Significance
During mitosis, Aurora B kinase plays a key role in ensuring that sister chromatids (each of the copies of a replicated chromosome) attach to different poles of the spindle to create a bipolar array of microtubules that allows correct distribution of the chromosomes. Aurora B deficiency leads to massive defects in chromosome segregation. Surprisingly, an increase in Aurora B activity also is deleterious for the cells and has been associated with various cancers. Here we demonstrate that in yeast an increase in Aurora B activity causes defects in chromosome segregation and spindle-assembly checkpoint activation by erroneously destabilizing even correct attachments of the chromosomes to the spindle, and also promotes premature collapse of the spindle midzone.
Keywords: Ipl1, Sli15, INCENP, CPC, SAC
Abstract
Aurora B kinase regulates the proper biorientation of sister chromatids during mitosis. Lack of Aurora B kinase function results in the inability to correct erroneous kinetochore–microtubule attachments and gives rise to aneuploidy. Interestingly, increased Aurora B activity also leads to problems with chromosome segregation, and overexpression of this kinase has been observed in various types of cancer. However, little is known about the mechanisms by which an increase in Aurora B kinase activity can impair mitotic progression and cell viability. Here, using a yeast model, we demonstrate that increased Aurora B activity as a result of the overexpression of the Aurora B and inner centromere protein homologs triggers defects in chromosome segregation by promoting the continuous disruption of chromosome–microtubule attachments even when sister chromatids are correctly bioriented. This disruption leads to a constitutive activation of the spindle-assembly checkpoint, which therefore causes a lack of cytokinesis even though spindle elongation and chromosome segregation take place. Finally, we demonstrate that this increase in Aurora B activity causes premature collapse of the mitotic spindle by promoting instability of the spindle midzone.
Problems with the partition of the replicated genome during mitosis can lead to aneuploidy, a hallmark of cancer cells (1). For appropriate distribution, chromosomes first must attach to the spindle. The presence of a single unattached chromosome triggers the spindle-assembly checkpoint (SAC), which restrains cell-cycle progression in metaphase (2). Additionally, each kinetochore of every pair of sister chromatids must be connected to opposite spindle poles (biorientation or amphitelic attachment). Aurora B kinase plays a pivotal role in this process (3). After replication, sister chromatids are bound together by ring-shaped cohesin complexes (4). Hence, when sister chromatids are bioriented, the pulling of the microtubules from both spindle poles generates tension. Aurora B detects the absence of tension resulting from erroneous kinetochore–microtubule (KT–MT) connections and destabilizes these connections by phosphorylating key components from the microtubule-binding site of the kinetochore (5–8), thereby generating an empty kinetochore that activates the SAC (5). Tension in the spindle resulting from the biorientation of the sister chromatids separates Aurora B from its substrates at the kinetochore, allowing the final stabilization of the KT–MT attachments (8, 9).
Aurora B, together with inner centromere protein (INCENP), borealin, and survivin, is part of the chromosomal passenger complex (CPC) (3), which controls key mitotic events by localizing to different structures during the cell cycle. After regulating proper chromosome attachment at the inner centromeres, the CPC later moves to the spindle midzone to regulate microtubule dynamics (10–14). The CPC also localizes to the division site, where it participates in the control of cytokinesis by regulating furrow ingression and abscission (3). Additionally, Aurora B and INCENP play a pivotal role in the NoCut pathway, an evolutionarily conserved checkpoint that delays the completion of cytokinesis until all chromosomes have cleared the cleavage plane so as to avoid chromosome breakage during abscission (15–17).
The absence of Aurora B activity leads to massive aneuploidy problems because of the inability to resolve incorrect KT–MT attachments. Interestingly, however, not only the absence of Aurora B but also its overexpression poses an important threat for cell viability, and increased levels of this kinase have been associated with various types of cancer (18–20). In mammals, elevated levels of Aurora B kinase cause defects in chromosome segregation and in the inhibition of cytokinesis (21), but little is known about the molecular mechanisms that underlie these phenotypes. Here we show that these problems can be recapitulated in Saccharomyces cerevisiae cells by increasing the expression of both the Aurora B and INCENP homologs [Increase in Ploidy 1 (Ipl1) and Synthetically Lethal with Ipl1 15 (Sli15), respectively]. Their increased expression leads to high levels of Aurora B activity and causes a dramatic loss of cell viability. Using this model, we further demonstrate that this increase in Aurora B activity causes defects in chromosome segregation by promoting continuous disruption of chromosome–microtubule attachments. This disruption leads to constitutive activation of the SAC, and this activation of the SAC explains the lack of cytokinesis after spindle elongation and chromosome segregation. Finally, we show that the elevated Aurora B activity in our budding yeast model also causes a premature collapse of the spindle because of the instability of the spindle midzone.
Results
Simultaneous Overexpression of Ipl1 and Sli15 Is Lethal for Yeast Cells.
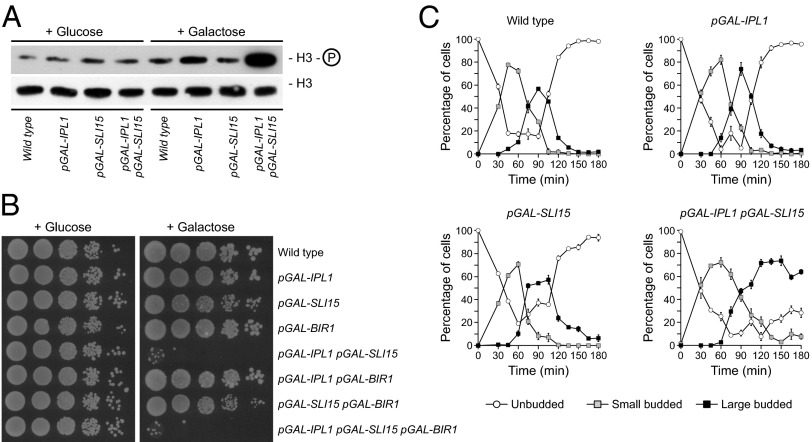
Despite the problems associated with Aurora B overexpression in mammalian cells (21), high levels of Ipl1 do not seem to interfere with cell viability in S. cerevisiae (22). This difference between mammalian and yeast cells has limited the molecular analysis of the consequences of increased Aurora B activity. Because the localization and activity of Aurora B depend on INCENP (3), we hypothesized that the lack of effect of Ipl1 overexpression in yeast cells could be explained if the levels of Sli15 were limiting, so that the excess of Aurora B could not be correctly activated and localized and therefore would not pose a threat to the cell’s viability. To test this notion, we generated cells carrying an additional copy of both IPL1 and SLI15 genes under the control of the galactose-inducible GAL1-10 promoter (pGAL). These cells showed simultaneous elevated expression of IPL1 and SLI15 in galactose-containing medium [89- and 110-fold increases in mRNA levels, respectively, as determined by quantitative RT-PCR (qRT-PCR)] and, more importantly, showed increased levels of histone H3 phosphorylation in serine 10 [a chromatin modification that depends on Aurora B activity (23)], as compared with wild-type cells or cells overexpressing only Ipl1 or Sli15 (Fig. 1A). Strikingly, increased levels of both Ipl1 and Sli15 led to a dramatic loss of cell viability (Fig. 1B). Importantly, overexpression of the survivin homolog Bir1 (24), another CPC component, together with Ipl1 or Sli15 overexpression did not cause an additive effect and was not able to rescue the lethality observed when Ipl1 and Sli15 were overexpressed (Fig. 1B).
Fig. 1.
Simultaneous overexpression of Ipl1 and Sli15 impairs cell viability. Wild-type (F955), pGAL-IPL1 (F256), pGAL-SLI15 (F953), pGAL-BIR1 (F1640), pGAL-IPL1 pGAL-SLI15 (F947), pGAL-IPL1 pGAL-BIR1 (F1644), pGAL-SLI15 pGAL-BIR1 (F1642), and pGAL-IPL1 pGAL-SLI15 pGAL-BIR1 (F1646) cells were grown at 25 °C in rich medium (yeast extract/peptone) with 2% raffinose (YPR). (A) Cells from the cultures grown in YPR were arrested in G1 with 5 μg/mL α-factor and were released into fresh rich medium with 2% galactose and 2% raffinose (YPRG). Thirty minutes before the release, 2% galactose was added to induce transcription from pGAL. α-Factor was added again 75 min after release to avoid cell-cycle reentry. Levels of H3-Ser10 phosphorylation and total H3 levels were determined by Western blot 60 min after the release. (B) Cell viability was analyzed by spotting 10-fold serial dilutions of cell cultures grown in YPR on yeast extract/peptone/dextrose (YPD) or YPRG plates, which then were incubated at 25 °C. (C) Cells were allowed to enter mitosis synchronously in YPRG as in A. Cell-cycle progression was determined by bud size. Percentages of unbudded and small and large budded cells are shown for each time point. Error bars indicate SD (n = 3).
Increased Levels of Ipl1 and Sli15 Cause Spindle Instability and Prevent Cytokinesis.
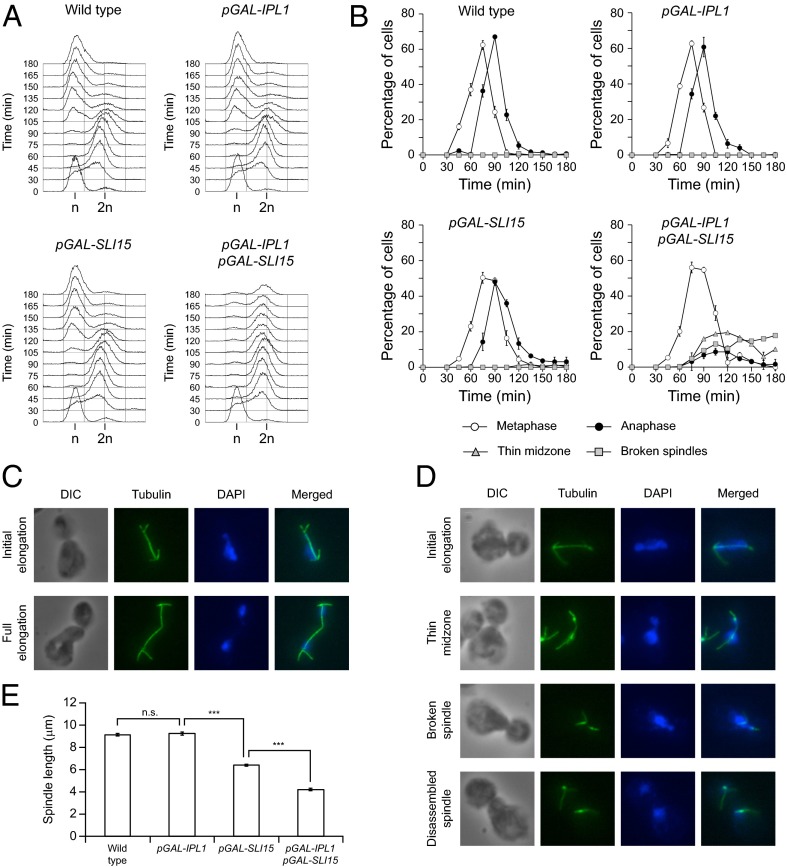
To analyze the effect of Ipl1 and Sli15 overexpression on cell-cycle progression, cells were allowed to enter mitosis in a synchronous manner after expression from pGAL was induced. Although cells overexpressing either Ipl1 or Sli15 progressed through mitosis as did wild-type cells, cells overexpressing both proteins apparently managed to enter the cell cycle without problems but failed to carry out cytokinesis and accumulated as large budded cells (Fig. 1C). FACS analysis showed that wild-type, pGAL-IPL1, and pGAL-SLI15 cells had fully replicated their DNA 60 min after they were released into galactose-containing medium and later exited mitosis and accumulated again as cells with a 1N DNA content (Fig. 2A). Increased Ipl1 and Sli15 levels did not seem to interfere with DNA replication, but, in accordance with our previous results, cells could not exit mitosis and finally accumulated with a 2N DNA content (Fig. 2A).
Fig. 2.
Cell-cycle progression after Ipl1 and Sli15 overexpression. Wild-type (F955), pGAL-IPL1 (F256), pGAL-SLI15 (F953), and pGAL-IPL1 pGAL-SLI15 (F947) cells were allowed to enter mitosis synchronously in YPRG as in Fig. 1A. (A) DNA content was determined by FACS analysis at the indicated time points. (B) Cell-cycle progression was analyzed by spindle (tubulin) and nuclear (DAPI) morphology. Percentages of metaphase and anaphase cells and of cells carrying spindles with a thin midzone and with broken spindles are shown for each time point. Error bars indicate SD (n = 3). (C and D) Representative images showing tubulin (green) and DAPI staining (blue) are presented for wild-type (F955) (C) and pGAL-IPL1 pGAL-SLI15 (F947) (D) cells. DIC, differential interference contrast. (E) Average maximum spindle length for each of the strains. Error bars indicate SEM (n = 150). Statistically significant (***P < 0.0001) or nonsignificant (n.s.) differences according to a two-tailed t test are shown.
We next analyzed spindle and nuclear morphology. Most wild-type cells displayed a typical short metaphase spindle ∼75 min after their release in galactose-containing medium, fully elongated the spindle in anaphase by 90 min, and finally disassembled the spindle and exited mitosis (Fig. 2 B and C and Movie S1). However, pGAL-IPL1 pGAL-SLI15 cells reached metaphase without any obvious problem but then experienced difficulties while trying to extend their spindles as chromosome segregation started (Fig. 2 B and D). As their elongation progressed, the spindles became thin, especially in the midzone, and finally collapsed (Fig. 2D and Movie S2). However, most of the cells managed finally to distribute their chromosomes between the mother and daughter cells, as demonstrated by the separated DNA masses (Fig. 2D). As a result, the average maximum spindle length was reduced from 9 μm in wild-type cells to about 4 μm when both Ipl1 and Sli15 were overexpressed (Fig. 2E). Spindle length was affected slightly by Sli15 overexpression (Fig. 2E), although in this case the spindle midzone did not seem to get thinner.
Overexpression of Ipl1 and Sli15 Leads to Severe Problems in Chromosome Segregation.
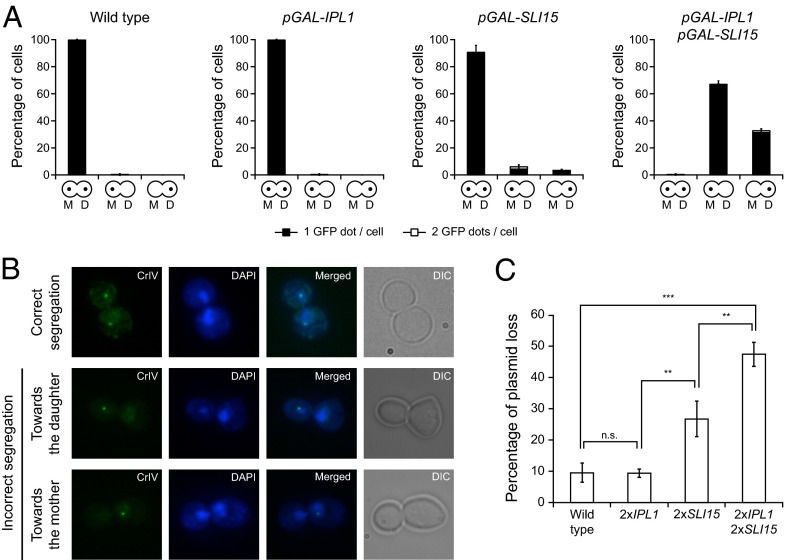
We also analyzed whether, as observed for mammalian cells (21), increased Aurora B activity leads to aneuploidy in yeast. To this end, we followed chromosome segregation using cells with chromosome 4 tagged with GFP at the centromere (CrIV-GFP) (25). In wild-type anaphase cells, two separate DNA masses of approximately equal size, each containing a fluorescent GFP dot, could be distinguished in the mother and daughter cells once the sister chromatids segregated toward opposite spindle poles (Fig. 3A). Although increased expression of only IPL1 or SLI15 did not affect chromosome segregation, simultaneous Ipl1 and Sli15 overexpression caused both sisters in most of the cells to segregate to the same spindle pole (Fig. 3 A and B). Unlike Ipl1-defective cells (26), pGAL-IPL1 pGAL-SLI15 cells showed a slight bias in the segregation of both sister chromatids toward the mother cell (Fig. 3A). This specific pattern of chromosome segregation, as well as the spindle instability and the lack of cytokinesis, depended on increased Ipl1 activity, because overexpression of both Sli15 and a kinase-dead Ipl1 (27) acted in a dominant-negative fashion, and cells behaved as mutants defective in Aurora B activity (Fig. S1). Finally, it is worth noting that even the duplication of both IPL1 and SLI15 genes under control of their own promoters generates a considerable increase in genome instability, as demonstrated by the elevated rate of plasmid loss in the cells (Fig. 3C).
Fig. 3.
Ipl1 and Sli15 overexpression causes defects in chromosome segregation. (A and B) Wild-type (F955), pGAL-IPL1 (F256), pGAL-SLI15 (F953), and pGAL-IPL1 pGAL-SLI15 (F947) cells carrying a GFP tag on chromosome 4 (CrIV-GFP) were allowed to enter mitosis synchronously in YPRG as in Fig. 1A. (A) Analysis of chromosome segregation using CrIV-GFP dots. The graphics below the bars indicate whether sister chromatids segregated correctly [yeast cells with a dot in the mother (M) and a dot in the daughter (D) cell] or cosegregated toward the mother (a dot only in the mother cell) or toward the bud (a dot only in the daughter cell). Black bars indicate that only one GFP dot could be detected per cell; white bars indicate that two GFP dots could be detected per cell. Error bars indicate SD (n = 3). (B) Representative images showing the different patterns of CrIV-GFP segregation (green) and DAPI staining (blue). (C) Plasmid-loss assay with wild-type cells (F496) and cells carrying two copies of IPL1 (2×IPL1; F1753), two copies of SLI15 (2×SLI15; F1754), or two copies of both genes (2×IPL1 2×SLI15; F2019) under the control of their own promoters and also carrying the pRS316 plasmid. The graph shows the median values and SDs of three fluctuation tests for each strain; each test was performed using six independent colonies. Statistically significant (***P < 0.001 and **P < 0.01) or nonsignificant (n.s.) differences according to a two-tailed t test are shown.
Increased Aurora B Activity Causes Spindle Instability by Impairing Ase1 Function.
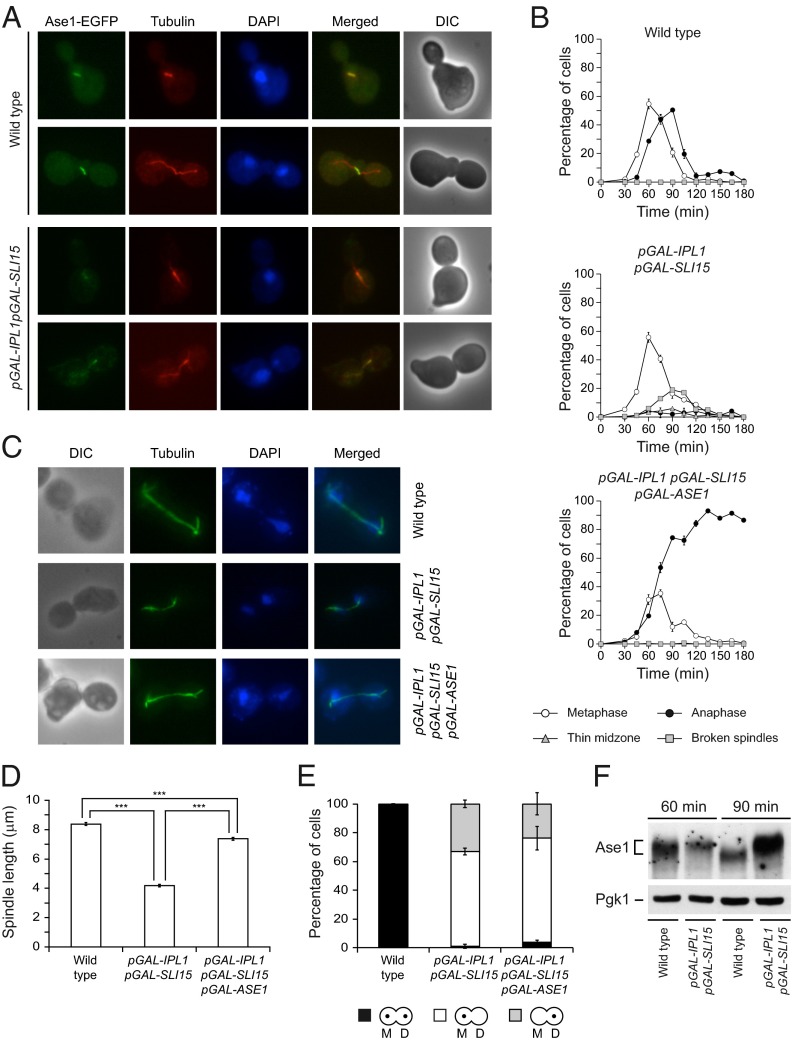
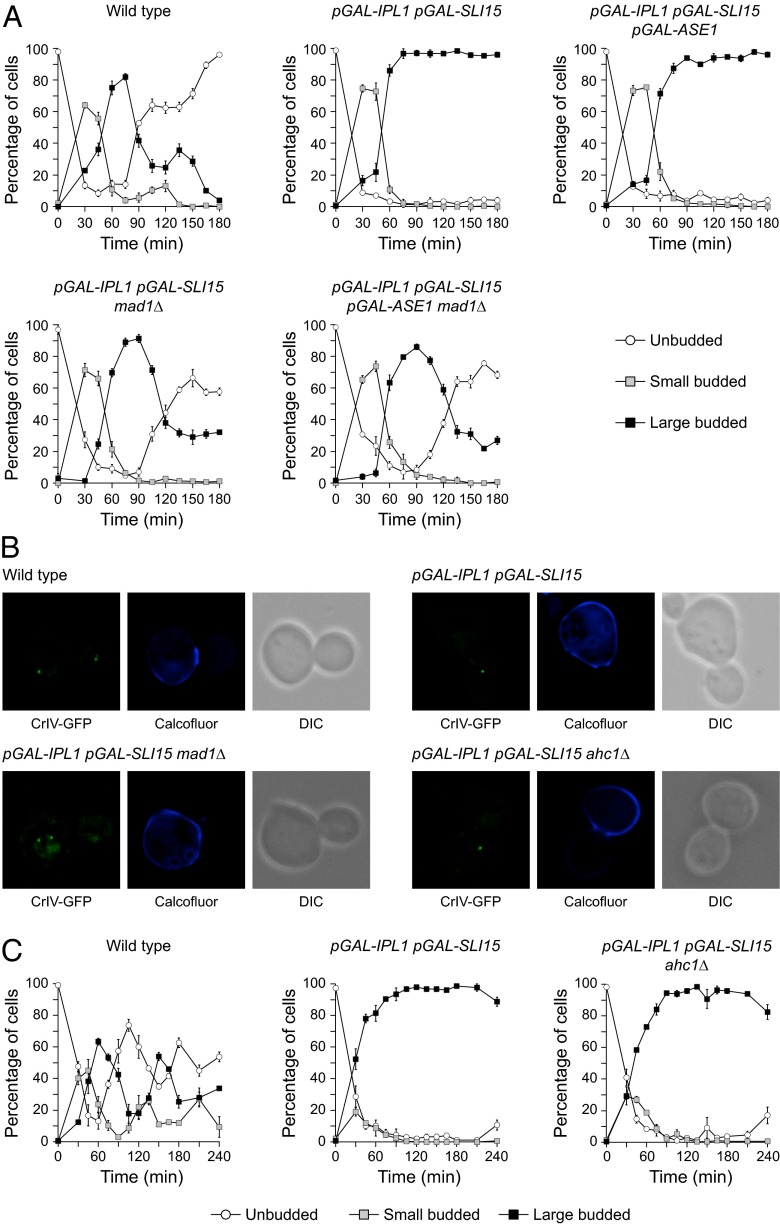
The defects in chromosome segregation caused by Ipl1 and Sli15 overexpression might be associated with the instability of the spindle. The high nuclear signal observed in cells overexpressing Ipl1 and Sli15 impeded the direct analysis of the localization of these proteins on the spindle, but the analysis of the localization of EGFP-tagged Bir1 indicated that the CPC still localized to the spindle in these cells (Fig. S2). Therefore the increase in Aurora B activity could cause premature spindle collapse by interfering with the function of microtubule-associated proteins. An interesting candidate is Ase1, a microtubule-bundling protein that is important for the stability of the spindle midzone (10). Intriguingly, the spindle collapses prematurely in ase1Δ cells (28), a phenotype that is similar to that observed after Ipl1 and Sli15 overexpression. Therefore we investigated whether localization of Ase1 is affected by increased Aurora B activity. In wild-type cells, Ase1-EGFP associated with the spindle in metaphase, and its localization was restricted to the midzone during anaphase (Fig. 4A). However, localization of Ase1-EGFP to the spindle was reduced during metaphase in cells overexpressing Ipl1 and Sli15, and it did not accumulate in the midzone as the spindle elongated (Fig. 4A). To test whether the spindle instability could be associated with defective Ase1 function, we next evaluated whether Ase1 overexpression could revert the premature spindle collapse caused by increased Aurora B activity. To this end, we placed an additional copy of ASE1 under the control of pGAL in our pGAL-IPL1 pGAL-SLI15 strain. Indeed, simultaneous Ase1 overexpression allowed cells with increased Ipl1 and Sli15 levels to elongate their spindles normally (Fig. 4 B and C). Accordingly, the maximum spindle length in pGAL-IPL1 pGAL-SLI15 pGAL-ASE1 cells was reduced only slightly as compared with wild-type cells (Fig. 4D). However, ASE1 overexpression did not alleviate other phenotypes associated with increased Ipl1 and Sli15 expression: The cells still displayed defects in chromosome segregation (Fig. 4E) and could not exit mitosis (Fig. 4B). Finally, we used SDS/PAGE to analyze whether Ase1 is hyperphosphorylated in the presence of high levels of Ipl1 and Sli15. Indeed, in these conditions Ase1 displayed a mobility shift that was abolished by phosphatase treatment (Fig. 4F and Fig. S2B); this finding suggests that increased Ipl1 and Sli15 levels interfere with Ase1 function by directly promoting its phosphorylation. Accordingly, expression of the ase1-5A allele, in which the five predicted Ipl1 consensus phosphorylation sites in Ase1 are mutated to alanine (10), reduced the percentage of cells with a debilitated spindle midzone and partially recovered the maximum spindle length after Ipl1 and Sli15 overexpression (Fig. S2C). In contrast to observations in cells overexpressing Ase1, the spindle finally collapsed in cells expressing Ase1-5A. However, Ipl1 still was able to phosphorylate the Ase1-5A protein in vitro (10), raising the possibility that the phospho-mutant still could be partially regulated by this kinase.
Fig. 4.
Spindle instability is associated with defective Ase1 function. (A) Wild-type (F1951) and pGAL-IPL1 pGAL-SLI15 (F1982) cells carrying Ase1-EGFP and Tub1-mCherry fusions were allowed to enter mitosis synchronously in YPRG as in Fig. 1A. Representative Ase1-EGFP (green), microtubules (red), DAPI (blue), merged, and DIC images are shown. (B–E) Wild-type (F955), pGAL-IPL1 pGAL-SLI15 (F947), and pGAL-IPL1 pGAL-SLI15 pGAL-ASE1 (F1735) cells were allowed to enter mitosis synchronously in YPRG as in Fig. 1A. (B) Cell-cycle progression was analyzed by spindle (tubulin) and nuclear (DAPI) morphology. Percentages of metaphase and anaphase cells and of cells displaying spindles with a thin midzone or with broken spindles are shown for each time point. Error bars indicate SD (n = 3). (C) Representative images showing tubulin (green) and DAPI staining (blue). (D) Average maximum spindle length. Error bars indicate SEM (n = 180). Statistically significant (***P < 0.0001) differences according to a two-tailed t test are shown also. (E) CrIV-GFP segregation was analyzed as in Fig. 3A. (F) Western blot shows Ase1-EGFP protein 60 and 90 min after release from α-factor in cells from A. Pgk1 was used as a loading control.
Increased Aurora B Activity Causes Destabilization of Amphitelic KT–MT Attachments.
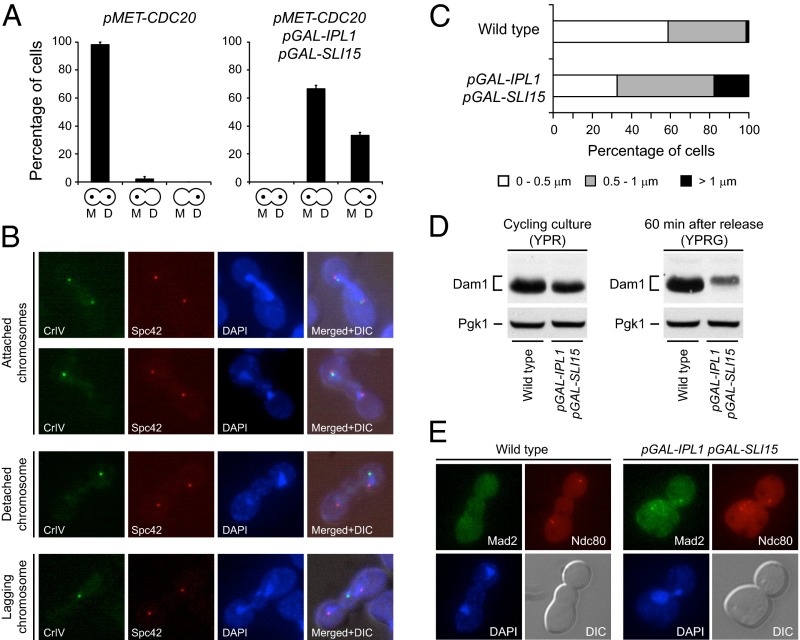
In the absence of tension in the spindle, phosphorylation of kinetochore components by Aurora B reduces their microtubule-binding affinity, thus generating an unattached kinetochore that activates the SAC (26, 29, 30). However, biorientation of sister chromatids separates Aurora B from its targets and stabilizes the KT–MT attachments (8, 9). Ipl1 and Sli15 overexpression could lead to the accumulation of the Aurora B homolog at the centromeres. In accordance with this notion, an analysis of Ipl1 localization by chromosome spreads showed that the Ipl1 signal at the centromeres is stronger after Ipl1 and Sli15 overexpression than in wild-type cells (Fig. S3A). This accumulation of Ipl1 at the centromeres could bring the kinase closer to its substrates than in wild-type cells, causing disruption of KT–MT attachments even when chromosomes are properly bioriented. A strong indication supporting this hypothesis is that the problems in chromosome segregation still were evident when Ipl1 and Sli15 were overexpressed in metaphase-arrested cells that subsequently were allowed to progress with the cell cycle, even though amphitelic attachments in these cells already had been established previously (Fig. 5A).
Fig. 5.
Increased Ipl1 and Sli15 expression causes continuous disruption of KT–MT attachments. (A) Analysis of chromosome segregation in pMET-CDC20 (F1670) and pMET-CDC20 pGAL-IPL1 pGAL-SLI15 (F1948) cells carrying CrIV-GFP. Cells were grown at 25 °C in synthetic complete (SC) medium with 2% raffinose and without methionine, were arrested in G1 with 5 μg/mL α-factor, and were released into SC with 2% raffinose and 8 mM methionine to impede CDC20 transcription and thus the metaphase-to-anaphase transition. Once cells were arrested in metaphase (2 h after release), 2% galactose was added to induce pGAL transcription, and 30 min later cells were released into SC with 2% raffinose and 2% galactose without methionine to allow cell-cycle progression. CrIV-GFP segregation was scored as in Fig. 3A, but no distinction was made between cells carrying one or two GFP dots. Error bars indicate SD (n = 3). (B and C) Wild-type (F1483) and pGAL-IPL1 pGAL-SLI15 (F1417) cells carrying CrIV-GFP and an mCherry-tagged version of Spc42 (a structural SPB component) were allowed to enter mitosis synchronously in YPRG as in Fig. 1A. (B) Representative images showing CrIV-GFP (green), Spc42-mCherry (red), and DAPI staining (blue), and merged images that also include DIC (merged+DIC) are presented. (C) Quantification of the average distance between the CrIV-GFP dot and the closest Spc42-mCherry signal (n = 150). (D) Wild-type (F2273) and pGAL-IPL1 pGAL-SLI15 (F2274) cells also expressing Dam1-3HA were allowed to enter mitosis synchronously in YPRG as in Fig. 1A. Western blot shows Pds1-3HA levels 60 min after release from α-factor. Pgk1 was used as a loading control. A Western blot displaying Dam1-3HA levels in cycling cultures of the same strains in YPR is shown as a reference. (E) Wild-type (F217) and pGAL-IPL1 pGAL-SLI15 (F1963) cells carrying Mad2-EGFP and Ndc80-CFP fusions were allowed to enter mitosis synchronously in YPRG as in Fig. 1A. Representative images showing Mad2-EGFP (green), Ndc80-CFP (red), DAPI staining (blue), and DIC are presented.
If KT–MT attachments were being disrupted continuously, the distance between the centromere of the chromosomes and the spindle pole bodies (SPBs; the equivalent of centrosomes in yeast) should increase after spindle elongation, because their disengagement from the microtubules would cause the chromosomes, which usually remain close to the SPBs, to be temporally separated from them. Indeed, Ipl1 and Sli15 overexpression increased the average distance between the centromere of chromosome 4 and the SPBs (Fig. 5 B and C). Remarkably, we even could detect pGAL-IPL1 pGAL-SLI15 cells in which CrIV-GFP was left behind as a lagging chromosome during spindle elongation (Fig. 5B). Dam1, a member of the DASH complex, is a key Ipl1 substrate in the kinetochore. Phosphorylation of Dam1 by Ipl1 weakens KT–MT attachments, and a dam1 S-to-D mutant that mimics constitutive Ipl1 phosphorylation of Dam1 shows evidence of lagging chromosomes (29). In agreement with our observations, although in wild-type cells Dam1 is dephosphorylated once the proper attachments are established during metaphase (Fig. 5D) (31), this protein is hyperphosphorylated in pGAL-IPL1 pGAL-SLI15 cells after transcription from the pGAL promoter is induced, as demonstrated by the mobility shift observed by SDS/PAGE gels that was abolished by phosphatase treatment (Fig. 5D and Fig. S3C). Interestingly, although histone H3 and Ase1 levels were not affected by the overexpression of Ipl1 and Sli15, the levels of Dam1 seemed to be reduced after the increase in Aurora B activity, possibly suggesting that the protein is less stable after Ipl1 phosphorylates it. Finally, further supporting the idea that chromosomes are constantly being detached, although all kinetochores are grouped together and visualized as a single fluorescent dot in wild-type anaphase cells expressing Ndc80 tagged with CFP as a kinetochore marker, multiple Ndc80-CFP foci could be detected in cells overexpressing Ipl1 and Sli15 after chromosome segregation (Fig. 5E).
Increased Ipl1 and Sli15 Expression Constitutively Activates the SAC.
If KT–MT attachments are constantly being destabilized, then the cells should be exposing unattached kinetochores continuously and thus permanently activating the SAC. One of the first steps in SAC activation is the localization of Mad2 to the unattached kinetochores (2). In wild-type metaphase cells, no Mad2-EGFP signal could be observed at the kinetochores once all the correct amphitelic attachments were established (Fig. S4A), nor could Mad2-EGFP be observed at the kinetochores in anaphase cells, as would be expected because the SAC is inactivated before this stage in the cell cycle (Fig. 5E). However, in agreement with our hypothesis, cells overexpressing Ipl1 and Sli15 displayed Mad2-EGFP foci that colocalized with the kinetochores during metaphase and, more strikingly, even after chromosome segregation and spindle elongation (Fig. 5E and Fig. S4A). Accordingly, and in contrast to wild-type cells, Ipl1 localization at the centromeres still could be observed in cells overexpressing Ipl1 and Sli15 that already had segregated their chromosomes (Fig. S3B).
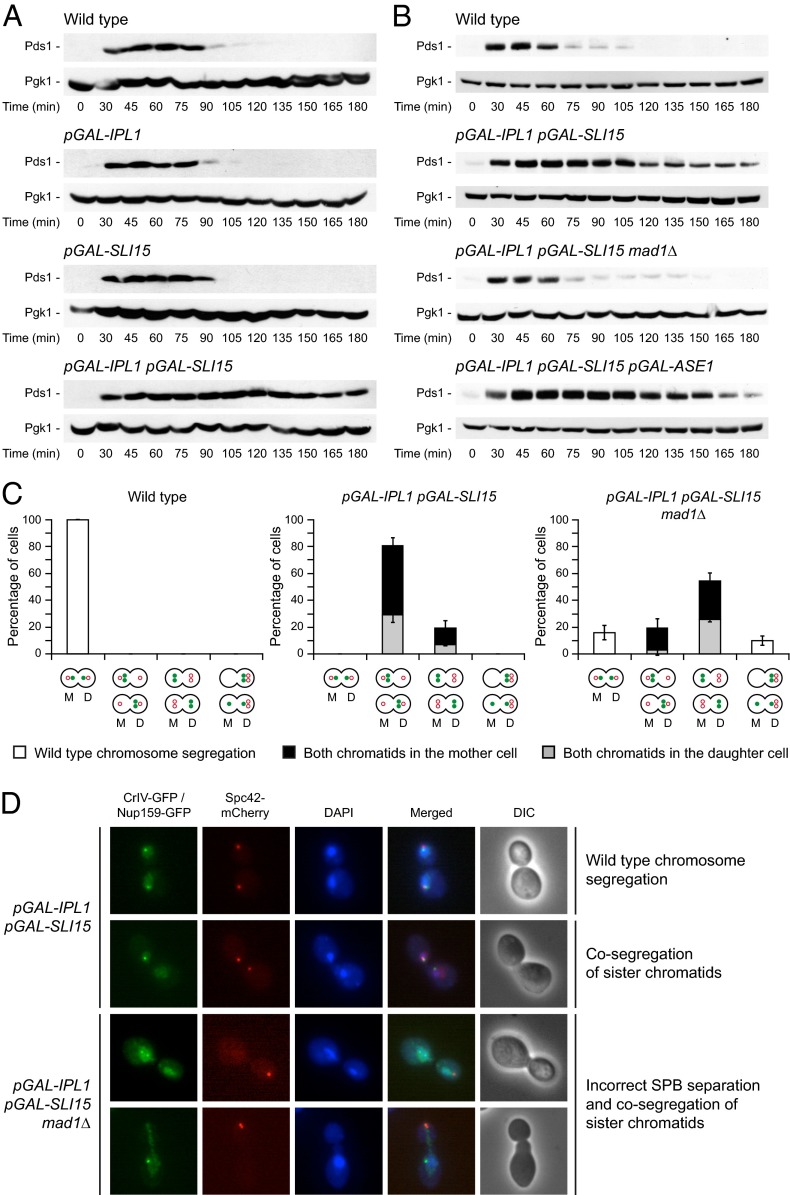
Activation of the SAC inhibits the anaphase-promoting complex (APC/C) by sequestering its coactivator Cdc20 (2). Therefore, we can easily determine whether the checkpoint is active by measuring the levels of securin (Pds1), which is targeted by the APC/C for its degradation at the metaphase-to-anaphase transition (32), and Clb2, because the APC/C also is responsible for Clb-CDK inactivation (2). According to our hypothesis, cells overexpressing Ipl1 and Sli15 did not degrade Pds1 and maintained high levels of Clb2 (Fig. 6A and Fig. S4B). Furthermore, deletion of MAD1, which encodes an essential SAC component, allowed Pds1 and Clb2 degradation after Ipl1 and Sli15 overexpression (Fig. 6B and Fig. S4B). However, SAC activation was not associated with the instability of the spindle, because Pds1 was not degraded in pGAL-IPL1 pGAL-SLI15 pGAL-ASE1 cells (Fig. 6B). Pds1 protects cohesin complexes from being degraded (4). Accordingly, a final indication of the constitutive activation of the SAC after Ipl1 and Sli15 overexpression is that both sister chromatids of CrIV remained bound when they cosegregated, and thus were visualized as a single GFP dot (Fig. 3B).
Fig. 6.
Ipl1 and Sli15 overexpression leads to constitutive SAC activation. (A and B) Wild-type (F144), pGAL-IPL1 (F1191), pGAL-SLI15 (F1196), pGAL-IPL1 pGAL-SLI15 (F980), pGAL-IPL1 pGAL-SLI15 mad1Δ (F1376), and pGAL-IPL1 pGAL-SLI15 pGAL-ASE1 (F1912) cells also expressing Pds1-3HA were allowed to enter mitosis synchronously in YPRG as in Fig. 1A. Western blots show Pds1-3HA levels at the indicated time points. Pgk1 was used as a loading control. (C) Analysis of CrIV and SPB segregation in wild-type (F1483), pGAL-IPL1 pGAL-SLI15 (F1417), and pGAL-IPL1 pGAL-SLI15 mad1Δ (F1927) cells carrying CrIV-GFP and a Spc42-mCherry fusion. Cells were allowed to enter mitosis synchronously in YPRG as in Fig. 1A. The graphics below the bars indicate the different patterns of CrIV-GFP (green closed circle) and SPB (red open circle) segregation included in each category. White bars indicate wild-type chromosome segregation; cosegregation of sister chromatids is represented by black bars [both sisters in the mother (M) cell] or gray bars [both sisters in the daughter (D) cell]. Error bars indicate SD (n = 3). (D) pGAL-IPL1 pGAL-SLI15 (F1562) and pGAL-IPL1 pGAL-SLI15 mad1Δ (F1811) cells carrying CrIV-GFP and also expressing Nup159-GFP and Spc42-mCherry fusions were allowed to enter mitosis synchronously in YPRG as in Fig. 1A. Representative images showing CrIV-GFP and Nup159-GFP (green), Spc42-mCherry (red), and DAPI staining (blue) and merged and DIC images are presented.
Although increased Ipl1 and Sli15 expression led to severe defects in chromosome segregation, most cells duplicated and segregated their SPBs correctly during mitosis (Fig. 6C). Intriguingly, MAD1 deletion caused a huge defect in SPB separation in these cells (Fig. 6 C and D). Despite the cosegregation of both SPBs, these cells showed separated DNA masses and carried out karyokinesis, as shown using DAPI and the Nup159 nucleoporin as a nuclear membrane marker (Fig. 6D). The most likely explanation for this observation is that activation of the SAC provides the cells with the additional time that is needed to set up the metaphase spindle properly in the presence of high levels of Aurora B activity. Ipl1 has been shown to play key roles in both spindle assembly and disassembly (10, 11, 13, 14). Therefore, spindle assembly also might be somewhat affected by the increase in Aurora B activity. Alternatively, increased Aurora B activity might interfere with the initial separation of the SPBs. Interestingly, Ipl1 activity has been shown to be required for maintenance of a tight association between duplicated SPBs during meiosis in budding yeast (33). In the absence of the SAC, cells cannot assemble the spindle correctly, both SPBs are dragged to the same cell, and most of the chromosomes become detached and are left behind because of the increased destabilization of KT–MT attachments. Accordingly, the DNA masses were greatly unequal in size in pGAL-IPL1 pGAL-SLI15 mad1Δ cells, and most of the DNA remained in the cell opposite the cell into which both SPBs were dragged (Fig. 6D). This defect in spindle assembly could not be rescued by ASE1 overexpression (Fig. S4C). In any case, and despite the problems with SPB separation, chromosome segregation defects still were evident in the few cells that managed to segregate the SPBs properly, as would be expected, given that abrogation of the checkpoint did not eliminate the problem that caused the aneuploidy (Fig. 6C). Additionally, the two CrIV sister chromatids could be visualized as separate dots (Fig. 6D), demonstrating that Pds1 was degraded and also that the DNA had been replicated previously.
SAC Inactivation Allows Cytokinesis in Cells Overexpressing Ipl1 and Sli15.
Although spindle elongation and chromosome segregation occurred, the cells that overexpressed Ipl1 and Sli15 did not carry out cytokinesis. However, as previously demonstrated, increased Aurora B activity causes constitutive SAC activation that might explain the absence of cytokinesis. Indeed, the abrogation of the SAC by the deletion of MAD1 allowed the cells with elevated Aurora B and INCENP levels to carry out cytokinesis and exit mitosis (Fig. 7A). Accordingly, a closed septum could be observed in the division plane before the separation of the daughter and mother cells (Fig. 7B).
Fig. 7.
Lack of cytokinesis is associated with activation of the SAC but not of the NoCut pathway. Wild-type (F1483 in A and F955 in B and C), pGAL-IPL1 pGAL-SLI15 (F1417 in A and F947 in B and C), pGAL-IPL1 pGAL-SLI15 ahc1Δ (F1700), pGAL-IPL1 pGAL-SLI15 pGAL-ASE1 (F1908), pGAL-IPL1 pGAL-SLI15 mad1Δ (F1927 in A and F1845 in B), and pGAL-IPL1 pGAL-SLI15 pGAL-ASE1 mad1Δ (F1910) cells were allowed to enter mitosis synchronously in YPRG as in Fig. 1A. (A and C) Percentages of unbudded and small and large budded cells are shown for each time point. Error bars indicate SD (n = 3). (B) Representative images showing CrIV-GFP (green), calcofluor staining (blue), and DIC.
The NoCut pathway inhibits abscission to prevent the cytokinesis machinery from damaging the DNA when the spindle prematurely breaks and the chromosomes are not properly segregated, and Ipl1 plays a key role in this checkpoint (15, 16). The presence of a functional NoCut pathway in SAC mutants (16) argues against the notion that Ipl1 and Sli15 overexpression inhibits the cytokinesis cascade by inducing hyperactivity of the NoCut pathway. Nevertheless, we evaluated whether increased Aurora B activity activated the NoCut pathway. Because the NoCut pathway is triggered in response to spindle midzone problems, and ASE1 deletion delays abscission in a NoCut-dependent manner (16), we evaluated whether stabilization of the spindle could allow cytokinesis to occur in cells with increased Aurora B activity. As expected, additional Ase1 overexpression did not allow cytokinesis to occur in cells with increased Ipl1 and Sli15 expression (Fig. 7A). Finally, deletion of AHC1, which encodes a scaffolding element of the ADA histone acetyltransferase [an essential NoCut component (15)], did not promote cytokinesis in cells with increased Ipl1 and Sli15 levels, and these cells still accumulated as large budded cells with an open septum (Fig. 7 B and C).
Discussion
Aurora B plays a key role in ensuring the correct biorientation of the sister chromatids during mitosis and therefore in preventing aneuploidy (26). Hence, its absence significantly impairs cell viability. Interestingly, elevated levels of this kinase also are detrimental for the cells (19, 21), and increased Aurora B expression has been linked to certain types of cancer (19). However, little is known about the molecular mechanisms leading to the defects associated with elevated Aurora B activity. A factor that has limited this analysis thus far is that the overexpression of Ipl1 does not seem to interfere with cell viability in S. cerevisiae, a model organism that has greatly facilitated our understanding of the roles of this kinase in cell-cycle regulation (22). Here, by simultaneously overexpressing Ipl1 and Sli15 in S. cerevisiae, we have developed a budding yeast model that has allowed the molecular dissection of the phenotypes determined by increased Aurora B activity.
Although it is evident how the lack of a protein that ensures proper KT–MT attachments can lead to massive defects during chromosome segregation, it is not obvious why its overexpression also can generate aneuploidy. In cells carrying the ipl1-321 allele, sister chromatids cosegregate preferentially into the bud at the restrictive temperature (26), because their inability to repair incorrect KT–MT attachments causes both sisters to be attached more frequently to the old SPB (the one that originally was in the mother cell), and in S. cerevisiae this SPB always migrates toward the bud (34). However, when Ipl1 and Sli15 are overexpressed simultaneously, sister chromatids preferentially cosegregate toward the mother cell. We suggest that, in this case, increased levels of active Aurora B accumulate at centromeres, determining the destabilization even of amphitelic chromosome attachments. In agreement with this hypothesis, Dam1, one of the key substrates targeted by Ipl1 to regulate KT–MT attachments, is hyperphosphorylated after the overexpression of Ipl1 and Sli15. Accordingly, although Aurora B is dispensable once the the proper KT–MT attachments are formed (26), the defects in chromosome segregation are still observed when Ipl1 and Sli15 are overexpressed after chromosome biorientation is established. Additionally, the average distance between the centromeres and the SPBs increases after spindle elongation in cells overexpressing Aurora B and INCENP homologs, probably reflecting a temporal disengagement of kinetochores from the spindle. We even observed lagging chromosomes that were left behind during spindle elongation. Interestingly, lagging chromosomes also are observed frequently in mammalian cells overexpressing AIM-1/Aurora B (21). This detachment of chromosomes also could explain why both sister chromatids are retained more frequently in the mother cell. Finally, and in agreement with KT–MT attachments being continuously disrupted, our results demonstrate that, although the lack of Ipl1 does not trigger the SAC (30), simultaneous overexpression of Ipl1 and Sli15 induces constitutive activation of this checkpoint. AIM-1/Aurora B overexpression in mammalian cells also activates the SAC (21), further demonstrating the validity of our model. Activation of the SAC inhibits the APC/C and therefore protects the cohesion of sister chromatids. This protection, however, creates an apparent paradox: If cohesins are not cleaved, why do cells overexpressing Ipl1 and Sli15 elongate their spindles and segregate their chromosomes? We believe that the constant detachment of KT–MT connections leads to spindle elongation because forces counteracting the activity of the motors that separate the SPBs are absent, as occurs in a cdc6 mutant (35); thus these cells continue their progression into mitosis until they reach an “anaphase-like prometaphase” state (36) in which they are physiologically still in metaphase but show an elongated spindle and distribute their chromosomes.
Increased Ipl1 and Sli15 expression also causes spindle instability, especially in the midzone. In cohesin-defective mutants arrested in metaphase, sister chromatids cannot be held together, and the cells attempt to elongate their spindles when the APC/C is still inactive and the proper conditions for spindle elongation are not yet established, thus debilitating the spindle midzone (37). In these cells, however, the inactivation of the SAC restores the normal timing of APC/CCdc20 activation and recovers the stability of the spindle (37), whereas the spindle defects in cells overexpressing Ipl1 and Sli15 are not rescued by MAD1 deletion. Therefore it is likely that the two situations are not similar. Instead, we suggest that increased Aurora B activity directly promotes premature spindle breakdown. Ipl1 plays an important role in spindle disassembly, and, accordingly, the ipl1-321 mutant delays spindle breakdown and shows a hyperelongated spindle at the restrictive temperature (13). Although our results suggest a centromeric accumulation of the CPC, the analysis of Bir1 localization shows that the complex still can localize to the spindle after Ipl1 and Sli15 overexpression. Once in the spindle, the CPC has been shown to regulate a number of microtubule-associated proteins, including Ase1 (10). Ase1 shows five consensus Ipl1 phosphorylation sites and can be phosphorylated in vitro by Aurora B (10). Our analysis of the phosphorylation status of Ase1 shows that this protein is hyperphosphorylated after overexpression of Ipl1 and Sli15 and that expression of the ase1-5A allele partially recovers the spindle defects caused by increased Aurora B activity. Furthermore, we have demonstrated that overexpression of Ase1 rescues the stability of the spindle midzone in cells with increased Ipl1 and Sli15 levels. Importantly, Ase1 overexpression cannot rescue the spindle defects observed in cohesin-defective mutants arrested in metaphase (Fig. S5). Therefore, we suggest that the premature breakdown of the spindle during its elongation in cells overexpressing Ipl1 and Sli15 is caused by a defective Ase1 function resulting from the hyperphosphorylation of this protein by the CPC.
One last phenotype associated with increased Aurora B activity in metazoans is the absence of cytokinesis even though chromosome segregation occurs (21, 38). We show that this phenotype also is seen in S. cerevisiae. Aurora B plays an important role in the NoCut pathway (15, 16). However, increased levels of Ipl1 and Sli15 do not seem to activate the NoCut pathway, and the lack of cytokinesis cannot be rescued by stabilization of the spindle midzone. Instead, our results demonstrate that the absence of cytokinesis in these cells is caused by the constitutive activation of the SAC. The NoCut pathway is still functional in SAC mutants (16), giving further evidence that this pathway is not activated despite the elevated Aurora B activity.
An interesting question is why increasing the expression of both Ipl1 and Sli15 in yeast is necessary to recapitulate the phenotypes associated to increased Aurora B activity in higher eukaryotes. Ipl1 is the only member of the Aurora family in S. cerevisiae, whereas in mammals there are three Aurora kinases—A, B, and C—that show different subcellular localization and functions (3). Ipl1 has additional functions in yeast that normally are carried out by Aurora A or C in mammals (10, 33, 39); thus it might be essential to additionally regulate Ipl1 activity by controlling Sli15 levels. A stricter regulation of Aurora B activity in higher eukaryotes could make the additional control of INCENP levels unnecessary. In any case, our results highlight the importance of the rigorous control of the expression of Aurora B and INCENP for cell viability and also may be helpful in providing new insights into how deregulation of Aurora B can promote genomic instability in higher eukaryotes. We have demonstrated that the duplication of both IPL1 and SLI15 genes considerably increases genomic instability in yeast cells as compared with the duplication of only one or the other of the two genes. Similarly, simultaneous elevated expression of INCENP could facilitate the tumorigenic process in cancer types associated with increased Aurora B levels. On the other hand, increased expression of both Aurora B and INCENP could be required specifically for tumor progression in particular cell types, as has been suggested previously (19).
Materials and Methods
Strains.
All strains are derivatives of W303 and are described in Table S1.
Plasmid Loss Assay.
Cells carrying the centromeric pRS316 plasmid, which allows expression of the URA3 gene, were grown on SC medium for 23 generations and then were diluted and plated on SC plates to determine the total number of cells. Subsequently, the colonies were replicated on SC plates without uracil to record the number of cells losing the pRS316 plasmid.
Immunolocalization and Fluorescence Microscopy.
Immunofluorescence was carried out as described in ref. 40. Anti-tubulin (Abcam) and anti-rat FITC (Jackson ImmunoResearch) antibodies were used at 1:250. Chromosome spreads were prepared as described in ref. 41. Anti-HA (HA.11; Covance) and anti-mouse Cy3 (Jackson ImmunoResearch) antibodies were used at 1:250. Microscope preparations were analyzed and imaged using a DM6000 microscope (Leica) equipped with a 100×/1.40 NA oil immersion objective lens, A4, L5, CFP, and TX2 filters, and a DF350 digital charge-coupled device camera (Leica). Pictures were processed with LAS AF (Leica) and ImageJ (http://rsbweb.nih.gov/ij/) software.
DAPI and Calcofluor Staining.
Samples were fixed for 15 min at room temperature in 2.5% (vol/vol) formaldehyde, washed twice with 0.1 M potassium phosphate buffer (pH 6.6), and resuspended in 0.1 M potassium phosphate buffer (pH 7.4). Calcofluor (Sigma) was used at 1 mg/mL For DAPI preparations, cells first were fixed for 10 min in 80% (vol/vol) ethanol and then were resuspended in 10 μg/mL DAPI. Microscope preparations were analyzed and imaged as for immunofluorescence.
FACS Analysis.
Cells were fixed in 70% (vol/vol) ethanol, incubated for 12 h in PBS with 1 mg/mL RNase A, and stained for 1 h with 5 mg/mL propidium iodide (Sigma). After sonication of the sample to separate single cells, DNA content was analyzed in a FACSCalibur flow cytometer (Becton Dickinson).
Western Blot Analysis.
Protein extracts were prepared using the trichloroacetic acid precipitation method described in ref. 42. Antibodies used are shown in Table S2. Protein signal was detected using the Western Bright ECL system (Advansta).
Supplementary Material
Acknowledgments
We thank Dr. S. Biggins for kindly providing yeast strains and Drs. F. Cortés-Ledesma, S. Picossi, and F. Prado, as well as members of the F.M.-C. laboratory, for critical reading of the manuscript. This work was supported by Junta de Andalucía Grant CVI-5806 and the European Fund for Economic and Regional Development of the European Union. F.M.-C. was the recipient of a contract from the Fifth Research Plan (VPPI) of the University of Seville. M.M.-B. received a Board for Advanced Studies Predoctoral Fellowship from the Spanish National Research Council.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408017111/-/DCSupplemental.
References
- 1.Siegel JJ, Amon A. New insights into the troubles of aneuploidy. Annu Rev Cell Dev Biol. 2012;28:189–214. doi: 10.1146/annurev-cellbio-101011-155807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 3.Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13(12):789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nasmyth K, Haering CH. Cohesin: Its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 5.Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8(1):78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- 6.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127(5):983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 7.DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127(5):969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 8.Welburn JP, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38(3):383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang E, Ballister ER, Lampson MA. Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient. J Cell Biol. 2011;194(4):539–549. doi: 10.1083/jcb.201103044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotwaliwale CV, Frei SB, Stern BM, Biggins S. A pathway containing the Ipl1/aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly. Dev Cell. 2007;13(3):433–445. doi: 10.1016/j.devcel.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereira G, Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302(5653):2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- 12.Douglas ME, Davies T, Joseph N, Mishima M. Aurora B and 14-3-3 coordinately regulate clustering of centralspindlin during cytokinesis. Curr Biol. 2010;20(10):927–933. doi: 10.1016/j.cub.2010.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buvelot S, Tatsutani SY, Vermaak D, Biggins S. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J Cell Biol. 2003;160(3):329–339. doi: 10.1083/jcb.200209018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodruff JB, Drubin DG, Barnes G. Mitotic spindle disassembly occurs via distinct subprocesses driven by the anaphase-promoting complex, Aurora B kinase, and kinesin-8. J Cell Biol. 2010;191(4):795–808. doi: 10.1083/jcb.201006028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendoza M, et al. A mechanism for chromosome segregation sensing by the NoCut checkpoint. Nat Cell Biol. 2009;11(4):477–483. doi: 10.1038/ncb1855. [DOI] [PubMed] [Google Scholar]
- 16.Norden C, et al. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 2006;125(1):85–98. doi: 10.1016/j.cell.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 17.Steigemann P, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136(3):473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 18.Tatsuka M, et al. Multinuclearity and increased ploidy caused by overexpression of the aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 1998;58(21):4811–4816. [PubMed] [Google Scholar]
- 19.Katayama H, Brinkley WR, Sen S. The Aurora kinases: Role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 2003;22(4):451–464. doi: 10.1023/a:1023789416385. [DOI] [PubMed] [Google Scholar]
- 20.Katayama H, et al. Mitotic kinase expression and colorectal cancer progression. J Natl Cancer Inst. 1999;91(13):1160–1162. doi: 10.1093/jnci/91.13.1160. [DOI] [PubMed] [Google Scholar]
- 21.Ota T, et al. Increased mitotic phosphorylation of histone H3 attributable to AIM-1/Aurora-B overexpression contributes to chromosome number instability. Cancer Res. 2002;62(18):5168–5177. [PubMed] [Google Scholar]
- 22.Biggins S, et al. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13(5):532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu JY, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102(3):279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 24.Li F, Flanary PL, Altieri DC, Dohlman HG. Cell division regulation by BIR1, a member of the inhibitor of apoptosis family in yeast. J Biol Chem. 2000;275(10):6707–6711. doi: 10.1074/jbc.275.10.6707. [DOI] [PubMed] [Google Scholar]
- 25.Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128(3):477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka TU, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108(3):317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 27.King EM, Rachidi N, Morrice N, Hardwick KG, Stark MJ. Ipl1p-dependent phosphorylation of Mad3p is required for the spindle checkpoint response to lack of tension at kinetochores. Genes Dev. 2007;21(10):1163–1168. doi: 10.1101/gad.431507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuyler SC, Liu JY, Pellman D. The molecular function of Ase1p: Evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J Cell Biol. 2003;160(4):517–528. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheeseman IM, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111(2):163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 30.Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15(23):3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keating P, Rachidi N, Tanaka TU, Stark MJ. Ipl1-dependent phosphorylation of Dam1 is reduced by tension applied on kinetochores. J Cell Sci. 2009;122(Pt 23):4375–4382. doi: 10.1242/jcs.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shirayama M, Tóth A, Gálová M, Nasmyth K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402(6758):203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- 33.Shirk K, Jin H, Giddings TH, Jr, Winey M, Yu HG. The Aurora kinase Ipl1 is necessary for spindle pole body cohesion during budding yeast meiosis. J Cell Sci. 2011;124(Pt 17):2891–2896. doi: 10.1242/jcs.086652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereira G, Tanaka TU, Nasmyth K, Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 2001;20(22):6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14(15):3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bajer AS. Functional autonomy of monopolar spindle and evidence for oscillatory movement in mitosis. J Cell Biol. 1982;93(1):33–48. doi: 10.1083/jcb.93.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Severin F, Hyman AA, Piatti S. Correct spindle elongation at the metaphase/anaphase transition is an APC-dependent event in budding yeast. J Cell Biol. 2001;155(5):711–718. doi: 10.1083/jcb.200104096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terada Y, et al. AIM-1: A mammalian midbody-associated protein required for cytokinesis. EMBO J. 1998;17(3):667–676. doi: 10.1093/emboj/17.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen J, Paris J, Willer M, Philippe M, Hagan IM. The S. pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J Cell Sci. 2001;114(Pt 24):4371–4384. doi: 10.1242/jcs.114.24.4371. [DOI] [PubMed] [Google Scholar]
- 40.Valerio-Santiago M, Monje-Casas F. Tem1 localization to the spindle pole bodies is essential for mitotic exit and impairs spindle checkpoint function. J Cell Biol. 2011;192(4):599–614. doi: 10.1083/jcb.201007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nairz K, Klein F. mre11S—a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 1997;11(17):2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Aquino KE, et al. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol Cell. 2005;19(2):223–234. doi: 10.1016/j.molcel.2005.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.