403–419 10.1104/pp.110.156000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquerré-Tugayé M. T. (1979). Cell surfaces in plant-microorganism interactions: I. A structural investigation of cell wall hydroxyproline-rich glycoproteins which accumulate in fungus-infected plants . Plant Physiol. 64 314–319 10.1104/pp.64.2.314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquerré-Tugayé M. T., Lafitte C., Mazau D., Toppan A., Touzé A. (1979). Cell surfaces in plant-microorganism interactions: II. Evidence for the accumulation of hydroxyproline-rich glycoproteins in the cell wall of diseased plants as a defense mechanism . Plant Physiol. 64 320–326 10.1104/pp.64.2.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanata W. I., Lee K. H., Son B. H., Yoo J. Y., Harmoko R., Ko K. S., et al. (2013). N-glycan maturation is crucial for cytokinin-mediated development and cellulose synthesis in Oryza sativa. Plant J. 73 966–979 10.1111/tpj.12087 [DOI] [PubMed] [Google Scholar]
- Farid A., Malinovsky F. G., Veit C., Schoberer J., Zipfel C., Strasser R. (2013). Specialized roles of the conserved subunit OST3/6 of the oligosaccharyltransferase complex in innate immunity and tolerance to abiotic stresses. Plant Physiol. 162 24–38 10.1104/pp.113.215509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkostefanakis S., Sedeek K. E. M., Raad M., Zaki M. S., Kalaitzis P. (2014). Virus induced gene silencing of three putative prolyl 4-hydroxylases enhances plant growth in tomato (Solanum lycopersicum). Plant Mol. Biol. 85 459–471 10.1007/s11103-014-0197-6 [DOI] [PubMed] [Google Scholar]
- Geshi N., Johansen J. N., Dilokpimol A., Rolland A., Belcram K., Verger S., et al. (2013). A galactosyltranferase acting on arabinogalactan protein glycans is essential for embryo development in Arabidopsis. Plant J. 76 128–137 10.1111/tpj.12281 [DOI] [PubMed] [Google Scholar]
- Gil G.-C., Velander W. H., Van Cott K. E. (2009). N-glycosylation microheterogeneity and site occupancy of an Asn-X-Cys sequon in plasma-derived and recombinant Protein C. Proteomics 9 2555–2567 10.1002/pmic.200800775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S., Hänsel U., Ziemann M., Pauly M. (2009). Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc. Natl. Acad. Sci. U.S.A. 106 14699–14704 10.1073/pnas.0905434106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S., Sharma V., Baidoo E. E. K., Keasling J. D., Scheller H. V., Pauly M. (2013). Arabinosylation of a Yariv-precipitable cell wall polymer impacts plant growth as exemplified by the Arabidopsis glycosyltransferase mutant ray1. Mol. Plant 6 1369–1372 10.1093/mp/sst029 [DOI] [PubMed] [Google Scholar]
- Hall Q., Cannon M. C. (2002). The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell 14 1161–1172 10.1105/tpc.010477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan H., Reis C. A., Bennett E. P., Mirgorodskaya E., Roepstorff P., Hollingsworth M. A., et al. (2000). The Lectin domain of UDP-N-acetyl-D-galactosamine: polypeptide N-acetylgalacto-saminyltransferase-T4 directs its glycopeptide specificities. J. Biol. Chem. 275 38197–38205 10.1074/jbc.M005783200 [DOI] [PubMed] [Google Scholar]
- Hawes M. C., Bengough G., Cassab G., Ponce G. (2003). Root caps and rhizosphere. J. Plant Growth Regul. 21 352–367 10.1007/s00344-002-0035-y [DOI] [Google Scholar]
- Hawes M. C., Gunawardena U., Miyasaka S., Zhao X. (2000). The role of root border cells in plant defense. Trends Plant Sci. 5 128–133 10.1016/S1360-1385(00)01556-9 [DOI] [PubMed] [Google Scholar]
- Held M. A., Tan L., Kamyab A., Hare M., Shpak E., Kieliszewski M. J. (2004). Di-isodityrosine is the intermolecular cross-link of isodityrosine-rich extensin analogs cross-linked in vitro. J. Biol. Chem. 279 55474–55482 10.1074/jbc.M408396200 [DOI] [PubMed] [Google Scholar]
- Hennet T. (2012). Diseases of glycosylation beyond classical congenital disorders of glycosylation. Biochim. Biophys. Acta 1820 1306–1317 10.1016/j.bbagen.2012.02.001 [DOI] [PubMed] [Google Scholar]
- Hieta R., Myllyharju J. (2002). Cloning and characterization of a low molecular weight prolyly 4-hydroxylase from Arabidopsis thaliana. J. Biol. Chem. 277 23965–23971 10.1074/jbc.M201865200 [DOI] [PubMed] [Google Scholar]
- Ioffe E., Stanley P. (1994). Mice lacking N-acetylglucosaminyltransferase I activity die at mid-gestation, revealing an essential role for complex or hybrid N-linked carbohydrates. Proc. Natl. Acad. Sci. U.S.A. 91 728–732 10.1073/pnas.91.2.728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P. A. P., Galinha C. I. R., Pereira C. S., Fortunato A., Soares N. C., Am ancio S. B. Q., et al. (2001). Rapid deposition of extension during the elicitation of grapevine callus cultures is specifically catalyzed by a 40-kilodalton peroxidase. Plant Physiol. 127 1065–1076 10.1104/pp.010192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Frank J., Kang C., Kajiura H., Vikram M., Ueda A., et al. (2008). Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. U.S.A. 105 5933–5938 10.1073/pnas.0800237105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski M. J., Lamport D. T. A. (1994). Extensins: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 5 157–172 10.1046/j.1365-313X.1994.05020157.x [DOI] [PubMed] [Google Scholar]
- Kieliszewski M. J., Lamport D. T. A., Cannon M. C. (2011). Hydroxyproline-rich glycoproteins: form and function. Annu. Plant Rev. 41 321–342 [Google Scholar]
- Kieliszewski M. J., Shpak E. (2001). Synthetic genes for the elucidation of glycosylation codes for arabinogalactan-proteins and other hydroxyproline-rich glycoproteins. Cell. Mol. Life Sci. 58 1386–1398 10.1007/PL00000783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivirikko K. I., Pihlajaniemi T. (1998). Collagen hydroxylases and the protein disulfide isomerase subunit of prolyl 4-hydroxylases. Adv. Enzymol. Relat. Areas Mol. Biol. 72 325–398 [DOI] [PubMed] [Google Scholar]
- Knoch E., Dilokpimol A., Tryfona T., Poulsen C. P., Xiong G., Harholt J., et al. (2013). A β-glucuronosyltransferase from Arabidopsis thaliana involved in biosynthesis of type II arabinogalactan has a role in cell elongation during seedling growth. Plant J. 76 1016–1029 10.1111/tpj.12353 [DOI] [PubMed] [Google Scholar]
- Lamport D. T. A., Kieliszewski M. J., Chen Y., Cannon M. C. (2011). Role of the extensin superfamily in primary cell wall architecture. Plant Physiol. 156 11–19 10.1104/pp.110.169011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin A., Imperiali B. (2011). The expanding horizons of asparagine-linked glycosylation. Biochemistry 50 4411–4426 10.1021/bi200346n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. J. D., Sakata Y., Mau S.-L., Pettolino F., Bacic A., Quatrano R. S., et al. (2005). Arabinogalactan proteins are required for apical cell extension in the moss Physcomitrella patens. Plant Cell 17 3051–3065 10.1105/tpc.105.034413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiter H., Mucha J., Staudacher E., Grimm R., Glössl J., Altmann F. (1999). Purification, cDNA cloning, and expression of GDP-L-Fuc:Asn-linked GlcNAc α-1,3-fucosyltransferase from mung beans. J. Biol. Chem. 274 21830–21839 10.1074/jbc.274.31.21830 [DOI] [PubMed] [Google Scholar]
- Léonard R., Costa G., Darrambide E., Lhernould S., Fleurat-Lessard P., Carlué M., et al. (2002). The presence of Lewis a epitopes in Arabidopsis thaliana glycoconjugates depends on an active α-4-fucosyltransferase gene. Glycobiology 12 299–306 10.1093/glycob/12.5.299 [DOI] [PubMed] [Google Scholar]
- Lerouge P., Cabanes-Macheteau M., Rayon C., Fischette-Lainé A.-C., Gomord V., Faye L. (1998). N-glycoprotein biosynthesis in plants: recent developments and future trends. Plant Mol. Biol. 38 31–48 10.1023/A:1006012005654 [DOI] [PubMed] [Google Scholar]
- Liang L., Basu D., Pattathil S., Xu W.-L., Venetos A., Martin S. L., et al. (2013). Biochemical and physiological characterization of fut4 and fut6 mutants defective in arabinogalactan-protein fucosylation in Arabidopsis. J. Exp. Bot. 64 5537–5551 10.1093/jxb/ert321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Faik A., Kieliszewski M. J., Tan L., Xu W. L., Showalter A. M. (2010). Identification and characterization of in vitro galactosyltransferase activities involved in the arabinogalactan protein glycosylation in tobacco and Arabidopsis. Plant Physiol. 154 632–642 10.1104/pp.110.160051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebminger E., Hüttner S., Vavra U., Fischl R., Schoberer J., Grass J., et al. (2009). Class I α-mannosidases are required for N-glycan pr 398 ocessing and root development in Arabidopsis thaliana. Plant Cell 21 3850–3867 10.1105/tpc.109.072363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J. B., Marth J. D. (2003). A genetic approach to mammalian glycan function. Annu. Rev. Biochem. 72 643–691 10.1146/annurev.biochem.72.121801.161809 [DOI] [PubMed] [Google Scholar]
- Matsui T., Takita E., Sato T., Kinjo S., Aizawa M., Sugiura Y., et al. (2011). N-glycosylation at noncanonical Asn-X-Cys sequences in plant cells. Glycobiology 21 994–999 10.1093/glycob/cwq198 [DOI] [PubMed] [Google Scholar]
- Metzler M., Gertz A., Sarkar M., Schachter H., Schrader J. W., Marth J. D. (1994). Complex asparagine-linked oligosaccharides are required for morphogenic events during post-implantation development. EMBO J. 13 2056–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet Y. A., Danna C. H., Clay N. K., Songnuan W., Simon M. D., Werck-Reichhart D., et al. (2010). Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22 973–990 10.1105/tpc.109.069658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. P., Fangel J. U., Willats W. G. T., Vivier M. A. (2014a). Pectic-β(1,4)-galactan, extensin and arabinogalactan–protein epitopes differentiate ripening stages in wine and table grape cell walls. Ann. Bot. 10.1093/aob/mcu053 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. P., Nguema-Ona E., Fagerström A., Fangel J. U., Willats W. G. T., Hugo A., et al. (2014b). Profiling the main cell wall polysac-charides of grapevine leaves using high throughput techniques. Carbohydr. Polym. 99 190–198 10.1016/j.carbpol.2013.08.013 [DOI] [PubMed] [Google Scholar]
- Motose H., Sugiyama M., Fukuda H. (2004). A proteoglycan mediates inductive interaction during plant vascular development. Nature 429 873–878 10.1038/nature02613 [DOI] [PubMed] [Google Scholar]
- Myllyharju J. (2003). Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 22 15–24 10.1016/S0945-053X(03)00006-4 [DOI] [PubMed] [Google Scholar]
- Nguema-Ona E., Andème-Onzighi C., Aboughe-Angone S., Bardor M., Ishii T., Lerouge P., et al. (2006). The reb1-1 mutation of Arabidopsis. Effect on the structure and localization of galactose-containing cell wall polysaccharides. Plant Physiol. 140 1406–1417 10.1104/pp.105.074997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguema-Ona E., Bannigan A., Chevalier L., Baskin T. I., Driouich A. (2007). Disruption of arabinogalactan proteins disorganizes cortical microtubules in the root of Arabidopsis thaliana. Plant J. 52 240–251 10.1111/j.1365-313X.2007.03224.x [DOI] [PubMed] [Google Scholar]
- Nguema-Ona E., Coimbra S., Vicré-Gibouin M., Mollet J.-C., Driouich A. (2012). Arabinogalactan-proteins in root and pollen tube cells: distribution and functional properties. Ann. Bot. 110 383–404 10.1093/aob/mcs143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguema-Ona E., Vicré-Gibouin M., Cannesan M.-A., Driouich A. (2013). Arabinoga-lactan-proteins in root microbe interactions. Trends Plant Sci. 18 440–449 10.1016/j.tplants.2013.03.006 [DOI] [PubMed] [Google Scholar]
- O’Neill M. A., Eberhard S., Albersheim P., Darvill A. G. (2001). Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294 846–849 10.1126/science.1062319 [DOI] [PubMed] [Google Scholar]
- Pagny S., Bouissonnie F., Sarkar M., Follet-Gueye M.-L., Driouich A., Schachter H., et al. (2003). Structural requirements for Arabidopsis β-1,2-xylosyltransferase activity and targeting to the Golgi. Plant J. 33 189–203 10.1046/j.0960-7412.2002.01604.x [DOI] [PubMed] [Google Scholar]
- Plancot B., Santaella C., Jaber R., Kiefer-Meyer M.-C., Follet-Gueye M.-L., Leprince J., et al. (2013). Deciphering the responses of root border-like cells of Arabidopsis thaliana and Linum usitatissimum to pathogen derived elicitors. Plant Physiol. 163 1583–1597 10.1104/pp.113.222356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Li Y., Gan J., Wang W., Zhang H., Liu Y., et al. (2013). OsDGL1, a homolog of an oligosaccharyltransferase complex subunit, is involved in N-glycosylation and root development in rice. Plant Cell Physiol. 54 129–137 10.1093/pcp/pcs159 [DOI] [PubMed] [Google Scholar]
- Qi W., Fong C., Lamport D. T. A. (1991). Gum arabic glycoprotein is a twisted hairy rope: new model based on O-galactosylhydroxyproline as a polysaccharide attachment site. Plant Physiol. 96 848–855 10.1104/pp.96.3.848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y., Egelund J., Gilson P. R., Houghton F., Gleeson P. A., Schultz C. J., et al. (2008). Identification of a novel group of putative Arabidopsis thaliana β-(1,3)-galactosyltransferases. Plant Mol. Biol. 68 43–59 10.1007/s11103-008-9351-3 [DOI] [PubMed] [Google Scholar]
- Saint-Jore-Dupas C., Nebenführ A., Boulaflous A., Follet-Gueye M.-L., Plasson C., Hawes C., et al. (2006). Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18 3182–3200 10.1105/tpc.105.036400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito F., Suyama A., Oka T., Yoko-o T., Matsuoka K., Jigami Y., et al. (2014). Identification of novel peptidyl serine α-galactosyltransferase alactosyltransferase gene family in plants. J. Biol. Chem. 289 20405–20420 10.1074/jbc.M114.553933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B., van Engelen F., van der Knaap E., van de Wiel C., van Kammen A., Bisseling T. (1990). Sequential induction of nodulin gene expression in the developing pea nodule. Plant Cell 2 687–700 10.1105/tpc.2.8.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz C. J., Rumsewicz M. P., Johnson K. L., Jones B. J., Gaspar Y. M., Bacic A. (2002). Using genomic resources to guide research directions. the arabinogalactan protein gene family as a test case. Plant Physiol. 129 1448–1463 10.1104/pp.003459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert G., Roberts K. (2007). The biology of arabinogalactan proteins. Annu. Rev. Plant Biol. 58 137–161 10.1146/annurev.arplant.58.032806.103801 [DOI] [PubMed] [Google Scholar]
- Showalter A. M. (2001). Arabinogalactan proteins: structure, expression and function. Cell. Mol. Life Sci. 58 1399–1417 10.1007/PL00000784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter A. M., Keppler B., Lichtenberg J., Gu D., Welch L. R. (2010). A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 153 485–513 10.1104/pp.110.156554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E., Leykam J. F., Kieliszewski M. J. (1999). Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-O-glycosylation codes. Proc. Natl. Acad. Sci. U.S.A. 96 14736–14741 10.1073/pnas.96.26.14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourrouille C., Marquet-Blouin E., D’Aoust M. A., Kiefer-Meyer M.-C., Seveno M., Pagny-Salehabadi S., et al. (2008). Down-regulated expression of plant-specific glycoepitopes in alfalfa. Plant Biotechnol. J. 6 702–721 10.1111/j.1467-7652.2008.00353.x [DOI] [PubMed] [Google Scholar]
- Strasser R. (2014). Biological significance of complex N-glycans in plants and their impact on plant physiology. Front. Plant Sci. 5:363 10.3389/fpls.2014.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R., Altmann F., Mach L., Glössl J., Steinkellner H. (2004). Generation of Arabidopsis thaliana plants with complex N-glycans lacking β-1,2-linked xylose and core α-1,3-linked fucose. FEBS Lett. 561 132–136 10.1016/S0014-5793(04)00150-4 [DOI] [PubMed] [Google Scholar]
- Strasser R., Bondili J. S., Vavra U., Schoberer J., Svoboda B., Glossl J., et al. (2007). A unique 1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing Lewis a structures in Arabidopsis thaliana. Plant Cell 19 2278–2292 10.1105/tpc.107.052985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R., Mucha J., Mach L., Altmann F., Wilson I., Glössl J., et al. (2000). Molecular cloning and functional expression of β-1, 2-xylosyltransferase cDNA from Arabidopsis thaliana. FEBS Lett. 472 105–108 10.1016/S0014-5793(00)01443-5 [DOI] [PubMed] [Google Scholar]
- Strasser R., Mucha J., Schwihla H., Altmann F., Glössl J., Steinkellner H. (1999a). Molecular cloning and characterization of cDNA coding for β-1,2N-acetylglucosaminyltransferase I (GlcNAc-TI) from Nicotiana tabacum. Glycobiology 9 779–785 10.1093/glycob/9.8.779 [DOI] [PubMed] [Google Scholar]
- Strasser R., Steinkellner H., Borén M., Altmann F., Mach L., Glössl J., et al. (1999b). Molecular cloning of cDNA encoding N-acetylglucosaminyltransferase II from Arabidopsis thaliana. Glycoconj. J. 16 787–791 10.1023/A:1007127815012 [DOI] [PubMed] [Google Scholar]
- Strasser R., Schoberer J., Jin C., Glossl J., Mach L., Steinkellner H. (2006). Molecular cloning and characterization of Arabidopsis thaliana Golgi α-mannosidase II, a key enzyme in the formation of complex N-glycans in plants. Plant J. 45 789–803 10.1111/j.1365-313X.2005.02648.x [DOI] [PubMed] [Google Scholar]
- Tan L., Eberhard S., Pattathil S., Warder C., Glushka J., Yuan C., et al. (2013). An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25 270–287 10.1105/tpc.112.107334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Showalter A. M., Egelund J., Hernandez-Sanchez A., Doblin M. S., Bacic A. (2012). Arabinogalactan-proteins and the research challenges for these enigmatic plant cell surface proteoglycans. Front. Plant Sci. 3:140 10.3389/fpls.2012.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Varnai P., Lamport D. T. A., Yuan C., Xu J., Qui F., et al. (2010). Plant O-hydroxyproline arabinogalactans are composed of repeating trigalactosyl subunits with short bifurcated side chains. J. Biol. Chem. 285 24575–24583 10.1074/jbc.M109.100149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M., Ross H., Mcrae D., Stewart D., Roberts I., Duncan G., et al. (2000). A potato α-glucosidase gene encodes a glycoprotein processing α-glucosidase II-like activity. Demonstration of enzyme activity and effects of down-regulation in transgenic plants. Plant J. 24 305–316 10.1046/j.1365-313x.2000.00873.x [DOI] [PubMed] [Google Scholar]
- Tiainen P., Myllyharju J., Koivunen P. (2005). Characterization of a second Arabidopsis thaliana prolyl-4-hydroxylase with distinct substrate specificity. J. Biol. Chem. 280 1142–1148 10.1074/jbc.M411109200 [DOI] [PubMed] [Google Scholar]
- Tryfona T., Liang H.-C., Kotake T., Tsumuraya Y., Stephens E., Dupree P. (2012). Structural characterization of Arabidopsis leaf arabinogalactan polysaccharides. Plant Physiol. 160 653–666 10.1104/pp.112.202309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryfona T., Theys T. E., Wagner T., Stott K., Keegstra K., Dupree P. (2014). Characterization of FUT4 and FUT6 α-(1-2)-Fucosyltransferases reveals that absence of root arabinogalactan fucosylation increases Arabidopsis root growth salt sensitivity. PLoS ONE 9:e93291 10.1371/journal.pone.0093291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulvskov P., Paiva D. S., Domozych D., Harholt J. (2013). Classification, naming and evolutionary history of glycosyltransferases from sequenced green and red algal genomes. PLoS ONE 8:e76511 10.1371/journal.pone.0076511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel A. J., Roberts K. (2002). Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. Plant J. 32 105–113 10.1046/j.1365-313X.2002.01406.x [DOI] [PubMed] [Google Scholar]
- Varki A. (1993). Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3 97–130 10.1093/glycob/3.2.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. (2011). Evolutionary forces shaping the Golgi glycosylation machinery: why cell surface glycans are universal to living cells. Cold Spring Harb. Perspect. Biol. 3:a005462 10.1101/cshperspect.a005462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Marth J. D., et al. (2009). Symbol nomenclature for glycan representation. Proteomics 9 5398–5399 10.1002/pmic.200900708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A., Lowe J. B. (2009). “Biological roles of glycans,” in Essentials of Glycobiology 2nd Edn Chap. 6 eds Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R.et al. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [PubMed] [Google Scholar]
- Velasquez M., Salter J. S., Dorosz J. G., Petersen B. L., Estevez J. M. (2012). Recent advances on the posttranslational modifications of EXTs and their roles in plant cell walls. Front. Plant Sci. 3:93 10.3389/fpls.2012.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez S. M., Ricardi M. M., Dorosz J. G., Fernandez P. V., Nadra A. D., Pol-Fachin L., et al. (2011). O-glycosylated cell wall proteins are essential in root hair growth. Science 332 1401–1403 10.1126/science.1206657 [DOI] [PubMed] [Google Scholar]
- Vicré M., Santaella C., Blanchet S., Gateau A., Driouich A. (2005). Root border-like cells of Arabidopsis. microscopical characterization and role in the interaction with rhizobacteria. Plant Physiol. 138 998–1008 10.1104/pp.104.051813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F., Spano T., Vlad D., Boudaher F., Ouelhadj A., Kalaitzis P. (2007). Arabidopsis prolyl-4 hydroxylases are differentially expressed in response to hypoxia, anoxia and mechanical wounding. Physiol. Plant. 130 471–483 10.1111/j.1399-3054.2007.00915.x [DOI] [Google Scholar]
- von Schaewen A., Frank J., Koiwa H. (2008). Role of complex N-glycans in plant stress tolerance. Plant Signal. Behav. 3 871–873 10.4161/psb.3.10.6227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schaewen A., Sturm A., O’Neill J., Chrispeels M. (1993). Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol. 102 1109–1118 10.1104/pp.102.4.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandall H. H., Irazoqui F., Tarp M. A., Bennett E. P., Mandel U., Takeuchi H., et al. (2007). The lectin domains of polypeptide GalNaC-transferases exhibit carbohydrate binding specificity for GalNaC. Lectin bind to GalNaC-glycopeptide substrates is required for high density GalNaC O-glycosylation. Glycobiology 17 374–387 10.1093/glycob/cwl082 [DOI] [PubMed] [Google Scholar]
- Wei G., Shirsat A. H. (2006). Extensin over-expression in Arabidopsis limits pathogen invasiveness. Mol. Plant Pathol. 7 579–592 10.1111/j.1364-3703.2006.00363.x [DOI] [PubMed] [Google Scholar]
- Wenderoth I., von Schaewen A. (2000). Isolation and characterization of plant N-acetyl glucosaminyltransferase I (GntI) cDNA sequences. Functional analyses in the Arabidopsis cgl mutant and in antisense plants .Plant Physiol. 123 1097–1108 10.1104/pp.123.3.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I. B., Rendić D., Freilinger A., Dumić J., Altmann F., Mucha J., et al. (2001a). Cloning and expression of cDNAs encoding α-1,3-fucosyltransferase homologues from Arabidopsis thaliana. Biochim. Biophys. Acta 1527 88–96 10.1016/S0304-4165(01)00151-9 [DOI] [PubMed] [Google Scholar]
- Wilson I. B., Zeleny R., Kolarich D., Staudacher E., Stroop C., Kamerling J., et al. (2001b). Analysis of Asn-linked glycans from vegetable foodstuffs: widespread occurrence of Lewis a, core α-1,3-linked fucose and xylose substitutions. Glycobiology 11 261–274 10.1093/glycob/11.4.261 [DOI] [PubMed] [Google Scholar]
- Wu Y., Williams M., Bernard S., Driouich A., Showalter A. M., Faik A. (2010). Functional identification of two non-redundant Arabidopsis α-(1,2)-fucosyltransferases specific to arabinogalactan proteins. J. Biol. Chem. 285 13638–13645 10.1074/jbc.M110.102715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D., Ma L., Šamaj J., Xu C. (2011). Immunohistochemical analysis of cell wall hydroxyproline-rich glycoproteins in the roots of resistant and susceptible wax gourd cultivars in response to Fusarium oxysporum f. sp.Benincasae infection and fusaric acid treatment. Plant Cell Rep. 30 1555–1569 10.1007/s00299-011-1069-z [DOI] [PubMed] [Google Scholar]
- Xie F., Williams A., Edwards A., Downie J. A. (2012). A plant arabinogalactan-like glycoprotein promotes a novel type of polar surface attachment by Rhizobium leguminosarum. Mol. Plant Microbe Interact. 25 250–258 10.1094/MPMI-08-11-0211 [DOI] [PubMed] [Google Scholar]
- Ye C. Y., Li T., Tuskan G. A., Tschaplinski T. J., Yang X. H. (2011). Comparative analysis of GT14/GT14-like gene family in Arabidopsis, Oryza, Populus, Sorghum and Vitus. Plant Sci. 181 688–695 10.1016/j.plantsci.2011.01.021 [DOI] [PubMed] [Google Scholar]
- Yuasa K., Toyooka K., Fukuda H., Matsuoka K. (2005). Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J. 41 81–94 10.1111/j.1365-313X.2004.02279.x [DOI] [PubMed] [Google Scholar]
- Zhou Y. H., Li S. B., Qian Q., Zeng D., Zhang M., Guo L., et al. (2009).BC10 a DUF266-containing and Golgi-located type II membrane protein, is required for cell wall biosynthesis in rice (Oriza sativa L.). Plant J. 57 446–462 10.1111/j.1365-313X.2008.03703.x [DOI] [PubMed] [Google Scholar]
- Zielinska D. F., Gnad F., Wiśniewski J. R., Mann M. (2010). Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141 897–907 10.1016/j.cell.2010.04.012 [DOI] [PubMed] [Google Scholar]