Significance
How body pattern evolves in nature remains largely unknown. Although recent progress has been made on the molecular basis of losing morphological features during adaptation to new environments (regressive evolution), there are few well worked out examples of how morphological features may be gained in natural species (constructive evolution). Here we use genetic crosses to study how threespine stickleback fish have increased their tooth number in a new freshwater environment. Genetic mapping and gene expression experiments suggest regulatory changes have occurred in the gene for a bone morphogenetic signaling molecule, leading to increased expression in the freshwater fish that have more teeth. Our studies suggest that changes in gene regulation may underlie both gain and loss traits during vertebrate evolution.
Keywords: Gasterosteus, polyphyodonty, craniofacial, adaptation, quantitative genetics
Abstract
Developmental genetic studies of evolved differences in morphology have led to the hypothesis that cis-regulatory changes often underlie morphological evolution. However, because most of these studies focus on evolved loss of traits, the genetic architecture and possible association with cis-regulatory changes of gain traits are less understood. Here we show that a derived benthic freshwater stickleback population has evolved an approximate twofold gain in ventral pharyngeal tooth number compared with their ancestral marine counterparts. Comparing laboratory-reared developmental time courses of a low-toothed marine population and this high-toothed benthic population reveals that increases in tooth number and tooth plate area and decreases in tooth spacing arise at late juvenile stages. Genome-wide linkage mapping identifies largely separate sets of quantitative trait loci affecting different aspects of dental patterning. One large-effect quantitative trait locus controlling tooth number fine-maps to a genomic region containing an excellent candidate gene, Bone morphogenetic protein 6 (Bmp6). Stickleback Bmp6 is expressed in developing teeth, and no coding changes are found between the high- and low-toothed populations. However, quantitative allele-specific expression assays of Bmp6 in developing teeth in F1 hybrids show that cis-regulatory changes have elevated the relative expression level of the freshwater benthic Bmp6 allele at late, but not early, stages of stickleback development. Collectively, our data support a model where a late-acting cis-regulatory up-regulation of Bmp6 expression underlies a significant increase in tooth number in derived benthic sticklebacks.
Understanding the developmental genetic basis of morphological evolution is a long-standing goal in biology (1, 2). Evolved morphological differences can be “loss” (regressive) traits, where morphological features are lost or reduced, or “gain” (constructive) traits, where morphological features are gained or increased. Although many of the traits best understood at the molecular level involve loss traits (1, 2), recent studies have begun to genetically dissect some evolved gain traits (3–5). However, whether gain traits have similar genetic architectures as loss traits and whether gain traits are also associated with cis-regulatory changes remains largely unknown.
Teeth are a classic vertebrate model system for studying morphological evolution, due to their excellent preservation in the fossil record. Teeth are also a classic vertebrate model system for organogenesis, because teeth, like many other organs, develop through reciprocal signaling interactions between epithelia and mesenchyme. Continuing efforts have produced a rich understanding of the genetic networks that orchestrate tooth morphogenesis in model systems (6). However, despite the wealth of knowledge about tooth evolution and development, we still know little about the number and type of genetic changes that accompany diversification of dental patterning during evolution.
Pharyngeal jaws and teeth, used during mastication in fish, are located in the posterior branchial segments in the fish’s throat (7, 8). In teleost fish, pharyngeal jaw patterning is an adaptive trait that covaries with diet and trophic niche (9). The rich phenotypic diversity of pharyngeal jaws and teeth in fish, coupled with the understanding of the genetic networks of tooth development from model organisms, offers an opportunity to understand the developmental genetic basis of evolved changes in tooth patterning.
The threespine stickleback (Gasterosteus aculeatus) fish has emerged as an excellent model system allowing for genetic dissection of evolutionary change in vertebrates (10). Sticklebacks have undergone an extensive adaptive radiation, independently colonizing thousands of freshwater lakes and creeks generated after widespread melting of glaciers at the end of the last ice age (11). The dietary shifts to larger prey accompanying freshwater adaptation have resulted in evolved changes in trophic morphology (12, 13). Despite striking morphological differences between marine and freshwater populations, hybrids are fertile, allowing forward genetic analysis of evolved differences. In several lakes, “species pairs” of benthic and limnetic stickleback morphs are found (13). In each of these lakes, a benthic species is adapted to feeding on macroinvertebrates in the littoral zone or deeper sediments. This derived diet differs from the diet of both the limnetic species and ancestral marine forms, both of which feed on smaller zooplankton. Benthic sticklebacks have evolved trophic adaptations matched for this specialized diet (13, 14). Here we describe evolved tooth gain, a heritable constructive increase in tooth number compared with ancestral marine fish, in a derived benthic stickleback population. We then apply quantitative genetics and developmental biology methods to begin to dissect the genetic and developmental basis of this evolved gain trait.
Results
Derived Benthic Fish Have Evolved Increases in Tooth Number.
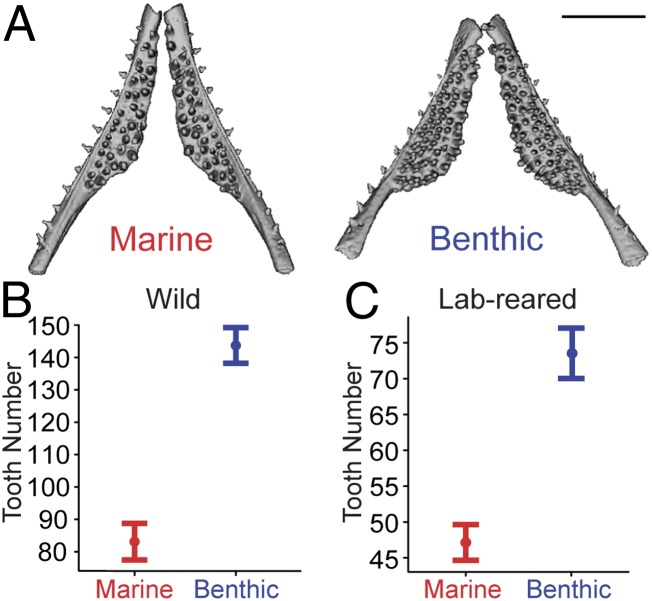
Because benthic sticklebacks have undergone an adaptive shift in diet, and because aspects of pharyngeal jaw patterning correlate with trophic niche in other species (9), we hypothesized that wild benthic fish have evolved changes in tooth patterning compared with ancestral marine fish. To test this hypothesis, we first quantified adult ventral pharyngeal tooth number from wild benthic fish from Paxton Lake, Canada (PAXB, hereafter called “benthic”) and an ancestral marine population from Rabbit Slough, Alaska (RABS, hereafter called “marine”). In these samples, the wild benthic population has an approximate twofold gain in tooth number compared with wild marine adults (Fig. 1 A and B and SI Appendix, Table S1). To determine whether this striking difference in tooth number is heritable, we quantified tooth number in adult fish from each population in a common laboratory-reared environment. The increased tooth number in benthic fish compared with marine fish is also seen in laboratory-reared stocks fed the same diet (Fig. 1C and SI Appendix, Table S1), showing that the tooth number differences have a large heritable component.
Fig. 1.
Heritable evolved tooth gain in derived benthic fish. (A) MicroCT images of wild adult stickleback ventral pharyngeal tooth plates of marine fish from Rabbit Slough, Alaska (Left) and benthic fish from Paxton Lake, Canada (Right). (Scale bar, 1 mm.) (B and C) Total ventral pharyngeal tooth number in wild (B) and laboratory-reared (C) adults shows benthic fish have significantly higher tooth counts (P = 8.8 × 10−7 and P = 4.9 × 10−7 in two-tailed t tests for wild and laboratory-reared, respectively). Error bars are SEM.
Evolved Changes in Tooth Patterning Occur Late in Development.
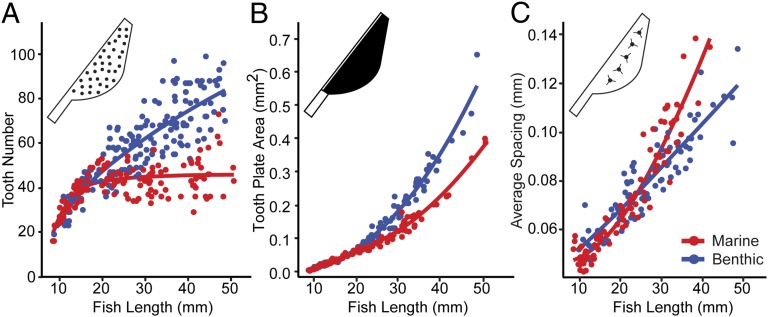
To examine when during development this evolved increase in tooth number appears, we generated a dense developmental time course of laboratory-reared fish from both marine and benthic populations and quantified tooth number (Fig. 2A). Tooth number between the two populations was not significantly different at early larval stages but began diverging when fish reached a total length of about 20 mm (Fig. 2A). Tooth number continued diverging after 20 mm, with benthic fish continuing to add new teeth while marine fish tooth number plateaus, resulting in the approximate twofold difference in tooth number seen in adults (Fig. 2A and SI Appendix, Table S2). The observed difference in tooth number between marine and benthic fish could arise either through an increase in the size of the tooth field and/or through an increased density of teeth in that field. We quantified tooth plate area (area of tooth-bearing portion of the fifth ceratobranchial bone) and average intertooth spacing throughout the developmental time courses. Relative to marine fish, in benthic fish tooth number and tooth plate area increased, while intertooth spacing decreased, with all three traits diverging late in development, after the 20-mm stage (Fig. 2 B and C and SI Appendix, Table S2). Thus, the late increase in tooth number in derived benthic fish is accompanied by at least two other late developmental patterning changes: an expansion of the tooth field and an increase in tooth density within that field. These increases in tooth number and tooth plate area and decreased intertooth spaces were also observed in F1 marine by benthic hybrid fish, showing that the benthic phenotypes are at least partially dominant (SI Appendix, Table S3).
Fig. 2.
Evolved differences in tooth number, area, and spacing appear late during development. Developmental time courses of laboratory-reared marine (red) and benthic (blue) fish of different total body lengths (x axis) for tooth number (A), tooth plate area (B), and tooth spacing (C). All three traits have diverged after 20-mm fish length.
Genome-wide Architecture of Evolved Changes in Tooth Number, Tooth Plate Area, and Spacing.
Previous work identified quantitative trait loci (QTL) controlling tooth number in a large genetic cross of 370 F2 fish derived from a Paxton benthic and a Japanese marine grandparent (15). Compared with Paxton benthic freshwater sticklebacks, Japanese marine sticklebacks, like Alaskan marine sticklebacks from the Rabbit Slough population, are low-toothed both in the wild and in the laboratory (SI Appendix, Table S1). To test whether tooth number, tooth plate area, and spacing are genetically separable traits, we measured tooth plate area and tooth spacing in 272 F2 progeny of the Japanese Marine × Paxton benthic intercross. In the F2 progeny, tooth plate area and spacing are significantly correlated with tooth number; however, tooth plate area and intertooth spacing are not correlated with each other (SI Appendix, Fig. S1). These results were robustly replicated in a Paxton benthic × Rabbit Slough marine F2 cross (SI Appendix, Fig. S1). Furthermore, a principal component analysis of these three tooth traits revealed that tooth plate area and spacing load orthogonally onto the first principal component (SI Appendix, Fig. S2), suggesting that the genetic controls of these tooth-patterning phenotypes are at least partially separable.
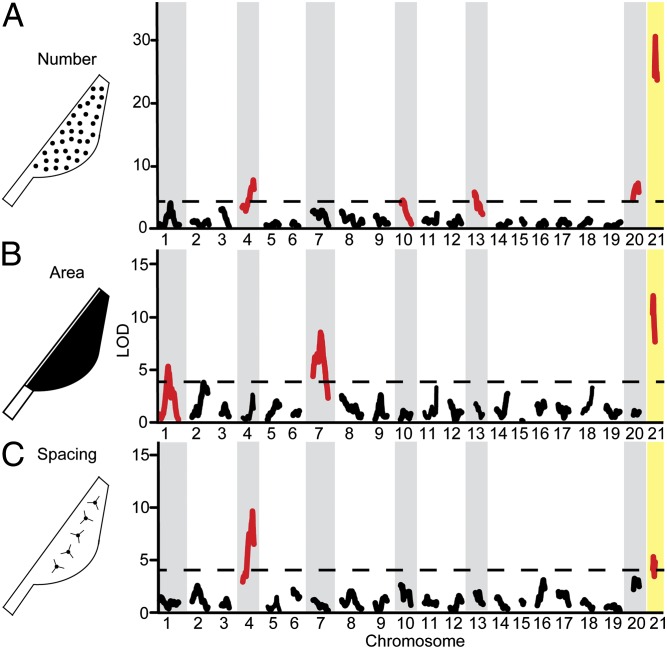
To further test whether benthic sticklebacks have evolved more teeth by modifying the genetic programs controlling tooth plate area and/or tooth spacing, we used genome-wide linkage mapping to map loci controlling these three traits (Fig. 3). All three tooth phenotypes have a strong genetic component, with different chromosome regions having effects on one, two, or all three phenotypes. The QTL with the largest effect on tooth number maps to chromosome 21, to a region where benthic alleles confer not only more teeth, but also larger tooth plate area and smaller intertooth spacing (Fig. 3 and SI Appendix, Table S4). Other significant QTL had specific effects on one or two, but not all three, tooth phenotypes. For example, a chromosome 4 tooth number QTL overlaps a large-spacing QTL but has no significant effect on tooth plate area. Conversely, QTL on chromosomes 1 and 7 control tooth plate area but had no significant effect on tooth number or spacing (Fig. 3 and SI Appendix, Table S4). Thus, the complex, polygenic architectures of tooth number, tooth plate area, and intertooth spacing are partially separable and include a large-effect QTL on chromosome 21 controlling all three traits.
Fig. 3.
Genome-wide linkage mapping for tooth number, tooth plate area, and tooth spacing in a marine × benthic F2 genetic cross. Genome-wide QTL mapping results for tooth number (A), tooth plate area (B), and intertooth spacing (C). All significant QTL are highlighted in red, and all chromosomes with significant effects on at least one tooth phenotype are shaded gray. The largest-effect tooth QTL on chromosome 21 is highlighted in yellow. The dashed line is the significance threshold of α = 0.05 determined by permutation tests (LOD scores of 4.1 for all three traits).
Bone Morphogenetic Protein 6 Maps Within the Major-Effect QTL Interval on Chromosome 21.
The largest-effect QTL controlling pharyngeal tooth patterning in this study maps to chromosome 21 and explains ∼30% of the variance in pharyngeal tooth number (Fig. 3A) (15). To test whether this large-effect tooth number QTL replicates in other wild-derived chromosomes from the Paxton lake benthic population and to ask when during development this QTL acts, we analyzed an additional F2 genetic cross at three different developmental stages: before the tooth number divergence in the time course, around the time of divergence, and an adult stage after tooth number diverged in the time course. The results show robust replication of the chromosome 21 tooth QTL at the two later time points. Indeed, the effects of the chromosome 21 tooth QTL get more significant as fish develop (SI Appendix, Fig. S3), mirroring the late-onset developmental appearance of the tooth number differences (Fig. 2A).
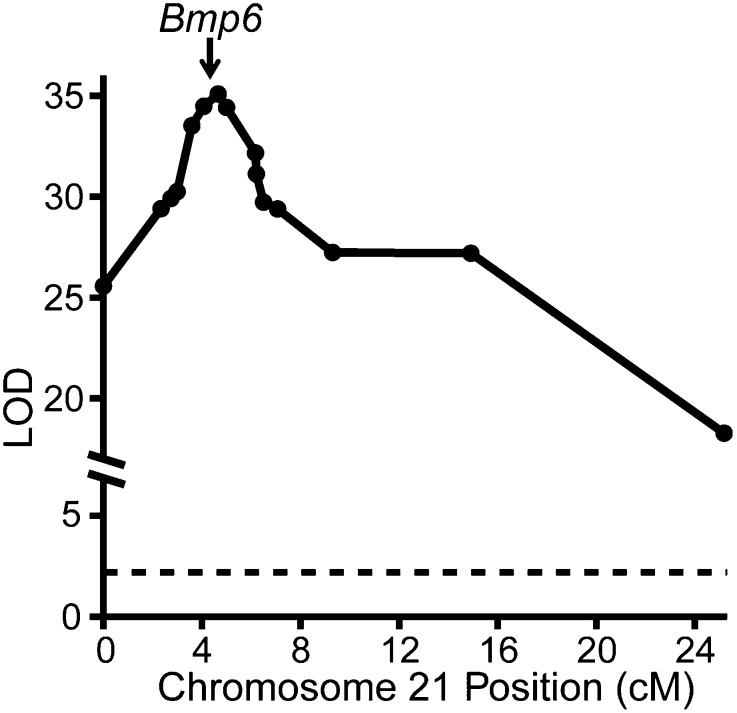
To begin to understand the molecular genetic basis of evolved tooth gain, we fine-mapped this chromosome 21 QTL. We first genotyped 1,004 additional F2 fish from the Paxton benthic × Japanese marine cross (16) to identify individuals with recombination events within the original chromosome 21 tooth QTL (15). This genotyping identified 91 recombinant F2 fish, which were then selectively phenotyped for tooth number. These data localized the QTL to a 2.6 cM, 2.56 Mb 1.5 logarithm of odds (LOD) candidate interval (Fig. 4) containing 59 predicted genes, including one outstanding candidate gene [Bone morphogenetic protein 6 (Bmp6)] and one additional gene (Tfap2a) whose mammalian ortholog also has documented tooth expression (http://bite-it.helsinki.fi/). During tooth development, the Bmp pathway plays an intimate role in specifying tooth number, shape, and size (17–22), strongly motivating Bmp6 as a candidate gene to underlie evolved tooth gain.
Fig. 4.
Fine mapping of the chromosome 21 tooth number QTL centers around Bmp6. Genotype–phenotype association for genetic markers (circles) across chromosome 21 (x axis). The position of Bmp6 is marked with the arrow, and the dashed line is the significance threshold of α = 0.05 determined by permutation tests (LOD score of 2.2).
Bmp6 Is Expressed in Developing Stickleback Teeth.
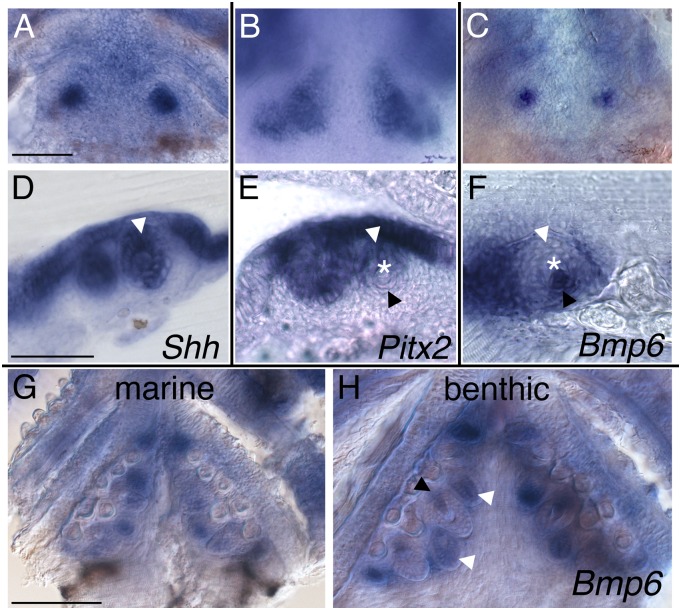
To test whether stickleback Bmp6 is expressed in developing teeth, we performed in situ hybridization on marine and benthic embryos and larvae (Fig. 5). We detected Bmp6 mRNA in developing stickleback tooth germs, in addition to mRNA of two known markers for tooth development in fish and mammals: Sonic hedgehog (Shh) and Pituitary homeobox 2 (Pitx2) (Fig. 5 A–C). In whole-mount embryos, Bmp6 expression at early tooth stages marked individual tooth germs similar to Shh and more restricted than Pitx2, which appeared to label the entire tooth-forming field (Fig. 5 A–C). Histological sections revealed that both Shh and Pitx2 were expressed in epithelial cells (Fig. 5 D and E), similar to the epithelial expression of these two genes in developing teeth in mice and other fish species (19, 23–26). In contrast, Bmp6 expression was dynamically detected in odontogenic epithelial and mesenchymal cells (Fig. 5F and SI Appendix, Fig. S4), similar to most other Bmp genes in fish and mice (19, 27–30). During tooth development at larval stages, Bmp6 showed complex but overall qualitatively similar expression patterns in marine and benthic fish (Fig. 5 G and H). Expression of Bmp6 in newly developing teeth persists throughout later stages, including in putative replacement tooth germs (SI Appendix, Fig. S5). This Bmp6 expression in developing teeth throughout embryonic and juvenile development supports the hypothesis that Bmp6 underlies the chromosome 21 tooth QTL. In contrast, no expression of Tfap2a was detected in developing teeth (SI Appendix, Fig. S6).
Fig. 5.
Bmp6 is expressed in developing stickleback teeth. Gene expression in developing benthic (A–F and H) and marine (G) stickleback teeth at 7.5 d postfertilization (dpf) (A–E) and 15 dpf (8 mm, F–H) revealed by in situ hybridization in whole-mount (A–C, G, and H) and 40-µm vibratome sections of comparably staged developing tooth germs (D–F and SI Appendix, Fig. S4). (A–F) Tooth markers Shh (A and D) and Pitx2 (B and E) are detected in the odontogenic epithelium, whereas Bmp6 is expressed dynamically in odontogenic epithelium early (C and H) and in odontogenic mesenchyme in newly ossifying teeth (F and H). (G and H) Bmp6 continues to be expressed in teeth later in development in both marine and benthic larvae. White arrowheads, odontogenic epithelium; asterisks, newly mineralized developing teeth; black arrowheads, odontogenic mesenchyme. (Scale bars, A–F = 50 μm, G and H = 100 µm.)
cis-Regulatory Changes Have Elevated Expression of the Benthic Bmp6 Allele During Tooth Development.
We sequenced the exons of Bmp6 in marine and benthic fish and found no nonsynonymous coding differences (SI Appendix, Fig. S7). To test for possible cis-acting regulatory differences in expression of marine and benthic alleles, we generated F1 hybrids between marine and benthic fish and used pyrosequencing assays to ask whether benthic and marine alleles made equal contributions to the overall level of Bmp6 mRNA expression in F1 hybrid tooth plates. Allele-specific expression assays allow for the precise quantification of cis-regulatory differences between the two chromosomes in the same cells of the same fish in an identical trans-acting environment (31). We tested for a cis-regulatory change in Bmp6 at three developmental time points, one before (larval), one during (juvenile), and one after (adult) the tooth number divergence. We detected no significant cis-regulatory difference in Bmp6 at an early larval stage before the tooth number divergence in the time course (Fig. 6). However, in both juveniles and adults, when tooth number differences are first being established and are further diverging between marine and benthic populations, we detected a highly significant allele-specific expression difference, with ∼1.4-fold up-regulation of Bmp6 expression from the benthic allele in F1 hybrid fish (Fig. 6). This significant up-regulation of Bmp6 at a later developmental stage mirrors both the late divergence in tooth number and the late-acting nature of the chromosome 21 QTL. These results support the hypothesis that a temporally regulated cis-regulatory difference in Bmp6 expression drives the difference in tooth number between benthic and marine sticklebacks.
Fig. 6.
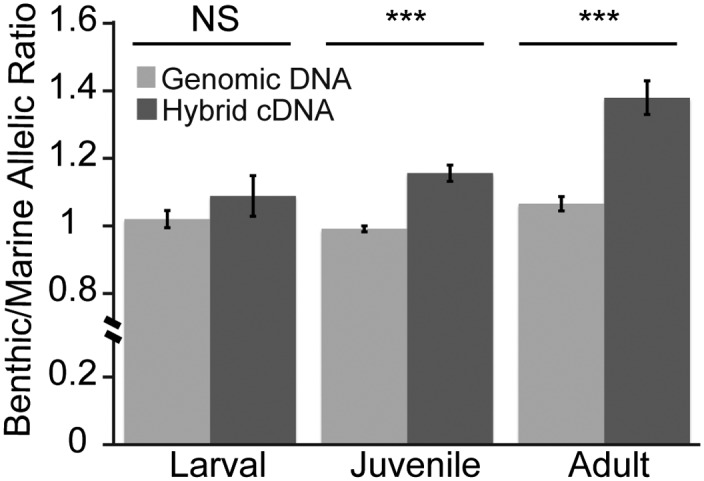
cis-regulatory up-regulation of the benthic allele of Bmp6 in late, not early, stages of tooth development. Shown are the ratios of benthic to marine alleles measured by pyrosequencing assays from either genomic DNA (light gray) or tooth plate cDNA (dark gray) from benthic × marine F1 hybrids at three different developmental stages. No significant difference in Bmp6 expression was detected between marine and benthic alleles at the larval stage, but at the juvenile and adult stage the benthic allele was significantly up-regulated (sample sizes and P values by the Wilcoxon signed rank test for early, juvenile, and adult are n = 12, P = 0.27; n = 18, P = 0.0003; and n = 13, P = 0.0005, respectively). Error bars are SEM.
Discussion
Our studies show that Paxton benthic freshwater sticklebacks have evolved major changes in tooth number, tooth plate area, and intertooth spacing that arise relatively late during development. Because sticklebacks, like most teleosts, retain the basal vertebrate condition of polyphyodonty (continuous tooth replacement) (32), the late divergence in tooth number could result from a change in the rate of the tooth regeneration program late in development, once the initial tooth pattern has been established. This late-forming increase in tooth number may match the time period when benthic fish begin to benefit from increased tooth number (i.e., perhaps wild benthic larvae do not normally begin exploiting a benthic diet until about 20–25 mm in length). Alternatively, developmental or genetic constraints may lead to late-forming divergence. For example, altering the tooth developmental program at earlier stages may lead to deleterious pleiotropic consequences, or available standing genetic variation might primarily affect late, not early, development.
Although our laboratory-reared data show that major differences in tooth number are maintained between marine and freshwater fish when reared in a common laboratory environment, tooth numbers in both populations are reduced in laboratory-reared fish compared with wild fish. Differences in chronological age likely contribute to this difference, because wild fish are likely at least 1 y old, whereas our laboratory-reared adults were 6 mo old. In addition, tooth number may be influenced by diet and rearing conditions, as has previously been reported in cichlids (33).
Previous quantitative genetic studies of stickleback pharyngeal tooth number revealed five QTL controlling ventral pharyngeal tooth number in a F2 genetic cross between an ancestral low-toothed Japanese marine fish and a derived high-toothed Paxton benthic freshwater fish (15). Our more detailed studies suggest that differences in total adult tooth number arise from a combination of several factors, including changes in the development programs controlling tooth number, the size of the tooth field, and the spacing of teeth within that field. This conclusion is supported by the statistical relationships between tooth number, area, and spacing in the F2 cross and by the genome-wide linkage mapping results of all three phenotypes. We have identified at least seven QTL that have significant effects on tooth number, tooth plate size, or tooth spacing. Different QTLs affect one, two, or three different tooth phenotypes (tooth number, tooth spacing, and tooth plate size), showing modular control of evolved changes in dental patterning.
In other fish, pharyngeal jaw patterning is correlated with dietary niche, likely due to adaptive advantages of different morphologies in feeding success on different diets (9). Because benthic fish are well described as having trophic specializations for eating benthos (13), we hypothesize that the evolved tooth gain in benthic sticklebacks is also an adaptive trait that has been selected during an ecological shift to a benthic diet. We note that of the seven tooth-patterning QTL, only three go in a direction that is concordant with the overall shift in tooth number in the parental populations (i.e., benthic alleles conferring more teeth) based on the developmental time courses. However, the QTL with the largest phenotypic effect on chromosome 21 does act in a direction that is consistent with the overall trend in tooth number in the parental populations (benthic allele conferring more teeth). Perhaps the smaller-effect QTL that have effects in the opposite direction result from chromosome 21’s effect overshooting the adaptive peak for tooth patterning in this recently evolved population, with other loci evolving to bring tooth patterning closer to the adaptive peak (34). The mixed direction of effects of benthic alleles could alternatively result from pleiotropy (35), with QTL controlling other adaptive benthic phenotypes that might secondarily affect tooth patterning. For example, the large-effect tooth-spacing QTL on chromosome 4 overlaps the Ectodysplasin (Eda) gene which controls adaptive reductions in armor plate patterning (36) and is also well known to affect vertebrate tooth patterning (37, 38). Interestingly, Eda also plays a role in the spacing of hair placodes and tooth cusps in mice (39, 40), making Eda an excellent candidate for underlying the tooth-spacing QTL on chromosome 4. A third possibility is that some or all of these tooth traits could be changing due to genetic drift occurring after freshwater colonization. As several other species pairs and hundreds of other freshwater populations with trophic modifications have been described (12, 13, 41–43), one test of adaptive significance will be to ask whether other derived benthic lake or creek freshwater stickleback populations have also evolved increases in tooth number. Molecular genetic identification of the tooth-patterning QTL that are segregating in the current cross, combined with population genetic tests of molecular variation surrounding causal loci, should also help distinguish these models.
To begin to study the molecular mechanisms behind evolved tooth gain, we fine-mapped the largest-effect tooth number QTL on chromosome 21. A previous study identified a cluster of QTL on chromosome 21 controlling several derived freshwater skeletal traits (15). This QTL cluster mapped near a large genomic inversion previously shown to display strong worldwide patterns of divergence between marine and freshwater populations (44), suggesting that multiple phenotypes may be controlled by linked genetic changes within the chromosome inversion. Interestingly, we find that the 1.5-LOD candidate interval for the chromosome 21 tooth QTL maps over 1.5 Mb from the inversion, strongly suggesting that the molecular changes driving tooth gain map outside the inverted region.
The new fine-mapped interval for the tooth QTL contains an excellent candidate gene, Bmp6. We show that Bmp6 is expressed in developing teeth in marine and benthic sticklebacks, has no predicted coding changes between populations, but has a late-onset cis-regulatory up-regulation in benthic fish. Because in other vertebrates, BMPs act as activators of tooth development (45), we hypothesize that the elevated Bmp6 expression observed in benthic sticklebacks contributes to their increased tooth number controlled by the chromosome 21 region. Bone Morphogenetic Proteins were originally identified based on their remarkable ability to induce ectopic bone when implanted at new sites in animals (46). Thus, increases in tooth plate area could also result from increased Bmp6 expression. The divergence in tooth number and Bmp6 cis-regulation at late, not early, developmental stages might reflect a heterochronic shift in the benthic population, where the benthic tooth development and replacement program is “stuck” in the early rapid tooth-generating phase observed in early larval stages in both marine and benthic fish. Although we parsimoniously favor the hypothesis that Bmp6 underlies the evolved differences in tooth number, tooth plate area, and intertooth spacing, we note that the fine mapping was only done for tooth number, so it is possible that other genes underlie the evolved changes in tooth plate area and intertooth spacing.
The use of BMP ligands as major drivers of morphological evolution in vertebrates is striking. BMP family members have been implicated in several vertebrate evolved traits: size and shape of the beak in Darwin’s finches, size and shape of the jaw in cichlids, jaw and skull variation in brachycephalic dogs, and avian feather patterning (47–50). Although based on a limited number of reported cases and possibly affected by ascertainment biases, this apparent reuse of the same signaling pathway across taxa may reflect a predisposition for Bmp genes to be used during morphological evolution, perhaps due to having complex, modular cis-regulatory architecture to generate evolutionary variation (51, 52).
Previous QTL mapping studies in sticklebacks have shown that major changes in pelvic hindfin development, armor plate formation, and body pigmentation are all due to alterations in key developmental signaling molecules and transcription factors (36, 53–55). In each of these previous cases, freshwater fish have evolved a major loss or reduction of skeletal structures that were originally present in marine ancestors. In all three cases, cis-regulatory changes are implicated, either directly (53, 54) or inferred (36). Here we show that a major gain in tooth number can also be genetically mapped to a relatively small number of chromosome regions. The QTLs with largest effects on tooth number control somewhat less of the overall variance than the previously identified QTL for armor plates, pelvis, and pigment (each of which controls 50% or more of the variance in the corresponding trait). Nevertheless, the overall effects of the tooth-patterning QTLs are still quite large compared with classical predictions of nearly infinitesimal effects for genetic changes underlying evolved differences in natural populations. Finally, our results with Bmp6 show that for both loss and gain traits, the chromosome regions with largest phenotypic effects show clear evidence of cis-acting regulatory changes in key developmental control genes. Although many more case studies will be needed to draw general conclusions, collectively, these studies suggest that similar general principles may underlie the evolution of both loss and gain traits and that regulatory changes in developmental control genes play an important role in both regressive and constructive evolution of the vertebrate skeleton.
Materials and Methods
Stickleback Husbandry.
Lab-reared fish were raised in 110-liter tanks under common conditions (3.5 g/l Instant Ocean salt, 0.4 mL/l NaHCO3) and fed live brine shrimp as larvae, then frozen daphnia, bloodworms, and Mysis shrimp as juveniles and adults. All experiments and field collections were done with the approval of the Institutional Animal Care and Use Committee from University of California, Berkeley, Stanford University, or the University of British Columbia.
QTL Mapping.
QTL mapping was done using R/qtl (56). To map QTL for adult tooth number, area, and spacing, we analyzed a subset (n = 272 fish) of a previously described (16) Paxton Benthic and Japanese Marine F2 cross. Two hundred seventy-five microsatellite markers were genotyped in each F2. Tooth number, area, and spacing were quantified in each F2. As all three traits were significantly correlated with fish total length, residuals from a linear regression were used for each of the three traits. See SI Appendix, SI Materials and Methods, for details of QTL mapping.
In Situ Hybridization.
Marine and benthic embryos and larvae were euthanized, fixed overnight in 4 g paraformaldehyde in 100 ml 1×PBS, then dehydrated and stored at −20 °C in methanol. For larvae older than 9 d postfertilization, ventral tooth plates were dissected after rehydration from methanol and before in situ hybridization. In situ hybridization was performed essentially as described (57) but with in situs done in tubes in a water bath not baskets and using a 2-d hybridization for older larval stages. For sections, whole-mount in situs were fixed overnight in 4 g paraformaldehyde in 100 ml 1× PBS, embedded in gelatin–albumin cross-linked with 1.75% glutaraldehyde, and sectioned at 40 µm on a Pelco 101 Vibratome Series 1000. Primer sequences for generating the clones used to make the Bmp6, Shh, Pitx2, and Tfap2a riboprobes are listed in SI Appendix.
Pyrosequencing of F1 Hybrids.
For allele-specific expression experiments, Paxton benthic freshwater fish were crossed with Rabbit Slough marine fish by in vitro fertilization to generate hybrid F1s. The bilateral pair of ventral pharyngeal tooth plates from each hybrid was dissected on ice from larval, juvenile, and adult stages (∼10–20 mm, n = 12; ∼25–40 mm, n = 18; and >40 mm in total length, n = 13, respectively). See SI Appendix for primer sequences used for RT-PCR and pyrosequencing and additional methods.
Supplementary Material
Acknowledgments
We thank Mike Bell and Brian Summers for generously providing wild adult RABS fish, Priscilla Erickson for generating some of the lab-reared PAXB fish, Andrew Glazer for generating the Shh probe, and Gareth Fraser and Peter Walentek for useful suggestions on sectioning. This work was supported in part by National Science Foundation Graduate Research Fellowships (to P.A.C. and N.A.E.); National Institutes of Health Genetics Training Grant 5T32GM007127 (to P.A.C.); an Achievement Rewards for College Scientists fellowship (to N.A.E.); a Canada Research Chair and grants from Natural Sciences and Engineering Research Council and the Canada Foundation for Innovation (to D.S.); an NIH Center for Excellence in Genomic Studies grant (5P50HG2568) and investigator position at the Howard Hughes Medical Institute (to D.M.K.); and a March of Dimes Basil O’Connor award, the Pew Charitable Trusts, and NIH R01-DE021475 (to C.T.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The DNA sequences reported in this paper have been deposited in the GenBank and Probe databases [accession nos. KM406380–KM406383 (Genbank); Pr032250564–Pr032250575, Pr032250589–Pr032250592 (Probe)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407567111/-/DCSupplemental.
References
- 1.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134(1):25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 2.Stern DL. Evolutionary developmental biology and the problem of variation. Evolution. 2000;54(4):1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- 3.Protas M, et al. Multi-trait evolution in a cave fish, Astyanax mexicanus. Evol Dev. 2008;10(2):196–209. doi: 10.1111/j.1525-142X.2008.00227.x. [DOI] [PubMed] [Google Scholar]
- 4.Wark AR, et al. Genetic architecture of variation in the lateral line sensory system of threespine sticklebacks. G3. 2012;2(9):1047–1056. doi: 10.1534/g3.112.003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Werner T, Koshikawa S, Williams TM, Carroll SB. Generation of a novel wing colour pattern by the Wingless morphogen. Nature. 2010;464(7292):1143–1148. doi: 10.1038/nature08896. [DOI] [PubMed] [Google Scholar]
- 6.Jernvall J, Thesleff I. Tooth shape formation and tooth renewal: Evolving with the same signals. Development. 2012;139(19):3487–3497. doi: 10.1242/dev.085084. [DOI] [PubMed] [Google Scholar]
- 7.Lauder GV. Functional design and evolution of the pharyngeal jaw apparatus in euteleostean fishes. Zool J Linn Soc-Lond. 1983;77(1):1–38. [Google Scholar]
- 8.Wainwright PC. Functional morphology of the pharyngeal jaw apparatus. In: Shadwick R, Lauder GV, editors. Biomechanics of Fishes. New York: Academic Press; 2005. pp. 77–101. [Google Scholar]
- 9.Muschick M, Indermaur A, Salzburger W. Convergent evolution within an adaptive radiation of cichlid fishes. Curr Biol. 2012;22(24):2362–2368. doi: 10.1016/j.cub.2012.10.048. [DOI] [PubMed] [Google Scholar]
- 10.Kingsley DM, Peichel CL. The molecular genetics of evolutionary change in sticklebacks. In: Ostlund-Nilsson S, Mayer I, Huntingford FA, editors. Biology of the Three-Spine Stickleback. Boca Raton: CRC Press; 2007. pp. 41–81. [Google Scholar]
- 11.Bell MA, Foster SA. 1994. The Evolutionary Biology of the Threespine Stickleback (Oxford Univ Press, Oxford, U. K.), 571 pp.
- 12.Caldecutt WJ, Bell MA, Buckland-Nicks JA. Sexual dimorphism and geographic variation in dentition of threespine stickleback, Gasterosteus aculeatus. Copeia. 2001;(4):936–944. [Google Scholar]
- 13.Schluter D, McPhail JD. Ecological character displacement and speciation in sticklebacks. Am Nat. 1992;140(1):85–108. doi: 10.1086/285404. [DOI] [PubMed] [Google Scholar]
- 14.McPhail JD. Ecology and evolution of sympatric sticklebacks (Gasterosteus): Evidence for a species pair in Paxton Lake, Texada Island, British Columbia. Can J Zool. 1992;70:361–369. [Google Scholar]
- 15.Miller CT, et al. Modular skeletal evolution in sticklebacks is controlled by additive and clustered quantitative trait Loci. Genetics. 2014;197(1):405–420. doi: 10.1534/genetics.114.162420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colosimo PF, et al. The genetic architecture of parallel armor plate reduction in threespine sticklebacks. PLoS Biol. 2004;2(5):E109. doi: 10.1371/journal.pbio.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andl T, et al. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131(10):2257–2268. doi: 10.1242/dev.01125. [DOI] [PubMed] [Google Scholar]
- 18.Bei M, Kratochwil K, Maas RL. BMP4 rescues a non-cell-autonomous function of Msx1 in tooth development. Development. 2000;127(21):4711–4718. doi: 10.1242/dev.127.21.4711. [DOI] [PubMed] [Google Scholar]
- 19.Fraser GJ, Bloomquist RF, Streelman JT. Common developmental pathways link tooth shape to regeneration. Dev Biol. 2013;377(2):399–414. doi: 10.1016/j.ydbio.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia S, et al. Roles of Bmp4 during tooth morphogenesis and sequential tooth formation. Development. 2013;140(2):423–432. doi: 10.1242/dev.081927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vainio S, Karavanova I, Jowett A, Thesleff I. Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell. 1993;75(1):45–58. [PubMed] [Google Scholar]
- 22.Wang Y, et al. BMP activity is required for tooth development from the lamina to bud stage. J Dent Res. 2012;91(7):690–695. doi: 10.1177/0022034512448660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bitgood MJ, McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol. 1995;172(1):126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- 24.Fraser GJ, Berkovitz BK, Graham A, Smith MM. Gene deployment for tooth replacement in the rainbow trout (Oncorhynchus mykiss): A developmental model for evolution of the osteichthyan dentition. Evol Dev. 2006;8(5):446–457. doi: 10.1111/j.1525-142X.2006.00118.x. [DOI] [PubMed] [Google Scholar]
- 25.Fraser GJ, Graham A, Smith MM. Conserved deployment of genes during odontogenesis across osteichthyans. Proc Biol Sci. 2004;271(1555):2311–2317. doi: 10.1098/rspb.2004.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mucchielli ML, et al. Mouse Otlx2/RIEG expression in the odontogenic epithelium precedes tooth initiation and requires mesenchyme-derived signals for its maintenance. Dev Biol. 1997;189(2):275–284. doi: 10.1006/dbio.1997.8672. [DOI] [PubMed] [Google Scholar]
- 27.Aberg T, Wozney J, Thesleff I. Expression patterns of bone morphogenetic proteins (Bmps) in the developing mouse tooth suggest roles in morphogenesis and cell differentiation. Dev Dyn. 1997;210(4):383–396. doi: 10.1002/(SICI)1097-0177(199712)210:4<383::AID-AJA3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 28.Fraser GJ, et al. An ancient gene network is co-opted for teeth on old and new jaws. PLoS Biol. 2009;7(2):e31. doi: 10.1371/journal.pbio.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Connell DJ, et al. A Wnt-bmp feedback circuit controls intertissue signaling dynamics in tooth organogenesis. Sci Signal. 2012;5(206):ra4. doi: 10.1126/scisignal.2002414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wise SB, Stock DW. Conservation and divergence of Bmp2a, Bmp2b, and Bmp4 expression patterns within and between dentitions of teleost fishes. Evol Dev. 2006;8(6):511–523. doi: 10.1111/j.1525-142X.2006.00124.x. [DOI] [PubMed] [Google Scholar]
- 31.Wittkopp PJ. Using pyrosequencing to measure allele-specific mRNA abundance and infer the effects of cis- and trans-regulatory differences. Methods Mol Biol. 2011;772:297–317. doi: 10.1007/978-1-61779-228-1_18. [DOI] [PubMed] [Google Scholar]
- 32.Huysseune A, Witten PE. Developmental mechanisms underlying tooth patterning in continuously replacing osteichthyan dentitions. J Exp Zoolog B Mol Dev Evol. 2006;306(3):204–215. doi: 10.1002/jez.b.21091. [DOI] [PubMed] [Google Scholar]
- 33.Huysseune A. Phenotypic plasticity in the lower pharyngeal jaw dentition of Astatoreochromis alluaudi (Teleostei: Cichlidae) Arch Oral Biol. 1995;40(11):1005–1014. doi: 10.1016/0003-9969(95)00074-y. [DOI] [PubMed] [Google Scholar]
- 34.Orr HA. The genetic theory of adaptation: A brief history. Nat Rev Genet. 2005;6(2):119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- 35.Rogers SM, et al. Genetic signature of adaptive peak shift in threespine stickleback. Evolution. 2012;66(8):2439–2450. doi: 10.1111/j.1558-5646.2012.01622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colosimo PF, et al. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005;307(5717):1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- 37.Aigler SR, Jandzik D, Hatta K, Uesugi K, Stock DW. Selection and constraint underlie irreversibility of tooth loss in cypriniform fishes. Proc Natl Acad Sci USA. 2014;111(21):7707–7712. doi: 10.1073/pnas.1321171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Häärä O, et al. Ectodysplasin regulates activator-inhibitor balance in murine tooth development through Fgf20 signaling. Development. 2012;139(17):3189–3199. doi: 10.1242/dev.079558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harjunmaa E, et al. On the difficulty of increasing dental complexity. Nature. 2012;483(7389):324–327. doi: 10.1038/nature10876. [DOI] [PubMed] [Google Scholar]
- 40.Mustonen T, et al. Ectodysplasin A1 promotes placodal cell fate during early morphogenesis of ectodermal appendages. Development. 2004;131(20):4907–4919. doi: 10.1242/dev.01377. [DOI] [PubMed] [Google Scholar]
- 41.Gross HP, Anderson JM. Geographic variation in the gillrakers and diet of european threespine sticklebacks, Gasterosteus Aculeatus. Copeia. 1984;(1):87–97. [Google Scholar]
- 42.Hagen DW, Gilbertson LG. Geographic variation and environmental selection in Gasterosteus Aculeatus L in Pacific Northwest, America. Evolution. 1972;26(1):32–51. doi: 10.1111/j.1558-5646.1972.tb00172.x. [DOI] [PubMed] [Google Scholar]
- 43.McPhail JD. Ecology and evolution of sympatric sticklebacks (Gasterosteus): Origin of the species pairs. Can J Zool. 1993;71(3):515–523. [Google Scholar]
- 44.Jones FC, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484(7392):55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kavanagh KD, Evans AR, Jernvall J. Predicting evolutionary patterns of mammalian teeth from development. Nature. 2007;449(7161):427–432. doi: 10.1038/nature06153. [DOI] [PubMed] [Google Scholar]
- 46.Reddi AH, Reddi A. Bone morphogenetic proteins (BMPs): From morphogens to metabologens. Cytokine Growth Factor Rev. 2009;20(5–6):341–342. doi: 10.1016/j.cytogfr.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 47.Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305(5689):1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 48.Albertson RC, Streelman JT, Kocher TD, Yelick PC. Integration and evolution of the cichlid mandible: The molecular basis of alternate feeding strategies. Proc Natl Acad Sci USA. 2005;102(45):16287–16292. doi: 10.1073/pnas.0506649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mou C, et al. Cryptic patterning of avian skin confers a developmental facility for loss of neck feathering. PLoS Biol. 2011;9(3):e1001028. doi: 10.1371/journal.pbio.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schoenebeck JJ, et al. Variation of BMP3 contributes to dog breed skull diversity. PLoS Genet. 2012;8(8):e1002849. doi: 10.1371/journal.pgen.1002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guenther C, Pantalena-Filho L, Kingsley DM. Shaping skeletal growth by modular regulatory elements in the Bmp5 gene. PLoS Genet. 2008;4(12):e1000308. doi: 10.1371/journal.pgen.1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kingsley DM. What do BMPs do in mammals? Clues from the mouse short-ear mutation. Trends Genet. 1994;10(1):16–21. doi: 10.1016/0168-9525(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 53.Chan YF, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327(5963):302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller CT, et al. cis-regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131(6):1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shapiro MD, et al. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428(6984):717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- 56.Broman KW, Sen S. A Guide to QTL Mapping with R/qtl. New York: Springer; 2009. [Google Scholar]
- 57.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3(1):59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.