Abstract
Peptide-mediated interactions, in which a short linear motif binds to a globular domain, play major roles in cellular regulation. An accurate structural model of this type of interaction is an excellent starting point for the characterization of the binding specificity of a given peptide-binding domain. A number of different protocols have recently been proposed for the accurate modeling of peptide-protein complex structures, given the structure of the protein receptor and the binding site on its surface. When no information about the peptide binding site(s) is a priori available, there is a need for new approaches to locate peptide-binding sites on the protein surface. While several approaches have been proposed for the general identification of ligand binding sites, peptides show very specific binding characteristics, and therefore, there is a need for robust and accurate approaches that are optimized for the prediction of peptide-binding sites.
Here we present PeptiMap, a protocol for the accurate mapping of peptide binding sites on protein structures. Our method is based on experimental evidence that peptide-binding sites also bind small organic molecules of various shapes and polarity. Using an adaptation of ab initio ligand binding site prediction based on fragment mapping (FTmap), we optimize a protocol that specifically takes into account peptide binding site characteristics. In a high-quality curated set of peptide-protein complex structures PeptiMap identifies for most the accurate site of peptide binding among the top ranked predictions. We anticipate that this protocol will significantly increase the number of accurate structural models of peptide-mediated interactions.
Keywords: protein peptide interactions, FFT sampling, binding site detection, mapping, PeptiDB
Introduction
Much of the maintenance of a cell is accomplished by communication between proteins. Such communication may involve complex layers of regulation that are mediated by local features, such as changes in local concentrations of the partners or / and post-translational modifications 1,2. These interactions are often weak and transient, and tuned so that the threshold of biological downstream response may easily be manipulated. Peptide – mediated interactions, in which a short, linear stretch binds to its protein receptor, are well suited for such transient binding, and therefore extensively used. In higher eukaryotes, up to 50% of known interactions between proteins are indeed mediated by peptides 3.
The classical peptide-protein interaction involves (1) a short motif that is often embedded within an unstructured region, and (2) a peptide-binding domain with a defined globular structure. This interaction may occur between two distinct proteins, or within a protein, and the very competition between cis and trans interactions is often the very step that regulates protein function 4.
One of the important sources of information about interactions is the structure of a protein-protein complex. This structure can be used as a starting point for the characterization and manipulation of an interaction. As an example, residues that are critical for an interaction may be identified using experimental or computational alanine scanning of interface residues 5–8. Abolishment of an interaction by mutation of these critical residues may help identify the functional role of this interaction 9. Finally, targeting of the interface of critical interactions by small molecules is gaining increasing importance in drug design, in addition to the traditional design of inhibitors of enzyme reactions 10,11.
While the number of experimentally solved structures is increasing, the fraction of protein complexes among these remains very low, around 10–20% 12. This calls for the development of approaches that identify a binding site on a protein structure, or even better model the structure of a complex from the free monomers. Indeed, the field of docking, in which the structure of a complex is modeled from the structures of the free components, has significantly improved over the last 2 decades (see this CAPRI issue for some of the latest improvements).
Identification of the binding site on a protein structure is a first step towards the generation of an accurate structural model of an interaction. If crucial residues that mediate the binding of two partners can be identified, this has two important effects: first of all, experiments can be directed towards those residues and the functional effect of an interaction may be studied. Secondly, docking approaches may be focused on a specific interface patch 13. For instance, we have previously developed a protocol that starting from a known binding site and an approximate peptide conformation within that site can accurately model the peptide-protein complex structure (FlexPepDock 14,15), even without any detailed knowledge of the peptide structure within the binding site (ab initio FlexPepDock 16). Thus, binding site identification allows to focus, and to intensify the search to relevant sites, rather than wasting time in a global full docking search, which can also result in additional false positives.
Limited approaches have been proposed to identify peptide binding sites on proteins (e.g. references 17–19). These use information both from the structures of the partners, as well as from the sequence.
PepSite identifies peptide binding sites on protein structures by searching for regions that match a spatial PSSM derived from known peptide binding protein receptor structures 17. As such, it can not only identify the location of the peptide binding site, but also suggests a sequence motif for the binding peptides. Consequently, information about the actual peptide-binding partners is also provided. Another recently published approach uses the BRIX database of interacting fragments to predict the structure of peptide-protein complexes starting from a peptide sequence and a solved receptor structure 19.
As for peptide binding sites, these existing methods perform well mainly on known binding sites, such as WW, SH3 and kinase domains, but less well on non-standard peptide-mediated interactions. Thus, new tools are needed to address this problem.
Here we suggest an approach based on the observation that protein functional sites, including peptide binding sites, also bind small organic molecules of various shapes and polarity, as observed by nucleic magnetic resonance (NMR) 20 and X-ray crystallography experiments 21. FTmap 22 is a direct computational analogue of the above experimental approaches. This protocol is based on the successful FFT-based docking protocol with statistical potential 23. We have recently reported application of this approach to ligand binding site identification 24 and druggable protein-protein interaction sites 25, demonstrating broad range commonality of principles of molecular recognition.
In this study, we have calibrated a protocol to detect peptide binding sites on protein structures. For this, we have adapted the mapping protocol to the identification of peptide binding sites. Two key differences distinguish peptide site prediction from ligand binding sites detection: (1) Prediction requires knowledge of the domain critical for peptide interaction: In contrast to proteins that bind ligands at one key site, multi-domain proteins can have multiple peptide regulatory sites on each of the domains and therefore focusing on one particular domain is recommended; (2) Peptides do not bind to inner buried sites of the proteins, which could well be ligand binding sites, and therefore these internal sites need to be removed. We show that with this tailored protocol, we can identify accurately the binding site of a peptide within the top-3 sites in 19/21 (90%) of a benchmark set of curated peptide-protein complexes. We validate the approach on a set of peptide-protein structures that were released after calibration of the protocol: for 7/9 (78%) of the structures we reliably identify the peptide-binding site. In addition to the robustness of our protocol, our results also highlight features that characterize specifically peptide-protein complex structures, and finally allows us to identify yet outstanding challenges in the modeling, and also the experimental identification of peptide-mediated interactions.
Methods
Compilation of training and test sets for protocol calibration
In a previous study we have studied the special character of peptide-protein complex structures. For this aim, we collected PeptiDB, a set of 103 peptide-protein complexes that represent a set of different known peptide-mediated interactions (less than 70% sequence identity among different protein receptors), and a more restricted set of 61 complexes in which no two proteins share the same fold (according to CATH classification) (See reference 26 and Table S1 therein).
In the present study, our aim is to identify the peptide binding site on the protein surface. In order to prevent bias to bound sites on a protein structure, we needed to verify that the surface accessible to the peptide for binding on the receptor is indeed not occupied by any other protein nor ligand. We therefore went over the original set and checked each protein-peptide interaction individually for the following features:
Availability of the free receptor structure. We filtered for cases in which the corresponding free conformation has been solved by x-ray crystallography at reasonable resolution (<2.7Å), for a protein with full (>98%) sequence identity.
No bound ligands or proteins on the surface of the receptor. We verified that no ligands are bound to the protein surface, in particular not to the binding sites (e.g., for the PeptiDB entry 1T4D (mdm-x), we could not find a corresponding structure of the free mdm2 molecule without any ligand that had been solved by crystallography, and therefore we removed it from our set).
No crystal contacts that stabilize the binding site. Inspection of the symmetry mates in the solved crystal structures revealed that in certain cases the binding site of the free conformation binds to a peptide stretch of a copy of the protein in the crystal lattice (see for example PeptiDB entry 1DDW, the evh1 domain in the homer protein). In these cases, the peptide-binding site might have been arranged in a similar way as the bound conformation, and this might bias our protocol.
Biological unit of protein. In order to simulate accurately the accessible surface to the peptide, we included in the dataset the biological unit of the protein, e.g. a homodimer (see for example PeptiDB entry 2DS5 of the CLPX protease Zn binding domain).
The resulting set for assessment is detailed in Table 1A. To assess the robustness of this protocol, we compiled an additional set by extracting peptide-protein complex structures that were released to the Protein Data Bank (PDB 12) after the development of PeptiMap (between the dates 1.1.2013–8.4.2013) and filtered according to the same guidelines as detailed above. Only protein receptor structures not structurally similar to the entries in the training set were retained (i.e. distinct CATH domain 27). This validation set is detailed in Table 1B.
Table I.
Benchmark and validation sets used for optimization and evaluation of PeptiMap peptide binding site identification
| (A) Initial Calibration Benchmark (Extracted from PeptiDB 26; n=21) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Name | bound | unbound | Peptide sequence | Cath domain | domains/multimer | treatment | rank of hit | Pepsite218 ranks |
| dystrophin (WW) | 1EG4A | 1EG3A_1 | NMTPYRSPPPYVPa | 2.20.70.10 (b; 2x1.10.238.10) | 1 of 4 different domains (47-84) | split: only first domain used | 2,3 | 6,8,10 |
| sh2a1 (SH2) | 1D4TA | 1D1ZA | KSLTIYAQVQK | 3.30.505.10 | 1 | - | ||
| lsb3 sla1 (SH3) | 1SSHA | 1OOTA | GPPPAMPARPT | 2.30.30.40 | 2 | 1-6 | ||
| erbb2 (PDZ) | 1MFGA | 2H3LA | EYLGLDVPV | 2.30.42.10 | 1 | - | ||
| wdr5 (WD40) | 2H9MA | 2H14A | ARTKQT | 2.120.10.80 | central hole not accessible to peptide | mask internal sites | 3 | 1,3,5,6 |
| usp7 | 2FOJA | 2F1WA | GARAHSS | NA* (β-sandwich) | 1,2 | 1-6 | ||
| cyclophilin | 1AWR | 2ALFA | HAGPIA | 2.40.100.10 | 1 | 1-10 | ||
| p97 N-glycanase | 2HPLA | 2HPJA | DDLYG | NA* (p97) | 1 | - | ||
| traf2 | 1CZY | 1CA4A | ace-PQQATDD | 2.60.210.10 | weak homotrimer (ABC) | use monomerc | 3 | - |
| i-ap1 | 1JD5A | 1JD4A | AIAYFIPD | 1.10.1170.10 | weak homodimer (AB) | use monomer | 1,2 | 4 |
| gga1 | 1JWG_AC | 1JWFA | DEDLLHI | 1.25.40.90 | weak homodimer (AC - in bound structure) | use monomer | 2 | - |
| ntf2 | 1GYB_AB | 1GY7_AB | FSF | 3.10.450.50 | tight homodimer | unit: 2 chains |
1 - |
- |
| Clpx | 2DS8_AB | 2DS5_AB | ALRVVK | NA* (clpx) | tight homodimer, 2 sites | unit: 2 chains |
1,2 | - |
| calpain small subunit | 1NX1A | 1ALVAB | DAIDALSSDFT | 1.10.238.10 | tight homodimer central hole not accessible to peptide | unit: 2 chainsc |
- | - |
| ap2 | 2VJ0A | 1B9KA_1 1B9KA_2 |
PKGWVTFE FEDNFVP |
2.60.40.1030 3.30.310.30 |
2 domains | split: both have peptide bound |
- 3 |
- - |
| pim1 kinase transferase domain | 2C3I | 2J2IB_2 | KRRRHPSG | (3.30.200.20) 1.10.510.10 | 2 domains | split: only second used | 2 | - |
| cdk2 cyclin | 2CCHB | 1H1RB | HTLKGRRLVFDN | 1.10.472.10 x2 | 2 same domains | do not split | 3 | 1,6 |
| trypsin | 2AGEX | 1UTNA | Sin-AAPR | 2.40.10.10 x2 | 2 same domains | do not split | 1 | 6 |
| Pcna | 1RXZ | 1RWZA | KSTQATLERWF | 3.70.10.10 | 2 same domains | do not split | 2 | - |
| Endothiapepsin | 1ER8 | 4APEA | PFHLLVY | 2.40.70.10 | 2 same domains | do not split | 1,2,3 | 1-6 |
| (B) Validation set (Compiled from recent PDB releases 1-4/2013; n=9) | ||||||||
| Name | bound | unbound | Peptide sequence | Cath domain | domains/multimer | treatment | rank of hit | Pepsite2 ranks |
| γ2 adaptin Ear domain | 2YMT | 4BCXA | EWGPWV | 2.60.40.1230 | 1 domain | 3 | - | |
| βcop WD40 | 4J73 | 3MKQA | EAKKLV | 2x 2.130.10.10 + additional domain (based on 1VYH) | 3 domains central hole not accessible to peptides | split, only first domain (2-300); site 1 masked | 2,3 | 2,3,5,6 |
| RADa | 4B3B | 4A74A | aceFHTA | 3.40.50.300 (based on 1PZN) | weak homodimer | use monomer | 2,3 | - |
| FKBP35 | 4ITZA | 3NI6A | sinALPFnit | 3.10.50.40 (based on 1KT0A) | weak homodimer | use monomer | 1 | 1-10 |
| jnk1 kinase transferase domain | 3VUH | 3ELJ | PKRPTTLNLF | (3.30.200.20); 1.10.510.10 | 2 domains | split: only second domain | - | 1-10 |
| HIF alpha | 4B7E | 2W0X | EVVKLLLEHGADVLAQD | 2-305; 1.10.287.1010 | 2 domains | split: only first domain | 1,3 | 1,4,7-10 |
| MLH1 mut alpha c-term | 4FMNA | 4E4WA | VRSKYFK | NA* | multidomain | too big without splittingc |
- No split information available |
- |
| demethylase | 4FWF | 4FWE | ARTMQTARKSTGG | (domain A02: 280-405 & 522-836, based on 2V1DA) | multidomain | split: only second domain | 1 | - |
| ck2 kinase | 4IB5 | 3BQC | GCRLYGFKIHGCG | 3.30.200.20; (1.10.510.10) | 2 domains | split: only first domain | 3 | - |
Peptide residues accurately mapped by fragments are highlighted in bold and underlined; Peptide residues with area covered by fragments are highlighted in bold but not underlined
XX: unassigned domains
Better prediction obtained with SCOP classification (1CA4: 1,2 instead of 3), or domain parser (1ALV)
No domain assignment available based on CATH
Definition of the functional unit that will be mapped
Similar to its experimental counterpart, Structure-Activity-Relationship (SAR)-NMR 28, our computational solvent mapping approach is aimed at targeting distinct “physiological units”, namely globular domains that form a stable unit and together act as a receptor. As an example, a tight dimer receptor will be defined as one physiological unit to map. On the other hand, when a protein is composed of distinct domains that can move one relative to the other, we expect each to act as an individual binding site to a peptide, and therefore each should be treated as a distinct physiological unit. Based on this rationale, we have devised the following rules by which we define the individual units that are separately mapped:
For “tight” multimers (ratio of buried surface area of the monomer >0.2, see for example 1GY7), the individual unit is defined as the full multimer.
If a protein consists of more than one domain, all of the same class (i.e. repeated domains), we do not split the protein into individual domains, but rather treat it as one individual unit.
If the protein consists of different domains, split these into individual domains and perform solvent mapping separately on the domain that is known to be critical for the interaction (if such information is not available, each domain should be screened separately).
Separation of receptor structure into distinct domains
In order to map the fragments onto individual domains, the protein receptor structure was decomposed into these domains based on CATH domain classification (v3.4). In cases where no CATH classification was available, we identified the most similar CATH domain using sequence alignments.
Detailed outline of protocol
The protocol consists of a series of steps described below. First, the “physiological unit” of the protein receptor is defined (as detailed above). The next steps (2–7) are identical to the FTsite protocol used to identify protein and ligand binding sites 24. The final steps (8–10) include the removal of sites involved in domain interactions, the merging of adjacent clusters, and finally the filter of internal sites not accessible to the peptide. Here we summarize the protocol shortly, and where appropriate highlight the changes specific to peptide binding site identification that were incorporated into PeptiMap. Steps new to PeptiMap are highlighted in bold.
Step 1
Selection of “physiological unit”. Decomposition is based on annotation of protein domains according to CATH (the rules for decomposition are outlined above and more detailed in the Results section).
Step 2
Grid-based sampling of the protein surface with FFT. All bound ligand water molecules and other ligands are removed prior to the calculations. We then sample the protein surface for 16 small molecule probe types 22. This is done using exhaustive sampling with grid-based Fast Fourier Transform (FFT) (109 docked probe positions). The best 2,000 poses with the lowest energies for each probe type are retained.
Step 3
Post-FFT clustering to discard spurious probe clusters. For each probe type, the 2000 retained poses are clustered using a simple greedy algorithm. We select the lowest energy pose as the center of the first cluster, and add all poses within 4Å center-to-center distance from it as cluster members 29. All clustered poses are removed, and we repeat the same steps to form the second and then the subsequent clusters until all poses are clustered. Clusters with less than 10 probes are removed, and the 6 largest clusters are retained for further analysis.
Step 4
Minimization and re-scoring. The energy of each retained protein-probe complex is minimized using the CHARMM 30 potential with the Analytic Continuum Electrostatic (ACE) model representing the electrostatics and solvation terms as implemented in version 27 of CHARMM. The algorithm uses the polar-hydrogen-only parameter set from version 19 of CHARMM. The energy minimization is performed using a limited memory Broyden–Fletcher–Goldfarb–Shannon (L-BFGS) method in which heavy atoms of the protein are held fixed, while the polar hydrogen atoms of the protein and all atoms of the probes are free to move. Poses with positive energies after minimization are discarded.
Step 5
Generating consensus clusters. Following the energy minimization we re-cluster the resulting probe poses. As in Step 2, we select the lowest energy pose as the center of the first cluster, but use 4 Å full-atom pairwise RMSD as the clustering radius. After all probes are clustered and clusters with less than 10 members are discarded, the clusters are ranked on the basis of the Boltzmann averaged energy, and the 6 lowest energy clusters are retained for every probe type. Consensus clusters are generated by grouping probe clusters with cluster centers within 4 Å. The centers of the resulting consensus clusters are fixed, and the probe clusters are re-distributed such that each cluster center is closer to the center of its own consensus cluster than to the center of any other consensus cluster. Consensus clusters that overlap with an integral element of the intact protein such as heme are discarded. A consensus cluster is considered to overlap with a co-factor if their volume overlap exceeds 80% of the consensus cluster.
Step 6
Ranking consensus clusters. The algorithm ranks the consensus clusters by the number of non-bonded contacts between the protein and all probes of the consensus cluster. A residue of the protein and a probe are considered to be in contact if any atom of the residue is less than 4 Å from any atom of the probe. A residue is considered to be in contact with a consensus cluster if it is in contact with any of its probes. After selecting the contact residues for a consensus cluster we reevaluate the number of contacts by adding also interactions with probes that are within 4 Å but are not part of the original consensus cluster. The resulting numbers are normalized using the overall number of contacts for all probes, and used for ranking the consensus clusters.
Step 7
Identification of putative peptide binding sites. To identify the putative binding site, the algorithm first selects the consensus cluster with the highest number of contacts. This cluster is then expanded by adding any neighboring consensus cluster if the center of any of its probe is closer than 3.5 Å to the center of any probe in the consensus cluster. The protein residues that are within 4 Å of the expanded consensus cluster constitute the top prediction of the binding site. The first consensus cluster is then removed, and the procedure is repeated using the next consensus cluster with the highest number of contacts to identify lower ranked predictions of the peptide-binding site.
Step 8
Discard sites located in non-accessible regions in the protein. Sites at the domain interface (in cases of proteins that were split into individual domains): If a putative site clashes significantly with secondary structures (α-helices or β-sheets) on the partner domain(s), the site is discarded. A putative site is discarded if the site is within 3.0 Å of one or more secondary structure-associated (helices or sheets) amino acids on partner domain(s) as defined by PyMol. This removes sites that are involved in domain-domain interactions.
Step 9
Expand final sites that are retained. Each of the sites is expanded by adding to the site probe cluster representatives that are not already part of the site but are closer than 4.0 Å (atom-atom distance) to probe cluster representatives that are already part of the site. Probe cluster representatives that are already members of other sites are not used and, therefore, the sites are only expanded and not co-joined.
Step 10
Remove inaccessible sites within the protein core: While the small molecules that are used to probe the protein surface may access internal cavities due to their small size, peptides are larger and therefore cannot reach these voids. At the same time such sites are attractive to probes since they provide much larger contact areas to a small molecule than the surface (and indeed they are good ligand binding sites). To adequately analyze peptide binding surface, such sites should therefore be excluded from mapping. In order to identify internal ligand binding sites, we building 100 rays uniformly covering a sphere 31 from the center of the site, and identify which of those contact the protein (a contact is defined if a ray passes within 2 Å from the center of any atom of the receptor). A binding pocket is considered internal if 80% of the rays contact the protein. Internal sites are masked and steps (2–10) are repeated.
Assessment of performance: criteria for binding site identification
Since peptide binding site identification criteria have not yet been established, we have employed accepted criteria used for ligand binding site identification 32. This criterion requires the geometric center of the predicted ligand-binding site to lie within 4.0 Å of any peptide atom. This allows easy comparison to other approaches. Residues mapped by PeptiMap are shown in bold in Table I, and residues for which side chain atoms have been accurately mapped at the same spatial position (i.e. within 2 Å).
Results
Compilation of curated benchmark set of peptide-protein complexes for peptide binding site prediction
In order to allow the objective assessment of peptide binding site prediction, we compiled a set of protein structures of a free receptor that do not contain any other molecule at the peptide binding site (i.e. no other bound ligands, and no crystal contacts, see Methods for more detail). We then parsed the structures into domains and identified their biological units. Based on these, we defined the receptor structure and the surface to mapped for peptide binding sites. The same procedure was repeated on a second set compiled after this calibration (see Methods). The total of 30 peptide-protein complexes are detailed in Table IA (benchmark of 21 interactions) & Table IB (additional validation set of 9 interactions).
We would like to note that while the optimization set, and more so the test set, are rather small, they are non-redundant and therefore do represent a wide variety of different peptide binding domains. More importantly, considerable efforts were made to verify that these sets are clean and not biased to any bound conformation. In a real-world scenario the available template structure might well bind a symmetry mate or a ligand in the binding site – thus PeptiMap is expected to perform even better.
Adaptation of FTsite to prediction of peptide binding sites
Computational fragment mapping can accurately identify small ligand binding sites 24, as well as locate druggable protein-protein interfaces 25. Peptides lie in between these two cases: In contrast to ligand binding sites, peptide binding sites tend to be more shallow and the pockets to be smaller. In contrast to protein binding sites, peptides tend to bind to smaller regions and with usually weaker affinity. We wanted to optimize PeptiMap to specifically identify peptide-binding sites (noting that such modifications might improve predictions of binding sites of larger ligands as well). Two major modifications were necessary to provide a robust protocol for peptide binding site location on protein structures, namely (1) the decomposition of protein structures into physiological, functional units, and (2) filtering out internal ligand binding sites not accessible to peptides.
(1) Decomposition of protein structures into physiological, functional units
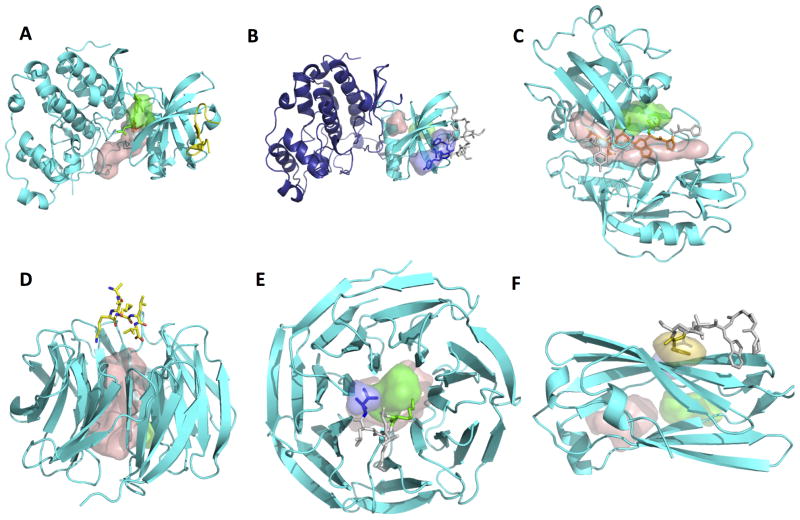
Assuming that a peptide will bind to an organized binding site, we decompose the protein structure into independent parts that represent the stable functional unit a peptide might encounter. Splitting a protein structure into individual domains prevents the identification of short-lived crevices at the boundary of the domain interface as peptide binding sites, and reduces the surface to be sampled (see Table I, and Figures 1A&B for the example of ck2 kinase, pdb ID 3BQC).
Figure 1. Examples of predictions of peptide binding sites using PeptiMap.
The same color-coding scheme is used in all figures: The receptor is shown as a cartoon colored in cyan, while the different predicted sites are shown in surface representation (top-ranking site – wheat; site 2 – green; site 3 - dark blue). The peptide is shown in stick representation and colored according to the predicted binding sites (residues that do not contact any predicted site are in light grey). Pdb IDs of the apo and bound structures are indicated in parentheses. Predictions were made on the apo structures; bound structures are for validation purposes only.
(A, B) Mapping on single domains improves peptide binding site prediction on ck2 kinase (apo: pdb ID 3BQC chain A): (A) mapping on the full structure identifies merely a known ligand binding site (pdb ID 3U4U), while (B) mapping on the n-terminal kinase domain only identifies the peptide binding site (pdb ID 4IB5; the transferase domain not mapped here is shown in dark blue). This highlights how implementation of segmentation into single domains may improve in particular peptide-binding site predictions (while small ligands can be identified also on the full structure).
(C) Example of accurate PeptiMap prediction of the peptide binding site covering a substantial part of the peptide: Endothiapepsin peptide binding site prediction identifies the peptide binding site of all but one residue (apo: pdb ID 4APE chain A; bound: 1ER8). (D, E) Masking of internal ligand binding sites improves peptide binding site prediction on coatomer b subunit. (D) In the original prediction, mapping of the receptor structure (pdb ID 3MKQ chain A) identifies mainly peptide-inaccessible sites within the whole of the WD40 domain, but not the peptide binding site (pdb ID 4J73). (E) When entrance into the inner cavity is blocked, PeptiMap identifies correctly the location of the peptide-binding site. (F) Example of target failed by Peptimap. In case of the AP-2 complex subunit alpha (CATH domain 2.60.40.1030; pdb ID 1B9K), the peptide binding site (from pdb ID 2VJ0) is not identified by the top 3 predictions, only by prediction ranked 4th (in yellow).
In contrast, tight homo-multimers (e.g. Clpx, pdb ID 2DS5), as well as repeated same domains within a protein (that in general also form tight interactions) are merged into one functional unit for mapping (e.g. Figure 1C that shows Endothiapepsin chain A that is composed of two identical domains according to the CATH classification).
(2) Filter out internal binding sites
Fragment mapping will identify among others also ligand-binding sites within proteins that are not accessible to the larger peptide ligands. Removal of such sites using ray tracing (see Methods section) improved predictions of peptide binding sites of WD40 domains that contain a hole at the center of the protein not accessible to peptides (Figures 1D & 1E show how ranking is significantly improved by masking inaccessible internal ligand binding sites in the protein cop b, pdb ID 3MKQ chain A), and similar results were also observed on additional cases of known internal ligand binding sites (results not shown).
PeptiMap accurately locates the peptide-binding site on most of the receptor structures in both the benchmark and validation sets, and compares very favorably to other approaches
Using this streamlined protocol, PeptiMap identified 10 out of 21 (50%) peptide binding sites in the benchmark as the top-ranking prediction. More importantly, it failed only in two cases to identify the site among the top-3 ranking predictions (19/21 success rate; 90%). In the independent subsequent validation, a lower performance was observed for top-ranking sites: 3/9 (33%) binding sites were top-ranked. However, again only two cases were not identified among the top-3 ranked predictions (7/9 successes; 78%). Details of performance for each of the proteins are given in Table I. These results demonstrate the general applicability of the protocol to a range of different peptide-protein complexes, involving many different folds as well as many different functional classes.
The predicted binding sites are in many cases very accurately mapped. Figure 1C shows a particularly successful prediction: for Endothiapepsin, the accurate location of six out of seven peptide residues is identified by PeptiMap. Overall, 1 to 6 peptide residue positions (median of 2) are identified by PeptiMap (these residues are highlighted for each peptide in Table I). It should be noted that this covers a significant part of the peptide residues that directly contact the receptor.
We compared our protocol with other available approaches. PepSite is the only approach that can be tested and validated via a server. The coverage of this approach is rather restricted (due to limited availability of enough structures to create the structural PSSMs). We ran PepSite (version2 18) on the same dataset in order to compare the two approaches. PepSite identified only 6 out of 21 peptide binding sites in the benchmark set within the top1 predicted sites (same results for top3 assessment). The corresponding performance for the validation set of 9 cases was 3 top-ranking predictions and 4 predictions ranked 1–3.
The recently published approach based on the BRIX database of interacting fragments 19 reports results for a set of protein-peptide complexes. Among these we could find predictions for two of the proteins assessed here: for p97 N-glycanase (2HPJ) this approach failed to identify the binding site, while an acceptable prediction was reported for PCNA (1RWZ), albeit not top-ranked.
Discussion and Conclusion
PeptiMap is a new, accurate and robust approach for peptide binding site detection on protein receptor surfaces. It is based on the successful computational fragment mapping approach previously applied to ligand site prediction 24 and the detection of druggable protein interactions 25. Overall the results presented here are promising and indicate that automated and efficient prediction of peptide binding sites on proteins is coming of age. Not only is the peptide binding site on the receptor surface identified for most of the cases among the top-3 ranking structures (26/30), but also the predictions are accurate and identify many of the important peptide binding residues. Furthermore, our results here suggest that PeptiMap clearly outperforms other available approaches, thanks to its general applicability and accurate prediction.
In the following we shortly assess the challenges that need to be addressed to further extend performance of PeptiMap and suggest several ways to do so, based on the constraints of this approach and the cases where it still fails (see e.g. Figure 1F for a peptide-binding site identified only by the prediction ranked 4th).
Possible strategies to improve Peptimap
Focused mapping: We have shown in several examples that once a site has been identified, focused local mapping can improve the coverage of the site, and provide general guidelines regarding peptide sequence preference 7,33.
Improved definition of functional units: Preliminary results indicate that performance can significantly be affected by how the functional units are defined, in particular how the individual domain boundaries are determined. While CATH-based domain definition provides overall adequate functional units, in some cases we noticed that different definitions can dramatically improve performance (see Table I). In particular for large proteins, when CATH provides no domain definition or does not split the protein into distinct domains, PeptiMap may fail to identify the peptide-binding site (e.g. pdb ID 4E4W chain A and 1ALV chain B, see Table I). In such cases, domain mapping based on alternative tools such as domainparser 34, or on visual inspection could define a subdomain that is useful for peptide binding site detection (results not shown).
Combination of PeptiMap with other approaches to characterize structure and specificity of peptide-protein interactions
Currently, PeptiMap predicts the location of peptide binding sites, but does not provide any information about actual structure of the peptide within this site, nor about possible sequences for a binding peptide. In order to proceed to a full model of the peptide-receptor complex, PeptiMap predictions could serve as input for peptide-protein docking protocols 13,35. We plan to incorporate PeptiMap into a scheme for FlexPepDock 14,16 that will allow for full ab initio prediction of peptide-protein complex structures starting from a given peptide sequence and a free receptor structure. In addition, while PeptiSite and the BRIX-based approach are less general, they do provide more information in their prediction, such as sequence-specific binding site identification, as well as an approximate structure of the peptide – protein complex. Therefore, combining information from different protocols that use rather complimentary approaches will ultimately improve our knowledge and understanding of peptide-mediated interaction and their structural basis.
Implications for globular protein-protein docking
In a previous study we have suggested that a significant fraction of globular protein-protein interactions is mediated by one linear, peptidic stretch that contributes most of the binding energy (e.g. a dominant loop at the interface) 36. Moreover, this study indicated that when bound to the protein partner, this peptide tends to adopt a structure that is very similar to the one adapted within the protein context. Consequently, PeptiMap could be used in a more general way to identify on a protein surface where such dominant peptidic stretches would bind. While FTmap was shown to identify druggable sites on protein interfaces 25, PeptiMap is expected to be very useful for the design of specific peptide-derived inhibitors of protein interactions.
This study demonstrates the strength of non-biased, ab initio prediction protocols for finding molecular recognition sites of peptides. The general applicability of such an approach will substantially contribute to improved characterization of a range of peptide-mediated interaction, and provides thus a good starting point for structure-based characterization of biological interaction and function. A server is under development to make PeptiMap generally available (and the PeptiMap software is freely available to academic users upon request).
Acknowledgments
This work was funded by the USA-Israel Binational Science Foundation, grant number 2009418 (jointly to DK & OSF); DK was also supported by NIH grants GM93147, GM064700, GM61687, NSF DBI1047082, and Russian Ministry of Education and Science (grant number 14.A18.21.1973); OSF was also supported by the Israel Science Foundation, founded by the Israel Academy of Science and Humanities (grant number 319/11) and by the European Research Council under the ERC Grant Agreement #310873.
References
- 1.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300(5618):445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 2.Akiva E, Friedlander G, Itzhaki Z, Margalit H. A dynamic view of domain-motif interactions. PLoS Comput Biol. 2012;8(1):e1002341. doi: 10.1371/journal.pcbi.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petsalaki E, Russell RB. Peptide-mediated interactions in biological systems: new discoveries and applications. Curr Opin Biotechnol. 2008;19(4):344–350. doi: 10.1016/j.copbio.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Van Roey K, Gibson TJ, Davey NE. Motif switches: decision-making in cell regulation. Curr Opin Struct Biol. 2012;22(3):378–385. doi: 10.1016/j.sbi.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Morrison KL, Weiss GA. Combinatorial alanine-scanning. Curr Opin Chem Biol. 2001;5(3):302–307. doi: 10.1016/s1367-5931(00)00206-4. [DOI] [PubMed] [Google Scholar]
- 6.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Sci STKE 2004. 2004;219:l2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 7.Zerbe BS, Hall DR, Vajda S, Whitty A, Kozakov D. Relationship between hot spot residues and ligand binding hot spots in protein-protein interfaces. J Chem Inf Model. 2012;52(8):2236–2244. doi: 10.1021/ci300175u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu X, Mitchell JC. KFC2: a knowledge-based hot spot prediction method based on interface solvation, atomic density, and plasticity features. Proteins. 2011;79(9):2671–2683. doi: 10.1002/prot.23094. [DOI] [PubMed] [Google Scholar]
- 9.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267(5196):383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 10.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450(7172):1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 11.London N, Raveh B, Schueler-Furman O. Druggable protein---protein interactions --- from hot spots to hot segments. Curr Opin Chem Biol. 2013 doi: 10.1016/j.cbpa.2013.10.011. under review. [DOI] [PubMed] [Google Scholar]
- 12.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, Zardecki C. The Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 61):899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 13.London N, Raveh B, Schueler-Furman O. Peptide docking and structure-based characterization of peptide binding: from knowledge to know-how. Curr Opin Struct Biol. 2013 doi: 10.1016/j.sbi.2013.07.006. In Press. [DOI] [PubMed] [Google Scholar]
- 14.Raveh B, London N, Schueler-Furman O. Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins. 2010;78(9):2029–2040. doi: 10.1002/prot.22716. [DOI] [PubMed] [Google Scholar]
- 15.London N, Raveh B, Cohen E, Fathi G, Schueler-Furman O. Rosetta FlexPepDock web server--high resolution modeling of peptide-protein interactions. Nucleic Acids Res. 2011;39(Web Server issue):W249–253. doi: 10.1093/nar/gkr431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raveh B, London N, Zimmerman L, Schueler-Furman O. Rosetta FlexPepDock ab-initio: simultaneous folding, docking and refinement of peptides onto their receptors. PLoS One. 2011;6(4):e18934. doi: 10.1371/journal.pone.0018934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petsalaki E, Stark A, Garcia-Urdiales E, Russell RB. Accurate prediction of peptide binding sites on protein surfaces. PLoS Comput Biol. 2009;5(3):e1000335. doi: 10.1371/journal.pcbi.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trabuco LG, Lise S, Petsalaki E, Russell RB. PepSite: prediction of peptide-binding sites from protein surfaces. Nucleic Acids Res. 2012;40(Web Server issue):W423–427. doi: 10.1093/nar/gks398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verschueren E, Vanhee P, Rousseau F, Schymkowitz J, Serrano L. Protein-Peptide Complex Prediction through Fragment Interaction Patterns. Structure. 2013;21(5):789–797. doi: 10.1016/j.str.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 20.Hajduk PJ, Huth JR, Fesik SW. Druggability indices for protein targets derived from NMR-based screening data. J Med Chem. 2005;48(7):2518–2525. doi: 10.1021/jm049131r. [DOI] [PubMed] [Google Scholar]
- 21.Mattos C, Ringe D. Locating and characterizing binding sites on proteins. Nature biotechnology. 1996;14(5):595–599. doi: 10.1038/nbt0596-595. [DOI] [PubMed] [Google Scholar]
- 22.Brenke R, Kozakov D, Chuang GY, Beglov D, Hall D, Landon MR, Mattos C, Vajda S. Fragment-based identification of druggable ‘hot spots’ of proteins using Fourier domain correlation techniques. Bioinformatics. 2009;25(5):621–627. doi: 10.1093/bioinformatics/btp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins. 2006;65(2):392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 24.Ngan CH, Hall DR, Zerbe B, Grove LE, Kozakov D, Vajda S. FTSite: high accuracy detection of ligand binding sites on unbound protein structures. Bioinformatics. 2012;28(2):286–287. doi: 10.1093/bioinformatics/btr651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozakov D, Hall DR, Chuang GY, Cencic R, Brenke R, Grove LE, Beglov D, Pelletier J, Whitty A, Vajda S. Structural conservation of druggable hot spots in protein-protein interfaces. Proc Natl Acad Sci U S A. 2011;108(33):13528–13533. doi: 10.1073/pnas.1101835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.London N, Movshovitz-Attias D, Schueler-Furman O. The structural basis of peptide-protein binding strategies. Structure. 2010;18(2):188–199. doi: 10.1016/j.str.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 27.Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, Sillitoe I, Yeats C, Thornton JM, Orengo CA. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35(Database issue):D291–297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274(5292):1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 29.Kozakov D, Clodfelter KH, Vajda S, Camacho CJ. Optimal clustering for detecting near-native conformations in protein docking. Biophys J. 2005;89(2):867–875. doi: 10.1529/biophysj.104.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. Journal of computational chemistry. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yershova A, Jain S, Lavalle SM, Mitchell JC. Generating Uniform Incremental Grids on SO(3) Using the Hopf Fibration. Int J Rob Res. 2010;29(7):801–812. doi: 10.1177/0278364909352700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang B, Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golden MS, Cote SM, Sayeg M, Zerbe BS, Villar EA, Beglov D, Sazinsky SL, Georgiadis RM, Vajda S, Kozakov D, Whitty A. Comprehensive experimental and computational analysis of binding energy hot spots at the NF-kappaB essential modulator/IKKbeta protein-protein interface. J Am Chem Soc. 2013;135(16):6242–6256. doi: 10.1021/ja400914z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, Xu D, Gabow HN. Protein domain decomposition using a graph-theoretic approach. Bioinformatics. 2000;16(12):1091–1104. doi: 10.1093/bioinformatics/16.12.1091. [DOI] [PubMed] [Google Scholar]
- 35.Trellet M, Melquiond AS, Bonvin AM. A unified conformational selection and induced fit approach to protein-peptide docking. PLoS One. 2013;8(3):e58769. doi: 10.1371/journal.pone.0058769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.London N, Raveh B, Movshovitz-Attias D, Schueler-Furman O. Can self-inhibitory peptides be derived from the interfaces of globular protein-protein interactions? Proteins. 2010;78(15):3140–3149. doi: 10.1002/prot.22785. [DOI] [PMC free article] [PubMed] [Google Scholar]