Significance
A modular 3D brain-like cortical tissue is constructed with silk protein-based scaffold and ECM composite and primary cortical neurons. This tissue responds in vitro with biochemical and electrophysiological outcomes, mimicking observations of brain homeostasis and mechanical injury responses.
Keywords: electrophysiology, connectivity, silk, scaffold, traumatic brain injury
Abstract
The brain remains one of the most important but least understood tissues in our body, in part because of its complexity as well as the limitations associated with in vivo studies. Although simpler tissues have yielded to the emerging tools for in vitro 3D tissue cultures, functional brain-like tissues have not. We report the construction of complex functional 3D brain-like cortical tissue, maintained for months in vitro, formed from primary cortical neurons in modular 3D compartmentalized architectures with electrophysiological function. We show that, on injury, this brain-like tissue responds in vitro with biochemical and electrophysiological outcomes that mimic observations in vivo. This modular 3D brain-like tissue is capable of real-time nondestructive assessments, offering previously unidentified directions for studies of brain homeostasis and injury.
The brain possesses extraordinary connectivity of neural networks. This complexity is evident at multiple levels of structural and functional hierarchy, including microcircuits dominated by neuronal clusters and larger distinctive regions of grey matter interconnected by white matter axon tracts. These features are highlighted in the Blue Brain project (1) and the Human Connectom Project (2) that aim to compile detailed information about connectivity at various levels and ultimately, reconstruct the human brain as a large-scale network. However, at the tissue level, the complex interconnectivity is masked by the distribution of neurons, such as in the stratified laminar layers of the neocortex. Although functionally related neurons generally group together (3, 4), boundaries of functional units cannot be readily revealed with phenotype markers, necessitating electrophysiological studies and correlative functional outcomes. It is, therefore, necessary to differentiate physical and functional associations of neuronal populations to unravel complex networks.
Three-dimensional tissue engineering could provide compartmentalized cultures of discrete and identifiable structures to emulate native tissues and thereby, provide insight into the complexities. By recreating cell–cell and cell–ECM interactions, 3D structures enable the formation of tissue-mimetic architectures and promote more realistic physiological responses than conventional 2D cultures (5). Toward this goal, multilayer lithography (6), 3D patterning of bulk structures (7), and 3D tissue printing (8) are used. These rationally designed structures have been generated for tissue engineering of the lung, liver, and kidney, for which the structure–function relationships are modular-based and well-defined. Recent advances in stem and progenitor cell technology have induced cells to differentiate into and produce tissue-appropriate cell compositions and ECM components and form biomimetic tissues with nascent functions, including the cerebral organoids (9). These technologies show self-organization capability of cells in tissue-mimetic environments, such as native tissue-derived decellularized scaffolds (10–13). However, densely packed brain tissue with an architecture defined by neuronal connectivity (14, 15) presents a unique challenge to define modular structures with specific functions. Rather than reconstructing a whole-brain network, we aimed at reducing the structural complexity to fundamental features that are relevant to tissue-level physiological functions.
Neural connectivity at the basic level, which includes segregated neuronal and axonal compartments, is particularly relevant for brain disorders, such as diffuse axonal injury in brain trauma (16, 17). However, ECM gel-based in vitro 3D systems have not yielded tissue-level functional assessments, possibly because of their inadequate mechanical properties and fast degradation compared with brain tissue. Here, we developed 3D compartmentalized neuronal cultures with silk fibroin-based biomaterials offering tunable mechanical properties, versatile structural forms, and brain and neural culture compatibility (18–21). This brain-like tissue provides rudimentary but relevant features of brain neural networks. The physiologically relevant and responsive 3D brain-like tissue also shows capability for the assessment of brain disorders, such as traumatic brain injury (TBI).
Results
Modular Design of Bioengineered Brain-Like Tissues.
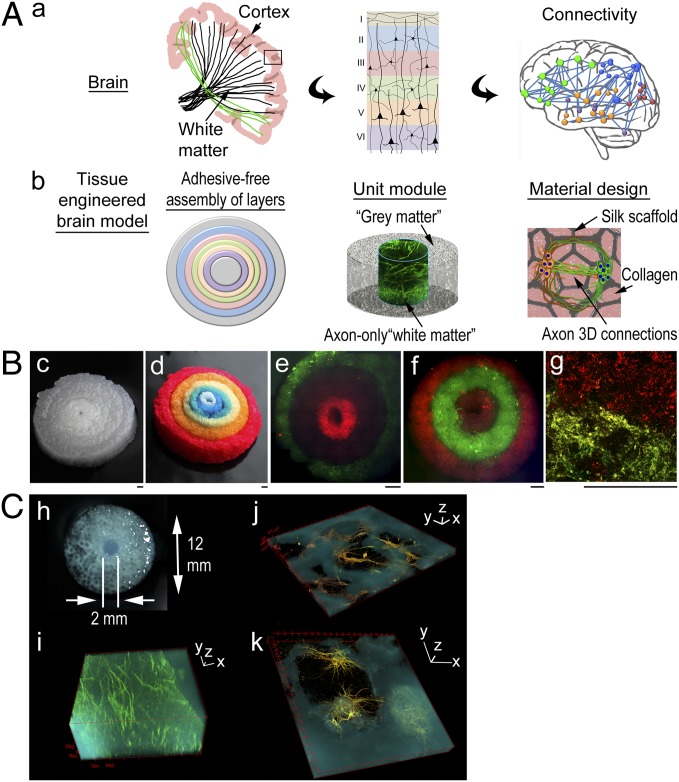
The approach consisted of a modular design of silk protein-based porous scaffolds. Fig. 1A shows the conceptual framework of the strategy. Fig. 1 A, a shows schematics of the targeted architectural features of the brain, including the cortex consisting of six laminar layers and white matter tracts [drawing based on the diffuse tensor imaging photographs in ref. 14] (Fig. 1 A, a, Left and Center) as well as microcircuitry (drawing based on the connectivity analysis described in ref. 15) (Fig. 1 A, a, Right). Fig. 1 A, b shows design concepts, including adhesive-free assembly of concentrically shaped donuts, the unit module consisting of neuron-rich grey matter regions and axon-only white matter regions, and the scaffold/collagen gel composite structure that supports the formation of axon connections in 3D.
Fig. 1.
A modular design of bioengineered brain-like cortical tissues. (A) Schematics of the conceptual framework of the strategy. (a) Targeted architectural features of the brain: (Left) the cortex and white matter tracks (drawing based on diffuse tensor imaging photographs in ref. 14), (Center) six laminar layers of the neocortex, and (Right) white matter connectivity in microcircuitries (drawing based on connectivity analysis described in ref. 15). (b) The design concepts: (Left) adhesive-free assembly of concentrically arranged layers, (Center) the unit module consisting of neuron-rich grey matter regions and axon-only white matter regions, and (Right) the material design of scaffold/collagen gel composite supporting axon connections in 3D. (B) 3D assembled tissue structures. (c and d) Assembly of six concentrically shaped donuts of silk scaffold [(c) original color; (d) dyed with food color]. (e–g) Each layer was seeded with different primary rat cortical neurons (live-stained with DiI in red and DiO in green) and assembled: (e and f) two three-layered constructs and (g) neurons at the interface. (Scale bar: 1 mm.) (C) A unit module. (h) The scaffold. (i–k) Three-dimensional view of confocal z stack of multichannel images of 3D brain-like tissues (fluorescently stained with axonal marker β3-tubulin in green and dendritic marker microtubule-associated protein-2 in red and superposed with inverted bright-field images of silk structure in cyan). (i) The center axon-only region. (j and k) The porous scaffold region. Note that the extensive neuronal coverage on the scaffold surface is obscured by the opaqueness of the scaffold material (cyan) compared with the porous region (dark) in the 3D images. (Scale bars: 3D axis, 100 µm.)
A jigsaw puzzle-like cutting process was developed to fabricate the modular structures and facilitate their assembly (Materials and Methods). The versatility of silk protein scaffolds allowed the modules to fit into self-supporting assembled structures in air or solution without additional bonding or mechanical reinforcements. Assembled six-layered structures are shown in Fig. 1B (the silk scaffold is shown in Fig. 1 B, c, and each layer of the scaffold dyed with a different food color is shown in Fig. 1 B, c). Each modular structure was populated with different neurons independently before assembly. Fig. 1 B, e and f shows two three-layered constructs, with neuronal populations differentiated by live staining with [1,1′-dioctadecyl-3,3,3′,3′-tetra-methylindo-carbocyanine perchlorate; diI-C18-(3)] (DiI) in red or [3,3′-dioctadecyloxa-carbocyanine perchlorate; diO-C18-(3)] (DiO) in green, with alternating dyes used in adjacent layers. Neurons at the interface (Fig. 1 B, g, layer I in red and layer II in green) showed the compartmentalization within the adjacent structures.
Unit modules were constructed to realize brain-like grey and white matter compartmentalization (Fig. 1C). The center region was comprised of millimeter-sized collagen gel matrix that was penetrated by dense long axons (Fig. 1 C, i). The neurons in the porous scaffold region organized as mininetworks around the pores (Fig. 1 C, j); within a pore, the neurons anchored to the scaffold surface and extended axons into the collagen gel-filled 3D space (Fig. 1 C, k).
Optimization of Structure, Matrix Components, and Mechanical Stiffness.
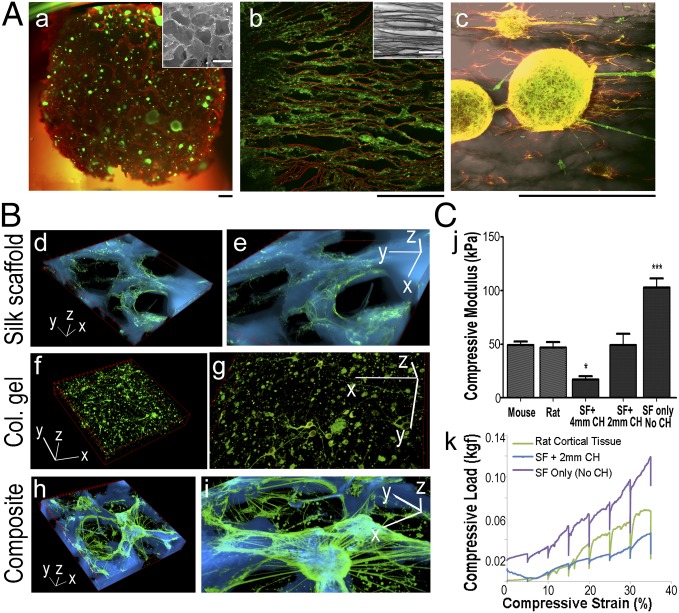
Based on the donut module approach, we optimized scaffold structure, matrix components, and mechanical stiffness to promote neuronal tissue growth (Fig. 2). Sponges of ∼500-µm pore size (previously characterized in ref. 22) were used (Fig. 2 A, a, Inset) to reduce diffusion distance (<200 µm) limitations in bulk biomaterial systems (23). The extensive porosity also allowed a more even distribution of dissociated neurons throughout the scaffolds. The size and orientation of the pores could be tuned with established processing strategies (24). Neuronal association with the scaffold surface was most pronounced on the aligned structures (Fig. 2 A, b, Inset). Cell processes arranged along the aligned scaffold features (Fig. 2 A, b shows live processes calcein acetoxymethyl-stained in green and silk material autofluoresced in red, and Fig. 2 A, c shows neurons stained for β3-tubulin in green, glial cells stained for GFAP in red, and silk material in dark gray).
Fig. 2.
Optimization of structure, matrix components, and mechanical stiffness for the brain-like tissue. (A) Structure. (a) Neurons (green) growing in silk sponges (red) with random pores (Inset; ∼500-µm diameter). (b and c) Neurons growing in silk sponges with aligned pores (b, Inset). (b) Live neuronal processes (green) growing along silk material structures (red). (c) Neurons (green) and glial cells (red) extend processes along aligned silk structures (dark gray). (Scale bar: 100 µm.) (B) Matrix components. (d and e) Silk scaffold only. (f and g) Collagen gel only. (h and i) Composite structures of silk scaffold infused with collagen gel. Fluorescence images of neurons (green) superposed with inverted bright-field images of silk structure (cyan). e, g, and i are zoomed-in views of d, f, and h, respectively. (Scale bars: 3D axis, 100 µm.) Col, collagen. (C) Mechanical stiffness measured by the confined compression test. SF, scaffold. (j) Compressive modulus of the composite structures with two different CH dimensions (2- and 4-mm CHs) compared with mouse and rat cortical tissues. Student t test. *P < 0.05; ***P < 0.001. (k) Representative load train traces.
The three-dimensional neural network formation required both mechanically stiff superstructure and a softer ECM gel matrix (Fig. 2B). Although silk material surfaces required a polylysine coating for cortical neuron adhesion, the material stiffness played a major role in promoting axon outgrowth (21). In silk scaffolds alone, the axons were confined to the convoluted 2D surfaces (Fig. 2 B, d and e and Fig. S1, time-evolved morphological changes). Conversely, ECM-like gels (such as collagen, fibrin, and Matrigel) supported neuronal growth but when used alone, did not support neural network formation; for example, in collagen gel-based cultures, neurons were individually dispersed (Fig. 2 B, f and g). In contrast, silk scaffold–ECM gel composite structures promoted axon connections in 3D as interconnected mininetworks through the pores (Fig. 2 B, h and i). Compared with the rapid degradation of fibrin gel and Matrigel in less than 1 wk, collagen gels survived in culture for months (Fig. S2). Therefore, collagen gels were used as the ECM component.
To mimic the mechanical properties of the brain ECM, confined compression tests were used to assess the compressive modulus (25, 26) of bioengineered tissues compared with mouse and rat cortical tissues of similar dimensions (Fig. 2C). The compressive modulus is proximate to the Young modulus in the confined compression test (Materials and Methods). By varying the center hole (CH) size (2 and 4 mm in diameter), the composite structure had different tissue modulus. Tissues with a 2-mm CH had a modulus of 49.1 ± 23.8 kPa (n = 6), similar to that of a mouse brain of 49.1 ± 6.7 kPa (n = 5) and a rat brain of 46.8 ± 8.9 kPa (n = 4). Tissues with a 4-mm CH had a significantly lower modulus of 17.1 ± 7.8 kPa (n = 7), and scaffolds without a CH had a significantly higher modulus of 102.8 ± 14.5 kPa (n = 4) than rodent brains (Fig. 2 C, j). In addition, scaffolds with a 2-mm CH showed similar viscoelastic behavior to rat brain tissue at strains <20% (Fig. 2 C, k). We chose this type of tissue structure for functional studies: silk sponges with ∼500-µm pore sizes, dimensions of 12- (for injury studies) or 5-mm diameters (for 96-well format) (Materials and Methods) × 2-mm height, and a CH of 2 mm in diameter seeded with cortical neurons and infused with a collagen gel.
Three-Dimensional Brain-Like Tissue Growth.
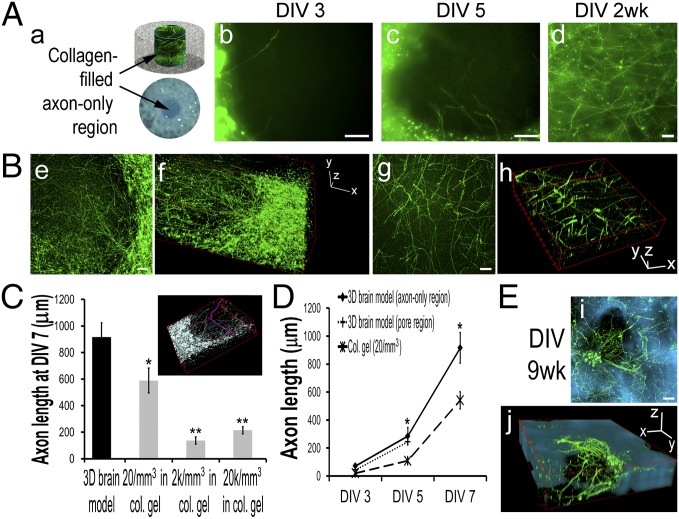
Axon growth was studied in the 3D brain-like tissue (Fig. 3). In the center white matter-like region (Fig. 3 A, a), axons penetrated into the collagen gel compartment by day 3 and traversed the 2-mm diameter gap by 2 wk (Fig. 3 A, b–d). The axon compartments were separated from scaffold-bound neuronal compartments (Fig. 3 B, e and f) and composed a cylindrical mass (2 mm in diameter by 1–2 mm in height) of pristine axon fibers (Fig. 3 B, g and h). Three-dimensional axon tracing (Materials and Methods and Fig. 3C, Inset, pink) showed that the axon length reached 916 ± 109 µm (n = 24/group) at day 7. This length was significantly longer than axons of 589 ± 94, 138 ± 28, and 215 ± 27 µm in the collagen gel-only control cultures with seeding densities of 20, 2,000, and 20,000 cells/mm3, respectively (n =32, 8, and 16, respectively) (Fig. 3C). Because by 2 wk, axon connections were established across the center region, it is unclear what the final length of the axons were and whether axons could grow longer in a larger compartment. Axon growth (Fig. 3D) showed an average growth rate of ∼106 and ∼100 µm/d by day 5 for the axon-only center and the scaffold region (n = 8/group), respectively, indicating homogeneous growth throughout the 3D brain-like tissue. Intact axonal networks were still present at 9 wk in vitro (Fig. 3E).
Fig. 3.
Three-dimensional brain-like tissue growth. (A) Axon growth in the collagen gel-filled center region. DIV, days in vitro. (a) Schematics. (b–d) Fluorescence images of (b) DIV3, (c) DIV5, and (d) 2-wk axons immunostained with β3-tubulin in green. (Scale bar: 100 µm.) (B) Three-dimensional view of confocal z stacks of DIV7 axons in the center region (e is the summed projection of f; g is the summed projection of h). (Scale bar: 100 µm.) (C) Axon length at DIV7 (Inset shows D axon tracing in pink) (Materials and Methods): comparison of 3D brain-like tissue with collage gel-based cultures with different cell densities. Student t test, collagen (col.) vs. 3D brain model. *P < 0.05; **P < 0.01. (D) Axon length growth. (E) Representative fluorescence images of neurons in a scaffold pore at DIV9 wk (β3-tubulin–stained in green superposed on silk structures in cyan; i is the summed projection of j). (Scale bar: 100 µm.)
Three-Dimensional Brain-Like Tissue Viability Depends on Neuron Density.
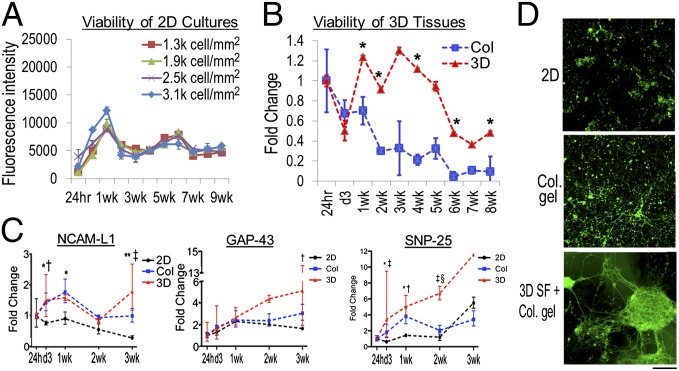
The 3D brain-like tissue growth was found to be affected by neuron densities. The axons showed a tendency to extend long distances over available space and form connections with neighboring neurons, consistent with other reports of hydrogel-based 3D neuronal systems (6). Two-dimensional cultures and collagen gel-based 3D cultures were used as control systems with cell numbers determined by DNA quantitation (Materials and Methods and Fig. S3). In both control systems, high-density cultures yielded more surviving neurons than low densities (Fig. 4A).
Fig. 4.
Three-dimensional brain-like tissue viability and gene expression. (A) Viability of 2D cultures measured with alamarBlue assay. Most cells died after 3 wk. (B) Viability of 3D brain-like tissues (red) and collagen (Col) gel-based cultures (blue) assayed with alamarBlue and expressed relative to 24-h levels. Student t test. *P < 0.05, 3D vs. Col. The two groups had similar cell numbers as determined from Fig. S3. (C) Expression of neural cell adhesion molecule L1 (NCAM-L1), growth-associated protein 43 (GAP-43), and synaptosomal-associated protein 25 (SNP-25) mRNA in 2D (black), collagen gel-based cultures (blue), and 3D brain-like tissues (red) over DIV3 wk relative to the 24-h expression. Asymmetric error bars show maximum and minimum fold change. Two-way ANOVA with Bonferroni posttests: 2D vs. Col: *P < 0.05; **P < 0.001; 2D vs. 3D: †P < 0.05; ‡P < 0.001; Col vs. 3D: §P < 0.05. (D) Representative fluorescence images of DIV21 neurons immunostained with β3-tubulin in green. (Scale bar: 100 µm.)
Because the initial seeding into the scaffold of the 3D brain-like tissue saturated the accessible surface area with neurons before being infused by the gel, the cell numbers in our system depended on scaffold surface area accessible to the cells. Indeed, we found that total cell numbers per scaffold were consistent, regardless of seeding variability (Fig. S3). When total seeding numbers were the same (∼400,000 cells/scaffold of 5-mm diameter × 2-mm height), the long-term viability of the 3D brain-like tissue showed more stable metabolic activity up to 5 wk and a more gradual decline thereafter compared with the consistent decrease in viability of the collagen gel-based system (Fig. 4B, Col).
Because of the high cell density in the 3D systems (∼8,000 cells/mm3) (SI Materials and Methods), it was difficult to discern tissue differences in viability with morphological features. However, gene expression showed that differences were manifested at mRNA levels early on during growth (Fig. 4C). The 3D brain-like tissues trended toward higher expression levels for neuronal adhesion (neural cell adhesion molecule L1), regenerative growth (growth-associated protein 43), and synaptogenesis (synaptosomal-associated protein 25) compared with 2D and collagen gel-based cultures (Fig. S4 shows primer design). At 3 wk, most neurons in 2D cultures had died, and large numbers of dead cells were retained within the collagen gel-based systems (Fig. 4D). In contrast, intact neuronal clusters and networks formed in the 3D brain-like tissues.
Three-Dimensional Brain-Like Tissue Electrophysiological Functions.
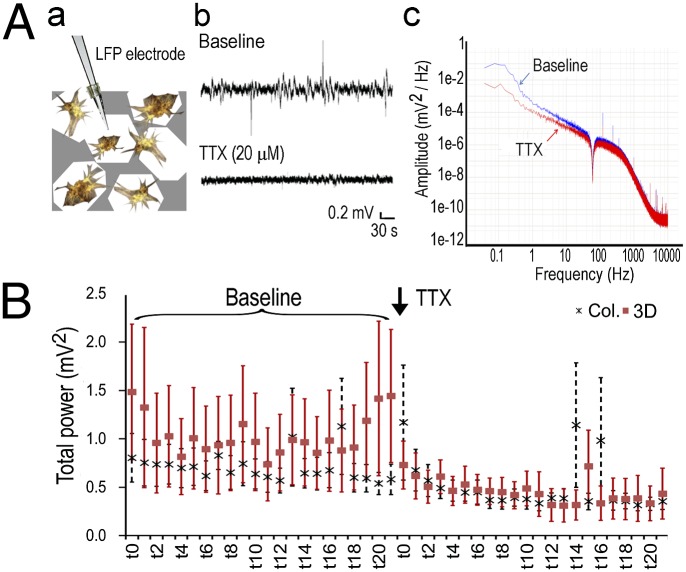
To determine the functional activity of the 3D brain-like tissues, local field potential (LFP) measurements were used based on an established setup for in vivo extracellular recording (27) (Fig. 5). The paired electrodes (50-µm tip and 250-µm separation) measured extracellular potential changes in the vicinity of a randomly selected neuronal cluster (Fig. 5 A, a, schematic). Tetradotoxin (TTX) treatment (20 µM) suppressed baseline activities of the 3D tissues across the power spectra from 0 to 50 Hz (Fig. 5 A, b and c). Power spectra of the signals obtained for 10-min baseline activities and 10-min TTX treatment were plotted in a 20-min trace with a 27-s window size (Fig. 5B). TTX blocked ∼50% (from 1.02 to 0.53 mV2) of the baseline activities of the 3D scaffold-based tissues compared with <30% decrease (from 0.69 to 0.50 mV2) of the collagen gel-based cultures (Fig. 5B, Col, n = 4; 3D, n = 10; 3D TTX vs. baseline: paired Student t test of measured power, P < 0.001; paired t test of normalized power against baseline average, P = 0.059). The time-evolved changes showed a more robust pharmacophysiological response of the 3D brain-like tissues than collagen gel-based cultures, although both systems had similar average cell densities.
Fig. 5.
Three-dimensional brain-like tissue electrophysiological activities. (A) LFP measurement. (a) Schematics. (b) Representative signal traces of baseline and after TTX (20 µM) treatment. (c) A representative power spectrum after fast Fourier transformation of raw signal traces. (B) Time-evolved changes of total power (millivolts2; 0–50 Hz) over a 20-min duration (10 min of baseline and 10 min of TTX treatment). Each segment (t0–t20) represents a 27-s window. Col, collagen.
Three-Dimensional Brain-Like Tissue Responses to TBI.
The similarity of the brain-like tissue mechanical properties with those of rodent brains suggested use as a model for mechanical injuries, such as TBI. In particular, the 3D brain-like tissue could provide real-time concurrent functional assessments that cannot be readily performed on patients or animals [for example, injury-induced surge of excitatory neurotransmitter Glu (28–31) and transient seizure activities post-TBI (32, 33)]. The 3D brain-like tissue allowed us to examine multiple end points postinjury, including cellular damage, electrophysiological activity, and neurochemical changes in real time (Fig. 6).
Fig. 6.
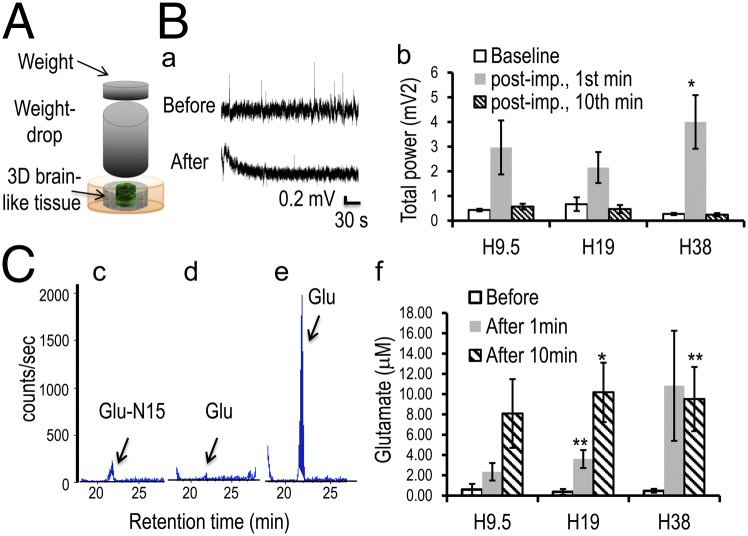
Three-dimensional brain-like tissue responses to TBI. (A) Schematics of the weight-drop impact injury model. (B) Injury-induced electrophysiological activity changes. (a) Representative signal traces of baseline (before) and after impact (after). The electrode was removed during injury and repositioned after an ∼2-min delay to avoid artifacts from the impact. (b) Total power of a 10-min baseline recording, the first 1-min postimpact, and minute 10 postimpact. Paired Student t test. *P < 0.05 vs. baseline. (C) Injury-triggered Glu release. Glu peaks (arrow) at a retention time of ∼21 min. (c–e) Representative LC/MS detection traces of (c) the internal control Glu-N15 and the Glu level (d) at the baseline and (e) after impact. (f) Quantification of Glu levels before and at 1 and 10 min after injury. Student t test. *P < 0.05; **P < 0.01 vs. before.
To induce TBI, we adopted the weight-drop model (34), in which force was delivered to the brain by dropping a blunt weight through a tube of controlled height to impact the skull. Using the 3D brain-like tissue to simulate the cortical tissue, 10.9-g weight drops from heights (Hs) of 9.5, 19, and 38 cm (H9.5, H19, and H38, respectively) were studied (Fig. 6A, schematics). Calculations based on the compressive modulus of 3D brain-like tissues estimated that these conditions would generate compression distances of 0.10, 0.15, and 0.22 mm, respectively, comparable with the ∼0.3-mm compression of the brain in vivo (34) (Materials and Methods).
Confocal images of the tissues immunostained for β3-tubulin showed graded neuronal damage corresponding to different heights of weight drop, confirming force-dependent injury responses (Fig. S5A). Impact-induced hyperactivity was observed using LFP measurements on the 3D brain-like tissues before and after injury (Fig. 6 B, a). At all three injury levels, there was a surge of activity that lasted for <2 min and then tapered to baseline levels of 691%, 322%, and 1,483%, respectively, for the first 1 min and returned to 132%, 69%, and 88%, respectively, of baseline at 10 min postinjury for H9.5, H19, and H38, respectively (n = 3, 5, and 3, respectively) (Fig. 6 B, b and Fig. S5B, time-evolved changes). Although currently, there are no reports on LFP measurements in in vivo TBI studies, the transient feature of the hyperactivity was similar to acute-phase EEG findings in TBI animal models (review in ref. 35).
To determine whether impact-induced hyperactivity was caused by excitatory neurotransmitter Glu release, which is thought to be a primary injury mechanism (28, 29), media samples before and after (1 and 10 min) impact on the 3D brain-like tissues were analyzed using tandem liquid chromatography (LC)/MS (Fig. 6C). Glu levels increased dramatically immediately after injury [Fig. 6 C, c shows the internal control compound Glu-N15, Fig. 6 C, d shows before impact, Fig. 6 C, e shows after impact (retention time ∼ 21 min), and Fig. S6A shows control assays]. Within 1 min, Glu levels reached to 2.3 ± 0.9, 3.6 ± 0.9, and 10.8 ± 5.4 μM for H9.5, H19, and H38, respectively (n = 6, 9, and 6 per group, respectively) from a negligible level of ∼0.5 ± 0.2 μM before injury (Fig. 6 C, f and Fig. S6 B–D, representative traces). In all three groups, Glu levels rose to ∼8–10 μM at 10 min after injury. The 1-min levels were similar to postinjury Glu levels in the rat brain immediately after TBI (29), and the severalfold increase was consistent with previous reports from animal studies (28, 30, 31). The time course, however, differed from in vivo studies showing Glu levels subsiding to preinjury levels by ∼5 min postinjury (29). This discrepancy may be ascribed to the lack of astrocyte-mediated Glu uptake (36) in this tissue model.
Discussion
The extraordinary connectivity of the brain’s neural network is evident at multiple levels and scales, including microcircuitry dominated by neuronal clusters, and compartmentalized grey and white matter regions. The complex network organization cannot be recapitulated in conventional 2D plate cultures or homogeneous ECM gel-based systems. Using a composite structure of silk protein-based porous scaffolds and ECM gels, a modular 3D brain-like tissue with brain-mimetic mechanical properties was developed to support compartmentalized neuronal network formation, long-term tissue growth, and electrophysiological functions and responses.
The 3D neuronal network formation was achieved by combining two types of biomaterials with different mechanical properties: a stiff scaffold to provide neuronal anchoring and a softer ECM gel matrix to permit axon penetration and connectivity in 3D. Selective material preferences provide the underlying principle for compartmentalizing the neurons from the axon connections (for example, at the microscale as mininetworks within pores of the scaffold and at a larger scale as grey and white matter-like architectures). We showed that the scaffolds provided an appropriate microenvironment for neural network formation, such as neuron anchoring, cellular compartments, and brain-like mechanical properties. In contrast, hydrogels permit cell reorganization without physical constraints, but neurons failed to form larger-sized networks in the collagen gel-only cultures. Additional structural features of the scaffold can further enhance neuronal organization, such as surface topology-directed alignment. As the mechanically dominant component of the composite structure, the control of the silk biomaterial stiffness allowed accommodation of other types of gels, while maintaining brain-like tissue elasticity. The use of silk in terms of tunable mechanics, slow degradation, hosting infused gels, and dimensional stability provided a suitable cortical niche to support the functions shown here; it is unknown if other biomaterials would generate similar outcomes.
This tissue model shows that neural network connectivity improves neuronal tissue viability, gene expression, and electrophysiological activities. In particular, the long-term viability up to at least 9 wk of the 3D scaffold-based cultures showed better performance than in collagen alone-based hydrogel cultures. The increased degree of neuronal clustering in the composite structure as opposed to disperse neurons in gels could provide enrichment of neurotrophic factors (37) and reciprocal synaptic transmission (38), therefore supporting sustained growth and synchronized activities. Additionally, the presence of 3D scaffolds can improve transport of oxygen and nutrients over time in contrast to gel-only matrices, where transport is reduced because of the collapse of the hydrogel structure during degradation. These features supported the viability of millimeter-sized tissues that was well beyond the diffusion limitations for gels (23). It is presently unclear whether the eventual decrease of the tissue viability may be caused by a lack of sustained neuronal activity (39). The long-term viability of this brain tissue system could be further extended by incorporating external biochemical and/or electrical stimulation to simulate the signaling environment in the brain and perfusion systems to improve diffusion.
The modular design provides a blueprint for more complex features of the brain. For example, the donut-shaped modules were assembled into concentric ring structures to simulate the laminar layers of the neocortex. The jigsaw puzzle-like cutting process could be used to fabricate other types of interlocking shapes to mimic specific tissue structures. Furthermore, the adhesive-free assembly process for constructing a self-supporting superstructure could be used to probe cell–cell interactions from different compartments. The proof of concept was shown by the microscopic contiguity of neuronal organization at the interface of two assembled silk donut structures. Although this model is rather basic, containing only primary neurons, other ECM components and soluble factors may be incorporated to support other types of cells, such as astrocytes and oligodendrocytes. Better-characterized neurons, such as cortical layer-specific neuron and/or glial cells, are needed to show functional interactions. Recent advances with pluripotent stem cell-based approaches have generated human cerebral tissue models (9, 40). Combining these region-specific brain cells with the bioengineering approach described here could provide a versatile and highly controllable platform to further extend the options of deriving brain-like tissue organization.
The capability of this 3D brain-like tissue for multimodal assessments, including structural, biochemical, genetic, and electrophysiological outcomes, makes it an attractive tissue with which to study a range of disease related to CNS disorders. Using TBI as an example, we showed robust tissue responses to physiologically relevant impact forces. The brain-like tissue model exhibited injury-induced Glu release and transient hyperactivity that are consistent with in vivo studies (28–33). Whereas real-time monitoring of electrophysiological activities of the brain is prohibitive with human or animal subjects, this model provides a unique opportunity for mapping out the temporal evolution of neurophysiological events and functional studies in vitro.
Materials and Methods
Silk Scaffold and ECM Gel Preparation.
Silk solution and porous scaffolds were prepared from Bombyx mori cocoons as described previously (19, 22). A biopsy punch with concentrically arranged rings was used to punch out layered donut shapes. These structures were separated to prepare for cell seeding and reassembled back into the original structure. The scaffolds were autoclaved and coated with poly-l-lysine (10 µg/mL). The scaffolds were immersed in concentrated neuron suspension (20–30 million cells/mL) for 24 h, washed extensively to remove unattached cells, and prepared for hydrogel infusion. Collagen gel was prepared from rat tail type I collagen I (3–4 mg/mL; BD Biosciences), 10× 199 media (Gibco), and 1 M NaOH by mixing at a ratio of 88:10:2. Fibrin gel was prepared by mixing fibrinogen (20 mg/mL; EMD Millipore) and thrombin (10 U/mL; Sigma) at a volume ratio of 2:5. Marigel was purchased from BD Biosciences. To make a scaffold–gel composite structure, the scaffolds were washed with ECM gels in liquid form to replace the media within the scaffold followed by incubation at 37 °C for 30 min before media immersion.
Primary Cortical Neuronal Culture.
The brain tissue isolation protocol was approved by the Tufts University Institutional Animal Care and Use Committee and complies with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (Institutional Animal Care and Use Committee B2011-45). Primary rat cortical neurons were isolated from embryonic day 18 Sprague–Dawley rats (Charles River). Cell densities are in SI Materials and Methods.
Confined Compression Test.
A mechanical tester (Instron 3366) was used for confined compression testing according to the manual (details in SI Materials and Methods). The aggregate modulus was based on the formula , where is the aggregate modulus, is the Young modulus (or elastic modulus), and is the Poisson ratio given as the ratio of the lateral deformation vs. the axial deformation. Given the small lateral deformation and negligible , the aggregate modulus was used to approximate the Young modulus.
Immunocytochemistry.
Antibodies included anti–β3-tubulin (rabbit, 1:500; Sigma), anti-GFAP (mouse, 1:1,000; rabbit, 1:500; Sigma), antimicrotubule-associated protein-2 (rabbit, 1;500; Sigma), and goat anti-mouse or rabbit Alexa 488 and 568 (1:250; Invitrogen) secondary antibodies. Standard immunocytochemistry methods were used (SI Materials and Methods).
3D Confocal Imaging and Image Analysis.
A Leica TCS SP2 (Leica Microsystems) confocal microscope and Fiji ImageJ with the Simple Neurite Tracer plugin were used for 3D image acquisition and measurements, respectively.
LFP Measurements.
LFPs were recorded with a custom-built recording probe consisting of two parallel tungsten electrodes (50-µm-diameter tip and 250-µm separation), and measurements were adapted from previous procedures (27) (SI Materials and Methods). Baseline signals were recorded for 10 min. A 10-µL droplet of 1 mM TTX (∼20 µM final concentration) was applied near the electrodes. After the signal stabilized in ∼1 min, recording was resumed for another 10 min. For the impact experiments, the electrode was removed during and repositioned after the weight drop to prevent damage from the impact. The postinjury recording contained an ∼2-min delay to exclude artifacts associated with reinsertion. The Clampfit (Molecular Devices) program was used for analysis. Power spectra were obtained by fast Fourier transformation average of a rectangular window over a time period of either 10 min (for cumulative effects) or 27 s (for time-dependent evolution analysis).
Impact Injury of 3D Cultures.
A custom-made weight-drop setup (a 10.9-g weight and a 15-mm i.d. tube for the height) was used for impact injury (modified from ref. 34). The test sample was immersed in artificial cerebrospinal fluid solution (1 mL) during the experiment. The sample was immediately removed after weight drop. Solution samples were collected (100 µL) before and after injury for liquid analysis. Calculation of compressive distance (x) was based on Newton’s gravitational formula ,where m is the weight (0.0109 kg), g is gravitational constant (9.91 m/s2), and h is the height (0.095, 0.19, and 0.38 m); k is the spring constant, which is given as the Young modulus (E) multiplied by the original area (A0) divided by the original length (L0) of the test sample.
Tandem LC/MS Analysis.
MS analysis of neurotransmitters was performed on a triple quadruple mass spectrometer (3200Q TRAP LC/MS/MS system; MDS Inc.) using an aminopropyl column (Luna NH2; Phenomenex) (41). The gradient method used solvent A, a 95:5 water/acetonitrile solution containing 20 mM ammonium acetate and 20 mM ammonium hydroxide (pH 9.5), and solvent B (acetonitrile). Solution samples were mixed with 1,000 ng 15N-labeled Glu (Sigma) and run with a flow rate of 150 µL/min with a gradient of t = 0 min, 85% (vol/vol) B; t = 15 min, 0% B; t = 28 min, 0% B; t = 30 min, 85% (vol/vol) B; and t = 40 min, 85% vol/vol B. Positive ionization mode was used to detect Glu (C5H10NO4+; Q1 mass = 148.14; Q3 mass = 84). Glu standards were used to obtain their characteristic retention time. For quantification, signals were integrated from peaks centered at the retention time. The ratio of the peak of the signal of interest over that of the internal control was calculated and converted to concentration.
Gene Expression Analysis.
RNA was extracted from 2D cultures (1 million cells/well of a six-well plate at ∼1,900 cell/mm2), collagen gel-based cultures (8,000 cells/mm3; 2 million cells/250 µL gel), and 3D scaffold gel composite cultures (5-mm diameter × 2-mm height) using an RNeasy Mini Kit (Qiagen Inc.). Standard protocols were used for RT-PCR analysis (SI Materials and Methods).
Cell Viability Assay.
AlamarBlue assay was used to assess cell viability (Invitrogen).
Statistical Analysis.
Data are means ± SEMs, except where otherwise noted. Analysis used Student t test, except where otherwise noted. For all tests, P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank the laboratory of Dr. Stephen Moss for providing embryonic rat brain tissues, Dr. Michael Whalen for discussing brain injury experiments, Antonio Varone and Dr. Kyongbum Lee for providing liquid chromatography/MS assistance, and Dr. Michaela Reagan and Dr. Biman Mandal for assisting with silk material processing. Special thanks to Donna B. Kaplan for inspiring this work. This work was funded by National Institutes of Health P41 Tissue Engineering Resource Center Grant EB002520.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 13682.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1324214111/-/DCSupplemental.
References
- 1.Hill SL, Wang Y, Riachi I, Schürmann F, Markram H. Statistical connectivity provides a sufficient foundation for specific functional connectivity in neocortical neural microcircuits. Proc Natl Acad Sci USA. 2012;109(42):E2885–E2894. doi: 10.1073/pnas.1202128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswal BB, et al. Toward discovery science of human brain function. Proc Natl Acad Sci USA. 2010;107(10):4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sperry RW. Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc Natl Acad Sci USA. 1963;50:703–710. doi: 10.1073/pnas.50.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imai T, et al. Pre-target axon sorting establishes the neural map topography. Science. 2009;325(5940):585–590. doi: 10.1126/science.1173596. [DOI] [PubMed] [Google Scholar]
- 5.Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7(3):211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 6.Gurkan UA, et al. Simple precision creation of digitally specified, spatially heterogeneous, engineered tissue architectures. Adv Mater. 2013;25(8):1192–1198. doi: 10.1002/adma.201203261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Odawara A, Gotoh M, Suzuki I. A three-dimensional neuronal culture technique that controls the direction of neurite elongation and the position of soma to mimic the layered structure of the brain. RSC Adv. 2013;3:23620–23630. [Google Scholar]
- 8.Mironov V, Boland T, Trusk T, Forgacs G, Markwald RR. Organ printing: Computer-aided jet-based 3D tissue engineering. Trends Biotechnol. 2003;21(4):157–161. doi: 10.1016/S0167-7799(03)00033-7. [DOI] [PubMed] [Google Scholar]
- 9.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen TH, et al. Tissue-engineered lungs for in vivo implantation. Science. 2010;329(5991):538–541. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song JJ, et al. Regeneration and experimental orthotopic transplantation of a bioengineered kidney. Nat Med. 2013;19(5):646–651. doi: 10.1038/nm.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hussein KH, et al. Fabrication of a biodegradable xenoantigen-free rat liver scaffold for potential drug screening applications. Transplant Proc. 2013;45(8):3092–3096. doi: 10.1016/j.transproceed.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Bonandrini B, et al. Recellularization of well-preserved acellular kidney scaffold using embryonic stem cells. Tissue Eng Part A. 2014;20(9-10):1486–1498. doi: 10.1089/ten.tea.2013.0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maddah M, Grimson WE, Warfield SK, Wells WM. A unified framework for clustering and quantitative analysis of white matter fiber tracts. Med Image Anal. 2008;12(2):191–202. doi: 10.1016/j.media.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingalhalikar M, et al. Sex differences in the structural connectome of the human brain. Proc Natl Acad Sci USA. 2014;111(2):823–828. doi: 10.1073/pnas.1316909110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith DH, Meaney DF, Shull WH. Diffuse axonal injury in head trauma. J Head Trauma Rehabil. 2003;18(4):307–316. doi: 10.1097/00001199-200307000-00003. [DOI] [PubMed] [Google Scholar]
- 17.Kraus MF, et al. White matter integrity and cognition in chronic traumatic brain injury: A diffusion tensor imaging study. Brain. 2007;130(Pt 10):2508–2519. doi: 10.1093/brain/awm216. [DOI] [PubMed] [Google Scholar]
- 18.Omenetto F, Kaplan D. From silk cocoon medical miracle. Scientists are crafting arteries, ligaments, circuitry and holograms from worm yarn. Sci Am. 2010;303(5):76–77. [PubMed] [Google Scholar]
- 19.Rockwood DN, et al. Materials fabrication from Bombyx mori silk fibroin. Nat Protoc. 2011;6(10):1612–1631. doi: 10.1038/nprot.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopkins AM, et al. Silk hydrogels as soft substrates for neural tissue engineering. Adv Funct Mater. 2013;23(41):5140–5149. [Google Scholar]
- 21.Tang-Schomer MD, et al. Film-based implants for supporting neuron-electrode integrated interfaces for the brain. Adv Funct Mater. 2014;24(13):1938–1948. doi: 10.1002/adfm.201303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nazarov R, Jin HJ, Kaplan DL. Porous 3-D scaffolds from regenerated silk fibroin. Biomacromolecules. 2004;5(3):718–726. doi: 10.1021/bm034327e. [DOI] [PubMed] [Google Scholar]
- 23.Rouwkema J, Rivron NC, van Blitterswijk CA. Vascularization in tissue engineering. Trends Biotechnol. 2008;26(8):434–441. doi: 10.1016/j.tibtech.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Wray LS, et al. A silk-based scaffold platform with tunable architecture for engineering critically-sized tissue constructs. Biomaterials. 2012;33(36):9214–9224. doi: 10.1016/j.biomaterials.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller K, Chinzei K. Constitutive modelling of brain tissue: Experiment and theory. J Biomech. 1997;30(11-12):1115–1121. doi: 10.1016/s0021-9290(97)00092-4. [DOI] [PubMed] [Google Scholar]
- 26.Mow VC, Huiskes R. Basic Orthopaedic Biomechanics & Mechano-Biology. Philadelphia: Lippincott Williams & Wilkins; 2005. p. xvi. [Google Scholar]
- 27.Schmitt LI, Sims RE, Dale N, Haydon PG. Wakefulness affects synaptic and network activity by increasing extracellular astrocyte-derived adenosine. J Neurosci. 2012;32(13):4417–4425. doi: 10.1523/JNEUROSCI.5689-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faden AI, Demediuk P, Panter SS, Vink R. The role of excitatory amino acids and NMDA receptors in traumatic brain injury. Science. 1989;244(4906):798–800. doi: 10.1126/science.2567056. [DOI] [PubMed] [Google Scholar]
- 29.Katayama Y, Becker DP, Tamura T, Hovda DA. Massive increases in extracellular potassium and the indiscriminate release of glutamate following concussive brain injury. J Neurosurg. 1990;73(6):889–900. doi: 10.3171/jns.1990.73.6.0889. [DOI] [PubMed] [Google Scholar]
- 30.Nilsson P, Hillered L, Pontén U, Ungerstedt U. Changes in cortical extracellular levels of energy-related metabolites and amino acids following concussive brain injury in rats. J Cereb Blood Flow Metab. 1990;10(5):631–637. doi: 10.1038/jcbfm.1990.115. [DOI] [PubMed] [Google Scholar]
- 31.Hinzman JM, et al. Diffuse brain injury elevates tonic glutamate levels and potassium-evoked glutamate release in discrete brain regions at two days post-injury: An enzyme-based microelectrode array study. J Neurotrauma. 2010;27(5):889–899. doi: 10.1089/neu.2009.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nilsson P, et al. Epileptic seizure activity in the acute phase following cortical impact trauma in rat. Brain Res. 1994;637(1-2):227–232. doi: 10.1016/0006-8993(94)91237-8. [DOI] [PubMed] [Google Scholar]
- 33.Bolkvadze T, Pitkänen A. Development of post-traumatic epilepsy after controlled cortical impact and lateral fluid-percussion-induced brain injury in the mouse. J Neurotrauma. 2012;29(5):789–812. doi: 10.1089/neu.2011.1954. [DOI] [PubMed] [Google Scholar]
- 34.Marmarou A, et al. A new model of diffuse brain injury in rats. Part I: Pathophysiology and biomechanics. J Neurosurg. 1994;80(2):291–300. doi: 10.3171/jns.1994.80.2.0291. [DOI] [PubMed] [Google Scholar]
- 35.Hunt RF, Boychuk JA, Smith BN. Neural circuit mechanisms of post-traumatic epilepsy. Front Cell Neurosci. 2013;7:89. doi: 10.3389/fncel.2013.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliet SH, Piet R, Poulain DA. Control of glutamate clearance and synaptic efficacy by glial coverage of neurons. Science. 2001;292(5518):923–926. doi: 10.1126/science.1059162. [DOI] [PubMed] [Google Scholar]
- 37.Korsching S. The neurotrophic factor concept: A reexamination. J Neurosci. 1993;13(7):2739–2748. doi: 10.1523/JNEUROSCI.13-07-02739.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Didier A, et al. A dendrodendritic reciprocal synapse provides a recurrent excitatory connection in the olfactory bulb. Proc Natl Acad Sci USA. 2001;98(11):6441–6446. doi: 10.1073/pnas.101126398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moody WJ, Bosma MM. Ion channel development, spontaneous activity, and activity-dependent development in nerve and muscle cells. Physiol Rev. 2005;85(3):883–941. doi: 10.1152/physrev.00017.2004. [DOI] [PubMed] [Google Scholar]
- 40.Mariani J, et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109(31):12770–12775. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bajad SU, et al. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A. 2006;1125(1):76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.