Abstract
Proteins that communicate signals from the cytoskeleton to the nucleus are prime targets for effectors of metastasis as they often transduce signals regulating adhesion, motility, and invasiveness. LIM domain proteins shuttle between the cytoplasm and the nucleus, and bind to partners in both compartments, often coupling changes in gene expression to extracellular cues. In this work, we characterize LIMD2, a mechanistically undefined LIM-only protein originally found to be overexpressed in metastatic lesions but absent in the matched primary tumor. LIMD2 levels in fresh and archival tumors positively correlate with cell motility, metastatic potential, and grade, including bladder, melanoma, breast, and thyroid tumors. LIMD2 directly contributes to these cellular phenotypes as shown by overexpression, knockdown, and reconstitution experiments in cell culture models. The solution structure of LIMD2 that was determined using nuclear magnetic resonance revealed a classic LIM-domain structure that was highly related to LIM1 of PINCH1, a core component of the integrin-linked kinase–parvin–pinch complex. Structural and biochemical analyses revealed that LIMD2 bound directly to the kinase domain of integrin-linked kinase (ILK) near the active site and strongly activated ILK kinase activity. Cells that were null for ILK failed to respond to the induction of invasion by LIMD2. This strongly suggests that LIMD2 potentiates its biologic effects through direct interactions with ILK, a signal transduction pathway firmly linked to cell motility and invasion. In summary, LIMD2 is a new component of the signal transduction cascade that links integrin-mediated signaling to cell motility/metastatic behavior and may be a promising target for controlling tumor spread.
Introduction
Defining the complex biology and the cascade of events that lead to metastatic spread of primary tumors, both locally and to distant sites, continue to be major unmet needs in cancer biology (1). Moreover, defining which molecular events in both the metastatic cascade and in the maintenance of tumor dormancy are targetable for therapeutic or preventative benefit is an even more daunting task. These results have defined molecules that control a large array of cellular phenotypes, including cell motility, cell–cell and cell–matrix interactions, and immune evasion (2, 3). Generally, it seems that rare metastatic variants appear stochastically in a genetically heterogeneous primary tumor, occurring quite early in tumor progression, and that normal developmental processes, such as epithelial–mesenchymal transition (EMT), mesenchymal-epithelial transition (MET), and angiogenic cascades, are often ectopically activated to achieve tumor spread (1, 4, 5). Both forward genetic and descriptive experimental approaches have been utilized to identify genetic and epigenetic determinants for metastatic capability, largely by selection and analysis of metastatic variants in populations, or by comparing the expression and mutation profiles of matched primary and metastatic lesions using the vast spectrum of “-omic” technologies currently popular (6–8). The finding of metastasis-associated antigens and transcriptional and/or genetic signatures in these comparisons has been a useful exercise, and may be useful in the clinic, but these approaches often fail to distinguish drivers of the metastatic phenotype from passenger/markers of the phenotype and, moreover, rarely led to specific mechanisms. In this study, we characterized the LIMD2 protein, which originally identified as highly and exclusively overexpressed in metastatic lesions but absent in matched normal tissue or primary tumor (9).
LIMD2 is a LIM-only domain protein that was identified as a biomarker for papillary thyroid carcinoma (PTC) lymph node metastasis (LNM) from molecular profiling of matched samples (9). LIMD2 was found to be highly expressed in LNM but absent from the primary tumor or normal thyroid tissue in matched patient PTC samples, suggesting that LIMD2 expression could provide an improved means of detecting potentially metastatic PTC cells during initial staging of a newly diagnosed carcinoma. In the human genome, there are 135 identifiable LIM-encoding sequences located within 58 genes. The LIM domain is organized as a tandem zinc-finger structure that functions as a modular protein-binding interface (Fig. 1A). LIM domain–containing proteins have diverse cellular roles such as regulators of gene expression, cyto-architecture, cell adhesion, cell motility, and signal transduction. LIM domain proteins are emerging as key molecules in a wide variety of human cancers (10). In particular, all members of the human LIM domain–only (LMO) proteins, LMO1 to LMO4, which are required for many normal developmental processes, are implicated in the onset or progression of several cancers, including T-cell leukemia, breast cancer, and neuroblastoma. Here, we report that LIMD2 regulates cell motility and is a novel effector of tumor progression via its role in the integrin-linked kinase (ILK) pathway.
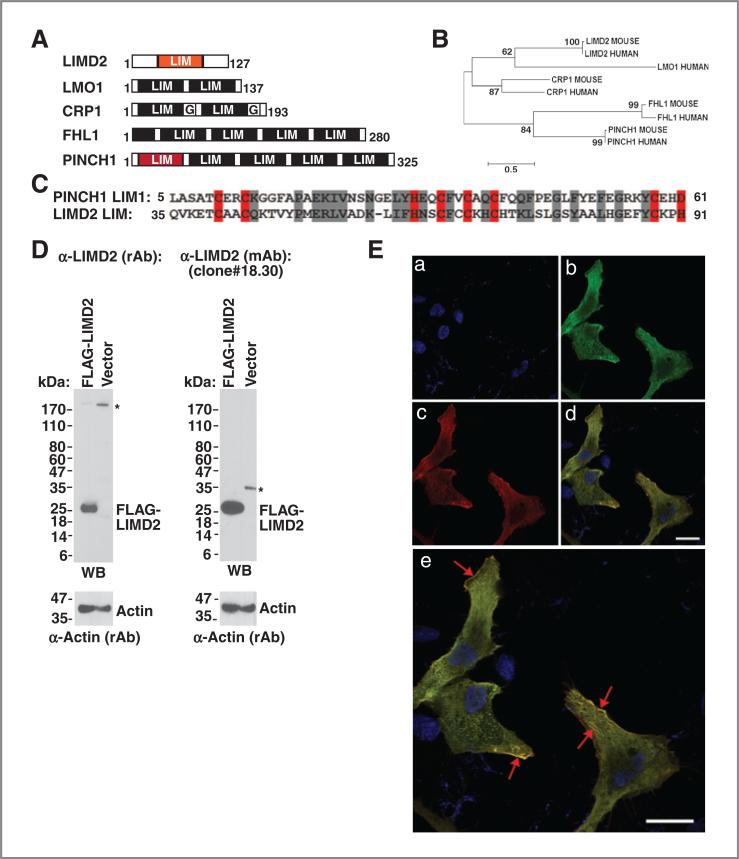
Figure 1.
A, LIMD2 and the LIM-only protein family. B, LIMD2 is most closely related to CRP1. C, the PINCH1-LIM1 and LIMD2 LIM domains are homologous. The zinc-chelating residues are highlighted in red; the conserved amino acids are highlighted in gray. D, antibodies robustly detect LIMD2 protein. *, nonspecific binding. E, TPC1 cells were transfected with myc-LIMD2 then fixed and stained with both anti-myc tag antibody (red) or anti-LIMD2 mAb (green). The cells were counterstained with DAPI to highlight the nucleus (blue). The red arrows indicate concentrations of Myc- and LIMD2 costaining signal, which is at the leading edge of the cells and also is present in streaks reminiscent of vinculin-containing adhesion plaques.
Materials and Methods
Plasmids, antibodies, cell culture, transfection, migration assays, soft-agar assays, GST-binding assays, immunofluorescence, and immunohistochemistry
The plasmids pET-28a(þ)-LIMD2, pGEX-4T1-LIMD2, pcDNA3.1-Flag-LIMD2, pcDNA3.1-Flag-LIMD1, and pcDNA3.1-Flag-AJUBA were constructed through PCR-based cloning (cloning details available on request). Sport6-LIMD2 (human) and pLKO.1-shLIMD2 (human) plasmids were purchased from Open Biosystems. pET-32-TT-ILK, 1–174 (His), and pCOLA-Duet-PINCH1-LIM1 (GST) plasmids are kindly provided by David Calderwood and Titus Boggon (11). Preparation of the His-LIMD2 and GST-LIMD2 fusion proteins were performed as described previously (12) and yielded soluble protein purified under native conditions. The DNA region encoding human LIMD2 amino acids 1 to 127 was isolated via PCR with synthetic oligonucleotides containing 5’ BamH1 and 3’ HindIII and the digested DNA fragment cloned into pQE30 vector at the corresponding restriction sites. This produced a protein that encoded the N-terminal vector-encoded peptide of MRGSHHHHHHGS-. Followed by AA1-127 of hLIMD2 followed by a stop codon. This was used to produce rabbit polyclonal antibody using standard techniques. For monoclonal antibody, the DNA region encoding LIMD2 (AA1-127) was isolated via PCR with synthetic oligonucleotides containing 5’ EcoRI and 3’ XhoI, and the digested DNA fragment cloned into pGEX4T-1 vector at the corresponding restriction sites. This produced a protein that encoded the N-terminal vector-encoded GST, followed by AA1-127 of hLIMD2 followed by a stop codon. The mAb used in this study was given the Wistar designation “LIMD2 clone #18.30” and was immunoglobulin G (IgG) iso-type. Both sources of crude antibodies were affinity purified using either the T-Gel or the SulfoLink Kits (Pierce) using full-length, native LIMD2 protein coupled to the resin.
HEK293, U2OS, MCF-7, MDA-MB-231, MDA-MB-435, and mouse fibroblast (ILKþ/þ and ILK −/− ) cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% FBS, 2 mmol/L l-glutamine, and penicillin (50 units/ mL)/streptomycin (50 μg/mL; P/S) at 37° C under 5% CO2 in a humidified chamber. MCF-10A mammary epithelial cells were maintained in DMEM:Ham's F12 containing 5% house serum, epidermal growth factor (10 ng/mL), hydrocortisone (0.5 mg/mL), cholera toxin (100 ng/mL), insulin (10 μg/mL), and P/S. Melanoma cell lines (Fom196-1, RGP-WM35, RGPWM3211, VGR-WM793, and Met1205 Lu) were maintained in Tu 2% medium containing 80% MCDB153, 20% Leibovitz's L-15, 2% FBS, insulin (bovine; 5 mg/mL), and 1.68 mmol/L CaCl2. Bladder cancer cell lines (T24 and RT4) were maintained in McCoys 5A containing 10% FBS, 1.5 mmol/L l- glutamine, 2.2 g/L sodium bicarbonate, and P/S. Bladder cancer cell lines (HT1376, UM-UC3, ScaBer, J82, and TCCSUP) were cultured in Eagle's Minimum Essential Medium containing 10% FBS, 2 mmol/L l-glutamine, 1.5 g/L sodium bicarbonate, and P/S. Thyroid cancer cell lines (XTC-1, FTC133, FTC236, FTC238, and TPC-1) were maintained in DMEM containing 10% FBS, thyroid-stimulating hormone (TSH; bovine; 0.01 units/mL), insulin (human; 10 μg/mL), P/S, and amphotericin B (250 ng/mL). Transient transfection in HEK293 and U2OS cells using lipofectamine were performed as described (13). Short hairpin RNA (shRNA)–mediated knockdown using lentiviral vectors was performed as described. The siRNAs directed against human LIMD2 were obtained from Open Biosystems. The infected cells were grown in puromycin selection medium for 1 week, and the pool was used for Western blotting and for functional assays. Whole cell extracts were prepared for Western blotting.
Cell migration assays were performed using standard trans-well chambers (8-μm pore size; Corning). The cells were serum starved for 4 hours before seeding for the transwell assay. After detachment with trypsin, the cells were washed gently with PBS and resuspended in serum-free medium. A 250 μL cell suspension (2 × 105 cells/mL) was added to the upper chamber. Triplicate wells were done for each assay. The complete medium was added to the bottom chamber. After overnight incubation, cells on the filters were fixed (glutaraldehyde) and stained with 0.5% toluidine blue. The number of migrated cells on each filter was quantified manually by counting all cells in 4 separate fields under × 10 magnification using a microscope. The mean number of cells in triplicate filters was calculated and is reported in each bar of the graph, ±SD for 1 experiment. Each experiment was repeated at least twice. T tests were performed and P values were obtained to compare the different cell lines used.
For immunofluorescence, TPC1 cells grown on sterile coverslips were transfected with pcDNA3.1 myc-LIMD2; 24 hours later the media was aspirated off and the cells were rinsed 2 times in 1 × PBS. The cells were fixed with 4% formaldehyde for 15 minutes at room temperature (RT). Then the cells were rinsed 3 times with 1 × PBS for 5 minutes each. The cells were permeated with 0.2% Triton X-100 for 5 minutes. The cells were rinsed 3 times with 1 × PBS for 5 minutes each. The cells were blocked for 1 hour in 5% goat serum. The cells were then incubated overnight at 4° C with the following dilutions of primary antibodies (made in 5% goat serum): 1:1,000 LIMD2 (mAb), 1:200 Myc-tag antibody (Cell Signaling Inc.). The cells were rinsed 3 times with 1 PBS. Then the cells were incubated with 1:1,000 dilution of secondary anti-mouse Alexa Fluor 594 and anti-rabbit Alexa Fluor 647 antibodies (diluted in 5% goat serum) for 30 minutes at RT (in the dark). The cells were rinsed 3 times with 1 × PBS for 5 minutes each. The cells were counterstained with DAPI for 2 minutes and rinsed 3 times in 1 × PBS for 5 minutes each. The coverslip was mounted on a glass slide with a drop of Prolong Gold anti-fade reagent and observed by confocal microscopy.
The soft agar assay was performed as described by the Wallert and Provost Lab. The base agar is composed of 0.5% agar, 1 × DMEM, and 10% FBS. The top agar is formed by mixing equal volume of 2 × DMEM, 20% FBS, 0.7% agar, and the cell suspension to give 1 × DMEM, 10% FBS, 0.35% agar, and 2 × 104 or 4 104 cells/well (6-well plate). The plates were incubated at 37° C in humidified incubator for 2 to 3 weeks. The cells were fed 2 times per week with cell culture media. The plates were stained with 0.5 mL of 0.005% Crystal Violet for 1 hour and the colonies in each well were quantified by counting 4 fields under 2 × magnification using a microscope. Each condition was done in triplicate and the average number of colonies per well is represented. Each experiment was repeated 3 times.
The GST association assays using in vitro transcribed and translated (IVT) protein was performed as described (12). Full-length, 35S-labeled ILK protein was used for binding to the bacterially produced GST fusion protein, and the association and washing buffer was 1 × radioimmunoprecipitation assay buffer. Standard IHC techniques were used to stain paraffin-embedded tissues, except that sections were subjected to antigen retrieval by pressure boiling in 10 mmol/L sodium citrate, 0.05% Tween 20, pH 6.0, for 3 minutes at 120° C. The sections were blocked with 1% normal horse serum for 30 minutes, incubated with αLIMD2 (mAb) overnight at 4° C (1:100 dilution of a 1 mg/mL IgG stock) and followed by 30 minutes incubation with horse anti-mouse biotinylated immunoglobulin (1:200). Subsequent steps followed the Vectastain Elite ABC Kit. The staining intensity and percentage of positive cells were scored using an ordinal scale blinded to the clinical and pathologic data. Staining intensity score was 0 for absent, 1 for faint, 2 for moderate, and 3 for strong. The percentage of positive cells score was 0 for 0%, 1 for less than 33%, 2 for ≥33% and ≤66%, and 3 for ≥66%.
Nuclear magnetic resonance spectroscopy
The hLIMD2 cDNA was cloned into pMAL-c5TEV (modified version of pMAL-c5X from NEB) and expressed in Escherichia coli BL21 (DE3). The cells were grown at 37° C until OD600 0.7 and then induced with 0.5 mmol/L of isopropylthio-β-galactoside at 25° C for 20 hours. The harvested cells were suspended in lysis buffer [20 mmol/L Tris pH 7.4, 200 mmol/L NaCl, 100 μmol/L ZnSO4, 0.2% IGEPAL, complete protease inhibitor (1 tablet per liter of the culture), 0.1 mg/mL lysozyme, 0.5 mmol/L phenylmethylsulfonyl fluoride, and 10 mmol/L β-mercaptoethanol]. The cells were disrupted by sonication, lysate was cleared by centrifugation at 18,000 rpm, and applied to amylose resin (NEB) equilibrated with the equilibrate buffer (20 mmol/L Tris pH 7.4, 200 mmol/L NaCl, 100 mmol/L ZnSO4, and 10 mmol/L b-mercaptoethanol) at 4° C for 1 hour. The resin was washed 5 times with 20 mL of equilibration buffer. MBP-fusion protein was eluted with 10 mmol/L maltose and cleaved by 100 μg of TEV protease per liter of the culture at 16° C overnight. The cleaved protein was subsequently purified by MonoS 4.6/100 PE and HiLoad 16/60 Superdex 75 pg [pre-equilibrated with 50 mmol/L phosphate pH 7.2, 100 mmol/L NaCl, and 100 μmol/L TCEP (Amersham Biosciences)]. Isotopically enriched LIMD2 was prepared from cells grown on minimal media (supplemented with 100 μmol/L of ZnSO4) containing 1 g/L of [15N] ammonium chloride with 2 g/L of either unlabeled glucose or [13C6] glucose (Cambridge Isotopes Laboratory). The ankyrin repeat domain (ARD; 1-174aa), kinase domains (182-452aa) of ILK were cloned into pET 32a (Invitrogen) and pGEX-6P1 (GE), respectively. Growth and induction were carried out as described above, except that the expression temperature was reduced to 15° C. The lysis buffer contained 50 mmol/L phosphate pH 7.2, 200 mmol/L NaCl, 10 mmol/L β-mercaptoethanol, and 10% glycerol. The protein used for nuclear magnetic resonance (NMR) was characterized by analytical ultracentrifugation and circular dichroism to determine its suitability for NMR.
Protein concentrations of LIMD2 (300–400 μmol/L) were used for NMR experiments for resonance assignment and structure calculation. The NMR spectra for assignment were collected in 90% H2O/10% D2O containing 50 mmol/L sodium phosphate, pH 7.2, 100 mmol/L NaCl, and 1 mmol/L TCEP at 600 MHz on a Varian INOVA spectrometer equipped with an HCN cold probe at 25° C. Backbone assignment was carried out using HNCACB and CBCA(CO)NH, whereas side chain assignments were determined using HCCH-TOCSY, CC(CO)NH, HBHA(CO)NH, as well as 2D (HB)CB(CGCD) HD and (HB) CB(CGCDCE)HE. The protonation state of the histidine residues was examined using a modified HMQC experiment (14). Distance restraints were derived from 15N-edited NOESY in 90% H2O/10% D2O, and 13C-edited NOESY and 2D NOESY in D2O. The spectra were processed by NMRPipe (15) and analyzed by SPARKY (16). The molecular dynamics based model of LIMD2–ILK–Parvin was obtained by AutoDOCK (17) analysis of fragmented LIMD2 structure against that of the pdb structure 3kmw (ILK/α-Parvin), rethreading the LIMD2 structure through the top 2 binding sites of the Autodock analysis and energy minimizations performed in water at a pH of 7.4.
The ILK kinase assay was performed as described previously (18) with modifications. ILK and LIMD2 were mixed and preincubated for 30 minutes at room temperature before addition of the substrate, myosin phosphatase target subunit 1 (MYPT1; Millipore). The amounts of ILK and MYPT1 were kept constant at 33.3 nmol/L (50 ng/20 μL reaction) and 417 nmol/L (500 ng/20 μL reaction), respectively, and increasing amounts of LIMD2 were added as indicated. Kinase reactions were initiated by the addition of ATP to a final concentration of 500 nmol/L. Reactions were carried out in 1 kinase reaction buffer (25 mmol/L Tris-HCl pH 7.5, 5 mmol/L β-glycerophosphate, 2 mmol/L dithiothreitol, 0.1 mmol/L Na3VO4, 10 mmol/L MgCl2) for 30 minutes at 30° C, terminated with the addition of sample buffer and proteins separated by SDS-PAGE. ILK and P-MYPT were assessed by Western blot analysis. MYPT concentration was assessed by Coomassie blue staining.
Results
Characterization of LIMD2 protein and production of αLIMD2 antibodies
LIMD2 belongs to the LIM-only family of LIM proteins, which contain between 1 and 5 LIM domains (Fig. 1A). A distance-based phylogenetic analysis of amino acid sequences among these LIM-only protein family members was performed using MEGA program (Fig. 1B). The LIMD2, CRP1, and LMO1 segregate from FHL1 and PINCH1, indicating that they are more similar to one another than they are to FHL1 and PINCH, which are more closely related, implying that there are shared ancestral relationships between the individual LIM domains (Fig. 1C; ref. 19). To define the targets and functions of LIMD2, rabbit polyclonal (rAb) and mouse monoclonal antibodies (mAb; Supplementary Fig. S1A) were produced and their specificity was determined. Both αLIMD2 (rAb) and αLIMD2 (mAb) detect the exogenous and endogenous LIMD2 protein in HEK293 and U2OS cell lines (Fig. 1D) but do not interact with AJUBA and LIMD1 (Supplementary Fig. S1B–S1D). By transient transfection, the Myc-LIMD2 protein [detected by both Myc (rAb) and LIMD2 (mAb)] is found predominantly in cytoplasm in a smooth granular distribution (Fig. 1E), a result confirmed by subcellular fractionation (data not shown). However, LIMD2 is concentrated in membrane ruffles and in streaks reminiscent of focal adhesion plaques (Fig. 1E). Interestingly, the protein is not excluded from the nucleus, consistent with many other reports of nucleocytoplasmic shuttling by LIM proteins. LIMD2 is robustly detected in formalin fixed, paraf-fin-embedded tissue specimens as a cytoplasmic protein, and no signal was seen in cells that lack LIMD2 (Supplementary Fig. S1E). Thus, these antibodies are specific and will be useful in examining the role of LIMD2 in cell motility and tumor metastasis.
LIMD2 transcript and protein levels correlate with malignant potential in breast, bladder, melanoma, and thyroid cancer cell lines and tumors
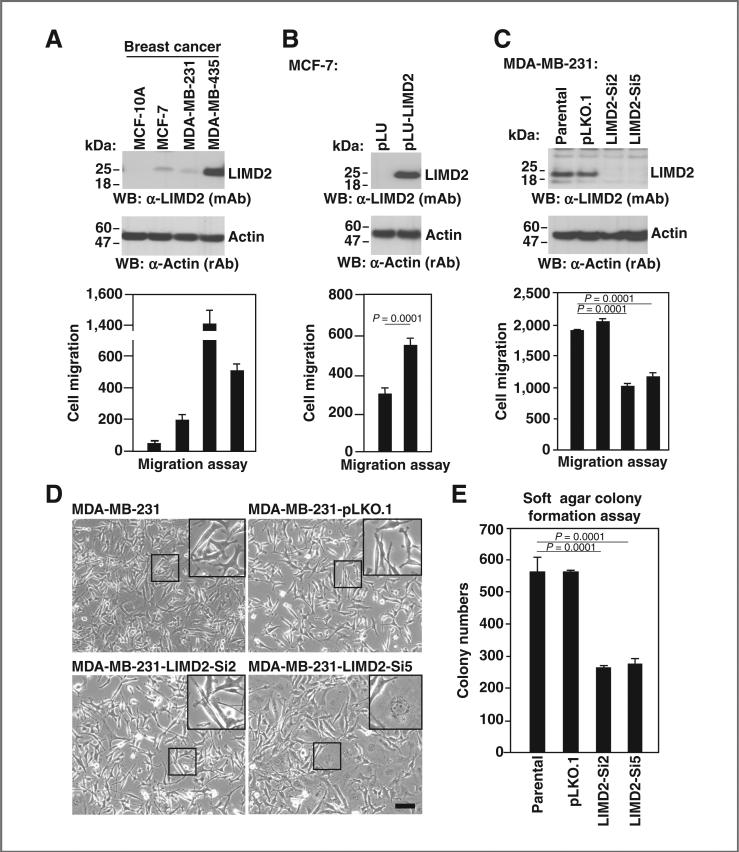
LIMD2 levels are well correlated with the higher degree of invasiveness among immortalized mammary epithelial cells (MCF-10A), nonmetastatic breast cancer cells (MCF-7), and breast cancer cells (MDA-MB-231 and MDA-MB-435; Fig. 2A). The LIMD2 RNA levels measured by quantitative PCR (qPCR) largely correlate with the protein levels (Supplementary Fig. S2A). MCF-7 cells expressing exogenous LIMD2 were obtained using lentivirus infection: this LIMD2 protein was readily detected by Western blot analysis (Fig. 2B, top). LIMD2 expression stimulated the migration of the MCF-7 cells compared with the control cell line (pLU vector control; Fig. 2B, bottom). Conversely, the knockdown of LIMD2 in MDA-MB-231 cells strongly impaired migration in these highly aggressive breast cancer cells (Fig. 2C). Restoration of wtLIMD2 expression in Si-5 knockdown MDA-MB-231 cells (using a LIMD2 cDNA with silent mutations in the coding sequence that render it resistant to the LIMD2-Si RNA) resulted in a regaining of the same migration ability as parental cells (Supplementary Fig. S3A and S3B), suggesting that the effects are because of LIMD2 expression and not to off-target effects. Depletion of LIMD2 strongly diminished colony formation in soft agar for all cell lines, but was efficiently restored by complementation by wtLIMD2 expression (Fig. 2E). Major differences in cell morphology between parental, control shRNA, LIMD2-Si2, and LIMD2-Si5 cells were also observed: the vector cells exhibited a spindle-like fibroblastic morphology, whereas the LIMD2-Si2 and LIMD2-Si5 cells showed an enlarged and flattened morphology (Fig. 2D). LIMD2 knockdown cells are enlarged and flattened as indicated with white arrows (Supplementary Figs. S3C and S4). LIMD2 overexpression restored the parental cells phenotype, showing a spindle-like fibroblastic morphology as indicated with black arrows (Supplementary Figs. S3C and S4). Thus, LIMD2 controls multiple hallmarks of oncogenic growth: anchorage-independent growth, migration, and altered cell morphology.
Figure 2.
A, breast cancer cell lines express endogenous LIMD2. B, ectopic expression of LIMD2 in MCF-7 cells. C, siRNA-mediated LIMD2 knockdown in MDA-MB-231 cells (the Si2 and Si5 are 2 independent cell clones). Migration potentials of each cell line. Data are presented as mean ± SD. T test was performed and P value is indicated. D, morphology of the cells. Bar, 100 μm. E, soft agar colony formation assays of each cell line.
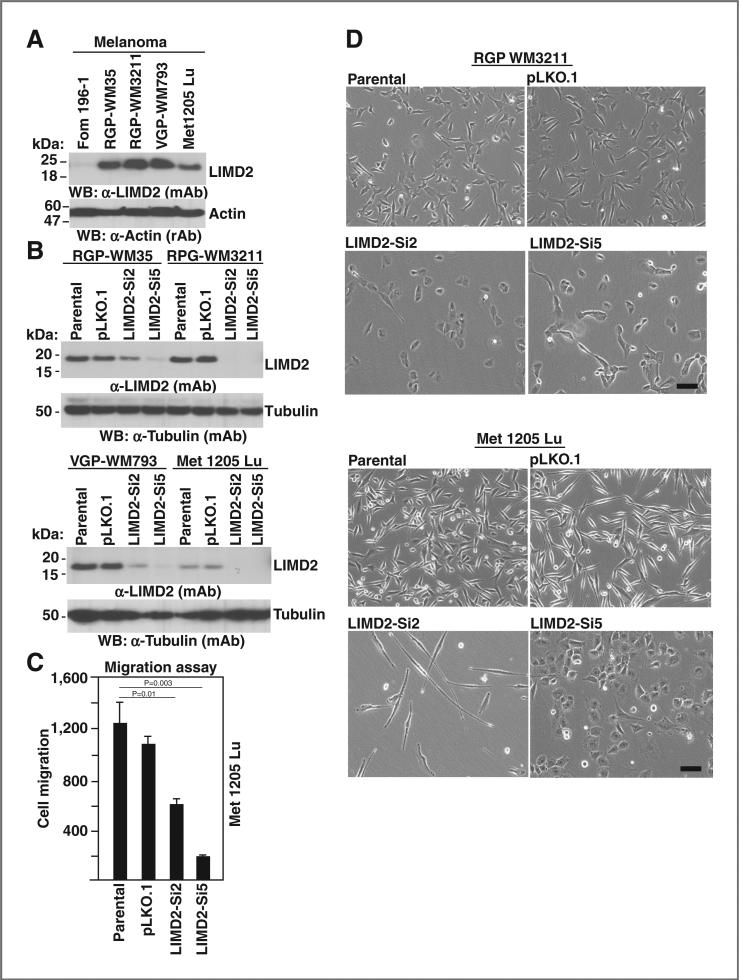
LIMD2 was analyzed in a series of melanoma [Fom196-1 cells (normal melanocytes), RGP-WM35, RGP-WM3211 cells (radial growth-phase melanoma), VGP-WM793 cells (vertical growth-phase melanoma), and Met1205 Lu cells (metastatic melanoma); Supplementary Fig. S2B]. LIMD2 protein was undetectable in normal melanocyte Fom196-1 cells, whereas the expression significantly increased in the RGP, VGP, and metastatic melanoma cells in a somewhat graded fashion (Fig. 3A). Following introduction of knockdown siRNA lentiviral vectors for LIMD2, protein levels and the migration rates were significantly reduced in all melanoma cell lines (Fig. 3B and C), whereas the cell morphology was widely variable (Fig. 3D). The RGP WM3211 cells with LIMD2 knockdown were flat compared with the spindle-shaped parental cells (Fig. 3D, RGP WM3211 panels). The metastatic melanoma cells (Met1205 Lu) morphology was changed dramatically showing a highly elongated shape reminiscent of neuronal cells, whereas the LIMD2-Si5 cells became flat and larger. Loss of LIMD2 significantly reduced Met1205 Lu cell migration (Fig. 3C).
Figure 3.
A, LIMD2 expression in a spectrum of melanoma cancer cells by Western blot analysis. B, knockdown of LIMD2. C, migration potentials of melanoma cells. The statistics were performed and P value is indicated. D, morphologic changes in melanoma cancer cells. Bars,100 μm.
In the series of bladder cancer cells [RT4, HT1376, UM-UC3, ScaBer, J82, TCCSUP, and T24 (listed from least to most aggressive)], LIMD2 levels coordinately increased with the known malignant characteristics of these cells (Supplementary Figs. S2C and S6A), with the more aggressive cell lines showing higher migration rates (Supplementary Fig. S6B and S6C). Ectopic LIMD2 expression in the more benign bladder cancer cell line RT4, which showed undetectable endogenous protein expression levels significantly increased migration potential (Supplementary Fig. S6D), indicating that the gain-of-function LIMD2 promotes bladder cancer cell migration. LIMD2 knockdown in T24 cells efficiently reduced LIMD2 protein levels to almost undetectable levels (Supplementary Fig. S6E), and these cells showed significant decreased numbers of colony formation in soft agar compared with the parental and nontargeted siRNA cells.
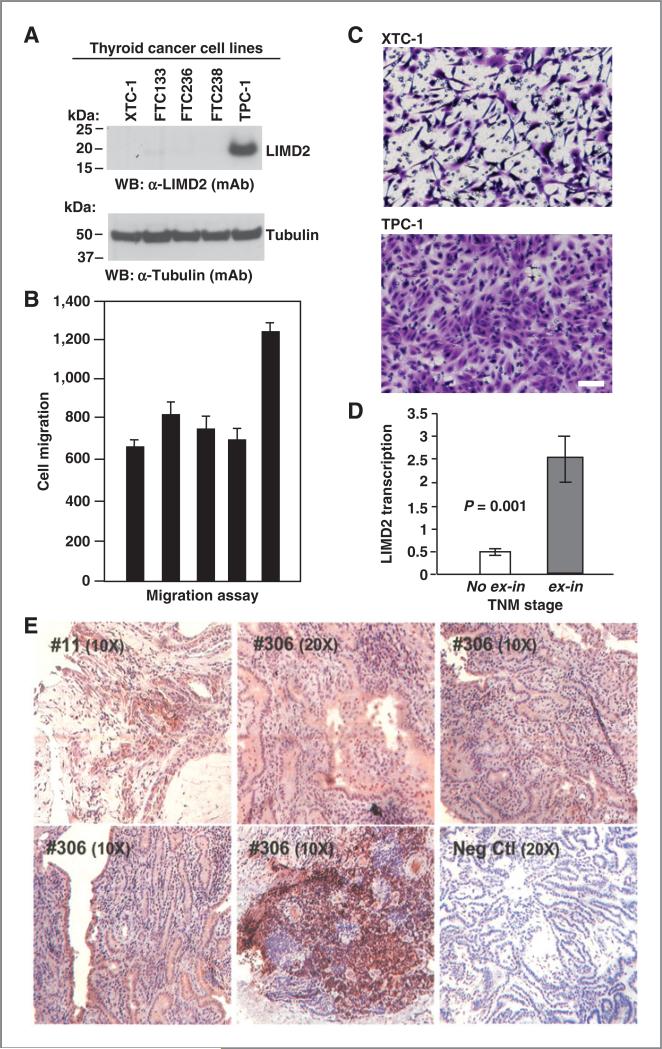
Because LIMD2 was identified from a papillary thyroid cancer metastasis, we evaluated LIMD2 expression in thyroid carcinoma cells lines: XTC-1, FTC133, FTC236, FTC238, and TPC-1. LIMD2 levels are elevated in the thyroid papillary cell line TPC-1, which is the most aggressive thyroid cancer cell line (Fig. 4A and Supplementary Fig. S2D) and shows the highest migration rate (Fig. 4B and C). We also evaluated LIMD2 expression in primary thyroid cancer specimens using qPCR. A total of 252 papillary thyroid cancer samples were evaluated for LIMD2 mRNA level by qPCR. We found that the LIMD2 mRNA level was significantly higher in papillary thyroid cancer with extrathyroidal invasion (P = 0.001; Fig. 4D and E). This highly significant finding further underscores the role of LIMD2 in invasion and metastasis processes.
Figure 4.
A, LIMD2 is highly expressed in thyroid cancer cells. B, migration potentials of the thyroid cancer cells. Data are presented as mean ± SD. C, images of thyroid carcinoma cell line XTC-1 and thyroid papillary carcinoma cell line TPC-1 taken from the migration assay. Bar, 100 mm. D, LIMD2 mRNA is higher in thyroid cancer samples with extrathyroidal invasion (n = 49). Total RNA was extracted from 252 papillary thyroid cancer samples using the TRIzol reagent (Invitrogen). TaqMan gene expression assay (Applied Biosystems) was used to quantitate LIMD2 mRNA expression levels. E, representative immunostaining results in papillary thyroid cancer samples using the aLIMD2 (mAb).
Structural studies into LIMD2 give insights into molecular function
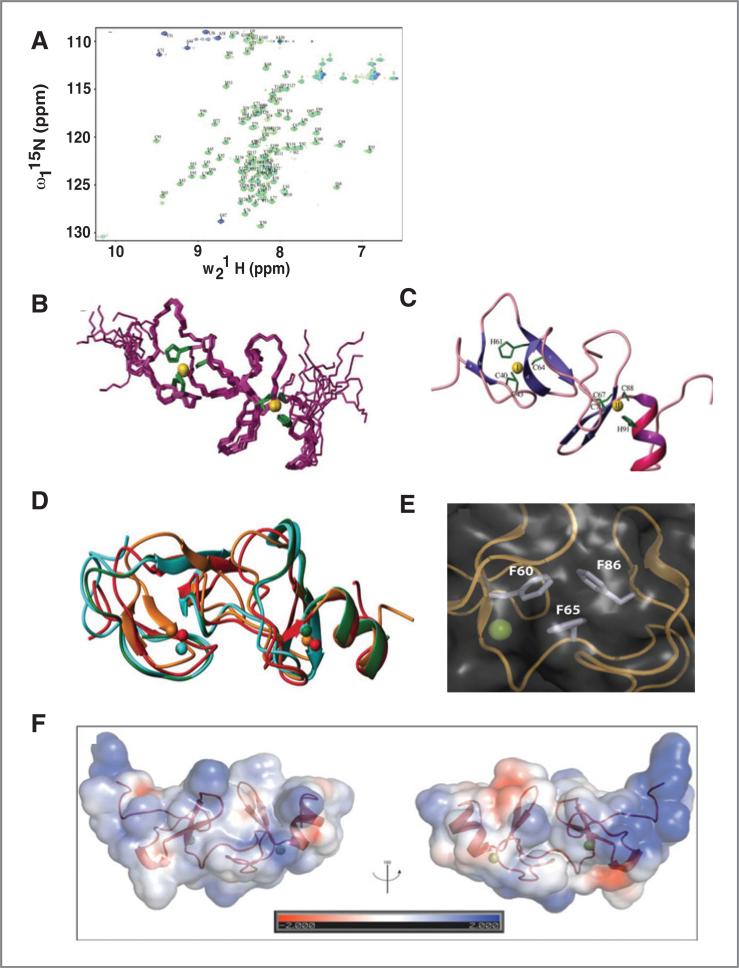
By analogy to the known functions of other LIM domains, LIMD2 likely binds to cellular proteins. We sought a structural biology solution to identify its targets by taking advantage of the fact that LIMD2 is well folded, globular, monomeric, and monodisperse in solution at high concentrations, making it suitable for NMR. Complete backbone assignments of the protein were determined (Fig. 5A), however analysis of NOESY spectra (13C- and 15N-edited NOESY-HSQC as well as 2DNOESY) indicated a lack of signals for the first 35 and last 23 amino acids of the protein consistent with these being unstructured. Consequently, only residues 33 to 104 were used for structure determination. This protein is composed of two treble clef fingers (19, 20) folded around two zinc atoms (Fig. 5B and C). ICP-EOS studies also indicate that there are 2.4 zinc molecules per LIMD2 domain. The zinc-ligated residues are Cys40, Cys43, His61, and Cys64 (CCHC) for site I, and Cys67, Cys70, Cys88, and His91 (CCCH) for site II. The first finger is composed of 2 antiparallel β-sheets with zinc-coordinated residues located on the loop connecting two strands in each antiparallel β-sheet. The second finger is composed of a long loop followed by an antiparallel β-sheet and ends in an approximately 2-turn helix. The zinc-ligating residues in the second site reside on the long loop and the beginning of a 2-turn helix (Fig. 5B). The Nδ1 atoms of the 2 histidines (His61 and His91) coordinate the zinc. The zinc-ligating residues comprise one of the key structural elements of LIMD2. In addition, structural analysis indicates that there is a small hydrophobic core likely involved in the stabilization of the protein, including the aromatic side chains of residues Phe60, Phe65, and Phe86, which form a hydrophobic cluster between the 2 zinc fingers and are buried (Fig. 5E).
Figure 5.
A, 1H-15N HSQC spectrum of LIMD2 with backbone assignment: BMRB accession number 18778. Purple represents aliased signals. B, superimposition of the 10 best structures calculated by ARIA (PDB accession code 2LZU) with an RMSD of 0.32 A for LIMD2 residues 39-96aa over backbone atoms. See Supplementary Table S1 for structure statistics. Zinc is colored in gold and side chains of the zinc-coordinated residues are shown in green. C, ribbon representation of LIMD2. D, alignment of LIMD2 (red) with PINCH1-LIM1 (cyan PDB code 3F6Q), PINCH2-LIM1 (green PDB code 3IXE), and PINCH1-LIM4 (orange PDB code 1NYP). E, side chains of F60, F65, and F86 from a LIMD2 hydrophobic core. F, electrostatic surface of LIMD2. The asymmetric charge distribution in LIMD2 is shown.
We assessed the structural similarity between LIMD2 and other reported LIM structures present in the PDB. The superposition of LIMD2 onto the 3-dimensional crystal structures of PINCH1-LIM1 (PDB code 3F6Q; ref. 11), and PINCH2-LIM1 (PDB code 3IXE; refs. 21 and 22) yielded an RMSD of 2.9 and 2.8 Å, respectively, for the backbone of the LIM consensus sequence (residues 39–96 in LIMD2). The secondary structure elements are well conserved, although the antiparallel β-sheet of the second finger of the LIMD2 adopts a flat conformation, and the c’ strand rotates around 90° along the axis of the sheet compared with other structures (Fig. 5C and D). The electrostatic surface of LIMD2 is very positive, as expected, given its pI of 8.7; however, the charge distribution is quite asymmetric, with both positive and hydrophobic patches (Fig. 5F). Comparison of LIMD2 to PINCH-LIM1 reveals that although the latter has a net negative charge (pI of 4.9), once again the charge distribution is asymmetric and contains hydrophobic patches (Supplementary Fig. S5). Similar to LIMD2, PINCH-LIM4 is a positively charged domain (pI 8.7); however, in contrast to LIMD2, the C-terminal helix is both negatively and positively charged on the corresponding faces based on alignment with the LIMD2 domain, respectively. The asymmetric charge distribution and positions of hydrophobic patches may supply various interfaces to accommodate specific binding proteins.
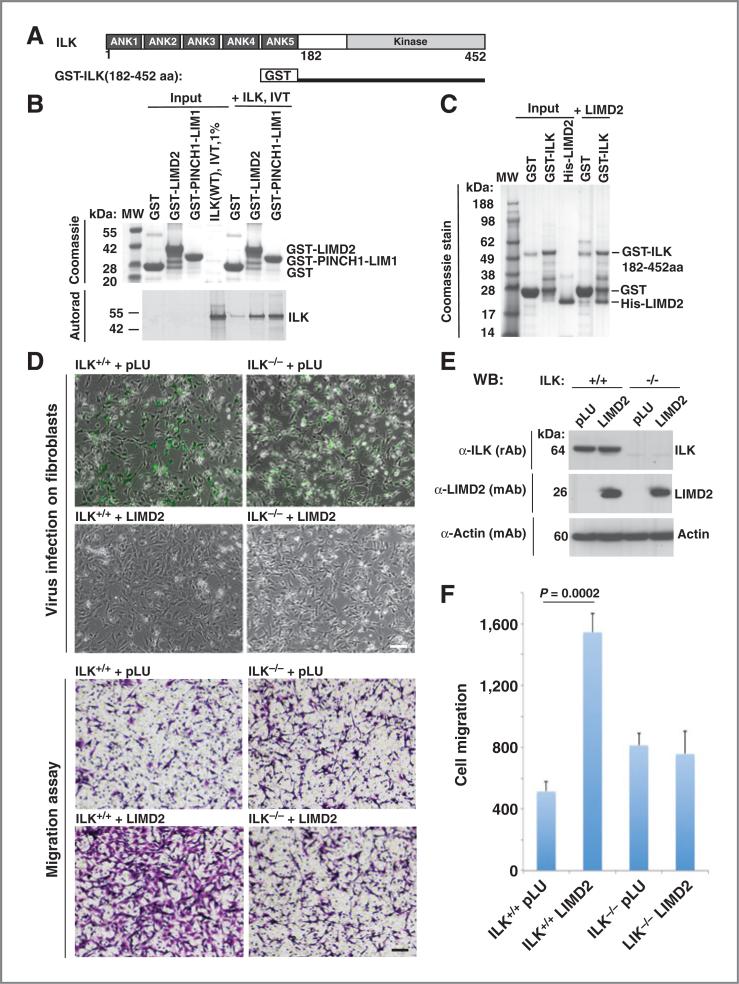
The LIMD2 structure was highly similar to the first LIM domain (LIM1) of PINCH1. PINCH1-LIM1 binds to the ARD of the ILK (11). ILK is a 452 amino acid, multidomain protein consisting of 5 N-terminal ankyrin repeats, and a C-terminal kinase domain (Figs. 6A and 8B). ILK is a core component of the IPP complex, which contains PINCH proteins and Parvin (Fig. 8B). The IPP complex binds to the C-terminal tails of β1-integrins, thus serving to communicate cell–cell and cell–matrix adhesion signals to the nucleus (23). To determine if PINCH1-LIM1 and LIMD2 bound to ILK, we used GST-LIMD2, GST-PINCH1-LIM1 and 35S-labeled ILK protein produced by IVT. Indeed, the GST-LIMD2 protein robustly binds to ILK under the same conditions as PINCH-LIM1; neither protein binds to GST alone (Fig. 6B). Identical results were observed for ILK and LIMD2 interaction using fully recombinant proteins, isolated from E. coli (Fig. 6C). These studies strongly suggest that ILK is a candidate target protein for binding by LIMD2. GST-PINCH1-LIM1 bound to the ARD robustly, as has been shown by other investigators (11), but the LIMD2 protein did not bind to the ILK-ARD (data not shown). Moreover, LIMD2 did not compete for PINCH1-LIM1 for binding to ILK (data not shown).
Figure 6.
LIMD2 and PINCH1 directly associate with ILK, LIMD2 affects cell migration through ILK. A, domain architecture of ILK. The N-terminus of ILK contains ankyrin repeat domain (ARD) and the C-terminus of ILK contains a kinase domain. B, ILK protein binds to GST-LIMD2 and GST-PINCH1-LIM1 but not GST alone. C, LIMD2 protein binds to GST-ILK (182-452aa). D, LIMD2 stimulates cell migration in ILKþ/þ fibroblast but not in ILK −/− fibroblast. Lentivirus pLU vector (GFP) and pLU-LIMD2 were infected on ILKþ/þ and ILK −/− fibroblast cells. Migration assay was performed on pLU and LIMD2 virus infected ILKþ/þ and ILK −/− fibroblast cells. Images were taken from the migrated cells. Bar, 100 μm. E, endogenous ILK protein were detected in ILKþ/þ fibroblast cells but not in ILK −/− cells by Western blot analysis using aILK (rAb). Exogenous LIMD2 proteins were detected in LIMD2 virus– infected fibroblast cells by Western blot analysis using aLIMD2 (mAb). F, ectopic LIMD2 expression promotes ILKþ/þ fibroblast cells migration. Three independent experiments were performed. Data, mean ± SD. P value is indicated.
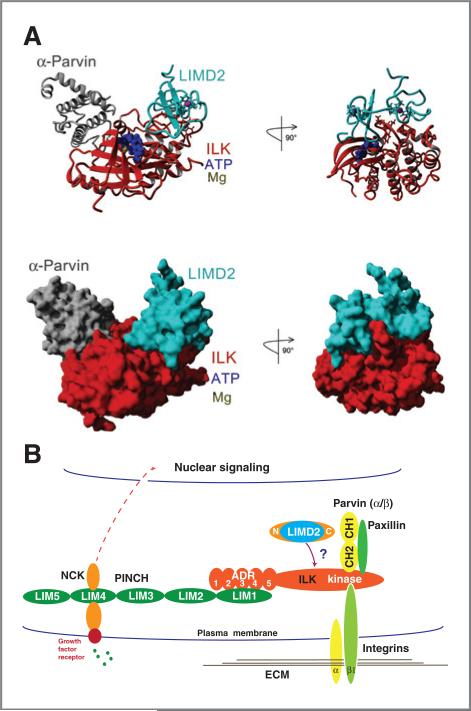
Figure 8.
A, computer molecular modeling of the LIMD2–ILK–aParvin protein complex. B, a proposed working model for LIMD2 functional roles in assembly of and normal function of the ILK–PINCH–Parvin complex in the integrin-linked signaling pathway.
To determine if endogenous cellular ILK is required for stimulation of migration by LIMD2, we used a matched set of fibroblasts that are either wild type for ILK (+/+) or have been rendered ILK null (−/−) via CRE-mediated excosopm pf floxed ILK alleles (24). Both of these cell types grow robustly in culture, show similar morphologies, similar doubling times, and are equally infectable by lentiviruses (Fig. 6D). The −/− cells lack ILK by Western blot analysis as expected (Fig. 6E). LIMD2 is robustly, and equally expressed in both cell types (Fig. 6E) when infected with lentiviruses. LIMD2 expression in ILKþ/þ cells stimulates cell migration almost 4-fold. However, there is no stimulation of migration by LIMD2 in cells lacking the −/− cells that lack ILK (Fig. 6F). Thus, ILK is likely a key effector for LIMD2 stimulated cell migration.
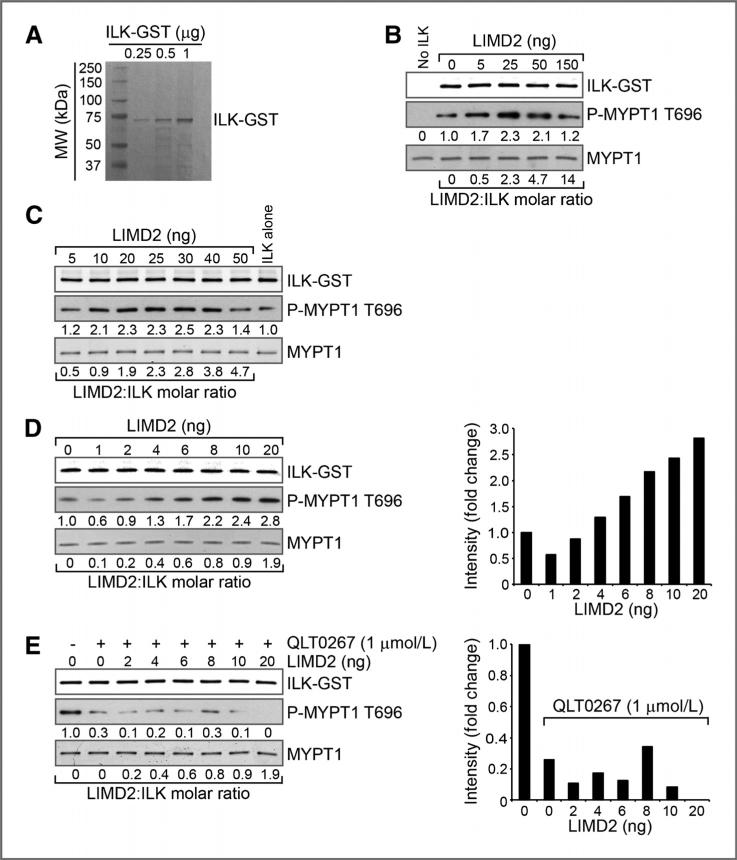
Because LIMD2 bound tightly to the region of ILK containing the kinase domain, we determined if LIMD2 could alter the kinase activity of ILK toward a model peptide substrate in vitro. Although there is considerable controversy in the literature about whether ILK is a kinase or a pseudo-kinase, the observation that baculovirus-expressed and purified protein contains this activity warranted a look at this question: we engaged the investigators who have championed this claim to perform these experiments. Baculovirus-produced full-length ILK (Fig. 7A) showed considerable kinase activity toward the MYPT peptide as has previously been shown (18). The addition of LIMD2 to the in vitro assay resulted in a dose-dependent increase in kinase activity, showing almost a 3-fold stimulation at the highest levels of LIMD2 added (Fig. 7B–D). This apparent activation of ILK by LIMD2 was abolished when the ILK small-molecule inhibitor kinase inhibitor QLT0267 was added to the reaction (Fig. 7E). Thus, binding of ILK by LIMD2 in vitro results in stimulation of the kinase activity exhibited by the recombinant ILK under these conditions. Together, these data strongly suggest that LIMD2 is a new positive regulator of ILK activity.
Figure 7.
LIMD2 increases ILK kinase activity in a dose-dependent manner. A, coomassie blue-stained polyacrylamide gel showing highly purified ILK–GST fusion protein used in the ILK kinase assay. B, dose-dependent phosphorylation of the ILK substrate myosin phosphatase target subunit 1 (MYPT1) on Thr696 in the presence of increasing concentrations of purified LIMD2. ILK and LIMD2 were mixed and preincubated for 30 minutes at room temperature before addition of the substrate. The amounts of ILK and MYPT1 were kept constant at 50 and 500 ng/reaction, respectively, and increasing amounts of LIMD2 were added as indicated. C, concentration curve similar to that shown in B, with LIMD2 concentrations increased stepwise from 5 to 50 ng. Reactions were carried out as described for B. D, concentration curve similar to that shown in C, with LIMD2 concentrations increased stepwise from 1 to 10 and 20 ng (molar ratio of 0.1–1.9). Reactions were carried out as described for B. Data in the bar graph are normalized to the signal observed in the presence of ILK alone. E, effect of the specific small molecule inhibitor of ILK, QLT0267, on the phosphorylation of MYPT by ILK-LIMD2. ILK-LIMD2 complexes were preincubated at the indicated concentrations as described above, and QLT0267 was added at a final concentration of 1.0 μmol/L just before initiation of the kinase reaction. Data in the bar graph are normalized to the signal observed in the presence of ILK alone.
Discussion
Characterization of the novel, metastasis-associated LIMD2 protein described herein supports the following conclusions: (i) LIMD2 is a small cytoplasmic protein whose endogenous expression levels correlate with the known malignant properties of cells derived from breast, melanoma, bladder, and thyroid cancers; (ii) ectopic expression or knockdown of LIMD2 stimulates or abrogates cell invasiveness, motility, and cell morphology consistent with tumor grade; (iii) primary human thyroid cancers that express LIMD2 show much greater extrathymic invasion; (iv) the LIMD2 structure is homologous to the PINCH1-LIM1 domain that binds to ILK; (v) both PINCH1-LIM1 and LIMD2 proteins bind to ILK directly but at nonoverlapping sites; (vi) LIMD2 stimulates the kinase activity of ILK, and endogenous ILK is required for the ability of LIMD2 to stimulate cell migration. Thus, in LIMD2, we have discovered a new target and function for a LIM-only protein in tumor progression using a combination of tumor biology and structural biology experimental approaches.
The finding of a direct connection between LIMD2 and the integrin signaling apparatus is wholly consistent with the promotion of cell migration and invasion seen in LIMD2-expressing cell lines and tumors. The multiprotein IPP complex (ILK–PINCH–Parvin) as a core complex links transmembrane integrin adhesion receptors to the actin cytoskeleton and intracellular signaling cascades (11, 25). The ILK localizes to sites of integrin-mediated cell adhesion (26, 25, 27). The presence of intrinsic catalytic activity as a kinase in ILK is still controversial, but the domain clearly binds to the CH2 domain of Parvin (28). Formation of the tripartite IPP complex is absolutely required for integrin signaling, migration, and invasion in both normal developmental and malignant programs. The PINCH1 LIM1 domain binds directly to the ARDs of ILK and stimulates integrin signaling (whereas the highly related PINCH2 protein binds to the ILK ARD and competes for PINCH1 binding; Figs. 22 and 29–31). Other proteins that bind to the core IPP complex, such as Nck, ERK1/2, GSK3β, and Rsu1, serve to remodel the IPP complex to respond to different growth factor signaling inputs emanating from the cell membrane (32) and largely control cell growth and differentiation pathways, most notably EMT. In addition to the direct effects of LIMD2 on ILK kinase function as described below, LIMD2 analogously may adapt the IPP complex to other as yet undefined inputs.
The finding that LIMD2 binds to ILK and promotes cell migration and invasion is, of course, inconsistent with it being a competitor for PINCH1 LIM1 binding, even though they share high structural similarity. That LIMD2 binds to ILK at a site distinct from PINCH1 is consistent with a model that LIMD2 may be a new positive regulator of the IPP complex (Fig. 8B). In fact, LIMD2 and PINCH1 likely bind simultaneously to ILK at widely dispersed sites. At first blush, it may seem surprising that LIMD2 and PINCH1 would target separate regions of ILK; however, given the vastly different charge distributions on the same structural scaffold (Fig. 5F), it is reasonable that these would bind different surfaces (Supplementary Fig. S5). In contrast, our data are consistent with binding of LIMD2 near the active site and thus directly influencing its enzyme activity. An in silico molecular dynamics–based docking analysis strongly suggests that LIMD2 binds to a site in ILK very near the potential active site but separate from the binding site for α-Parvin (Fig. 8A). It is noteworthy that when ILK is coex-pressed with α-Parvin in bacteria, the complex is enzymatically dead and in fact α-Parvin is an inhibitor of kinase activity when added to in vitro ILK kinase assays. Thus, α-Parvin and LIMD2 may serve as negative and positive regulators (respectively) of kinase activity by binding the 2 different lobes of the kinase domain, which border the active site cleft (Fig. 8A). The model also suggests that LIMD2 forms a well-defined pocket/groove bordered by the 2 zinc-binding fingers, which directly interact with an active site loop in ILK: this pocket may be a promising target for small molecule–based inhibitor design.
Like LIMD2, almost all components of the IPP complex have been observed to be deregulated in human cancer (32) and are largely correlated with the later metastatic phenotype. PINCH1 expression is highly correlated with tumor grade and invasiveness in gliomas, gastric cancer, and squamous cell carcinoma, suggesting that LIMD2 overexpression may phenocopy PINCH1 overexpression (33). It may be that optimal activation of ILK requires binding of both PINCH1 and LIMD2. The negative regulator of IPP, PINCH2, is often silenced by methylation in tumors. Moreover, the PINCH1 LIM domain 5 binds directly to an altered form of the Ras suppressor, Rsu1, thereby directly participating in Ras-mediated transformation (33). Thus, IPP deregulation is a common denominator among tumors arising from different lineages, which supports the observations that LIMD2 expression occurs in multiple tumor types.
Does LIMD2 function as a classic “adaptor” protein as has been demonstrated in other LIM-only proteins? One of the best examples of this function is the LIM-only protein LMO2, which uses its 2 LIM domains to bind the GATA1 and TAL1 transcription factors in the nucleus (34, 35). In this model, each LIM domain in LMO2 is specific for each partner. Similar specificity for protein-protein interactions are demonstrated by the individual LIM domains in Paxillin, TRIP6, Ajuba, and of course PINCH1, whose 5 LIM domains are likely specialized for different partners. Thus, LIM domain proteins with 2 or more LIM domains function as true scaffolds to combinatorially assemble multiprotein complexes (10). The case of LIMD2 is problematic to propose as a classic scaffold in that it encodes only a single LIM domain. However, the unstructured NH2- and COOH-terminal protein sequence in LIMD2 may provide this function. The pronounced charge differences observed on each “face” of the LIMD2 LIM domain may indicate that the LIM domain itself can provide separate surfaces for protein–protein interactions. It is formally possible that LIMD2 functions as a positive regulator of the IPP complex by simply using its single LIM domain to compete for a negative regulator of ILK. LIMD2 may also directly modulate the activity of the kinase domain of ILK. It is noteworthy that LIMD2 has not been observed in multiple proteomics–based approaches to the composition of the ILK complex and the proteins, which inhabit focal adhesion plaques (36). This is surprising as our preliminary analyses of LIMD2 distribution in cells by confocal microscopy show it to partially overlap with focal adhesion streaks, and the fact that LIMD2 seems to bind ILK directly and with considerable affinity using recombinant proteins. An ongoing dynamic approach using video-microscopy of genetically tagged fluorescent proteins should address this.
Does LIMD2 carry out multiple independent functions in different subcellular compartments? Many multi-LIM domain proteins shuttle between the cytoplasm and nucleus. These studies were initiated by the pioneering work of Beckerle showing that the LIM domain protein paxillin rapidly shuttled between the cytoplasm and the nucleus and had key regulatory roles in both compartments (19, 37). LIMD2 may function like its closest evolutionary homologue, CRP1. CRP1 is a 2 LIM protein that shuttles between the cytoplasm and nucleus. In the nucleus it binds a complex of the SRF and GATA transcription factors, whereas in the cytoplasm, CRP1 binds to α-actinin and zyxin to reorganize the cytoskeleton (38). The Ajuba LIM domain protein illustrates a similar pathway highly relevant to cancer progression. Our laboratory has shown that the 3 LIM domain protein Ajuba binds to the SNAG repression domain in the snail/slug transcription factors and plays a key role in the EMT program during tumor progression by repressing E-cadherin and other genes (39, 40). However, when in the cytoplasm, Ajuba is found at cell adhesion junctions and binds to CTNNA, F-actin, and GRB2. It is noteworthy that PINCH1 can be found in both the cytoplasm and the nucleus, and this location has prognostic value in primary human tumors (33).
In summary, the study of LIMD2 as a marker identified in a simple profiling study of primary and metastatic tumors has yielded a new player in the ILK signaling pathway. It is conceivable that small molecule approaches to disrupting LIMD2 binding to its partners could influence the metastatic process.
Supplementary Material
Acknowledgments
The authors thank F. Keeney (Wistar) for help with the imaging studies and The Wistar Institute Cancer Center Shared Facilities. They also thank R. Fassler and colleagues for the gift of the ILK −/− cells and D. Calderwood and T. Boggon (Yale University) for ILK and PINCH1 reagents.
Grant Support
Work in the Rauscher laboratory is supported by NIH grants CA129833, CA010815, CA163761, and DOD-BCRP W81XWH-11-1-0494, The Samuel Waxman Cancer Research Foundation, Susan G. Komen for the Cure, and The Noreen O'Neill Foundation for Melanoma Research. K.L.B. Borden holds a Canada Research Chair and is supported by NIH grants NIH RO1-80728 and NIH 98571.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
Conception and design: H. Peng, M. Talebzadeh-Farrooji, M.J. Osborne, Z. Hou, M. He, E. Kebebew, K.L.B. Borden, F.J. Rauscher
Development of methodology: H. Peng, M. Talebzadeh-Farrooji, M.J. Osborne, Z. Hou, M. He, M. Herlyn, A.J. Caton, C.S. Paterno, Q. Huang, S. Dedhar
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): M. Talebzadeh-Farrooji, M.J. Osborne, P.C. McDonald, J. Karar, Z. Hou, M. He, E. Kebebew, W. Fredericks, B. Malkowicz, A.S. Carolin, E. Skordalakes, Q. Huang, K.L.B. Borden, F.J. Rauscher
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): H. Peng, M. Talebzadeh-Farrooji, M.J. Osborne, J.W. Prokop, P.C. McDonald, M. He, E. Kebebew, T. Orntoft, D.W. Speicher, Q. Huang, S. Dedhar, K.L.B. Borden, F.J. Rauscher
Writing, review, and/or revision of the manuscript: H. Peng, M. Talebzadeh-Farrooji, M.J. Osborne, J.W. Prokop, P.C. McDonald, J. Karar, M. He, E. Kebebew, W. Fredericks, B. Malkowicz, D.W. Speicher, S. Dedhar, K.L.B. Borden, F.J. Rauscher
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M. Talebzadeh-Farrooji, M. He, T. Orntoft, M. Herlyn, F.J. Rauscher
Study supervision: M. Talebzadeh-Farrooji, S. Dedhar, K.L.B. Borden, F.J. Rauscher
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 2.Sethi N, Kang Y. Unravelling the complexity of metastasis—molecular understanding and targeted therapies. Nat Rev Cancer. 2011;11:735–48. doi: 10.1038/nrc3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott J, Kuhn P, Anderson AR. Unifying metastasis—integrating intravasation, circulation and end-organ colonization. Nat Rev Cancer. 2012;12:445–6. doi: 10.1038/nrc3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunter K. Host genetics influence tumour metastasis. Nat Rev Cancer. 2006;6:141–6. doi: 10.1038/nrc1803. [DOI] [PubMed] [Google Scholar]
- 5.Alderton GK. Metastasis: epithelial to mesenchymal and back again. Nat Rev Cancer. 2013;13:3. doi: 10.1038/nrc3428. [DOI] [PubMed] [Google Scholar]
- 6.Gumireddy K, Huang Q. Identification of metastasis genes by a functional genomics approach. Cell Cycle. 2010;9:423. doi: 10.4161/cc.9.3.10678. [DOI] [PubMed] [Google Scholar]
- 7.Gumireddy K, Li A, Gimotty PA, Klein-Szanto AJ, Showe LC, Katsaros D, et al. KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer. Nat Cell Biol. 2009;11:1297–304. doi: 10.1038/ncb1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–84. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 9.Cerutti JM, Oler G, Michaluart P, Jr, Delcelo R, Beaty RM, Shoemaker J, et al. Molecular profiling of matched samples identifies biomarkers of papillary thyroid carcinoma lymph node metastasis. Cancer Res. 2007;67:7885–92. doi: 10.1158/0008-5472.CAN-06-4771. [DOI] [PubMed] [Google Scholar]
- 10.Matthews JM, Lester K, Joseph S, Curtis DJ. LIM-domain-only proteins in cancer. Nat Rev Cancer. 2013;13:111–22. doi: 10.1038/nrc3418. [DOI] [PubMed] [Google Scholar]
- 11.Chiswell BP, Zhang R, Murphy JW, Boggon TJ, Calderwood DA. The structural basis of integrin-linked kinase-PINCH interactions. Proc Natl Acad Sci U S A. 2008;105:20677–82. doi: 10.1073/pnas.0811415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng H, Begg GE, Harper SL, Friedman JR, Speicher DW, Rauscher FJ., 3rd Biochemical analysis of the Kruppel-associated box (KRAB) transcriptional repression domain. J Biol Chem. 2000;275:18000–10. doi: 10.1074/jbc.M001499200. [DOI] [PubMed] [Google Scholar]
- 13.Peng H, Ivanov AV, Oh HJ, Lau YF, Rauscher FJ., 3rd Epigenetic gene silencing by the SRY protein is mediated by a KRAB-O protein that recruits the KAP1 co-repressor machinery. J Biol Chem. 2009;284:35670–80. doi: 10.1074/jbc.M109.032086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelton JG, Torchia DA, Meadow ND, Roseman S. Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli, using two-dimensional heteronuclear NMR techniques. Protein Sci. 1993;2:543–58. doi: 10.1002/pro.5560020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 16.Goddard TD, Kneller DG. SPARKY. University of California; San Francisco, CA: 2003. [Google Scholar]
- 17.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maydan M, McDonald PC, Sanghera J, Yan J, Rallis C, Pinchin S, et al. Integrin-linked kinase is a functional Mn2+-dependent protein kinase that regulates glycogen synthase kinase-3β (GSK-3β) phosphorylation. PLoS ONE. 2010;5:e12356. doi: 10.1371/journal.pone.0012356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–31. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 20.Grishin NV. Treble clef finger—a functionally diverse zinc-binding structural motif. Nucleic Acids Res. 2001;29:1703–14. doi: 10.1093/nar/29.8.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiswell BP, Stiegler AL, Razinia Z, Nalibotski E, Boggon TJ, Calder-wood DA. Structural basis of competition between PINCH1 and PINCH2 for binding to the ankyrin repeat domain of integrin-linked kinase. J Struct Biol. 2010;170:157–63. doi: 10.1016/j.jsb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velyvis A, Vaynberg J, Yang Y, Vinogradova O, Zhang Y, Wu C, et al. Structural and functional insights into PINCH LIM4 domain-mediated integrin signaling. Nat Struct Biol. 2003;10:558–64. doi: 10.1038/nsb938. [DOI] [PubMed] [Google Scholar]
- 23.Rooney N, Streuli CH. How integrins control mammary epithelial differentiation: a possible role for the ILK-PINCH-Parvin complex. FEBS Lett. 2011;585:1663–72. doi: 10.1016/j.febslet.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 24.Sakai T, Li S, Docheva D, Grashoff C, Sakai K, Kostka G, et al. Integrin-linked kinase (ILK) is required for polarizing the epiblast, cell adhesion, and controlling actin accumulation. Genes Dev. 2003;17:926–40. doi: 10.1101/gad.255603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hannigan G, Troussard AA, Dedhar S. Integrin-linked kinase: a cancer therapeutic target unique among its ILK. Nat Rev Cancer. 2005;5:51–63. doi: 10.1038/nrc1524. [DOI] [PubMed] [Google Scholar]
- 26.Legate KR, Montanez E, Kudlacek O, Fassler R. ILK, PINCH and parvin: the tIPP of integrin signalling. Nat Rev Mol Cell Biol. 2006;7:20–31. doi: 10.1038/nrm1789. [DOI] [PubMed] [Google Scholar]
- 27.Wu C. PINCH, N(i)ck and the ILK: network wiring at cell-matrix adhesions. Trends Cell Biol. 2005;15:460–6. doi: 10.1016/j.tcb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Hannigan GE, McDonald PC, Walsh MP, Dedhar S. Integrin-linked kinase: not so 'pseudo' after all. Oncogene. 2011;30:4375–85. doi: 10.1038/onc.2011.177. [DOI] [PubMed] [Google Scholar]
- 29.Fukuda K, Gupta S, Chen K, Wu C, Qin J. The pseudoactive site of ILK is essential for its binding to a-Parvin and localization to focal adhesions. Mol Cell. 2009;36:819–30. doi: 10.1016/j.molcel.2009.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velyvis A, Yang Y, Wu C, Qin J. Solution structure of the focal adhesion adaptor PINCH LIM1 domain and characterization of its interaction with the integrin-linked kinase ankyrin repeat domain. J Biol Chem. 2001;276:4932–9. doi: 10.1074/jbc.M007632200. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Chen K, Tu Y, Velyvis A, Yang Y, Qin J, et al. Assembly of the PINCH-ILK-CH-ILKBP complex precedes and is essential for localization of each component to cell-matrix adhesion sites. J Cell Sci. 2002;115:4777–86. doi: 10.1242/jcs.00166. [DOI] [PubMed] [Google Scholar]
- 32.Persad S, Dedhar S. The role of integrin-linked kinase (ILK) in cancer progression. Cancer Metastasis Rev. 2003;22:375–84. doi: 10.1023/a:1023777013659. [DOI] [PubMed] [Google Scholar]
- 33.Kovalevich J, Tracy B, Langford D. PINCH: more than just an adaptor protein in cellular response. J Cell Physiol. 2011;226:940–7. doi: 10.1002/jcp.22437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–44. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, et al. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29:232–42. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo JC, Han X, Hsiao CT, Yates JR, 3rd, Waterman CM. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for b-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol. 2011;13:383–93. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hervy M, Hoffman L, Beckerle MC. From the membrane to the nucleus and back again: bifunctional focal adhesion proteins. Curr Opin Cell Biol. 2006;18:524–32. doi: 10.1016/j.ceb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Chang DF, Belaguli NS, Iyer D, Roberts WB, Wu SP, Dong XR, et al. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev Cell. 2003;4:107–18. doi: 10.1016/s1534-5807(02)00396-9. [DOI] [PubMed] [Google Scholar]
- 39.Hou Z, Peng H, White DE, Negorev DG, Maul GG, Feng Y, et al. LIM protein Ajuba functions as a nuclear receptor corepressor and negatively regulates retinoic acid signaling. Proc Natl Acad Sci U S A. 2010;107:2938–43. doi: 10.1073/pnas.0908656107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou Z, Peng H, White DE, Wang P, Lieberman PM, Halazonetis T, et al. 14-3-3 binding sites in the snail protein are essential for snail-mediated transcriptional repression and epithelial-mesenchymal differentiation. Cancer Res. 2010;70:4385–93. doi: 10.1158/0008-5472.CAN-10-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.