Significance
Polysaccharide monooxygenases (PMOs) are recently discovered extracellular fungal and bacterial enzymes that are able to cleave the recalcitrant polysaccharides cellulose and chitin. We describe the discovery of a new family of fungal PMOs that act on starch based on bioinformatic, biochemical, and spectroscopic studies on NCU8746, a representative starch-active PMO from Neurospora crassa. The data support a proposed enzymatic mechanism and show that NCU08746 shares evolutionarily conserved features with previously reported PMOs. This discovery extends the currently known PMO family, suggesting the existence of a PMO superfamily with a much broader range of substrates. Starch-active PMOs provide an expanded perspective on studies of starch metabolism and may have potential in the food and starch-based biofuel industries.
Keywords: copper enzymes, oxygen activation, CBM20
Abstract
The recently discovered fungal and bacterial polysaccharide monooxygenases (PMOs) are capable of oxidatively cleaving chitin, cellulose, and hemicelluloses that contain β(1→4) linkages between glucose or substituted glucose units. They are also known collectively as lytic PMOs, or LPMOs, and individually as AA9 (formerly GH61), AA10 (formerly CBM33), and AA11 enzymes. PMOs share several conserved features, including a monocopper center coordinated by a bidentate N-terminal histidine residue and another histidine ligand. A bioinformatic analysis using these conserved features suggested several potential new PMO families in the fungus Neurospora crassa that are likely to be active on novel substrates. Herein, we report on NCU08746 that contains a C-terminal starch-binding domain and an N-terminal domain of previously unknown function. Biochemical studies showed that NCU08746 requires copper, oxygen, and a source of electrons to oxidize the C1 position of glycosidic bonds in starch substrates, but not in cellulose or chitin. Starch contains α(1→4) and α(1→6) linkages and exhibits higher order structures compared with chitin and cellulose. Cellobiose dehydrogenase, the biological redox partner of cellulose-active PMOs, can serve as the electron donor for NCU08746. NCU08746 contains one copper atom per protein molecule, which is likely coordinated by two histidine ligands as shown by X-ray absorption spectroscopy and sequence analysis. Results indicate that NCU08746 and homologs are starch-active PMOs, supporting the existence of a PMO superfamily with a much broader range of substrates. Starch-active PMOs provide an expanded perspective on studies of starch metabolism and may have potential in the food and starch-based biofuel industries.
Polysaccharide monooxygenases (PMOs) are enzymes secreted by a variety of fungal and bacterial species (1–5). They have recently been found to oxidatively degrade chitin (6–8) and cellulose (8–14). PMOs have been shown to oxidize either the C1 or C4 atom of the β(1→4) glycosidic bond on the surface of chitin (6, 7) or cellulose (10–12, 14), resulting in the cleavage of this bond and the creation of new chain ends that can be subsequently processed by hydrolytic chitinases and cellulases. Several fungal PMOs were shown to significantly enhance the degradation of cellulose by hydrolytic cellulases (9), indicating that these enzymes can be used in the conversion of plant biomass into biofuels and other renewable chemicals.
There are three families of PMOs characterized thus far: fungal PMOs that oxidize cellulose (9–12) (also known as GH61 and AA9); bacterial PMOs that are active either on chitin (6, 8) or cellulose (8, 13) (also known as CBM33 and AA10); and fungal PMOs that oxidize chitin (AA11) (7). Sequence homology between these three families is very low. Nevertheless, the available structures of PMOs from all three families reveal a conserved fold, including an antiparallel β-sandwich core and a highly conserved monocopper active site on a flat protein surface (Fig. 1A) (2, 6, 7, 9, 10, 15–17). Two histidine residues in a motif termed the histidine brace coordinate the copper center. The N-terminal histidine ligand binds in a bidentate mode, and its imidazole ring is methylated at the Nε position in fungal PMOs (Fig. 1A).
Fig. 1.
(A) Representative overall and active site structures of fungal PMOs (PDB ID code 2YET) (10). (B) Structure of cellulose (18, 19). Chitin also contains β(1→4) linkages and has similar crystalline higher order structure to cellulose. (C) Model structure of amylopectin (23–25). Hydrogen bonds are shown with green dashed lines.
Considering the conserved structural features, it is not surprising that the currently known PMOs act on substrates with similar structures. Cellulose and chitin contain long linear chains of β(1→4) linked glucose units and N-acetylglucosamine units, respectively (Fig. 1B). The polymer chains form extensive hydrogen bonding networks, which result in insoluble and very stable crystalline structures (18–21). PMOs are thought to bind to the substrate with their flat active site surface, which orients the copper center for selective oxidation at the C1 or C4 position (6, 16, 22). Some bacterial chitin-binding proteins are cellulose-active PMOs (8, 13, 14), further suggesting that the set of PMO substrates is restricted to β(1→4) linked polymers of glucose and glucose derivatives.
Here, we report on the identification of new families of PMOs that contain several key features of previously characterized PMOs, but act on substrates different from cellulose or chitin. A member of one of these novel families of PMOs, NCU08746, was shown to oxidatively cleave amylose, amylopectin, and starch. We designate the NCU08746 family as starch-active PMOs. Both amylose and amylopectin contain linear chains of α(1→4) linked glucose, whereas the latter also contains α(1→6) glycosidic linkages at branch points in the otherwise α(1→4) linked polymer. Unlike cellulose and chitin, amylose and amylopectin do not form microcrystals; instead, they exist in disordered, single helical, and double helical forms (23–27) (see Fig. 1C for example). Starch exists partially in nanocrystalline form, but lacks the flat molecular surfaces as those found in chitin and cellulose. The discovery of starch-active PMOs shows that this oxidative mechanism of glycosidic bond cleavage is more widespread than initially expected.
Results
Identification of New Fungal PMO Families.
All currently known fungal PMOs are predicted to be secreted via the canonical signal peptide-triggered pathway and contain several conserved features, including an absolutely conserved N-terminal histidine residue, a second conserved histidine residue, and the N/Q/E-X-F/Y motif. Using SignalP 4.0 (28), we found 777 N. crassa sequences with predicted signal peptides. Among these predicted secreted proteins, 49 were predicted to contain an N-terminal histidine residue. For 21 of these 49 proteins, the N-terminal histidine was absolutely conserved in all identified fungal homologs. The second histidine residue and the N/Q/E-X-F/Y motif in each of these 21 sequences were identified by using multiple sequence alignments with homologous sequences found in public databases. Table 1 shows N. crassa proteins with these PMO-conserved features. The majority of these proteins are cellulose active PMOs (in light blue; formerly known as GH61) (3, 11, 12). Three sequences belonging to a family annotated as chitinases are also shown (in aqua); a member of this family was recently characterized as a chitin-active PMO (7). The N terminus of NCU08746 (in yellow) shares weak homology with chitin binding domains and a C-terminal carbohydrate binding module 20 (CBM20) (29). The other sequences in Table 1 with unknown substrates are not annotated.
Table 1.
Predicted N. crassa PMOs, based on secreted proteins with a N-terminal histidine residue
 |
Cel, cellulose; Chi, chitin; MS, the first two residues of mature sequence; SP, the last three residues of signal peptide; St, starch; Sub, substrate; U, unknown. Colors represent different PMO families as determined by sequence homology.
Fig. 2 shows the domain architecture and the consensus sequence of NCU08746 and homologs. Thirty sequences of this family were found with an iterative protein database search by using a Hidden Markov Model algorithm (30). They are all found in the genomes of fungi in the Ascomycota phylum. Unlike chitin- and cellulose-active PMOs, the NCU08746 family has very high sequence identity. The N-terminal domain suggested by this sequence alignment contains 204–233 amino acids; this finding is inconsistent with its original annotation. Together with the biochemical characterization reported herein, we designate this catalytic domain as a starch-active PMO domain.
Fig. 2.
(A) Common domain architecture of 30 predicted starch-active PMOs from different fungal species. Eighteen have the CBM20 domain. (B) Consensus sequence logo representing the putative catalytic domain. Asterisks indicate the absolutely conserved residues also found in cellulose-active PMOs and chitin-active PMOs.
Expression, Purification, and Verification of NCU08746.
NCU08746 was obtained by using a previously reported recombinant expression method in N. crassa (22) using the primers listed in SI Appendix, Table S1. This expression method yields recombinant PMOs indistinguishable from the corresponding natively purified enzymes. NCU08746 was purified by using an amylose resin column, followed by a size exclusion column. SDS/PAGE analysis of the fractions from the amylose resin column shows that NCU08746 has relatively strong affinity for amylose (SI Appendix, Fig. S1), supporting a functional role for the predicted C-terminal CBM20 domain. Furthermore, MS/MS analysis of the trypsin-digested purified NCU08746 yielded nearly 50% sequence coverage, including both the N termini and C termini, confirming the predicted sequence of this protein. The mass spectrometry data are consistent with a methylated N-terminal histidine (SI Appendix, Fig. S2 and Table S2), a feature previously observed for PMOs natively purified from or recombinantly expressed in fungi (10, 11, 22).
NCU08746 Activity Assays and Product Identification.
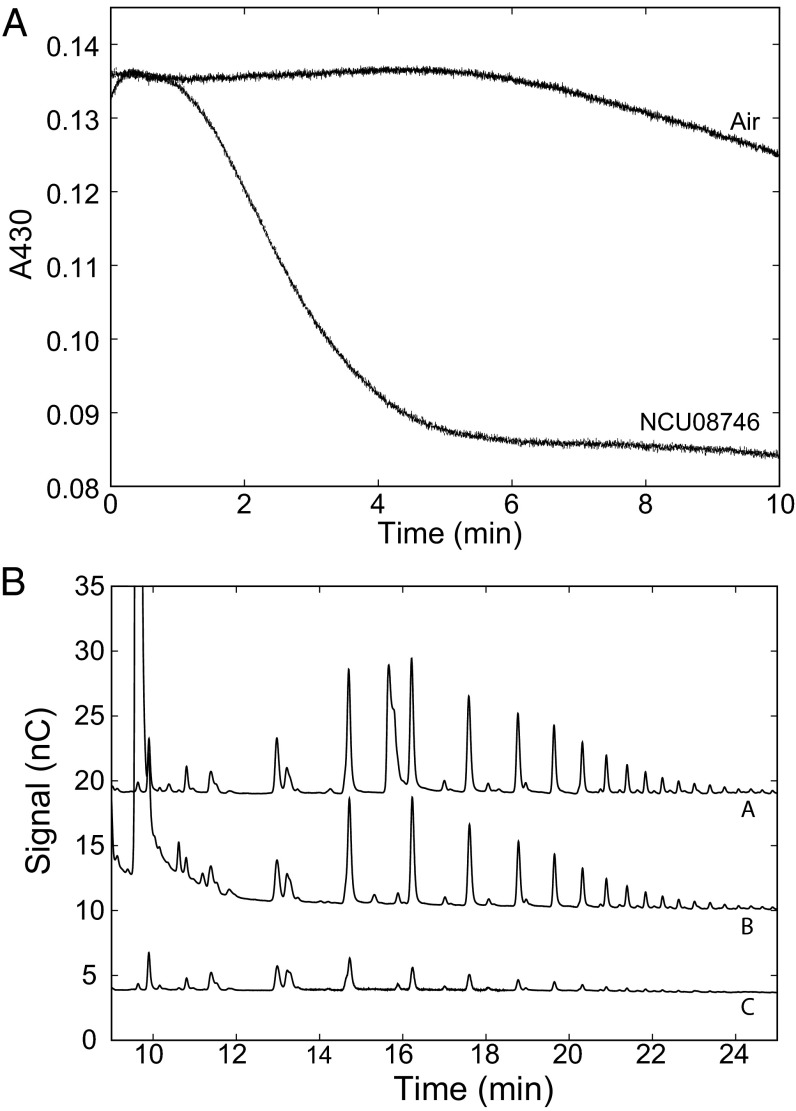
Fig. 3 shows the high performance anion exchange chromatographic (HPAEC) traces of NCU08746 assays under various conditions. The reaction with amylopectin depends on reductant (ascorbic acid) and oxygen. When both ascorbic acid and oxygen were present, the chromatogram exhibits a set of new peaks (trace C) that are not observed in the absence of either reductant or oxygen (SI Appendix, Fig. S3). The same set of peaks is also found in the assays with corn starch or amylose (SI Appendix, Figs. S4–S6), but not in the assays with phosphoric acid swollen cellulose (PASC) or chitin (Fig. 3, traces D and E, respectively). These results show clearly that NCU08746 specifically cleaves starch substrates via an oxygen-dependent mechanism.
Fig. 3.
Activity assays of NCU08746. Assays contained 5 μM NCU08746 with 2 mM ascorbic acid and atmospheric oxygen. (A and B) Maltodextrins (1–7 units) and soluble portion of amylose (average molecular mass ∼2.8 kDa), respectively, oxidized with Lugol's solution. (C–E) Assays with 50 mg/mL amylopectin, 5 mg/mL PASC, and 50 mg/mL chitin, respectively. The assays were carried out in 50 mM sodium acetate buffer at pH 5.0 and 42 °C.
The products of NCU08746 assays were identified by comparison with synthetic standards via HPAEC and mass spectrometry analysis. The product peaks of NCU08746 overlay with the peaks of malto-aldonic acids (Fig. 3, traces A and B), but not with those of maltodextrins (SI Appendix, Fig. S6). The mass spectrum of the assay with amylose exhibits a series of peaks 16 amu higher than the corresponding maltodextrin peaks (SI Appendix, Table S3), which is consistent with the presence of an additional oxygen atom. Together, the HPAEC and mass spectrometry data indicate that NCU08746 oxidizes the C1 position of starch substrates.
Copper Is the Native Metal Cofactor of NCU08746.
NCU08746 reconstituted with excess copper(II) sulfate and purified with size exclusion chromatography contained approximately one copper atom per protein molecule based on inductively coupled plasma optical emission spectrometry (ICP-OES) analysis (SI Appendix, Table S4). The addition of one equivalent of CuSO4 to the assays of holo NCU08746 had no effect on activity (SI Appendix, Figs. S4 and S5). Treatment of holo NCU08746 with 10 mM EDTA resulted in a partially apo form containing ∼0.12 copper atom per protein molecule. This form was assayed with the addition of one equivalent of various second-row metal salts. Among the metal ions tested, only Cu(II) increased the oxidative activity of this partially apo NCU08746 (SI Appendix, Fig. S7). This data supports the notion that copper is the native metal cofactor of NCU08746.
NCU08746 Accepts Electrons from Cellobiose Dehydrogenase.
Because NCU08746 shares several conserved features with cellulose-active PMOs, it was expected that the starch-active PMO would have a biological redox partner equivalent to that of cellulose-active PMOs, cellobiose dehydrogenase (CDH). Because such a redox partner has not been identified, we investigated the possibility of electron transfer from Myceliophthora thermophila CDH-2 (MtCDH-2) to NCU08746. CDH uses flavin adenine dinucleotide and heme cofactors to rapidly oxidize cellodextrins, including cellobiose, cellotriose, and cellotetraose, whereas activity on glucose and maltose is minimal (31). Oxidation of reduced MtCDH-2, obtained by incubation with cellobiose, was measured as the decrease in absorbance at 430 nm of the reduced heme cofactor. This oxidation occurs slowly in the presence of atmospheric oxygen but was significantly enhanced in the presence of NCU08746 (Fig. 4A), indicating that NCU08746 is an efficient electron acceptor for MtCDH-2. Subsequently, NCU08746 activity assays on amylopectin were carried out with MtCDH-2 as the electron donor. In the presence of MtCDH-2 only, NCU08746 exhibited very weak activity on amylopectin (Fig. 4B, trace C), which is consistent with the low activity of CDH on glucose and maltodextrins (31). In the presence of MtCDH-2 and cellobiose, NCU08746 activity (Fig. 4B, trace B) was comparable to that in the presence of excess ascorbic acid (Fig. 4B, trace A). Together, these results indicate that CDH can serve as an electron donor for NCU08746 catalysis.
Fig. 4.
(A) Effect of NCU08746 (5 μM) on the rate of oxidation of MtCDH-2 (1 μM) incubated with 6 μM cellobiose at room temperature. (B) NCU08746 (5 μM) activity assays on amylopectin (50 mg/mL) with 2 mM ascorbic acid (a), with 0.5 μM MtCDH-2 and 5 mM cellobiose (b), and with 0.5 μM MtCDH-2 only (c).
Active Site Characterization of Cu(II)-NCU08746.
Because NCU08746 contains two domains with a flexible linker region, it is not readily amenable to crystallization. Thus, the copper active site was characterized by using X-ray absorption spectroscopy, which provides information on the local structure of the copper center. The Cu K-edge X-ray absorption near edge spectrum (XANES) of Cu(II)-NCU08746 is typical of a five- or six-coordinate copper(II) species containing oxygen/nitrogen ligands, which exhibits no sizable 1s→3d transition or very weak 1s→4p transition features (SI Appendix, Fig. S8) (32).
Extended X-ray absorption fine structure (EXAFS) data and the resulting Fourier transform are shown in Fig. 5. The Fourier transform exhibits a strong inner shell feature at r′ ∼1.5 Å, a second shell feature at r′ ∼2.3 Å, and third shell features between r′ = 2.3 and 3.7 Å. The third shell features in the Fourier transform correspond to the double-humped feature centered at ∼4 Å−1 in the EXAFS spectrum, which arises from the multiple scattering paths of several imidazole moieties as found in the spectra of many other metalloproteins and model complexes (33–37). Fitting progress is described in SI Appendix, Table S5. The best fit includes 4 Cu-O/N paths at 1.97 Å, 1 Cu-O/N path at 2.22 Å, 1 Cu-O/N path at 2.42 Å, 2 Cu-C paths at 3.23 Å, and paths from 2.2 imidazole moieties (SI Appendix, Table S5). This result indicates that the copper center of NCU08746 contains two or three histidine ligands.
Fig. 5.
k3-weighted EXAFS data of Cu(II)-NCU08746 and its Fourier transform. The boxes highlight the features that arise from the outer-shell atoms of the imidazole ligands. Best fit parameters are provided in SI Appendix, Table S5 (Fit 9).
Discussion
Bioinformatic, biochemical, and spectroscopic studies reported here show that NCU08746 represents a member of a new family of PMOs that cleave starch. Sequence analysis reveals that this family contains the conserved histidine residues that are expected to form the histidine brace motif, and the motif N/Q/E-X-F/Y that contains the active site tyrosine residue in PMOs (Fig. 1). MS/MS analysis supports the presence of an N-terminal methylhistidine residue in NCU08746. Activity assays indicate that copper is the native metal cofactor of this enzyme. ICP analysis shows that it contains one Cu atom per protein molecule. Cu K-edge EXAFS analysis indicates that purified NCU08746 contains a copper(II) center with two or three histidine ligands. These features closely resemble those of the active sites in chitin- and cellulose-active PMOs (Fig. 1A) (2, 6, 7, 9, 10, 15–17).
We propose a structure for the active site of NCU08746 similar to that shown in Fig. 1A in which the elongated hexacoordinate copper(II) site contains two histidine ligands. The coordination number is consistent with both the XANES features and EXAFS analysis. The coordination sphere deduced from EXAFS fitting contains 4 O/N ligands at 1.97 Å from the copper center and two additional O/N ligands at longer distances, 2.22 and 2.42 Å, which is consistent with the elongated octahedral geometry as predicted by the Jahn–Teller effect. The uncertainty in the EXAFS-derived coordination number does not allow us to rule out a third histidine ligand. There are three conserved histidine residues throughout the NCU08746 family, which are also observed in fungal cellulose-active PMOs (9, 10, 15–17, 22). In fungal cellulose-active PMOs, the N-terminal histidine and the second histidine, around position 70–100, form the histidine brace (22); the third histidine, near the end of the catalytic domain, is involved in a conserved hydrogen bonding network at the active site (9, 10, 15–17). The length of the catalytic domain and the distribution of the three conserved histidine residues in starch-active PMOs are similar to those of cellulose-active PMOs, suggesting that starch-active PMOs likely adopt a homologous structural arrangement of the conserved histidine residues. Further mutagenesis and structural studies are needed to gain more detailed information about the copper center of starch-active PMOs.
NCU08746 requires both oxygen and a source of electrons. As shown, ascorbic acid is an efficient electron donor for the NCU08746 reaction. It is possible that oligosaccharide dehydrogenases or maltose dehydrogenases can serve as biological electron donors to starch-active PMOs; however, they have not been well characterized (38, 39). CDH, which is widely accepted as the biological partner of cellulose-active PMOs, can donate electrons to NCU08746 as efficiently as ascorbic acid. This result suggests that NCU08746 and other family members may possess some protein structural features similar to those in cellulose-active PMOs that allow the transfer of electrons from CDH to the copper center. It is also possible that CDH is the biological partner of starch-active PMOs. In the wild, fungi routinely encounter food sources containing mixed polysaccharides, and coexpression of enzymes that cleave diverse substrates would allow for parallel degradation pathways. Transcriptomic and proteomic analysis of the N. crassa secretome on different growth substrates would shed light on the potential biological relationship between CDH and starch-active PMOs.
Proposed starch degradation steps by NCU08746 are shown in Fig. 6, analogous to that of cellulose-active PMOs and chitin-active PMOs. NCU08746 is shown here to oxidize the C1 position in both amylose and amylopectin. For amylopectin, it is unclear whether only the α(1→4) bond is cleaved or whether NCU08746 can attack both α(1→4) and α(1→6) linkages. In addition, although C4 and C6 oxidized products are not detected for NCU08746, oxidation at these positions is relevant to bond cleavage and may be carried out by other starch-active PMOs. C4 oxidation is known to occur with some cellulose-active PMOs (11, 12, 14, 22, 40) and has recently been implicated in fungal chitin-active PMOs (7).
Fig. 6.
Proposed starch degradation steps by NCU08746 involving the cleavages of α(1→4) (Top and Middle) and α(1→6) (Center and Bottom) linkages via hydroxylation at the C1 position.
The discovery and characterization of starch-active PMOs shows that the bioinformatic approach described above can be used to identify new PMO families. There are two more uncharacterized potential PMO families listed in Table 1, whose substrates might be different from chitin, cellulose, and starch. This finding suggests the existence of a PMO superfamily with a much broader range of substrates. Unlike the uniform microcrystalline structure of chitin and cellulose (Fig. 1B), the higher order structure of starch is complicated. Starch is a mixture of amylose and amylopectin with the relative ratio varying depending on the source. Amylose contains α(1→4) glycosidic linkages and exists in disordered, single helical, and double helical forms. Amylopectin contains α(1→6) bonds in addition to α(1→4) bonds, which creates branches (Fig. 1C). Double helices can form between amylose and amylopectin in starch, which further complicates the structure (23–25). Recently, a cellulose-active PMO from N. crassa (22) was found to act on soluble cellodextrins (40) and hemicelluloses that contain β(1→4) linked glucose and glucose derivative units (41). The crystalline surface has thus not been required for all previously identified PMOs; however, β(1→4) linkages of glucose and glucose derivatives have been essential. Our finding emphasizes that PMOs can act on substrates with higher order structures and glycosidic linkages beyond those found in cellulose and chitin.
Our results show that NCU08746 oxidatively cleaves starch substrates. There are no other enzymes with such activity reported thus far to our knowledge. Oligosaccharide dehydrogenases (38) and maltose dehydrogenases (39) are capable of oxidizing the reducing ends of a number of oligosaccharides, including maltodextrins. However, these enzymes have not been reported to cleave glycosidic bonds. Starch-active PMOs introduce an expanded perspective on starch degradation and metabolism. Starch is known to be degraded mainly by α-amylase and glucoamylase, two efficient hydrolytic enzymes used in the food and starch-based biofuel industries. In industrial hydrolysis of starch to glucose, the first step is the swelling of starch granules with heat (90–100 °C) and water (26). In this step, the radially ordered structure of starch is disrupted; starch becomes amorphous and amenable to hydrolysis by amylases. Because fungi cannot perform this high temperature pretreatment, they may need other means to overcome the resistance of starch granule. One such mean would be the secretion of a complex mixture of amylolytic enzymes. Analogous to the role of chitin- and cellulose-active PMOs in the degradation of their respective crystalline substrates, starch-active PMOs may play a key role in the efficient degradation of raw starch.
Materials and Methods
Materials.
Amylopectin from maize (Sigma; 11020), amylose with average molecular mass of 2.8 kDa (AM2.8, TCI Chemicals; A0846), and corn starch (Sigma; S4126), and chitin (Sigma, C9752) were washed with water by centrifugation several times before the addition to assays. Phosphoric acid swollen cellulose was prepared from Avicel PH101 (Sigma; 11365) as described (42). Metal salts were purchased from various sources with analytical grade purity. Cellobiose dehydrogenase from Myceliophthora thermophila (MtCDH-2) was purified as reported (11). Standard glucose and maltodextrins with degrees of polymerization (DP) of 2–7 were purchased from Sigma. Higher DP soluble maltodextrins were obtained by suspending 100 mg/mL AM2.8 in water and removing the pellet by centrifugation. Malto-aldonic acids were prepared by oxidation of maltodextrins with Lugol’s solution by using a method described (12).
Identification of New PMOs.
All Neurospora crassa proteins were input into SignalP 4.0 program (28). Predicted sequences with an N-terminal histidine residue were compared with orthologs found in protein databases. Potential PMOs were identified as those sequences having a second conserved histidine residue and an N/Q/E-X-F/Y motif. Structural prediction was also performed by using Phyre2 (43).
Expression and Purification.
NCU08746 was expressed in N. crassa as reported (22) by using the primers in SI Appendix, Table S1. Five- to 10-L filtered culture was concentrated to ∼100 mL, desalted to 10 mM sodium acetate buffer at pH 5.0 (buffer A), and mixed with 10 mM EDTA overnight. The EDTA-treated sample was subsequently loaded onto a preequilibrated amylose resin column (10 mL). After sample injection, the column was washed with 15 column volumes of buffer A. Protein was eluted with 10 column volumes of 50 mM maltose in buffer A. Fractions containing NCU08746 were pooled and mixed with 100 μM CuSO4 overnight. Subsequently, the CuSO4-treated sample was then concentrated to ∼5 mL and purified further with a size exclusion column (Superdex 75; GE Healthcare). To obtain apo NCU08746, the reconstitution with CuSO4 was replaced with another EDTA treatment.
Protein MS/MS Analysis.
Trypsin-digested purified NCU08746 was analyzed with nano LC MS/MS at the Scripps Center for Metabolomics and Mass Spectrometry of The Scripps Research Institute.
Inductively Coupled Plasma Optical Emission Spectrometry Analysis.
ICP-OES analysis of 27 elements was performed at the Research Analytical Laboratory (RAL) at the University of Minnesota–Twin Cities. The samples were prepared according to the instructions from RAL.
Activity Assays.
Assays were carried out in 100 μL of total volume by using 96-well plates, which were shaken at 1,000 rpm and 42 °C for 4 h using a microplate shaker (Fisher, Model 9454FIMPUSA). All assays contained 50 mg/mL substrate suspension, except for assays with PASC that contained 5 mg/mL substrate. Typical assays contained 5 μM holo NCU08746, 2 mM ascorbic acid, and 50 mM sodium acetate at pH 5.0. Ascorbic acid was substituted by 0.5 μM MtCDH-2 in some assays. Cellobiose (5 mM) was added to an assay with MtCDH-2. For assays with partially apo NCU08746, 5 μM metal salts were added.
CDH Oxidation Assays.
The rate of oxidation was measured by monitoring the decrease in the absorption at 430 nm of the reduced heme domain. Immediately before data collection, 1 μM as-isolated MtCDH-2 in 50 mM sodium acetate at pH 5.0 was mixed with 6 μM cellobiose. To test the effect of NCU08746 on the rate of CDH oxidation, 5 μM NCU08746 was added to the CDH solution before mixing with cellobiose. All of the assays were carried out at room temperature and in the presence of atmospheric oxygen.
Product Analysis.
Assay samples were mixed with one volume of 0.2 M NaOH and analyzed with HPAEC by using a DIONEX ICS-5000 system and a reported gradient (11, 12). The synthetic malto-aldonic acids were run in the same manner. Product analysis with LC-MS was carried out at the Energy Biosciences Institute as described (11).
X-Ray Absorption Spectroscopy.
Cu(II)-NCU08746 was prepared in 100 mM Mes buffer at pH 5.0, concentrated, and mixed with 20% (vol/vol) glycerol to obtain a final concentration of 1.28 mM enzyme. The sample was transferred to X-ray absorption spectroscopy (XAS) solution sample holder. XAS data were collected at Beamline X3B of National Synchrotron Light Source at Brookhaven National Laboratory on frozen samples by using a He Displex cryostat (sample T ∼20 K). The energy scale was internally calibrated by using a Cu metal reference foil (Eref = 8,979 eV). To minimize photoreduction of the sample, eight first scans were collected on eight different sample spots (0.9 mm × 5.8 mm). Standard procedures were applied to reduce, calibrate, and average data by using EXAFSPAK (44). EXAFS fitting was performed by using the OPT function of EXAFSPAK. Theoretical scattering paths were calculated with FEFF8.4 (45) by using the model shown in SI Appendix, Fig. S9. The r and σ2 values of all significant single and multiple scattering paths of the imidazole moiety were linked together in the fit.
Supplementary Material
Acknowledgments
We thank Stefan Bauer at the Energy Biosciences Institute, University of California, Berkeley for help with LC/MS analysis and the M.A.M. group members for experimental support and helpful discussion. XAS data were collected at Beamline X3B of the National Synchrotron Light Source at Brookhaven National Laboratory, which was supported by National Institutes of Health Grant P30-EB009998 and the US Department of Energy under Contract DE-AC02-98CH10886. This work was supported by the Energy Biosciences Institute and The Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408090111/-/DCSupplemental.
References
- 1.Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VGH. Novel enzymes for the degradation of cellulose. Biotechnol Biofuels. 2012;5(1):45–56. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hemsworth GR, Davies GJ, Walton PH. Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr Opin Struct Biol. 2013;23(5):660–668. doi: 10.1016/j.sbi.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Tian C, et al. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc Natl Acad Sci USA. 2009;106(52):22157–22162. doi: 10.1073/pnas.0906810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yakovlev I, et al. Substrate-specific transcription of the enigmatic GH61 family of the pathogenic white-rot fungus Heterobasidion irregulare during growth on lignocellulose. Appl Microbiol Biotechnol. 2012;95(4):979–990. doi: 10.1007/s00253-012-4206-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berka RM, et al. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat Biotechnol. 2011;29(10):922–927. doi: 10.1038/nbt.1976. [DOI] [PubMed] [Google Scholar]
- 6.Vaaje-Kolstad G, et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330(6001):219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 7.Hemsworth GR, Henrissat B, Davies GJ, Walton PH. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat Chem Biol. 2014;10(2):122–126. doi: 10.1038/nchembio.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forsberg Z, et al. Comparative study of two chitin-active and two cellulose-active AA10-type lytic polysaccharide monooxygenases. Biochemistry. 2014;53(10):1647–1656. doi: 10.1021/bi5000433. [DOI] [PubMed] [Google Scholar]
- 9.Harris PV, et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: Structure and function of a large, enigmatic family. Biochemistry. 2010;49(15):3305–3316. doi: 10.1021/bi100009p. [DOI] [PubMed] [Google Scholar]
- 10.Quinlan RJ, et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci USA. 2011;108(37):15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phillips CM, Beeson WT, Cate JHD, Marletta MA. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem Biol. 2011;6(12):1399–1406. doi: 10.1021/cb200351y. [DOI] [PubMed] [Google Scholar]
- 12.Beeson WT, Phillips CM, Cate JHD, Marletta MA. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J Am Chem Soc. 2012;134(2):890–892. doi: 10.1021/ja210657t. [DOI] [PubMed] [Google Scholar]
- 13.Forsberg Z, et al. Cleavage of cellulose by a CBM33 protein. Protein Sci. 2011;20(9):1479–1483. doi: 10.1002/pro.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsberg Z, et al. Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases. Proc Natl Acad Sci USA. 2014;111(23):8446–8451. doi: 10.1073/pnas.1402771111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karkehabadi S, et al. The first structure of a glycoside hydrolase family 61 member, Cel61B from Hypocrea jecorina, at 1.6 A resolution. J Mol Biol. 2008;383(1):144–154. doi: 10.1016/j.jmb.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Beeson WT, 4th, Phillips CM, Marletta MA, Cate JHD. Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure. 2012;20(6):1051–1061. doi: 10.1016/j.str.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu M, et al. Crystal structure and computational characterization of the lytic polysaccharide monooxygenase GH61D from the Basidiomycota fungus Phanerochaete chrysosporium. J Biol Chem. 2013;288(18):12828–12839. doi: 10.1074/jbc.M113.459396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishiyama Y, Langan P, Chanzy H. Crystal structure and hydrogen-bonding system in cellulose Ibeta from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc. 2002;124(31):9074–9082. doi: 10.1021/ja0257319. [DOI] [PubMed] [Google Scholar]
- 19.Nishiyama Y, Sugiyama J, Chanzy H, Langan P. Crystal structure and hydrogen bonding system in cellulose I(α) from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc. 2003;125(47):14300–14306. doi: 10.1021/ja037055w. [DOI] [PubMed] [Google Scholar]
- 20.Nishiyama Y, Noishiki Y, Wada M. X-ray structure of anhydrous β-chitin at 1 Å resolution. Macromolecules. 2011;44(4):950–957. [Google Scholar]
- 21.Sikorski P, Hori R, Wada M. Revisit of α-chitin crystal structure using high resolution X-ray diffraction data. Biomacromolecules. 2009;10(5):1100–1105. doi: 10.1021/bm801251e. [DOI] [PubMed] [Google Scholar]
- 22.Vu VV, Beeson WT, Phillips CM, Cate JHD, Marletta MA. Determinants of regioselective hydroxylation in the fungal polysaccharide monooxygenases. J Am Chem Soc. 2014;136(2):562–565. doi: 10.1021/ja409384b. [DOI] [PubMed] [Google Scholar]
- 23.Pérez S, Bertoft E. The molecular structures of starch components and their contribution to the architecture of starch granules: A comprehensive review. Starke. 2010;62(8):389–420. [Google Scholar]
- 24.Popov D, et al. Crystal structure of A-amylose: A revisit from synchrotron microdiffraction analysis of single crystals. Macromolecules. 2009;42(4):1167–1174. [Google Scholar]
- 25.Imberty A, Chanzy H, Pérez S, Buléon A, Tran V. The double-helical nature of the crystalline part of A-starch. J Mol Biol. 1988;201(2):365–378. doi: 10.1016/0022-2836(88)90144-1. [DOI] [PubMed] [Google Scholar]
- 26.Oates CG. Towards an understanding of starch granule structure and hydrolysis. Trends Food Sci Technol. 1997;8(11):375–382. [Google Scholar]
- 27.Buléon A, Colonna P, Planchot V, Ball S. Starch granules: Structure and biosynthesis. Int J Biol Macromol. 1998;23(2):85–112. doi: 10.1016/s0141-8130(98)00040-3. [DOI] [PubMed] [Google Scholar]
- 28.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 29.Christiansen C, et al. The carbohydrate-binding module family 20—diversity, structure, and function. FEBS J. 2009;276(18):5006–5029. doi: 10.1111/j.1742-4658.2009.07221.x. [DOI] [PubMed] [Google Scholar]
- 30.Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henriksson G, Johansson G, Pettersson G. A critical review of cellobiose dehydrogenases. J Biotechnol. 2000;78(2):93–113. doi: 10.1016/s0168-1656(00)00206-6. [DOI] [PubMed] [Google Scholar]
- 32.Sarangi R. X-ray absorption near-edge spectroscopy in bioinorganic chemistry: Application to M-O2 systems. Coord Chem Rev. 2013;257(2):459–472. doi: 10.1016/j.ccr.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vu VV, Makris TM, Lipscomb JD, Que L., Jr Active-site structure of a β-hydroxylase in antibiotic biosynthesis. J Am Chem Soc. 2011;133(18):6938–6941. doi: 10.1021/ja201822v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pellei M, et al. Nitroimidazole and glucosamine conjugated heteroscorpionate ligands and related copper(II) complexes. Syntheses, biological activity and XAS studies. Dalton Trans. 2011;40(38):9877–9888. doi: 10.1039/c1dt10486a. [DOI] [PubMed] [Google Scholar]
- 35.Sanyal I, Karlin KD, Strange RW, Blackburn NJ. Chemistry and structural studies on the dioxygen-binding copper-1,2-dimethylimidazole system. J Am Chem Soc. 1993;115(24):11259–11270. [Google Scholar]
- 36.Costello A, Periyannan G, Yang K-W, Crowder MW, Tierney DL. Site-selective binding of Zn(II) to metallo-β-lactamase L1 from Stenotrophomonas maltophilia. J Biol Inorg Chem. 2006;11(3):351–358. doi: 10.1007/s00775-006-0083-z. [DOI] [PubMed] [Google Scholar]
- 37.D’Angelo P, et al. X-ray absorption investigation of a unique protein domain able to bind both copper(I) and copper(II) at adjacent sites of the N-terminus of Haemophilus ducreyi Cu,Zn superoxide dismutase. Biochemistry. 2005;44(39):13144–13150. doi: 10.1021/bi050925x. [DOI] [PubMed] [Google Scholar]
- 38.Tessema M, et al. Oligosaccharide dehydrogenase-modified graphite electrodes for the amperometric determination of sugars in a flow injection system. Anal Chem. 1997;69(19):4039–4044. doi: 10.1021/ac970127f. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi Y, Horikoshi K. Purification and properties of NAD+-dependent maltose dehydrogenase produced by alkalophilic Corynebacterium sp. No. 93-1. Biochim Biophys Acta. 1980;614(2):256–265. doi: 10.1016/0005-2744(80)90215-6. [DOI] [PubMed] [Google Scholar]
- 40.Isaksen T, et al. A C4-oxidizing lytic polysaccharide monooxygenase cleaving both cellulose and cello-oligosaccharides. J Biol Chem. 2014;289(5):2632–2642. doi: 10.1074/jbc.M113.530196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agger JW, et al. Discovery of LPMO activity on hemicelluloses shows the importance of oxidative processes in plant cell wall degradation. Proc Natl Acad Sci USA. 2014;111(17):6287–6292. doi: 10.1073/pnas.1323629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang YHP, Cui J, Lynd LR, Kuang LR. A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: Evidence from enzymatic hydrolysis and supramolecular structure. Biomacromolecules. 2006;7(2):644–648. doi: 10.1021/bm050799c. [DOI] [PubMed] [Google Scholar]
- 43.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 44.George GN, Pickering IJ. EXAFSPAK: A Suite of Computer Programs for Analysis of X-Ray Absorption Spectra. Stanford, CA: Stanford Synchrotron Radiation Laboratory, Stanford Linear Accelerator Center, Stanford University; 2000. http://ssrl.slac.stanford.edu/∼george/exafspak/manual.pdf. Accessed January 1, 2014. [Google Scholar]
- 45.Rehr JJ, Mustre de Leon J, Zabinsky SI, Albers RC. Theoretical x-ray absorption fine structure standards. J Am Chem Soc. 1991;113(14):5135–5140. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.