Abstract
Understanding, modeling, and predicting the impact of global change on ecosystem functioning across biogeographical gradients can benefit from enhanced capacity to represent biota as a continuous distribution of traits. However, this is a challenge for the field of biogeography historically grounded on the species concept. Here we focus on the newly emergent field of functional biogeography: the study of the geographic distribution of trait diversity across organizational levels. We show how functional biogeography bridges species-based biogeography and earth science to provide ideas and tools to help explain gradients in multifaceted diversity (including species, functional, and phylogenetic diversities), predict ecosystem functioning and services worldwide, and infuse regional and global conservation programs with a functional basis. Although much recent progress has been made possible because of the rising of multiple data streams, new developments in ecoinformatics, and new methodological advances, future directions should provide a theoretical and comprehensive framework for the scaling of biotic interactions across trophic levels and its ecological implications.
Keywords: biodiversity, functional trait, predictive ecology
Biogeography is the study of the distribution of species and ecosystems across space and time and of the underlying biotic and abiotic factors, mechanisms, and processes (1). Biogeography is central to synthesizing small- and large-scale patterns of species' responses to global environmental change and providing a window to assess the importance of earth and evolutionary history, as well as changing biotic and abiotic factors that underlie the current distribution of taxa. Historically, biogeography has been rooted in the species concept. Biologists long ago built global maps of species’ ranges and species diversity patterns, which allowed scientists to develop, test, and validate prominent ecological theories at the root of biogeography (2, 3). Biogeography, however, has largely developed separately from ecosystem ecology and earth system biology, yet we argue that one of the great challenges of 21st century biogeography is to provide theoretical baselines and tools for the understanding and prediction of ecosystem responses to environmental changes in terms of species composition and biogeochemical cycles (water, carbon, nutrients, and energy).
For the last several decades, new directions in biogeography have been aided by statistical and computational advances (4, 5), and new species distribution models have provided maps of priority areas for conservation and projections of the effects of global change on species diversity patterns for the next century (6–9). Nonetheless, predictions of how ecological communities respond to past and projected future climates have increasingly been challenged (10) due to the limitations of species-based approaches, including conceptual and technical difficulties in incorporating species interactions, dispersal limitations, and species’ adaptations into predictive models (11–16). Indeed, if we treat species as qualitative entities, then we need to understand each and every one. In contrast, trait-based biogeography can help model species interactions, dispersal ability, and physiological tolerance more simply and generically (17). More precisely, if we array species along some continuous trait axes (18), then knowing cross-species relationships (19), e.g., between plant physiological tolerance to drought and diameter of xylem vessel (20), can help forecast the change in species composition of ecological communities in response to variation in abiotic conditions like water availability.
Contrary to species-based biogeography, earth science models directly address changes in biogeochemical cycles at a global scale but using a simplistic representation of biodiversity in most cases (21–24): few plant functional types (PFT), characterized by mean ecophysiological characteristics, are used per biome in models dedicated to simulate and forecast the consequences of global changes on biogeochemical cycling (24). However, in biodiversity–ecosystem functioning research (25–27), the taxonomic composition of an ecosystem is identified as a key driver of ecosystem functioning. More recently, the functional component of biodiversity, i.e., the diversity of forms and functions, has been recognized as the missing link between biodiversity patterns and biogeochemical cycles (28–30) and perhaps is a core driver of ecosystem services (31, 32). Given these insights from ecosystem ecology, there is a growing consensus on the need for a better representation of biodiversity—in particular the composition of species, forms, and functions—in earth science models (21, 23, 24), as stressed in this issue by Reichstein et al. (33).
In this Introduction, and in the related special feature, to bridge historical species-based biogeography with earth systems science, we argue that the time is ripe to advance functional biogeography. We define functional biogeography as the analysis of the patterns, causes, and consequences of the geographic distribution of the diversity of form and function—namely, trait diversity. Indeed treating the biota as a continuous distribution of traits appears pivotal if one is expected to model the biosphere when there are literally millions of separate species. Describing and explaining the global distribution of forms and functions is a long-standing goal for ecologists (34–36). Such attempts have been synthetized under a variety of terms including macrophysiology (19, 37–39). The emergence of functional biogeography pursues this goal but also aims at linking biogeographical patterns of trait diversity to biogeographical patterns of species diversity, ecosystem functioning, and services because of the integration of concepts and methodologies from multiple fields (Fig. 1). Advancing functional biogeography becomes timely with the rise of a predictive era in ecology and the acceleration of societal demand regarding the evaluation and forecast of past, current, and future ecosystem services.
Fig. 1.
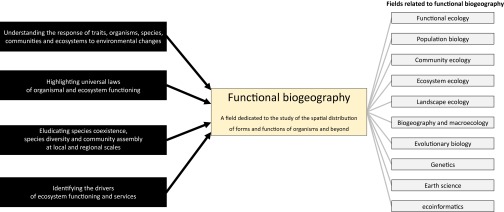
Functional biogeography: an emergent field at the crossroads of several science fields. Functional biogeography calls for knowledge from multiple fields to answer questions related to the distribution of forms and functions of organisms, populations, communities, ecosystems, and biomes across spatial scales.
Biogeography Beyond Species: Functional Traits as a Common Currency Across Biological Organizational Levels and Taxonomic Groups
We posit that a useful approach to functional biogeography is to use a common currency, namely functional traits. Functional traits are morphological, physiological, phenological, or behavioral features measured on organisms that can ultimately be linked to their performance (40). In trait-based ecology, it has been shown that it is possible to aggregate functional traits measured on organisms to explain the functioning of populations, communities, ecosystems, and beyond. For instance, at the community level, the use of community-weighed means (CWM, i.e., the average trait value of a community accounting for effects of species abundance within communities) (40, 41) is a promising tool to accurately predict ecosystem functions such as plant primary productivity (41, 42). As a whole, functional traits can be implemented into more or less complex integrative functions (e.g., CWM is a simple integrative function) to scale up from organs to higher organizational levels including ecosystems and biomes (40, 43, 44). Trait-based approaches have also been extensively used to describe the diversity of forms and functions within a study unit—often termed functional diversity sensu lato—using different distance metrics (e.g., variance based) (45–55) and how it scales spatially (56–61).
Variation in functional traits within species is also key. For instance, in this special feature, Reich et al. (62) highlight huge intraspecific variation in gymnosperm needle traits with latitude across the vast boreal domain. They note how this variation fits with trait economic theory and model the impacts of trait variation on carbon cycling across the world’s boreal forests. They validate the model output against independent data. Studying phenotypic and genotypic variation within species across space (e.g., within the range of species) represents one primary aspect of functional biogeography (63, 64). Future efforts to quantify both intraspecific phenotypic and genetic variation within species’ ranges should bring interesting insights into the eco-evolutionary drivers of species’ distributions and help bridge functional ecology, spatial ecology, genetics, and evolutionary biology (65) (Fig. 1). The question of accounting for intraspecific variation in cross-species trait-based studies has been extensively discussed for the last 5 years (52, 66, 67). The importance of accounting for intraspecific variation at macroecological scales, when interspecific variations are expected to be large, is still debated (21, 67–69).
A functional trait approach has now been rapidly and extensively developed in plant ecology, aided by the development of standardized protocols and methodologies (70, 71). More recently, similar initiatives have been initiated for microbes and animals (17, 72–74). These initiatives will favor cross-taxonomic group comparisons of trait diversity patterns from local to global and pave the road of an integrated, comprehensive framework for the understanding of ecology at large spatial and temporal scales. In this special feature, Whittaker et al. (75) are able to provide new insights in the field of island biogeography by building functional diversity–area relationships, in complement to species richness–area relationships and compare them across taxonomic groups (here between beetles and spiders). The analysis of congruence of trait diversities across groups is also timely for better targeting conservation areas, quantifying and explaining multitropic networks and evaluating ecosystem services (76). Interestingly, in this special feature, Kembel et al. (77) demonstrate how phyllosphere bacterial communities impact the performance of plants and the structure of plant communities in a Neotropical forest. The authors propose that the phyllosphere bacterial diversity is a key component of plant functioning and should be considered as a plant functional trait, hence extending our current definition of functional traits (40). This study sheds light on the ecological role of the extended phenotype, still largely ignored in trait-based ecology; in particular, its importance for the adaption of organisms and for the interactions among them. More broadly, such pioneering studies are a step forward in the quantification of species interactions (including mutualism, competition, facilitation, predation), which remains a priority question in community ecology and biogeography. Identifying the traits involved in species interactions (interaction traits hereafter) is challenging but pivotal for several approaches including network analyses. Considering the interacting communities (the phyllosphere bacterial communities) as traits of other organisms (the plants) is one promising step in that direction.
From Patterns to Predictions
Describing Trait Diversity Across Space.
We still lack knowledge of the response of organismal traits to environmental changes for most kingdoms, including plants, which is, however, a prerequisite to make trait-based ecology a predictive science (Fig. 2). The first trait–environment relationships were established from local studies (78, 79), but site-dependent effects and the restricted range of environmental conditions covered in most studies question the robustness of the results. A functional biogeography approach to this question is a relevant alternative (80). Indeed, examining how species and their functionalities vary geographically can be useful because many of the same drivers of change that occur at every single site on earth have already varied across time and across space. The study from Reich et al. (81) is a notable example. Using a large global biomass dataset, the authors provide biogeographically explicit relationships between biomass partitioning in trees and temperature, elucidating a long-standing ecological question about the variation of plant biomass allocation with increasing stressful conditions.
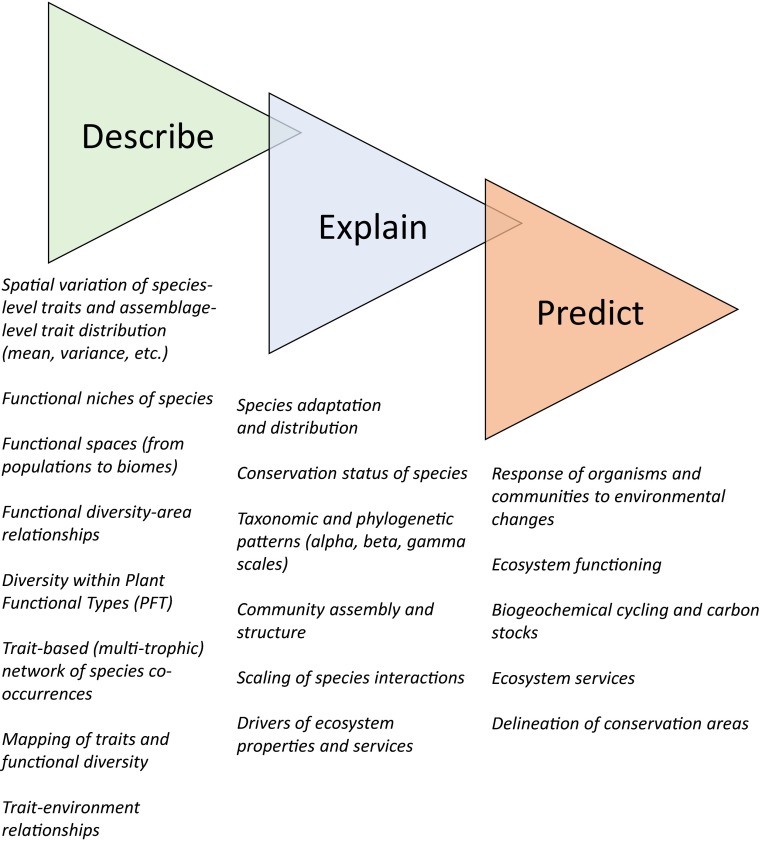
Fig. 2.
The three tenets of functional biogeography: describe, explain, predict. The first tenet of functional biogeography is to describe the distribution of forms and functions along environmental gradients and across spatial scales. The second is to use this information to explain the geographic distribution of organisms, biodiversity (notably species and phylogenetic diversity) patterns, and ecosystem processes and services. The third is to predict their responses to environmental changes using trait-based predictive functions and models.
The establishment of robust trait–environment relationships will help achieve one core goal of functional biogeography: to place measures of the functions of biodiversity on a map (58, 60, 82). Such functional maps are indeed the bases of many questions in functional biogeography and derived fields (57). In this special feature, Bennie et al. (83) elegantly map a core behavioral trait in mammals: the diel time partitioning. They show that the biogeography of this trait is under the influence of thermal constraints but also artificial lighting and other anthropogenic activities. Such a finding is important for improving the modeling of mammals’ distribution. Beyond macrophysiological implications on the biogeography of species, mapping the diversity of organismal functions is also central to the quantification of ecosystem functioning and services. Here, van Bodegom et al. (84) show that trait data are now available worldwide to map core traits pertaining to plant functioning (leaf mass per area, stem-specific density, and seed mass in their study) at a global scale. Further, they demonstrate that the spatial variation of these three traits alone explains a large part of observed vegetation types, thus linking trait distributions to dynamic global vegetation models (DGVMs). Ultimately such maps can help refine earth science and land surface models in a more continuous manner, i.e., by replacing the spatial distribution of plant functional types by continuous maps of functional traits as input information in those models (33) provided that data on traits and species occurrences are available (Fig. 3).
Fig. 3.

General chart for functional biogeography studies using a combination of heterogeneous data sources. Combing organismal traits, species occurrences and abundances, and environmental information allows exploration of the response of organisms, populations, communities, and beyond to environmental changes. In combination with (measured, estimated, modeled, simulated) information about ecosystem properties, one can examine the abiotic and biotic drivers of ecosystem functioning (single processes and multifunctionality) and services and provide predictive outcomes. Although these approaches are increasingly used, the incorporation of time (e.g., biogeography of time series, functional trajectories of ecosystems) and a systematic functional analysis of multitrophic networks are currently lacking.
Finally, the supposed link between functional traits and resource availabilities offers new insights into the physiological limits of the biogeographical distribution of species and the delineation of their fundamental and realized niches. In this issue, Stahl et al. (85) successfully show that three key functional traits—seed mass, wood density and plant height—explain species' range limits of a continental flora. Maps of functional traits could also be considered as new environmental layers for species distribution modeling (86, 87). Such analyses are a step forward in the definition and quantification of the functional niches of species (18, 88).
Explaining Species Coexistence, Species Diversity, and Community Assembly at Local to Regional and Continental Scales.
Functional biogeography can shed light on local species assembly processes and the structure of communities. The idea that local processes depend on regional ones is not new in biogeography (89, 90), but the rising of systematic and standardized descriptions of trait diversity across scales can accelerate quantification of ecological properties and processes and the test of ecological theories on patterns of species diversity (Fig. 3). For instance, delineating the functional trait space of a region is a way to evaluate the niche space available in this region (18, 86), which should subsequently help explain local species diversity patterns (86, 91, 92). In this special feature, Lamanna et al. (86) recast several biodiversity theories for species richness gradients in terms of variation in functional diversity. As a result, they are able to uniquely assess several trait-based hypotheses for the latitudinal gradient of species richness but from a trait perspective. Based on a large botanical and trait database for woody species in the New World, they find that species occupy a larger functional space in the temperate zone in comparison with the tropics. This finding contradicts several theories that have suggested that there are a greater number of species in the tropics due to larger trait or niche space. In the future we expect more and more powerful tests of biodiversity theories and the elucidation of the biogeographical patterns of species richness by using new botanical and animal trait databases worldwide (Fig. 3). These efforts will help solidify the development of trait-based biogeography theory.
Functional traits are increasingly used in community ecology to detect and quantify the processes that shape ecological communities (52, 93, 94). With a biogeography perspective, this approach can be applied to understand the spatial scaling of assembly processes across broad gradients. In particular, the scaling of species interactions is the source of one of the most intense debates in biogeography (3, 14, 95–98). Indeed understanding when and how local biotic interactions influence the biogeographical distribution of species is crucial for species distribution modeling (13, 15). Further, the intensity of species interactions (competition in particular) has long been supposed to vary along environmental and latitudinal gradients (e.g., less competition expected in harsh environments and/or higher latitude), which could explain the latitudinal patterns of species richness (3, 95). Mapping traits directly involved in species interactions (82, 99) can help us to understand the scaling of species interactions. For instance, one can expect past intense competition in biogeographical areas characterized by greater variance in interaction traits (58) as a result of limiting similarity processes (100).
Elucidating the Drivers of Ecosystem Functioning and Services Across Space.
Functional biogeography offers a unique window to explain the variation of ecosystem functioning at large scales (33). Elucidating the drivers of ecosystem functioning at this scale is of high priority for policymakers from the perspective of modulating the impacts of human-driven global changes and adjusting conservation policies (101). Past functional biogeography exercises successfully linked the mean functional characteristics of ecosystems to resource cycling (102), but the functional structure of communities, i.e., the distribution of trait values within communities, was not fully characterized. More recent advances in functional ecology have provided tools to quantify both mean functional characteristics (e.g., CWM) and functional dispersion (e.g., Rao’s entropy) within ecosystems. Today it is urgent to disentangle effects of both facets of trait diversity in driving ecosystem functioning (103, 104). In particular, functional dispersion could be considered as a proxy of the result of biotic interactions (see above); thus, its quantification can provide insights into the importance of species interactions in the modulation of ecosystem functioning.
Latitudinal gradients, often considered as natural laboratories (105), are particularly appropriate tools for functional biogeography exercises and are of particular relevance to the analysis of large-scale patterns in trait diversity and how it is related to observed or inferred ecosystem processes. In the next few years, relating the functional structure of ecosystems to ecosystem functional features inferred from remote sensing outputs will be particularly valuable to provide continuous maps of functional traits, as well as highly resolved proxies for ecosystem functioning (Fig. 3), as revealed by recent successful attempts (106, 107).
Collectively, a better understanding and quantification of the functional drivers of ecosystem functioning across biogeographical gradients will help refine plant functional types, or eliminate their need entirely, for earth science modeling (24). Practically, providing well-resolved continuous maps of functional traits is a first important step in that direction (84).
The same line of reasoning can be applied to ecosystem services given that ecosystem functions are the biological bases of ecosystem services. Interestingly, as shown by Lamarque et al. (108), it is possible to evaluate ecosystem services bundles and tradeoffs using a trait-based empirical model. Using mountain grasslands in the French Alps as a case study, the authors are able to disentangle the direct and indirect effects of climate and land use on ecosystem services, including water quality, aesthetic value, and fodder quality and quantity, through a scenario-based study.
Toward Predictions in a Changing World.
Because functional biogeography links organismal functions to their environment, it is theoretically possible to predict the response of organisms, communities, and ecosystems to environmental changes from functional traits (43, 44, 109). Ultimately, it appears possible to predict bundles of ecosystem services based on theoretical and empirical knowledge and the mapping of different facets of functional diversity (108, 110).
The stacking of different maps (e.g., taxonomic, phylogenetic functional diversity, carbon stocks, ecosystem productivity, aesthetic values, socio-ecological mapping of species’ favorable habitats) (Fig. 3) will help refine conservation areas because there is little reason for a perfect overlapping of species, functional, and phylogenetic rarity (111). For instance, in this special feature, Mouillot et al. (112) show that the functional vulnerability—defined as a potential decrease of functional diversity following species loss—of fish fauna on tropical reefs can be high in areas characterized by high species diversity. Conversely, functional redundancy is discovered in low species diversity areas. Together, this suggests that conserving hotspots of species diversity is not sufficient if one also aims at targeting the functional insurance of biota.
The Rise of Specific Tools and Resources
Functional biogeography will advance faster if large heterogeneous datasets and computing tools are combined and brought to bear in a spatially explicit context (Fig. 3). These datasets are now available or will emerge soon. They encompass a global coverage of geo-referenced species occurrences (113–115), the functional characteristics of species (21, 73, 116, 117), and more and more precise environmental layers (118, 119). Ecoinformatics accelerated the development of datasets structured in a complex and consistent fashion by the means of ontologies for ecology (120, 121), methods for model–data integration (122, 123), and the calculation of the n-dimensional hypervolume (55, 86). Computing and mathematical science are developing approaches to spatial data mining and the extrapolation of information from incomplete datasets (124). For instance, using Bayesian statistics, it will be possible to fill gaps in trait databases for a given trait and a given species with information from other traits and other species (125, 126). Similarly, in a spatial context, missing information in a grid cell can be filled based on information contained in surrounding grids (124).
Future Perspectives
Functional biogeography should help improve our understanding of the biogeographical patterns of species diversity and, ultimately, may allow us to predict the consequences of global changes for ecosystem functions and services. However, the development of expectations specific to functional biogeography is in its infancy. Indeed, although functional biogeography is inherently process based, theory is still lacking. To make functional biogeography a predictive science, it will be important to include the eco-evolutionary dynamics of organisms, communities, ecosystems, and beyond (10). Some recent findings suggest a tight linkage between functional traits and demography (127), which represents a promising step in the development of an integrative and dynamical theory to functional biogeography.
Another priority avenue is to provide insights into trophic or food web models, first by providing maps of functions of primary producers. To do so, it is urgent to better characterize traits across taxonomic groups and their interrelationships (32, 128), as well as to identify and quantify interaction traits. Rapid progress has been made in ecoinformatics, but global coverage of functional traits, even for plants, is still sparse. Until greater coverage is achieved, it will be impossible to achieve an accurate estimation of trait diversity at a global scale and to account for some facets of trait diversity, including functional rarity. Further efforts in ecoinformatics should also provide relevant ontologies, including trait ontologies that are proving themselves indispensable for functional biogeography exercises.
A functional perspective to biogeography can easily integrate intraspecific trait variation to assess the importance of local adaptation and phenotypic plasticity in the modulation of large-scale processes. Nevertheless, although recent debates in functional ecology have advanced our understanding of the importance of intraspecific variation at local scales (52, 66, 129, 130), it is urgent to evaluate its importance at large scales. It has been assumed that species turnover and interspecific variations predominate over intraspecific variation at large scales (66), but empirical tests are scarce and mostly come from the plant kingdom (68, 69). Further, there are some specific areas of functional biogeography where intraspecific variation should be accounted for, in particular when characterizing the functional space of a given species (18) and its phenotypic variation within its distribution range (63). A better evaluation of intraspecific trait variation within species’ ranges, in complement to a quantification of genetic variation, should bring novel insights into the eco-evolutionary drivers of species’ biogeography and functional niches.
Acknowledgments
C.V. was supported by a Marie Curie International Outgoing Fellowship within the 7th European Community Framework Program (DiversiTraits Project 221060). B.J.E. was supported by National Science Foundation Macrosystems Award DBI-1065861. This study was partly conducted as a part of the Botanical Information and Ecology Network (BIEN) working group supported by the National Centre for Ecological Analysis and Synthesis (NCEAS), a center funded by the NSF (Grant EF 0553768); the University of California, Santa Barbara; and the state of California. The BIEN Working Group was also supported by iPlant (NSF Grant DBI-0735191). This research was also supported by the French Foundation for Research on Biodiversity (FRB; www.fondationbiodiversite.fr) in the context of the Centre de Synthèse et d'Analyse sur la Biodiversité (CESAB) project Assembling, analysing and sharing data on plant functional diversity to understand the effects of biodiversity on ecosystem functioning: a case study with French Permanent Grasslands (DIVGRASS). This study was inspired by the TRY Initiative on plant traits (try-db.org) supported by International Geosphere-Biosphere Programme and DIVERSITAS.
References
- 1.Brown J, Lomolino M. Biogeography. Sunderland, MA: Sinauer Associates; 1998. [Google Scholar]
- 2.Brown J. Macroecology. Chicago: Univ of Chicago Press; 1995. [Google Scholar]
- 3.Pianka E. Latitudinal gradients in species diversity: A review of concepts. Am Nat. 1966;100(910):33–46. [Google Scholar]
- 4.Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecol Modell. 2006;190(3-4):231–259. [Google Scholar]
- 5.Thuiller W, Lafourcade B, Engler R, Araújo MB. BIOMOD - A platform for ensemble forecasting of species distributions. Ecography. 2009;32(3):369–373. [Google Scholar]
- 6.Thomas CD, et al. Extinction risk from climate change. Nature. 2004;427(6970):145–148. doi: 10.1038/nature02121. [DOI] [PubMed] [Google Scholar]
- 7.Thuiller W, Lavorel S, Araújo MB, Sykes MT, Prentice IC. Climate change threats to plant diversity in Europe. Proc Natl Acad Sci USA. 2005;102(23):8245–8250. doi: 10.1073/pnas.0409902102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Araújo MB, Alagador D, Cabeza M, Nogués-Bravo D, Thuiller W. Climate change threatens European conservation areas. Ecol Lett. 2011;14(5):484–492. doi: 10.1111/j.1461-0248.2011.01610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellard C, et al. 2014. Hotspot hotness: Biodiversity hotspot vulnerability to global changes. Glob Ecol Biogeogr, in press.
- 10.Lavergne S, Mouquet N, Thuiller W, Ronce O. Biodiversity and climate change: Integrating evolutionary and ecological responses of species and communities. Annu Rev Ecol Evol Syst. 2010;41:321–350. [Google Scholar]
- 11.Soberón J. Grinnellian and Eltonian niches and geographic distributions of species. Ecol Lett. 2007;10(12):1115–1123. doi: 10.1111/j.1461-0248.2007.01107.x. [DOI] [PubMed] [Google Scholar]
- 12.Thuiller W, et al. Predicting global change impacts on plant species distributions: Future challenges. Perspect Plant Ecol Evol Syst. 2008;9(3-4):137–152. [Google Scholar]
- 13.Boulangeat I, Gravel D, Thuiller W. Accounting for dispersal and biotic interactions to disentangle the drivers of species distributions and their abundances. Ecol Lett. 2012;15(6):584–593. doi: 10.1111/j.1461-0248.2012.01772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kissling W, et al. Towards novel approaches to modelling biotic interactions in multispecies assemblages at large spatial extents. J Biogeogr. 2012;39(12):2163–2178. [Google Scholar]
- 15.Thuiller W, et al. A road map for integrating eco-evolutionary processes into biodiversity models. Ecol Lett. 2013;16(Suppl 1):94–105. doi: 10.1111/ele.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wisz MS, et al. The role of biotic interactions in shaping distributions and realised assemblages of species: Implications for species distribution modelling. Biol Rev Camb Philos Soc. 2013;88(1):15–30. doi: 10.1111/j.1469-185X.2012.00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green JL, Bohannan BJ, Whitaker RJ. Microbial biogeography: From taxonomy to traits. Science. 2008;320(5879):1039–1043. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- 18.Violle C, Jiang L. Towards a trait-based quantification of species niche. J Plant Ecol-UK. 2009;2(2):87–93. [Google Scholar]
- 19.Reich PB. The world-wide 'fast-slow' plant economics spectrum: A traits manifesto. J Ecol. 2014;102(2):275–301. [Google Scholar]
- 20.Craine JM, et al. Global diversity of drought tolerance and grassland climate-change resilience. Nature Clim Change. 2013;3:63–67. [Google Scholar]
- 21.Kattge J, et al. TRY: A global database of plant traits. Glob Change Biol. 2011;17(9):2905–2935. [Google Scholar]
- 22.Boulangeat I, et al. Improving plant functional groups for dynamic models of biodiversity: At the crossroads between functional and community ecology. Glob Change Biol. 2012;18(11):3464–3475. doi: 10.1111/j.1365-2486.2012.02783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verheijen LM, et al. Impacts of trait variation through observed trait-climate relationships on performance of an Earth system model: A conceptual analysis. Biogeosciences. 2013;10:5497–5515. [Google Scholar]
- 24.Wullschleger SD, et al. Plant functional types in Earth system models: Past experiences and future directions for application of dynamic vegetation models in high-latitude ecosystems. Ann Bot (Lond) 2014;114(1):1–16. doi: 10.1093/aob/mcu077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naeem S, Duffy JE, Zavaleta E. The functions of biological diversity in an age of extinction. Science. 2012;336(6087):1401–1406. doi: 10.1126/science.1215855. [DOI] [PubMed] [Google Scholar]
- 26.Cardinale BJ. Towards a general theory of biodiversity for the Anthropocene. Elem Sci Anth. 2013;1:000014. [Google Scholar]
- 27.Cardinale BJ, et al. Biodiversity loss and its impact on humanity. Nature. 2012;486(7401):59–67. doi: 10.1038/nature11148. [DOI] [PubMed] [Google Scholar]
- 28.Chapin FS, 3rd, et al. Consequences of changing biodiversity. Nature. 2000;405(6783):234–242. doi: 10.1038/35012241. [DOI] [PubMed] [Google Scholar]
- 29.Wardle DA, et al. Ecological linkages between aboveground and belowground biota. Science. 2004;304(5677):1629–1633. doi: 10.1126/science.1094875. [DOI] [PubMed] [Google Scholar]
- 30.Diaz S, et al. 2007. Terrestrial Ecosystems in a Changing World, The IGBP Series, eds Canadell J, Pataki D, Pitelka L (Springer, Berlin)
- 31.Cadotte MW, Carscadden K, Mirotchnick N. Beyond species: Functional diversity and the maintenance of ecological processes and services. J Appl Ecol. 2011;48(5):1079–1087. [Google Scholar]
- 32.Lavorel S. Plant functional effects on ecosystem services. J Ecol. 2013;101(1):4–8. [Google Scholar]
- 33.Reichstein M, Bahn M, Mahecha MD, Kattge J, Baldocchi DD. Linking plant and ecosystem functional biogeography. Proc Natl Acad Sci USA. 2014;111:13697–13702. doi: 10.1073/pnas.1216065111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whittaker R. Communities and Ecosystems. New York: Macmillan; 1975. [Google Scholar]
- 35.Schimper A. Planzengeographie auf Physiologischer Grundlage. Bonn, Germany: G. Fisher; 1898. [Google Scholar]
- 36.Harrison S, et al. Ecophysiological and bioclimatic foundations for a global plant functional classification. J Veg Sci. 2010;21(2):300–317. [Google Scholar]
- 37.Chown SL, Gaston KJ, Robinson D. Macrophysiology: Large-scale patterns in physiological traits and their ecological implications. Funct Ecol. 2004;18(2):159–167. [Google Scholar]
- 38.Chown SL, Gaston KJ. Macrophysiology for a changing world. Proc Biol Sci. 2008;275(1642):1469–1478. doi: 10.1098/rspb.2008.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaston KJ, et al. Macrophysiology: A conceptual reunification. Am Nat. 2009;174(5):595–612. doi: 10.1086/605982. [DOI] [PubMed] [Google Scholar]
- 40.Violle C, et al. Let the concept of trait be functional! Oikos. 2007;116(5):882–892. [Google Scholar]
- 41.Garnier E, et al. Plant functional markers capture ecosystem properties during secondary succession. Ecology. 2004;85(4):2630–2637. [Google Scholar]
- 42.Reich PB. Key canopy traits drive forest productivity. Proc Biol Sci. 2012;279(1736):2128–2134. doi: 10.1098/rspb.2011.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lavorel S, Garnier E. Predicting changes in community composition and ecosystem functioning from plant traits: Revisiting the Holy Grail. Funct Ecol. 2002;16(5):545–556. [Google Scholar]
- 44.Suding K, et al. Scaling environmental change through the community-level: A trait-based response-and-effect framework for plants. Glob Change Biol. 2008;14(5):1125–1140. [Google Scholar]
- 45.Petchey O, Hector A, Gaston K. How do different measures of functional diversity perform? Ecology. 2004;85(3):847–857. [Google Scholar]
- 46.Mouillot D, et al. Niche overlap estimates based on quantitative functional traits: A new family of non-parametric indices. Oecologia. 2005;145(3):345–353. doi: 10.1007/s00442-005-0151-z. [DOI] [PubMed] [Google Scholar]
- 47.Mason NWH, Mouillot D, Lee WG, Wilson JB. Functional richness, functional evenness and functional divergence: The primary components of functional diversity. Oikos. 2005;111(1):112–118. [Google Scholar]
- 48.Cornwell WK, Schwilk LD, Ackerly DD. A trait-based test for habitat filtering: Convex hull volume. Ecology. 2006;87(6):1465–1471. doi: 10.1890/0012-9658(2006)87[1465:attfhf]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 49.Villéger S, Mason NW, Mouillot D. New multidimensional functional diversity indices for a multifaceted framework in functional ecology. Ecology. 2008;89(8):2290–2301. doi: 10.1890/07-1206.1. [DOI] [PubMed] [Google Scholar]
- 50.Schleuter D, Daufresne M, Massol F, Arguillier C. A user's guide to functional diversity indices. Ecol Monogr. 2010;80(3):469–484. [Google Scholar]
- 51.Mouchet M, Villéger S, Mason N, Mouillot D. Functional diversity measures: An overview of their redundancy and their ability to discriminate community assembly rules. Funct Ecol. 2010;24(4):867–876. [Google Scholar]
- 52.Violle C, et al. The return of the variance: Intraspecific variability in community ecology. Trends Ecol Evol. 2012;27(4):244–252. doi: 10.1016/j.tree.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 53.De Bello F, Carmona C, Mason N, Sebastia M-T, Leps J. Which trait dissimilarity for functional diversity: Trait means or trait overlap? J Veg Sci. 2013;24(5):807–819. [Google Scholar]
- 54.Cadotte MW, Albert C, Walker S. The ecology of differences: IIntegrating evolutionary and functional distances. Ecol Lett. 2013;16(10):1234–1244. doi: 10.1111/ele.12161. [DOI] [PubMed] [Google Scholar]
- 55.Blonder B, Lamanna C, Violle C, Enquist B. The n-dimensional hypervolume. Glob Ecol Biogeogr. 2014;23(5):595–609. [Google Scholar]
- 56.Devictor V, et al. Spatial mismatch and congruence between taxonomic, phylogenetic and functional diversity: The need for integrative conservation strategies in a changing world. Ecol Lett. 2010;13(8):1030–1040. doi: 10.1111/j.1461-0248.2010.01493.x. [DOI] [PubMed] [Google Scholar]
- 57.Swenson NG, Weiser MD. Plant geography upon the basis of functional traits: An example from eastern North American trees. Ecology. 2010;91(8):2234–2241. doi: 10.1890/09-1743.1. [DOI] [PubMed] [Google Scholar]
- 58.Swenson NG, et al. The biogeography and filtering of woody plant functional diversity in North and South America. Glob Ecol Biogeogr. 2012;21(8):798–808. [Google Scholar]
- 59.Freschet GT, et al. Global to community scale differences in the prevalence of convergent over divergent leaf trait distributions in plant assemblages. Glob Ecol Biogeogr. 2011;20(5):755–765. [Google Scholar]
- 60.Newbold T, Butchart SHM, Sekercioğlu CH, Purves DW, Scharlemann JPW. Mapping functional traits: comparing abundance and presence-absence estimates at large spatial scales. PLoS ONE. 2012;7(8):e44019. doi: 10.1371/journal.pone.0044019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stahl U, et al. Whole-plant trait spectra of North American woody plant species reflect fundamental ecological strategies. Ecosphere. 2013;4:art128. [Google Scholar]
- 62.Reich PB, Rich RL, Lu X, Wang Y-P, Oleksyn J. Biogeographic variation in evergreen conifer needle longevity and impacts on boreal forest carbon cycle projections. Proc Natl Acad Sci USA. 2014;111:13703–13708. doi: 10.1073/pnas.1216054110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banta JA, et al. Climate envelope modelling reveals intraspecific relationships among flowering phenology, niche breadth and potential range size in Arabidopsis thaliana. Ecol Lett. 2012;15(8):769–777. doi: 10.1111/j.1461-0248.2012.01796.x. [DOI] [PubMed] [Google Scholar]
- 64.McKown AD, et al. Geographical and environmental gradients shape phenotypic trait variation and genetic structure in Populus trichocarpa. New Phytol. 2014;201(4):1263–1276. doi: 10.1111/nph.12601. [DOI] [PubMed] [Google Scholar]
- 65.Guillot G, Renaud S, Ledevin R, Michaux J, Claude J. A unifying model for the analysis of phenotypic, genetic, and geographic data. Syst Biol. 2012;61(6):897–911. doi: 10.1093/sysbio/sys038. [DOI] [PubMed] [Google Scholar]
- 66.Albert CH, Grassein F, Schurr FM, Vieilledent G, Violle C. When and how should intraspecific trait variability be considered in plant ecology? Persp Plant Ecol Evol. 2011;13(3):217–225. [Google Scholar]
- 67.Albert C, et al. On the importance of intraspecific variability for the quantification of functional diversity. Oikos. 2012;121(1):116–126. [Google Scholar]
- 68.Kazakou E, et al. Are trait-based species' rankings consistent across datasets and spatial scales? J Veg Sci. 2014;25(1):235–247. [Google Scholar]
- 69.Cordlandwehr V, et al. Do plant traits retrieved from a database accurately predict on-site measurements? J Ecol. 2013;101(3):662–670. [Google Scholar]
- 70.Cornelissen JHC, et al. A handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Aust J Bot. 2003;51:335–380. [Google Scholar]
- 71.Pérez-Harguindeguy N, et al. New handbook for standardised measurement of plant functional traits worldwide. Aust J Bot. 2013;61(3):167–234. [Google Scholar]
- 72.Pey B, et al. 2014. A new conceptual framework for soil invertebrate trait-based approaches in ecology. Basic Appl Ecol 15:194–206.
- 73.Homburg K, Homburg N, Schäfer F, Schuldt A, Assmann T. Carabids.org - A dynamic online database of ground beetle species traits (Coleoptera, Carabidae) Insect Conserv Diver. 2014;7(3):195–205. [Google Scholar]
- 74.Krause S, et al. Front Microbiol. 2014. Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Whittaker RJ, et al. Functional biogeography of oceanic islands and the scaling of functional diversity in the Azores. Proc Natl Acad Sci USA. 2014;111:13709–13714. doi: 10.1073/pnas.1218036111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lavorel S, et al. A novel framework for linking functional diversity of plants with other trophic levels for the quantification of ecosystem services. J Veg Sci. 2013;24(5):942–948. [Google Scholar]
- 77.Kembel SW, et al. Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc Natl Acad Sci USA. 2014;111:13715–13720. doi: 10.1073/pnas.1216057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Poorter H, Niinemets U, Poorter L, Wright IJ, Villar R. Causes and consequences of variation in leaf mass per area (LMA): A meta-analysis. New Phytol. 2009;182(3):565–588. doi: 10.1111/j.1469-8137.2009.02830.x. [DOI] [PubMed] [Google Scholar]
- 79.Poorter H, Niinemets U, Walter A, Fiorani F, Schurr U. A method to construct dose-response curves for a wide range of environmental factors and plant traits by means of a meta-analysis of phenotypic data. J Exp Bot. 2010;61(8):2043–2055. doi: 10.1093/jxb/erp358. [DOI] [PubMed] [Google Scholar]
- 80.Moles AT, et al. Which is a better predictor of plant traits: Temperature or precipitation? J Veg Sci. 2014;25(5):1167–1180. [Google Scholar]
- 81.Reich PB, et al. Temperature drives global patterns in forest biomass distribution in leaves, stems, and roots. Proc Natl Acad Sci USA. 2014;111:13721–13726. doi: 10.1073/pnas.1216053111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moles AT, et al. Putting plant resistance traits on the map: A test of the idea that plants are better defended at lower latitudes. New Phytol. 2011;191(3):777–788. doi: 10.1111/j.1469-8137.2011.03732.x. [DOI] [PubMed] [Google Scholar]
- 83.Bennie JJ, Duffy JP, Inger R, Gaston KJ. Biogeography of time partitioning in mammals. Proc Natl Acad Sci USA. 2014;111:13727–13732. doi: 10.1073/pnas.1216063110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Bodegom PM, Douma JC, Verheijen LM. A fully traits-based approach to modeling global vegetation distribution. Proc Natl Acad Sci USA. 2014;111:13733–13738. doi: 10.1073/pnas.1304551110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stahl U, Reu B, Wirth C. Predicting species' range limits from functional traits for the tree flora of North America. Proc Natl Acad Sci USA. 2014;111:13739–13744. doi: 10.1073/pnas.1300673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lamanna C, et al. Functional trait space and the latitudinal diversity gradient. Proc Natl Acad Sci USA. 2014;111:13745–13750. doi: 10.1073/pnas.1317722111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kearney M, Porter W. Mechanistic niche modelling: Combining physiological and spatial data to predict species’ ranges. Ecol Lett. 2009;12(4):334–350. doi: 10.1111/j.1461-0248.2008.01277.x. [DOI] [PubMed] [Google Scholar]
- 88.Ackerly DD, Cornwell WK. A trait-based approach to community assembly: partitioning of species trait values into within- and among-community components. Ecol Lett. 2007;10(2):135–145. doi: 10.1111/j.1461-0248.2006.01006.x. [DOI] [PubMed] [Google Scholar]
- 89.Ricklefs RE. Community diversity: Relative roles of local and regional processes. Science. 1987;235(4785):167–171. doi: 10.1126/science.235.4785.167. [DOI] [PubMed] [Google Scholar]
- 90.Ricklefs RE. A biogeographical perspective on ecological systems: Some personal reflections. J Biogeogr. 2011;38(11):2045–2056. [Google Scholar]
- 91.Ricklefs RE, O'Rourke K. Aspect diversity in moths: A temperate-tropical comparison. Evolution. 1975;29(2):313–324. doi: 10.1111/j.1558-5646.1975.tb00211.x. [DOI] [PubMed] [Google Scholar]
- 92.Ricklefs RE. Species richness and morphological diversity of passerine birds. Proc Natl Acad Sci USA. 2012;109(36):14482–14487. doi: 10.1073/pnas.1212079109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McGill BJ, Enquist BJ, Weiher E, Westoby M. Rebuilding community ecology from functional traits. Trends Ecol Evol. 2006;21(4):178–185. doi: 10.1016/j.tree.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 94.Weiher E, et al. Advances, challenges and a developing synthesis of ecological community assembly theory. Philos Trans R Soc Lond B Biol Sci. 2011;366(1576):2403–2413. doi: 10.1098/rstb.2011.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pianka E. Latitudinal gradients in species diversity. Trends Ecol Evol. 1989;4(8):223–223. [Google Scholar]
- 96.Wiens JJ. The niche, biogeography and species interactions. Philos Trans R Soc Lond B Biol Sci. 2011;366(1576):2336–2350. doi: 10.1098/rstb.2011.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Araujo MB, Rozenfeld A. The geographic scaling of biotic interactions. Ecography. 2014;37(5):406–415. [Google Scholar]
- 98.Schemske DW. In: Speciation and Patterns of Diversity. Butlin RK, Bridle JR, Schluter D, editors. Cambridge, UK: Cambridge Univ Press; 2009. pp. 220–239. [Google Scholar]
- 99.Moles AT. Dogmatic is problematic: Interpreting evidence for latitudinal gradients in herbivory and defence. Ideas Ecol Evol. 2013;6(1):1–4. [Google Scholar]
- 100.MacArthur RH, Levins R. The limiting similarity, convergence and divergence of coexisting species. Am Nat. 1967;101(971):377–385. [Google Scholar]
- 101.Thomas CD, et al. Reconciling biodiversity and carbon conservation. 2013;16(S1):39–47. doi: 10.1111/ele.12054. [DOI] [PubMed] [Google Scholar]
- 102.Schulze ED, Kelliher FM, Korner C, Lloyd J, Leuning R. Relationships among maximum stomatal conductance, ecosystem surface conductance, carbon assimilation rate, and plant nitrogen nutrition: A global ecology scaling exercise. Annu Rev Ecol Syst. 1994;25:629–660. [Google Scholar]
- 103.Díaz S, et al. Incorporating plant functional diversity effects in ecosystem service assessments. Proc Natl Acad Sci USA. 2007;104(52):20684–20689. doi: 10.1073/pnas.0704716104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ricotta C, Moretti M. CWM and Rao’s quadratic diversity: A unified framework for functional ecology. Oecologia. 2011;167(1):181–188. doi: 10.1007/s00442-011-1965-5. [DOI] [PubMed] [Google Scholar]
- 105.De Frenne P, et al. Latitudinal gradients as natural laboratories to infer species' responses to temperature. J Ecol. 2013;101(3):784–795. [Google Scholar]
- 106.Silman MR. Functional megadiversity. Proc Natl Acad Sci USA. 2014;111(16):5763–5764. doi: 10.1073/pnas.1402618111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Asner GP, et al. Amazonian functional diversity from forest canopy chemical assembly. Proc Natl Acad Sci USA. 2014;111(15):5604–5609. doi: 10.1073/pnas.1401181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lamarque P, Lavorel S, Mouchet M, Quétier F. Plant trait-based models identify direct and indirect effects of climate change on bundles of grassland ecosystem services. Proc Natl Acad Sci USA. 2014;111:13751–13756. doi: 10.1073/pnas.1216051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chown SL, Hoffmann AA. Ecophysiological forecasting for environmental change adaptation. Funct Ecol. 2013;27(4):930–933. [Google Scholar]
- 110.Lavorel S, et al. Using plant functional traits to understand the landscape distribution of multiple ecosystem services. J Ecol. 2011;99(1):135–147. [Google Scholar]
- 111.Winter M, Devictor V, Schweiger O. Phylogenetic diversity and nature conservation: Where are we? Trends Ecol Evol. 2013;28(4):199–204. doi: 10.1016/j.tree.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 112.Mouillot D, et al. Functional over-redundancy and high functional vulnerability in global fish faunas of tropical reefs. Proc Natl Acad Sci USA. 2014;111:13757–13762. doi: 10.1073/pnas.1317625111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Guralnick RP, Hill AW, Lane M. Towards a collaborative, global infrastructure for biodiversity assessment. Ecol Lett. 2007;10(8):663–672. doi: 10.1111/j.1461-0248.2007.01063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jetz W, McPherson JM, Guralnick RP. Integrating biodiversity distribution knowledge: Toward a global map of life. Trends Ecol Evol. 2012;27(3):151–159. doi: 10.1016/j.tree.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 115.Schaminée J, Hennekens S, Chytry M, Rodwell J. Vegetation-plot data and databases in Europe: An overview. Preslia. 2009;81:173–185. [Google Scholar]
- 116.Dell AI, Pawar S, Savage VM. Trait database for size and temperature dependence of species interactions. Ecology. 2013;94(5):1205–1206. [Google Scholar]
- 117.Wilman H, et al. EltonTraits 1.0: Species-level foraging attributes of the world's birds and mammals. Ecology. 2014;95(7):2027–2027. [Google Scholar]
- 118.Tuanmu M, Jetz W. A global 1km consensus land cover product for biodiversity and ecosystem modeling. Glob Ecol Biogeogr. 2014;23(9):1031–1045. [Google Scholar]
- 119.Weigelt P, Jetz W, Kreft H. Bioclimatic and physical characterization of the world’s islands. Proc Natl Acad Sci USA. 2013;110(38):15307–15312. doi: 10.1073/pnas.1306309110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jones MB, Schildhauer M, Reichman OJ, Bowers S. The new Bioinformatics: Integrating ecological data from the gene to the biosphere. Annu Rev Ecol Evol Syst. 2006;37:519–544. [Google Scholar]
- 121.Madin JS, Bowers S, Schildhauer MP, Jones MB. Advancing ecological research with ontologies. Trends Ecol Evol. 2008;23(3):159–168. doi: 10.1016/j.tree.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 122.Kattge J, et al. A generic structure for plant trait databases. Methods Ecol Evol. 2011;2(2):202–213. [Google Scholar]
- 123.Wiser SK, et al. Veg-X: An exchange standard for plot-based vegetation data. J Veg Sci. 2011;22(4):598–609. [Google Scholar]
- 124.Shekhar S, Zhang P, Huang Y, Vatsavai R. 2003. Data Mining: Next Generation Challenges and Future Directions, eds Kargupta H, Anupam J (AAAI Press, Palo Alto, California)
- 125.Shan H, et al. 2012. Gap Filling in the Plant Kingdom—Trait Prediction Using Hierarchical Probabilistic Matrix Factorization (Edinburgh, UK), Vol. 12, pp 1303–1310.
- 126.Swenson NG. Phylogenetic imputation of plant functional trait databases. Ecography. 2014;37(2):105–110. [Google Scholar]
- 127.Adler PB, et al. Functional traits explain variation in plant life history strategies. Proc Natl Acad Sci USA. 2014;111(2):740–745. doi: 10.1073/pnas.1315179111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hillebrand H, Matthiessen B. Biodiversity in a complex world: Consolidation and progress in functional biodiversity research. Ecol Lett. 2009;12(12):1405–1419. doi: 10.1111/j.1461-0248.2009.01388.x. [DOI] [PubMed] [Google Scholar]
- 129.Jung V, Violle C, Mondy C, Hoffmann L, Muller S. Intraspecific variability and trait-based community assembly. J Ecol. 2010;98(5):1134–1140. [Google Scholar]
- 130.Albert CH, et al. A multi-trait approach reveals the structure and the relative importance of intra- vs. interspecific variability in plant traits. Funct Ecol. 2010;24(6):1192–1201. [Google Scholar]