Abstract
It is becoming increasingly clear that there are unique sets of miRNAs that have distinct governing roles in several aspects of both innate and adaptive immune responses. In addition, new tools allow selective modulation of the expression of individual miRNAs, both in vitro and in vivo. Here, we summarize recent advances in our understanding of how miRNAs drive the activity of immune cells, and how their modulation in vivo opens new avenues for diagnostic and therapeutic interventions in multiple diseases, from immunodeficiency to cancer.
Keywords: microRNA, immunotherapy, tumor immunology, adaptive immunity
Introduction
MicroRNAs (miRNAs) are 18-25 nucleotides long encoded RNAs that are processed by the enzyme Dicer from precursor stem loop RNA hairpins. In non-germinal human cells, four Ago proteins with different expression profiles and substrate specificities provide a platform for the recruitment of other proteins. Ago variants bind to a “guide” RNA strand that becomes unwound from the original double-stranded miRNA structure (Kaya and Doudna, 2012). Interactions occur in such a manner that the “seed” region of this ssRNA has its Watson-and-Crick base edges exposed to complementary sequences that are typically found in the 3’ untranslated region of target mRNAs (Schirle and MacRae, 2012). Translation of hundreds of target genes is therefore finely modulated by a single miRNA via mRNA degradation or translational repression, which (directly and indirectly) has dramatic effects in the acquisition of specific phenotypes in multiple cell types.
While the consequences of the dramatic differences in the expression of multiple miRNAs in cancerous cells vs. their healthy counterparts are now being recurrently reported, the role of miRNAs in physiological and pathological immune responses appears to be different and at least equally complex (Sempere and Conejo-Garcia, 2012). Several lines of evidence suggest that miRNAs (and perhaps other non-coding RNAs) are particularly important in regulating immune responses in both healthy and disease states: Firstly, several miRNAs are selectively expressed by hematopoietic cells, and some of them (e.g., miR-142) are expressed at significantly higher levels, compared to other sequences. Secondly, immune responses result from rapid phenotypic changes in leukocytes. These particularly plastic cell types utilize fine-tuning mechanisms of genomic regulation to avoid disrupting the delicate balance between effective protection and autoimmunity. Thirdly, emerging evidence indicates that changes in a single miRNA sequence are sufficient to induce the activation or the acquisition of regulatory properties by multiple immune cell types (Contreras and Rao, 2012; Josefowicz et al., 2012; Li et al., 2007; Lodish et al., 2008; Rodriguez et al., 2007; Shen et al., 2012).
In this review, we will summarize some recent advances into the immunobiology of miRNAs, and how this field may open new avenues for diagnostic and therapeutic interventions in multiple diseases, from immunodeficiency to cancer.
Hematopoietic-specific miRNAs
The first line of evidence about the critical role of miRNAs in the orchestration of immune responses is the selective expression of at least two miRNAs (miR-142 and miR-223) in hematopoietic cells under steady-state conditions. In addition, several other miRNAs are expressed in hematopoietic cells at significantly higher levels, compared to other cell types (e.g., miR-181 and miR-155; http://www.mirz.unibas.ch/cloningprofiles/). Furthermore, multiple miRNAs (termed “immunomiRs”) are predicted to preferentially target immune genes (Asirvatham et al., 2008). These miRNAs are crucial regulators of early hematopoiesis and lineage commitment (Chen et al., 2004). Correspondingly, Dicer knockout mice show severe defects in hematopoietic development, which affects all leukocyte compartments (Alemdehy et al., 2012; Koralov et al., 2008; Xu et al., 2012). miR-181a, for instance, is critical for the development of virtually all lymphocyte subsets (Cichocki et al., 2011; Li et al., 2007; Spierings et al., 2011), while miR-155, miR-17~92 and miR-150 participate in the differentiation of B and T lymphocytes (Garzon and Croce, 2008). Consistently, these miRNAs also play pivotal roles as oncogenes and as tumor suppressor genes in hematological malignancies, through various mechanisms that are incompletely understood (Jiang et al., 2012; Sandhu et al., 2012; Schwind et al., 2010).
Among the several leukocyte-specific miRNAs, miR-142 is one of the most abundant sequences in the T and B lymphoid lineages (Chen et al., 2004; Wu et al., 2007), as well as in the entire myeloid compartment (Chen et al., 2004). Perhaps due to its critical role in the development, survival and/or activity of many immune cells, relatively little is known about the functions of miR-142. It has been recently reported that, along with miR-29a, miR-142-3p directly silences the cyclin T2 gene, preventing the release of hypophosphorylated retinoblastoma and therefore promoting monocytic differentiation (Wang et al., 2012). Correspondingly, a decrease in miR-142-3p levels is observed in blasts from acute myeloid leukemia (Lv et al., 2012; Wang et al., 2012), while miR-142-3p is translocated in human B cell malignancy (Robbiani et al., 2009).
In physiological responses in lineage-committed immune cells, miR-142-3p controls IL-6 production by LPS-stimulated dendritic cells (DCs), and dampens miR-142-3p activity reducing endotoxin-induced mortality (Sun et al., 2011). This particular strand regulates the production of cAMP by targeting adenylyl cyclase 9 in regulatory T cells, thus mitigating their suppressor activity (Huang et al., 2009). Further supporting a role for miR-142-3p in regulating T cell activity, CD4 T cells in patients with systemic lupus erythematosus show reduced expression of both (miR-142-3p/5p) strands, which promotes T cell activation and eventually leads to B cell hyperstimulation (Ding et al., 2012). In contrast, miR-142-5p is overexpressed in peripheral blood in patients undergoing acute transplant rejection (Anglicheau et al., 2009). Although the biological effects of these changes are unclear, the levels of both miR-142-3p and miR-142-5p are dramatically decreased in TCR-activated naïve CD8 T cells, but their expression becomes partially restored in memory lymphocytes (Wu et al., 2007). Taken together, these results point to a role for miR-142 in promoting T cell quiescence, but more mechanistic studies are needed to clarify its complicated immunobiology.
In addition, miR-223 is selectively expressed in neutrophils and macrophages. Manipulation of miR-223 profoundly affects hematopoiesis (O’Connell et al., 2012), but understanding its functional role has been complicated due to conflicting results in different experimental systems. It was initially thought that miR-223 promoted the differentiation of myeloid progenitors into granulocytes, based on ectopic expression studies in human leukemic cell lines (Fazi et al., 2007). However, miR-223 knockout mice have increased counts of granulocyte progenitors in the bone marrow and hypermature neutrophils in the circulation (Johnnidis et al., 2008). Further studies unveiled an elegant mechanism, whereby C/EBPα and NFI-A compete for binding to the miR-223 promoter. During differentiation, C/EBPα replaces NFI-A, which leads to the up-regulation of miR-223, which in turns silences NFI-A (Fazi et al., 2005; Lindsay, 2008). Thus, an autoregulatory loop controls miR-223 expression, which promotes granulocytic differentiation. Furthermore, deletion of Mef2c, a direct target of miR-223 involved in determining cell lineage choice between B lymphocytes and myeloid cells (Stehling-Sun et al., 2009), results in a partial correction of granulocyte counts, although these granulocytes maintain an activated phenotype.
Besides its role in granulocytes, miR-223 is required for regulating erythrocyte development by targeting FBXW7 (Xu et al., 2010). In lymphocytes, miR-223 may be important for the effector function of NK and CD8 T cells, because granzyme B is as a direct target of miR-223 (Fehniger et al., 2010).
miRNAs are critical for the function of antigen-presenting cells
Besides miRNAs that are mostly exclusively found in hematopoietic cells in healthy adults, there are multiple miRNA sequences differentially expressed in some leukocyte subsets, or that preferentially target a set of transcripts that are important for the functions of a lineage of immune cells, including myeloid leukocytes. For instance, miR-143 and miR-145 are preferentially or selectively expressed by neutrophils (Allantaz et al., 2012), while miR-378 is only expressed in monocytes (Allantaz et al., 2012).
There are other miRNA sequences that are crucial for the orchestration of adaptive immunity. Among the miRNAs that are most relevant for the function of antigen-presenting cells, miR-146 and miR-155, along with miR-125b, respond to the activation of pattern recognition receptors, including Toll-Like Receptors (TLRs). miR-155 in particular is up-regulated in macrophages and DCs in response to endotoxin (Rodriguez et al., 2007), TNFα (Tili et al., 2007), or a synergistic cocktail of CD40 and TLR3 agonists (Cubillos-Ruiz et al., 2012; Scarlett et al., 2009). All these signals elicit DC activation and promote effective antigen presentation to T cells. miR-155 is also the only miRNA substantially up-regulated by primary macrophages stimulated with a TLR3 agonist plus interferon-beta (O’Connell et al., 2007; Rodriguez et al., 2007). Interestingly, DCs matured in the absence of miR-155 can express MHC–II and costimulatory molecules at levels comparable to those seen in identically treated matured wild-type DCs, but miR-155-defficient DCs fail to activate T cells, consistent with defective antigen presentation (Rodriguez et al., 2007). Correspondingly, mice that are deficient for miR-155 are also severely immunodeficient (Rodriguez et al., 2007; Thai et al., 2007). As commented below, we have also demonstrated that supplementing miR-155 in (miR-155low) tumor-associated regulatory DCs (Cubillos-Ruiz et al., 2012) induces genome-wide transcriptional changes that transform them from an immunosuppressive to an immunostimulatory cell type (Cubillos-Ruiz et al., 2012). Confirmed direct targets of miR-155 include immunosuppressive PU.1 (Vigorito et al., 2007), SOCS1 (Androulidaki et al., 2009) and CD200 (Cubillos-Ruiz et al., 2012). They also included C/epb-β, a regulator of the immunosuppressive environment created by aberrant myelopoiesis in established tumors (Marigo et al.), along with multiple mediators of transforming growth factor-beta (Tgf-β) signaling pathway, including Tgfβ1, Smad1, Smad6 and Smad7 (Rai et al., 2010). We finally demonstrated that Ccl22, which recruits regulatory T cells to the tumor microenvironment (Curiel et al., 2004), is also down-regulated in the presence of enhanced miR-155 activity (Cubillos-Ruiz et al., 2012). miR-155 is therefore the paradigm of a stimulatory “immunomiR”, and perhaps the most obvious target for therapeutic interventions aimed to modulate miRNA expression. Remarkably, pro-inflammatory intracellular factors released from damaged cells that serve as danger signals or ligands (such as High Mobility Group B1 (HMGB1)), do not appear to up-regulate miR-155, but alter the expression of other miRNAs such as miR-34c and miR-214 (Unlu et al., 2012).
Besides miR-155, miR-146 also participates in TLR signaling (Sonkoly et al., 2008) and is up-regulated in macrophages and DCs upon NF-κB nuclear translocation (Taganov et al., 2006). However, unlike miR-155, miR-146a inhibits NF-κB signaling, thus acting as an anti-inflammatory mediator. Consistent with a role as an inhibitor of excessive inflammation, miR146a is significantly decreased in leukocytes from systemic lupus erythematosus patients (Tang et al., 2009). Furthermore, miR-146a plays a negative regulatory role in the development of myeloid cells, so that mice deficient in miR-146a display an excessive proliferation of leukocytes of the myeloid lineage (Boldin et al., 2011). These effects appear to result from the up-regulation of the M-CSF receptor, although CSFR1 is not a direct target of miR-146a (Boldin et al., 2011).
Of note, miRNAs that regulate the function of myeloid leukocytes are particularly relevant for therapeutic targeting, due to the spontaneous endocytic activity of these leukocytes, which allows systemic or local delivery of RNA mimetics in the form of micro- or nano-particles.
miRNAs are critical for the function of lymphocytes
Among the miRNAs that are known to be most critical for the development of the T cell compartment, perhaps the most investigated is miR-181. Increasing miR-181a expression in mature T cells augments the sensitivity to peptide antigens and prevents the abrogation of cytotoxic function by downregulating multiple phosphatases. This increases the accumulation of specific phosphorylated intermediates and therefore enhances the T cell’s response to T cell receptor signaling (Li et al., 2007). These targets include DUSP6, and it has been recently proposed that a decline in miR-181a expression with age impairs TCR sensitivity by enhancing DUSP6 activity (Li et al., 2012). In addition, miR-181a promotes the deletion of particular clones with moderate affinity by modulating the threshold of TCR activation during thymic selection (Ebert et al., 2009).
Independent of its effects on hematopoietic development, the overall effects of miR-181 (e.g., activation vs. unresponsiveness) on mature, peripheral effector T cells are less clear (unpublished observations). It has been published, however, that functional miRNAs may not be absolutely required for the initial activation of effector T cells, although they are essential for their survival and functions (Zhang and Bevan, 2010). In CD8 T cells, a limited set of miRNAs (miR-16, miR-21, miR-142-3p, miR-142-5p, miR-150, miR-15b and let-7f) are expressed at significantly higher levels at any stage of activation/differentiation, and, together, they account for ~60% of all miRNAs (Wu et al., 2007). With the exception of miR-21, all of these miRNAs are significantly down-regulated upon TCR-mediated T cell activation, and their expression levels are restored back to intermediate levels when these stimulated T cells acquire a memory phenotype (Wu et al., 2007). In contrast, miR-21, a major inhibitor of Th1 polarization that targets IL12 and IFN-γ (Lu et al., 2011), is up-regulated in stimulated and memory lymphocytes, when compared to their naïve counterparts.
Furthermore, the four members of the miR-29 family have been identified as important regulators of lymphocyte activities. Thus, miR-29 suppresses immune responses by directly targeting IFN-γ in NK cells, CD4 and CD8 T lymphocytes (Ma et al., 2011). In addition, the transcription factors T-bet and Eomes, which also induce IFN-γ, are regulated by miR-29. Thus, miR-29 regulates helper T cell differentiation by repressing multiple target genes (Steiner et al., 2011).
As a paradigmatic immunostimulatory miRNA, miR-155 is also required for the function of T lymphocytes (Rodriguez et al., 2007). This, combined with the inability of miR-155-deficient DCs to activate T cells, contributes to explain the severe immunodeficiency observed in miR-155 knockout mice (Rodriguez et al., 2007; Thai et al., 2007), and occurs at multiple levels. Firstly, miR-155 is required for T helper cell differentiation and, subsequently, an optimal T cell-dependent antibody response (Thai et al., 2007). In addition, miR-155-deficient T cells show impaired expansion in response to CD3/CD28 activation and secrete less IFN-γ and IL-17 than their wild-type counterparts (Oertli et al., 2011). In contrast, by targeting SOCS1, miR-155 enhances Treg, but also Th17 differentiation (Yao et al., 2012). Nevertheless, an overall activating role is supported by the fact that miR-155 promotes autoimmune inflammation by enhancing inflammatory T cell development (O’Connell et al., 2010), while decreasing its expression ameliorates murine autoimmune encephalomyelitis (Murugaiyan et al., 2011).
In addition to its effects on T cells, miR-181 also promotes the activation of cells of the B lymphocyte lineage, and the level of miR-181a expression inversely correlates with overall survival and progression-free survival in B-cell lymphoma patients (Alencar et al., 2011). miR-155 also has corresponding effects on B cells, and Eμ-miR-155 transgenic mice have aberrant B cell proliferation and leukemia through dysregulation of the BCL6 transcriptional machinery (Sandhu et al., 2012).
Several other miRNAs are also critical for B cell development and functionality, including miR-17~92. Thus, mice deficient in miR-17~92 show higher expression of Bim, which promotes apoptosis and subsequently inhibits B cell development at the pro-B to pre-B transition (Ventura et al., 2008). Correspondingly, transgenic mice with increased expression of miR-17~92 develop lymphoproliferative disease and autoimmunity (Xiao et al., 2008).
Finally, dysregulation of miR-29 has also been associated with the pathogenesis of indolent B-CLL (Santanam et al., 2010).
In regards to miRNAs that regulate the function of myeloid leukocytes, miRNAs that govern T cell functions, in particular, can be ectopically and stably supplemented with, for instance, viral vectors. These lymphocytes, with enhanced effector or regulatory functions, could then be adoptively transferred and exert their activity in vivo at, for instance, tumor locations or sites of inflammatory damage, respectively.
miRNAs in anti-tumor immunity: The role of miR-155
Changes in the miRNA expression profile in tumors vs. healthy tissue have been the focus of hundreds of publications in recent years. However, epigenetic regulation of the function of the immune cells that participate in anti-tumor immunity and infiltrate the tumor microenvironment has received comparatively little attention. However, the crucial role of hematopoietic cells in both promoting malignant progression and eliminating nascent tumors - or at least adding immune pressure against established malignancies - is today beyond doubt among immunologists. T cell infiltration is significantly associated with better outcomes in cancer patients (Clemente et al., 1996; Galon et al., 2006; Yoshimoto et al., 1993; Zhang et al., 2003), and T cell based immunotherapies have already produced impressive clinical results (Dudley et al., 2002; Kalos et al., 2011; Porter et al., 2011). In addition, blocking immunosuppressive checkpoints such as CTLA4 and the PD-1/PD-L1 axis unleashes immune cells to regain control of advanced tumors (Pardoll and Drake, 2012; Topalian et al., 2012). Modulating the expression of the miRNAs that govern the immunosuppressive activity of certain microenvironmental leukocyte subsets (e.g., myeloid cells), or the activity of those miRNAs that enhance the activity of anti-tumor effector lymphocytes, could represent a valuable option for the design of novel therapeutic interventions. Supporting the feasibility of this proposition, we have recently documented the potential of transforming the phenotype of tumor-associated DCs in ovarian cancer-bearing hosts, by delivering polyplexes in the nanometer scale (Cubillos-Ruiz et al., 2009a) that carry synthetic miR-155 (Cubillos-Ruiz et al., 2012). Thus, tumor-associated myeloid cells (mostly macrophages and regulatory DCs) typically express very low levels of miR-155 and, accordingly, phagocytize tumor antigens but rather than eliciting protective immunity promote tolerance (Cubillos-Ruiz et al., 2010a; Huarte et al., 2008; Nesbeth et al., 2009; Nesbeth et al., 2010; Scarlett et al., 2009; Scarlett et al., 2012). However, the antigen-processing capacity of these leukocytes can be promoted in vivo and in situ, simply by supplementing miR-155 in an accessible microenvironment such as the peritoneal cavity (e.g., in ovarian cancer). Enhancing the activity of this miRNA resulted in down-regulation of multiple immunosuppressive genes, including Socs1, PU.1, TGFβ and Cebpβ, which transformed tumor DCs from an immunosuppressive to an immunostimulatory cell type. Non-viral miR-155 delivery therefore emerges as a transient, safe and effective intervention that is ready for clinical testing.
It is important to note, however, that dysregulation of miR-155 expression has also been associated with enhanced malignant potential. miR-155 is overexpressed in lymphomas and acute myeloid leukemia (Xiao and Rajewsky, 2009), and stable overexpression of miR-155 in hematopoietic progenitors causes a myeloproliferative disorder (O’Connell et al., 2008). In addition, constitutive (sustained) overexpression of miR-155 in the B cell lineage induces pre-B cell proliferation and promotes B cell malignancies (Costinean et al., 2006). Nevertheless, our experimental results demonstrate that the abundant phagocytes in the tumor microenvironment selectively incorporate miRNA mimetic compounds as long as these are complexed within micro- or nano-particles. In addition, their expression should be transient in the absence of viral vectors, and thereforemuch safer. Correspondingly, transient increase of miR-155 in lineage-committed (terminal and short-lived) myeloid cells never resulted in secondary tumors in our experimental systems (Cubillos-Ruiz et al., 2012). In fact, oncogenesis and adaptive immunity typically utilized common pathways because robust adaptive immune responses require rapid expansion of leukocytes. Finally, although miR-155 in solid tumors has been repeatedly assumed to be overexpressed in tumor cells, our recent optimization of a new detection technique in a panel of breast, colorectal, lung, pancreas, and prostate carcinomas showed that miR-155 is predominantly confined to immune cells in the tumor microenvironment (Sempere et al., 2010).
miRNAs in autoimmunity and transplantation
While multiple mechanisms of tolerance typically converge in the tumor microenvironment of virtually all solid tumors, miRNAs governing immune responses play an opposite role in pathological conditions involving excessive and harmful immune responses. Correspondingly, opposite expression levels of miR-155 have been critically involved in multiple inflammatory and autoimmune conditions (Shen et al., 2012). For instance, the contribution of excessive miR-155 activity to the pathogenesis of systemic lupus erythematosus has been mechanistically demonstrated. Thus, miR-155 and, interestingly, miR-155*, co-operatively promote type I IFN production by human plasmacytoid DCs.
In a different setting of deleterious immunity - acute graft-versus-host disease - miR-155 expression was (expectedly) up-regulated in T cells, in both mice and humans (Ranganathan et al., 2012). In addition, mice receiving miR-155-deficient donor lymphocytes had markedly reduced lethal acute graft-versus-host disease. Most importantly, down-regulating miR-155 expression with antagomiRs decreased the severity of rejection and increased survival in experimental models. miR-155 therefore emerges as a target for therapeutic intervention also in transplantation, in this case through systemic delivery of antagonists.
In addition, several studies have reported the potential of miRNA changes in peripheral blood cells and allograft biopsies as biomarkers in solid organ transplantation (Anglicheau et al., 2009; Sui et al., 2008). Besides the recurrent involvement of miR-155, hematopoietic-specific miRNAs (miR-142-5p and miR-223) were found by Anglicheau and colleagues to be overexpressed in both biopsies and peripheral leukocytes, although phytohaemagglutinin activation appears to decrease miR-223 expression (Anglicheau et al., 2009). However, Sui and colleagues reported a very different set of differently expressed miRNAs in acute rejection after renal transplantation, which includes preferential up-regulation of miR-125a and miR-629, and preferential down-regulation of miR-324-3p and miR-611 (Sui et al., 2008).
Role of miRNAs during HIV infection
As stated above, miRNAs play critical roles in regulating several developmental, physiological and pathological processes that involve different cellular subsets of the immune system. Thus, it is not surprising that these small RNAs could also participate in enhancing or inhibiting the course of T cell-targeting viral infections such as HIV. Bioinformatic analyses first predicted multiple binding sites for host miRNAs on various regions of the HIV genome (Hariharan et al., 2005), suggesting that cellular miRNAs could directly target HIV transcripts to modulate the infectious life cycle. Functional experiments have confirmed that HIV mRNAs interact with the RNA induced silencing complex (RISC) and that disrupting cellular P body structures enhances HIV viral production and infectivity (Nathans et al., 2009). Specifically, miR-29a expressed by T cells is capable of repressing HIV replication by targeting the viral 3’UTR (Nathans et al., 2009). In addition, it has been reported that miR-29 family members can directly target HIV nef in order to inhibit viral replication (Ahluwalia et al., 2008). Interestingly, the miR-29 family has recently been shown to regulate the production of interferon gamma in T cells by directly targeting the Tbx21 and Ifng genes (Steiner et al., 2011), indicating that miR-29 could also participate during the course of HIV infection by altering the proportions of circulating or tissue-specific Th1, Th2 and Th17 cells, which may play different roles during the course of infection (Ancuta et al., 2010). Nevertheless, it remains elusive whether miR-29 plays a protective role during in vivo infection and if HIV can counteract the effects of miR-29 by reducing its expression in T cells during acute or chronic infection in patients.
miR-28, miR-125b, miR-150, miR-223 and miR-382 have been demonstrated to play a crucial role in HIV infection by promoting viral latency in resting T cells (Huang et al., 2007). While anti-retroviral therapy has greatly reduced mortality in HIV-infected individuals, it is becoming increasingly clear that this treatment does not completely eradicate the virus. In fact, HIV can efficiently establish latent infection in a small proportion of resting T cells by stably integrating its proviral DNA into the host genome, a mechanism that restricts production of HIV proteins and that consequently enables the virus to evade immune responses and anti-retroviral drugs. Interestingly, this cluster of miRNAs is preferentially expressed in resting T cells compared with activated T cells, and has also been described to directly target the HIV 3’UTR. Loss-of-function experiments using a cocktail of inhibitors targeting all five miRNAs demonstrated viral re-activation in T cells isolated from HIV-infected patients treated with anti-retrovirals, indicating that these cellular miRNAs control viral protein expression during latency to promote an efficient HIV reservoir, and that a panel of miRNA inhibitors such as antagomirs or locked nucleic acids (LNAs) could be used in the future for therapeutic purposes.
Of note, host miRNAs can also influence HIV replication by targeting cellular factors that are utilized by the virus to undergo productive infection. Seminal studies by Triboulet and colleagues first demonstrated that the miR-17~92 cluster directly targets expression of the histone acetyltransferase PCAF, a cellular cofactor required for Tat activation and HIV replication during infection (Triboulet et al., 2007). Importantly, HIV markedly represses expression of miR-17/92 in vitro in T cells as a novel pathogenic mechanism to promote viral replication. However, it remains unknown whether members of the miR-17~92 cluster are also downregulated in T cells from HIV-infected individuals and if the levels of miR-17~92 in T cells from these patients correlate with CD4 counts or plasma viral loads. Strikingly, HIV infection can also trigger upregulation of several miRNAs (Triboulet et al., 2007), but a role for HIV-induced cellular miRNAs during infection and progression to AIDS via T cell depletion has not been identified.
The let-7 family represents another example of miRNAs that can directly influence immune responses to HIV. It has been reported that HIV infection rapidly triggers IL-10 expression and secretion by T cells, a process that correlates with reduced expression of let-7 family members (Swaminathan et al., 2012). Interestingly, the Il10 3’UTR is directly targeted by let-7 miRNAs and their expression is dramatically downregulated in CD4 T cells isolated from patients infected with HIV, compared with uninfected individuals (Swaminathan et al., 2012). Thus, the decrease in let-7 miRNAs in HIV-infected CD4 T cells may provide the virus with an important survival advantage via upregulation of immunosuppressive IL-10.
Finally, it has been demonstrated that a single nucleotide polymorphism (SNP) 35 kb upstream of HLA-C associates with control of HIV, and with levels of HLA-C transcripts and cell-surface expression (Kulkarni et al., 2011). Elegant work by Carrington and colleagues has demonstrated that variations within HLA-C 3’UTR regulate binding of miR-148 to its target site, thus resulting in lower surface expression of alleles that bind this miRNA and higher expression of HLA-C alleles that escape post-transcriptional regulation (Kulkarni et al., 2011). Strikingly the 3’ UTR variant also associates strongly with control of HIV, likely contributing to the effects of genetic variation encoding the peptide-binding region of the HLA class I loci.
miRNA supplementation/inhibition: The delivery platform
The crucial role of miRNAs in the orchestration of immune responses in many pathological conditions opens new avenues for therapeutic interventions, which can be based on augmenting or dampening the activity of one or more immunomiRs.
As commented above, our results demonstrate the feasibility of delivering transiently expressed but functional immunostimulatory miRNAs using a non-viral approach. Tumor-associated phagocytes are ideal targets for the use of nanomaterials due to their unforced enhanced endocytic activity, which allows preferential (if not selective) engulfment of polyplexes and lipoplexes carrying functional miRNA mimetic compounds. In our experimental systems, we tested a variety of antibody-targeted and untargeted lipoplexes and polymers to complex synthetic miR-155. Eventually, superior results were observed with the use of polyethylenimines (PEI), a biocompatible polymer currently in clinical trials that stabilizes double-stranded RNA and forms nanocomplexes that are avidly taken up by phagocytic cells (Cubillos-Ruiz et al., 2009a; Cubillos-Ruiz et al., 2009b; Cubillos-Ruiz et al., 2010b). Importantly, we have never been able to preferentially target epithelial tumor cells with nanomaterials (antibody-targeted or not), primarily due to poor cellular uptake by cancerous cells, compared to surrounding myeloid leukocytes. Therefore, rather than aiming to correct changes in miRNA expression in tumor cells, we propose that modulating the activity of immunomiRs in immune cells is a much more feasible approach, which also results in dramatic therapeutic effects (Cubillos-Ruiz et al., 2012).
In this context, tumors that are reachable for intratumoral administration, or diseases such as ovarian cancer, which are accessible through intraperitoneal injection, are ideal for modulation of miRNA expression. While increasing or diminishing the expression of crucial miRNAs in other inflammatory and autoimmune conditions is also feasible, RNA delivery typically results in activation of multiple TLRs and other immune sensors, which will boost additional responses a non-specific manner independently of sequence specificity (Cubillos-Ruiz et al., 2012; Cubillos-Ruiz et al., 2009a). It should be noted, however, that DNA (not RNA)-carrying, PEI-based nanocomplexes were also recently found to induce a rapid (and non-specific) increase in type I Interferons (IFNs) when they were, as expected, selectively taken-up by phagocytes in vivo (Huang et al., 2012). However, the authors propose that IFN type I receptor signaling eventually lead to IDO-mediated Treg activation and, correspondingly, regulatory outcomes. Because we have shown opposite (immunostimulatory) non-specific effects upon delivery of PEI-RNA polyplexes, it is likely that the systemic effects depend on target cells, which vary with different pathological conditions. The same platform could be therefore theoretically used to deliver antagomiRs or to supplement inhibitory miRNAs in autoimmunity and transplantation, but more mechanistic studies are needed.
In the context of HIV, recent advances in the generation of humanized mice have made possible the study and manipulation of HIV infection in vivo in a small animal model. Interestingly, various groups have reported the optimization of T cell-targeting nanoparticles to systemically deliver siRNA in vivo in HIV-infected humanized mice ((Kim et al., 2010)). Therefore, these nanocomplexes could also be used to encapsulate and deliver miRNA mimicking reagents (for miRNA supplementation) or antagomiR/LNA oligonuclotides for inhibiting aberrant miRNA dysregulation in T cells during infection.
miRNA supplementation/inhibition: Functional miRNA preparations
Besides the vehicle used for in vivo delivery of miRNA mimetics, the key element for success is the optimization of synthetic RNA that can be functionally active when processed by the host and recapitulate the activities of endogenous miRNAs. In certain populations, this can be achieved through ectopic expression with viral vectors. For instance, immunostimulatory miRNAs could be theoretically overexpressed in ex vivo primed tumor-reactive T cells, which could be further used for adoptive transfer. However, in vivo delivery of miRNA mimetics to tissue resident cells does not allow this approach.
Some commercial reagents are available from various suppliers but they are expensive, have not been tested in vivo, and the sequence and structure is not disclosed. However, we have recently reported a platform for the design and cost-effective production of custom synthetic miRNAs. Our optimization studies demonstrated that siRNA-like preparations consisting of two perfectly matching RNA strands that include the “guide” sequence of endogenous miR-155 do not recapitulate the activity of this miRNA. In contrast, 25-30 nt-long Dicer substrates (Kim et al., 2005), synthesized as RNA duplexes that include a bulged structure are much more effectively loaded into different Ago variants (referentially Ago2). They are alsodetected in vivo as overexpressed, functionally processed miR-155 at higher levels (Cubillos-Ruiz et al., 2012). Importantly, both reagents include the same core region (the mature sequence of miR-155), and in both cases thermodynamic criteria favored 5’ antisense strand uptake into RISC.
These dramatic differences in functionality, critical for the design of mimetic compounds for other sequences, are likely attributable to several issues: Firstly, it has been recently demonstrated that the overall structure of substrate RNA, including the position of an internal bulge, affects the position of Dicer cleavage (Gu et al., 2012). Secondly, the inclusion of one or more internal bulges, as in the endogenous sequence, reduces the thermostability of the RNA duplex, thus facilitating the “activation” (unwinding and upload of the guide strand into the RISC complex) of the synthetic miRNA. Thirdly, in the absence of asymmetrical bulges at particular positions, Dicer makes canonical cleavages 21 bp away from the 5′ end of the guide strand (19 bp away from the passenger strand) (Park et al., 2011). However, the position of loop (likely mimicked by terminal DNA nucleotides in Dicer substrates used as synthetic miR-155), also influences the position of Dicer cleavage. In fact, a “loop-counting rule” has been recently proposed, whereby Dicer cleaves precisely when it is able to recognize a single-stranded RNA sequence either from the loop region or internal bulge at a fixed distance (two nucleotides) relative to the site of cleavage.
Whatever the reasons are, the introduction of an internal bulge is critical for the functionality of miRNA mimicking sequences, and should be considered for future applications.
Besides supplementing miRNA activity, nucleic acids can be delivered in vivo and in vitro for specific silencing of overexpressed endogenous miRNAs in certain pathological conditions (Krutzfeldt et al., 2005). Chemically engineered oligonucleotides, termed ‘antagomiRs’, have been specifically optimized for this use, and proven to be effective upon intravenous administration. Therefore, the same platform used for augmenting the expression of selected miRNAs could be theoretically used to silence miRNAs with opposite activities, thus providing alternative tools for therapeutic interventions.
Finally, ectopic expression of “sponges” - tandem repetition of transcribed complementary sequences that “soak up” endogenous miRNAs (Ebert et al., 2007) – can be stably achieved ex vivo in cells such as lymphocytes, which could then be adoptively transferred.
Multiple viral and non-viral approaches are therefore available for in vivo modulation of miRNAs in immune cells, and therefore for immunotherapeutic interventions that should be both effective and mechanistically informative.
Figure 1. An internal bulge is critical for the functionality of miRNA mimicking sequences.
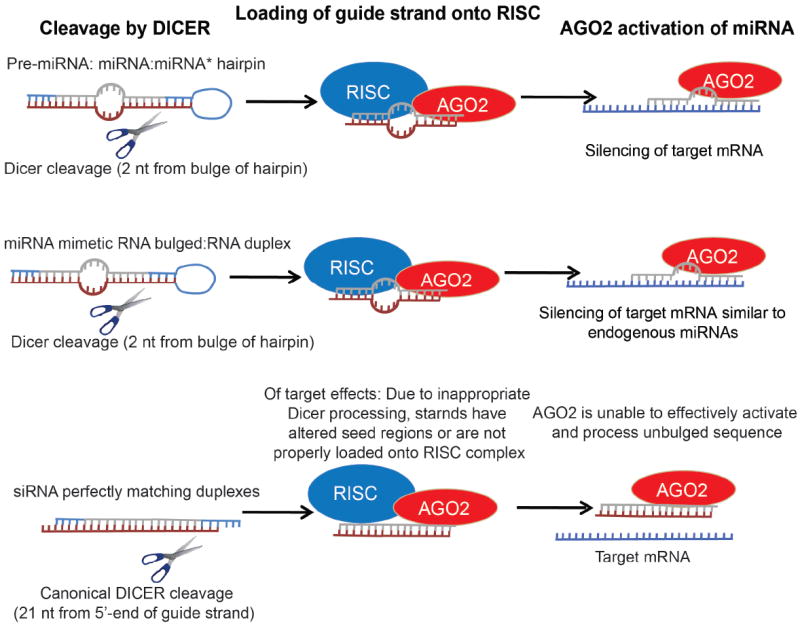
RNA duplexes that include a bulged structure, but not siRNA-like preparations, recapitulate the activity of endogenous miRNAs. A hypothetical model explaining these differential effects is depicted.
Figure 2. Peritoneal delivery of immunomiR-carrying polyplexes in ovarian cancer patients elicits anti-tumor immunity.
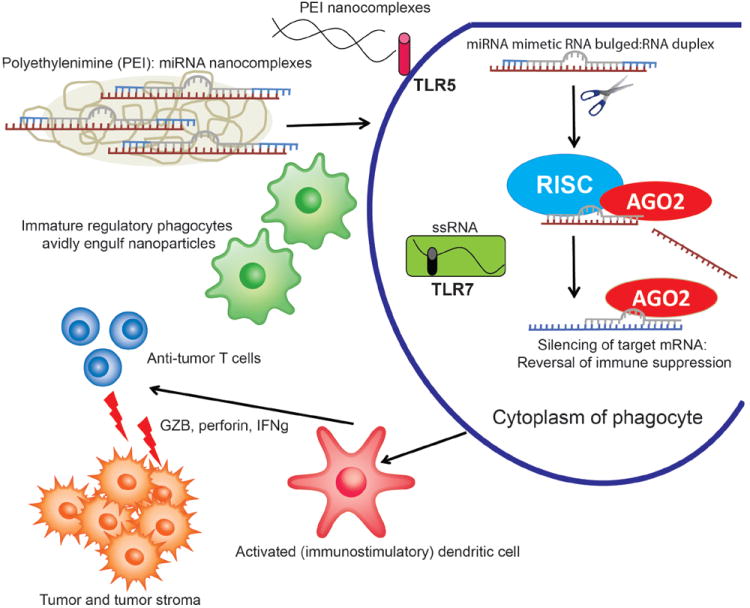
Intraperitoneally-injected nanoparticles encapsulating synthetic miR-155 are preferentially taken-up by regulatory phagocytes at ovarian cancer locations. Non-specific activation of TLR5 and TLR7, combined with silencing of immunosuppressive targets of endogenous immunomiRs elicits the activation of tumor-associated regulatory dendritic cells, transforming them from an immunosuppressive to an immunostimulatory cell type. In situ activated dendritic cells promote the expansion and function of anti-tumor T cells by effectively presenting tumor antigens engulfed in the tumor microenvironment.
References
- Ahluwalia JK, Khan SZ, Soni K, Rawat P, Gupta A, Hariharan M, Scaria V, Lalwani M, Pillai B, Mitra D, et al. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology. 2008;5:117. doi: 10.1186/1742-4690-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemdehy MF, van Boxtel NG, de Looper HW, van den Berge IJ, Sanders MA, Cupedo T, Touw IP, Erkeland SJ. Dicer1 deletion in myeloid-committed progenitors causes neutrophil dysplasia and blocks macrophage/dendritic cell development in mice. Blood. 2012;119:4723–4730. doi: 10.1182/blood-2011-10-386359. [DOI] [PubMed] [Google Scholar]
- Alencar AJ, Malumbres R, Kozloski GA, Advani R, Talreja N, Chinichian S, Briones J, Natkunam Y, Sehn LH, Gascoyne RD, et al. MicroRNAs are independent predictors of outcome in diffuse large B-cell lymphoma patients treated with R-CHOP. Clin Cancer Res. 2011;17:4125–4135. doi: 10.1158/1078-0432.CCR-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allantaz F, Cheng DT, Bergauer T, Ravindran P, Rossier MF, Ebeling M, Badi L, Reis B, Bitter H, D’Asaro M, et al. Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression. PLoS ONE. 2012;7:e29979. doi: 10.1371/journal.pone.0029979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancuta P, Monteiro P, Sekaly RP. Th17 lineage commitment and HIV-1 pathogenesis. Curr Opin HIV AIDS. 2010;5:158–165. doi: 10.1097/COH.0b013e3283364733. [DOI] [PubMed] [Google Scholar]
- Androulidaki A, Iliopoulos D, Arranz A, Doxaki C, Schworer S, Zacharioudaki V, Margioris AN, Tsichlis PN, Tsatsanis C. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–231. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglicheau D, Sharma VK, Ding R, Hummel A, Snopkowski C, Dadhania D, Seshan SV, Suthanthiran M. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci U S A. 2009;106:5330–5335. doi: 10.1073/pnas.0813121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol Immunol. 2008;45:1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, Garcia-Flores Y, Luong M, Devrekanli A, Xu J, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, Miller JS. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol. 2011;187:6171–6175. doi: 10.4049/jimmunol.1100835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente CG, Mihm MC, Jr, Bufalino R, Zurrida S, Collini P, Cascinelli N. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer. 1996;77:1303–1310. doi: 10.1002/(SICI)1097-0142(19960401)77:7<1303::AID-CNCR12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Contreras J, Rao DS. MicroRNAs in inflammation and immune responses. Leukemia. 2012;26:404–413. doi: 10.1038/leu.2011.356. [DOI] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Ruiz JR, Baird JR, Tesone AJ, Rutkowski MR, Scarlett UK, Camposeco-Jacobs AL, Anadon-Arnillas J, Harwood NM, Korc M, Fiering SN, et al. Reprogramming tumor-associated dendritic cells in vivo using microRNA mimetics triggers protective immunity against ovarian cancer. Cancer Res. 2012;72:1683–1693. doi: 10.1158/0008-5472.CAN-11-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Ruiz JR, Engle X, Scarlett UK, Martinez D, Barber A, Elgueta R, Wang L, Nesbeth Y, Durant Y, Gewirtz AT, et al. Polyethylenimine-based siRNA nanocomplexes reprogram tumor-associated dendritic cells via TLR5 to elicit therapeutic antitumor immunity. J Clin Invest. 2009a;119:2231–2244. doi: 10.1172/JCI37716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Ruiz JR, Fiering S, Conejo-Garcia JR. Nanomolecular targeting of dendritic cells for ovarian cancer therapy. Future Oncol. 2009b;5:1189–1192. doi: 10.2217/fon.09.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Ruiz JR, Martinez D, Scarlett UK, Rutkowski MR, Nesbeth YC, Camposeco-Jacobs AL, Conejo-Garcia JR. CD277 is a Negative Co-stimulatory Molecule Universally Expressed by Ovarian Cancer Microenvironmental Cells. Oncotarget. 2010a;1:329–328. doi: 10.18632/oncotarget.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Ruiz JR, Rutkowski M, Conejo-Garcia JR. Blocking ovarian cancer progression by targeting tumor microenvironmental leukocytes. Cell Cycle. 2010b;9:260–268. doi: 10.4161/cc.9.2.10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- Ding S, Liang Y, Zhao M, Liang G, Long H, Zhao S, Wang Y, Yin H, Zhang P, Zhang Q, et al. Decreased microRNA-142-3p/5p expression causes CD4+ T cell activation and B cell hyperstimulation in systemic lupus erythematosus. Arthritis Rheum. 2012;64:2953–2963. doi: 10.1002/art.34505. [DOI] [PubMed] [Google Scholar]
- Dudley ME, Wunderlich JR, Robbins PF, Yang JC, Hwu P, Schwartzentruber DJ, Topalian SL, Sherry R, Restifo NP, Hubicki AM, et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298:850–854. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert PJ, Jiang S, Xie J, Li QJ, Davis MM. An endogenous positively selecting peptide enhances mature T cell responses and becomes an autoantigen in the absence of microRNA miR-181a. Nat Immunol. 2009;10:1162–1169. doi: 10.1038/ni.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, Diverio D, Ammatuna E, Cimino G, Lo-Coco F, et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Fehniger TA, Wylie T, Germino E, Leong JW, Magrini VJ, Koul S, Keppel CR, Schneider SE, Koboldt DC, Sullivan RP, et al. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res. 2010;20:1590–1604. doi: 10.1101/gr.107995.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15:352–358. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang Y, Huang Y, Zhang F, Valdmanis PN, Kay MA. The Loop Position of shRNAs and Pre-miRNAs Is Critical for the Accuracy of Dicer Processing In Vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan M, Scaria V, Pillai B, Brahmachari SK. Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun. 2005;337:1214–1218. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- Huang B, Zhao J, Lei Z, Shen S, Li D, Shen GX, Zhang GM, Feng ZH. miR-142-3p restricts cAMP production in CD4+CD25- T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep. 2009;10:180–185. doi: 10.1038/embor.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- Huang L, Lemos HP, Li L, Li M, Chandler PR, Baban B, McGaha TL, Ravishankar B, Lee JR, Munn DH, et al. Engineering DNA nanoparticles as immunomodulatory reagents that activate regulatory T cells. J Immunol. 2012;188:4913–4920. doi: 10.4049/jimmunol.1103668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte E, Cubillos-Ruiz JR, Nesbeth YC, Scarlett UK, Martinez DG, Buckanovich RJ, Benencia F, Stan RV, Keler T, Sarobe P, et al. Depletion of dendritic cells delays ovarian cancer progression by boosting antitumor immunity. Cancer Res. 2008;68:7684–7691. doi: 10.1158/0008-5472.CAN-08-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Huang H, Li Z, Li Y, Wang X, Gurbuxani S, Chen P, He C, You D, Zhang S, et al. Blockade of miR-150 Maturation by MLL-Fusion/MYC/LIN-28 Is Required for MLL-Associated Leukemia. Cancer Cell. 2012;22:524–535. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalos M, Levine BL, Porter DL, Katz S, Grupp SA, Bagg A, June CH. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011;3:95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya E, Doudna JA. Biochemistry. Guided tour to the heart of RISC. Science. 2012;336:985–986. doi: 10.1126/science.1223549. [DOI] [PubMed] [Google Scholar]
- Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- Kim SS, Peer D, Kumar P, Subramanya S, Wu H, Asthana D, Habiro K, Yang YG, Manjunath N, Shimaoka M, et al. RNAi-mediated CCR5 silencing by LFA-1-targeted nanoparticles prevents HIV infection in BLT mice. Mol Ther. 2010;18:370–376. doi: 10.1038/mt.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kulkarni S, Savan R, Qi Y, Gao X, Yuki Y, Bass SE, Martin MP, Hunt P, Deeks SG, Telenti A, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472:495–498. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Yu M, Lee WW, Tsang M, Krishnan E, Weyand CM, Goronzy JJ. Decline in miR-181a expression with age impairs T cell receptor sensitivity by increasing DUSP6 activity. Nat Med. 2012;18:1518–1524. doi: 10.1038/nm.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- Lu TX, Hartner J, Lim EJ, Fabry V, Mingler MK, Cole ET, Orkin SH, Aronow BJ, Rothenberg ME. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J Immunol. 2011;187:3362–3373. doi: 10.4049/jimmunol.1101235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv M, Zhang X, Jia H, Li D, Zhang B, Zhang H, Hong M, Jiang T, Jiang Q, Lu J, et al. An oncogenic role of miR-142-3p in human T-cell acute lymphoblastic leukemia (T-ALL) by targeting glucocorticoid receptor-alpha and cAMP/PKA pathways. Leukemia. 2012;26:769–777. doi: 10.1038/leu.2011.273. [DOI] [PubMed] [Google Scholar]
- Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- Marigo I, Bosio E, Solito S, Mesa C, Fernandez A, Dolcetti L, Ugel S, Sonda N, Bicciato S, Falisi E, et al. Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity. 32:790–802. doi: 10.1016/j.immuni.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J Immunol. 2011;187:2213–2221. doi: 10.4049/jimmunol.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbeth Y, Scarlett U, Cubillos-Ruiz J, Martinez D, Engle X, Turk MJ, Conejo-Garcia JR. CCL5-mediated endogenous antitumor immunity elicited by adoptively transferred lymphocytes and dendritic cell depletion. Cancer Res. 2009;69:6331–6338. doi: 10.1158/0008-5472.CAN-08-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbeth YC, Martinez DG, Toraya S, Scarlett UK, Cubillos-Ruiz JR, Rutkowski MR, Conejo-Garcia JR. CD4+ T cells elicit host immune responses to MHC class II- ovarian cancer through CCL5 secretion and CD40-mediated licensing of dendritic cells. J Immunol. 2010;184:5654–5662. doi: 10.4049/jimmunol.0903247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertli M, Engler DB, Kohler E, Koch M, Meyer TF, Muller A. MicroRNA-155 is essential for the T cell-mediated control of Helicobacter pylori infection and for the induction of chronic Gastritis and Colitis. J Immunol. 2011;187:3578–3586. doi: 10.4049/jimmunol.1101772. [DOI] [PubMed] [Google Scholar]
- Pardoll D, Drake C. Immunotherapy earns its spot in the ranks of cancer therapy. J Exp Med. 2012;209:201–209. doi: 10.1084/jem.20112275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5’ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai D, Kim SW, McKeller MR, Dahia PL, Aguiar RC. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc Natl Acad Sci U S A. 2010;107:3111–3116. doi: 10.1073/pnas.0910667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan P, Heaphy CE, Costinean S, Stauffer N, Na C, Hamadani M, Santhanam R, Mao C, Taylor PA, Sandhu S, et al. Regulation of acute graft-versus-host disease by microRNA-155. Blood. 2012;119:4786–4797. doi: 10.1182/blood-2011-10-387522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, McBride KM, Klein IA, Stone G, Eisenreich TR, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu SK, Volinia S, Costinean S, Galasso M, Neinast R, Santhanam R, Parthun MR, Perrotti D, Marcucci G, Garzon R, et al. miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Emu-miR-155 transgenic mouse model. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1213764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santanam U, Zanesi N, Efanov A, Costinean S, Palamarchuk A, Hagan JP, Volinia S, Alder H, Rassenti L, Kipps T, et al. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proc Natl Acad Sci U S A. 2010;107:12210–12215. doi: 10.1073/pnas.1007186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlett UK, Cubillos-Ruiz JR, Nesbeth YC, Martinez DG, Engle X, Gewirtz AT, Ahonen CL, Conejo-Garcia JR. In situ stimulation of CD40 and Toll-like receptor 3 transforms ovarian cancer-infiltrating dendritic cells from immunosuppressive to immunostimulatory cells. Cancer Res. 2009;69:7329–7337. doi: 10.1158/0008-5472.CAN-09-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlett UK, Rutkowski MR, Rauwerdink AM, Fields J, Escovar-Fadul X, Baird J, Cubillos-Ruiz JR, Jacobs AC, Gonzalez JL, Weaver J, et al. Ovarian cancer progression is controlled by phenotypic changes in dendritic cells. J Exp Med. 2012;209:495–506. doi: 10.1084/jem.20111413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwind S, Maharry K, Radmacher MD, Mrozek K, Holland KB, Margeson D, Whitman SP, Hickey C, Becker H, Metzeler KH, et al. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:5257–5264. doi: 10.1200/JCO.2010.29.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Conejo-Garcia JR. Modulation of cancer progression by tumor microenvironmental leukocyte-expressed microRNAs. In: Biswas Subhra K., editor. Tumor Microenvironment and Myelomonocytic Cells. Rijeka, Croatia: InTech Open Access Publisher; 2012. pp. 221–54. [Google Scholar]
- Sempere LF, Preis M, Yezefski T, Ouyang H, Suriawinata AA, Silahtaroglu A, Conejo-Garcia JR, Kauppinen S, Wells W, Korc M. Fluorescence-based codetection with protein markers reveals distinct cellular compartments for altered MicroRNA expression in solid tumors. Clin Cancer Res. 2010;16:4246–4255. doi: 10.1158/1078-0432.CCR-10-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen N, Liang D, Tang Y, de Vries N, Tak PP. MicroRNAs-novel regulators of systemic lupus erythematosus pathogenesis. Nat Rev Rheumatol. 2012 doi: 10.1038/nrrheum.2012.142. [DOI] [PubMed] [Google Scholar]
- Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs and immunity: Novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008 doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Spierings DC, McGoldrick D, Hamilton-Easton AM, Neale G, Murchison EP, Hannon GJ, Green DR, Withoff S. Ordered progression of stage-specific miRNA profiles in the mouse B2 B-cell lineage. Blood. 2011;117:5340–5349. doi: 10.1182/blood-2010-10-316034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehling-Sun S, Dade J, Nutt SL, DeKoter RP, Camargo FD. Regulation of lymphoid versus myeloid fate ‘choice’ by the transcription factor Mef2c. Nat Immunol. 2009;10:289–296. doi: 10.1038/ni.1694. [DOI] [PubMed] [Google Scholar]
- Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, Matloubian M, Blelloch R, Ansel KM. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity. 2011;35:169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui W, Dai Y, Huang Y, Lan H, Yan Q, Huang H. Microarray analysis of MicroRNA expression in acute rejection after renal transplantation. Transpl Immunol. 2008;19:81–85. doi: 10.1016/j.trim.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Sun Y, Varambally S, Maher CA, Cao Q, Chockley P, Toubai T, Malter C, Nieves E, Tawara I, Wang Y, et al. Targeting of microRNA-142-3p in dendritic cells regulates endotoxin-induced mortality. Blood. 2011;117:6172–6183. doi: 10.1182/blood-2010-12-325647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Suzuki K, Seddiki N, Kaplan W, Cowley MJ, Hood CL, Clancy JL, Murray DD, Mendez C, Gelgor L, et al. Differential regulation of the Let-7 family of microRNAs in CD4+ T cells alters IL-10 expression. J Immunol. 2012;188:6238–6246. doi: 10.4049/jimmunol.1101196. [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, de Vries N, Tak PP, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, Activity, and Immune Correlates of Anti-PD-1 Antibody in Cancer. N Engl J Med. 2012 doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- Unlu S, Tang S, Wang E, Martinez I, Tang D, Bianchi ME, Zeh HJ, 3rd, Lotze MT. Damage associated molecular pattern molecule-induced microRNAs (DAMPmiRs) in human peripheral blood mononuclear cells. PLoS ONE. 2012;7:e38899. doi: 10.1371/journal.pone.0038899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XS, Gong JN, Yu J, Wang F, Zhang XH, Yin XL, Tan ZQ, Luo ZM, Yang GH, Shen C, et al. MicroRNA-29a and microRNA-142-3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood. 2012;119:4992–5004. doi: 10.1182/blood-2011-10-385716. [DOI] [PubMed] [Google Scholar]
- Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA profiling of naive, effector and memory CD8 T cells. PLoS ONE. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Guo K, Zeng Q, Huo J, Lam KP. The RNase III enzyme Dicer is essential for germinal center B-cell formation. Blood. 2012;119:767–776. doi: 10.1182/blood-2011-05-355412. [DOI] [PubMed] [Google Scholar]
- Xu Y, Sengupta T, Kukreja L, Minella AC. MicroRNA-223 regulates cyclin E activity by modulating expression of F-box and WD-40 domain protein 7. J Biol Chem. 2010;285:34439–34446. doi: 10.1074/jbc.M110.152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Ma YL, Liang W, Li HH, Ma ZJ, Yu X, Liao YH. MicroRNA-155 Modulates Treg and Th17 Cells Differentiation and Th17 Cell Function by Targeting SOCS1. PLoS ONE. 2012;7:e46082. doi: 10.1371/journal.pone.0046082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto M, Sakamoto G, Ohashi Y. Time dependency of the influence of prognostic factors on relapse in breast cancer. Cancer. 1993;72:2993–3001. doi: 10.1002/1097-0142(19931115)72:10<2993::aid-cncr2820721022>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, Makrigiannakis A, Gray H, Schlienger K, Liebman MN, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348:203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- Zhang N, Bevan MJ. Dicer controls CD8+ T-cell activation, migration, and survival. Proc Natl Acad Sci U S A. 2010;107:21629–21634. doi: 10.1073/pnas.1016299107. [DOI] [PMC free article] [PubMed] [Google Scholar]


