Significance
Microtubules are dynamic protein filaments assembled from tubulin subunits, which play a key role for cell division. Ligands that target microtubules and affect their dynamics belong to the most successful classes of chemotherapeutic drugs against cancer by inhibiting cell proliferation. Here we have analyzed three structurally unrelated drugs that destabilize microtubules, using X-ray crystallography. The data reveal a new tubulin-binding site for these drugs, which renders their mechanism of action distinct from that of other types of microtubule assembly inhibitors. Similar key interactions with tubulin are observed for all three ligands, thus defining a common pharmacophore. Our results offer an opportunity for the rational design of potent tubulin modulators for the development of more efficient cancer therapies.
Keywords: drug mechanism, microtubule-targeting agents, X-ray crystallography
Abstract
The recent success of antibody–drug conjugates (ADCs) in the treatment of cancer has led to a revived interest in microtubule-destabilizing agents. Here, we determined the high-resolution crystal structure of the complex between tubulin and maytansine, which is part of an ADC that is approved by the US Food and Drug Administration (FDA) for the treatment of advanced breast cancer. We found that the drug binds to a site on β-tubulin that is distinct from the vinca domain and that blocks the formation of longitudinal tubulin interactions in microtubules. We also solved crystal structures of tubulin in complex with both a variant of rhizoxin and the phase 1 drug PM060184. Consistent with biochemical and mutagenesis data, we found that the two compounds bound to the same site as maytansine and that the structures revealed a common pharmacophore for the three ligands. Our results delineate a distinct molecular mechanism of action for the inhibition of microtubule assembly by clinically relevant agents. They further provide a structural basis for the rational design of potent microtubule-destabilizing agents, thus opening opportunities for the development of next-generation ADCs for the treatment of cancer.
Microtubule-targeting agents such as the taxanes and the vinca alkaloids represent a successful class of anticancer drugs (1). Vinblastine, for example, is a microtubule-destabilizing agent (MDA) that is widely used in combination therapy for the treatment of childhood and adult malignancies (2). The broad clinical application of MDAs, however, is hampered by their severe adverse effects (3). This problem has been very recently addressed by the use of antibody–drug conjugate (ADC) approaches, which have revived interest in the development of highly potent MDAs for therapeutic use (4–6).
For several important MDAs, the molecular mechanism of action on tubulin and microtubules has so far remained elusive. Rhizoxin, for example, is a potent MDA that has been investigated in phase 2 clinical trials, but for reasons poorly understood, it has demonstrated only very limited clinical efficacy (7). At the molecular level, it is well established that rhizoxin interferes with the binding of vinblastine to tubulin; however, the exact location of its binding site has been a matter of debate (8–10). Interestingly, biochemical and mutagenesis data suggest that the structurally unrelated MDA maytansine (9, 11), which is part of an ADC that was recently approved by the FDA for the treatment of advanced breast cancer (11, 12), and the phase 1 drug PM060184 (13, 14) (Fig. 1A) share a common tubulin-binding site with rhizoxin (9, 13, 14). These two latter drugs have also been reported to interfere with the binding of vinblastine; however, as for rhizoxin, the exact binding sites and modes of action of maytansine and PM060184 have not been elucidated (9, 14–16).
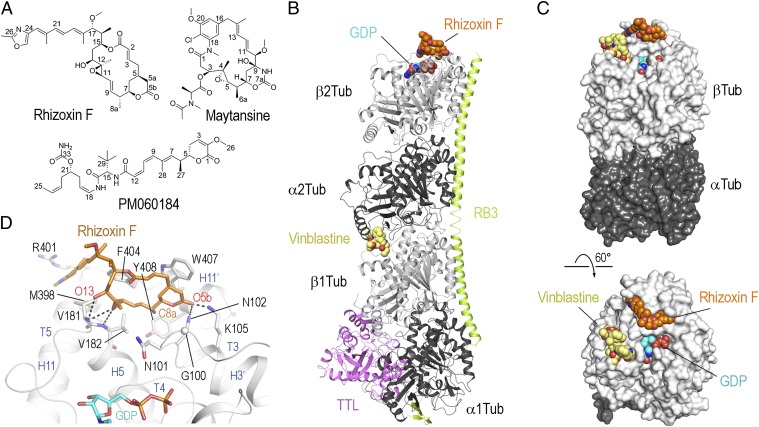
Fig. 1.
Structure of the tubulin–rhizoxin F complex. (A) Chemical structures of rhizoxin F, maytansine, and PM060184. (B) Overall view of the T2R-TTL–rhizoxin F complex. Tubulin (gray), RB3 (light green), and TTL (violet) are shown in ribbon representation; the MDA rhizoxin F (orange) and GDP (cyan) are depicted in spheres representation. As a reference, the vinblastine structure (yellow, PDB ID no. 1Z2B) is superimposed onto the T2R complex. (C) Overall view of the tubulin–rhizoxin F interaction in two different orientations. The tubulin dimer with bound ligand (α-tubulin-2 and β-tubulin-2 of the T2R-TTL–rhizoxin F complex) is shown in surface representation. The vinblastine structure is superimposed onto the β-tubulin chain to highlight the distinct binding site of rhizoxin F. All ligands are in sphere representation and are colored in orange (rhizoxin F), cyan (GDP), and yellow (vinblastine). (D) Close-up view of the interaction observed between rhizoxin F (orange sticks) and β-tubulin (gray ribbon). Interacting residues of β-tubulin are shown in stick representation and are labeled.
To establish the exact tubulin-binding site of rhizoxin, maytansine, and PM060184 and to clarify their specific interactions with the protein, we have investigated the structures of the corresponding ligand–tubulin complexes by X-ray crystallography. Our data reveal a new tubulin-binding site and pharmacophore for small molecules, and binding to this site is associated with a distinct molecular mechanism for the inhibition of microtubule formation.
Results and Discussion
A New Tubulin-Binding Site for Structurally Diverse MDAs.
We initially sought to investigate the molecular mechanism of action of rhizoxin. To provide insight into the binding mode of rhizoxin with tubulin, we soaked crystals of a protein complex composed of two αβ-tubulin (T2), the stathmin-like protein RB3 (R), and tubulin tyrosine ligase (TTL; the complex is denoted T2R-TTL) (17, 18) with the natural rhizoxin variant 2,3-desepoxy rhizoxin (19) [referred to as “rhizoxin F” from here on (20); Fig. 1A] and determined its tubulin-bound structure by X-ray crystallography at 2.0-Å resolution (Fig. 1B and Table S1 and Fig. S1A). The overall structure of tubulin in the tubulin–rhizoxin F complex superimposed well with the one obtained in the absence of the ligand (17) (rmsd, 0.124 Å over 354 Cα atoms). This result suggests that binding of the compound does not affect the global conformation of the tubulin, although we cannot exclude that the ligand may affect the conformation of the protein in its free state. More important, rhizoxin F was found to bind to a site on β-tubulin that is distinct from what is commonly referred to as the vinca domain, which is targeted by vinblastine (21–23) (Fig. 1C). As shown in Fig. 1D, this pocket is adjacent to the guanine-nucleotide binding site and is shaped by hydrophobic and polar residues of helices H3′, H11, and H11′, as well as the loops S3-H3′ (T3-loop), S5-H5 (T5-loop), and H11-H11′ of β-tubulin.
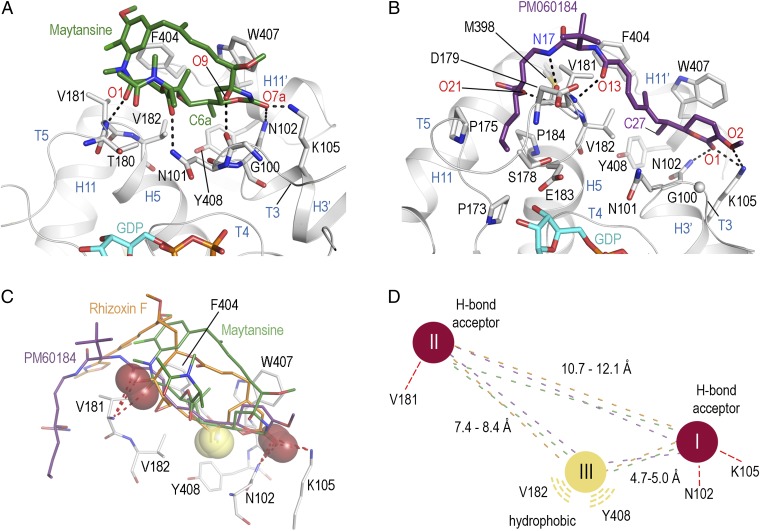
It has been previously suggested that the structurally distinct MDAs maytansine (9, 11) and PM060184 (13, 14) (Fig. 1A) share a common tubulin-binding site with rhizoxin (9, 13, 14); however, both drugs were also reported to interfere with the binding of vinblastine (9, 14–16). To clarify the exact binding site of maytansine and PM060184 on tubulin and their mode of interaction with the protein, we solved tubulin structures in complex with either drug at 2.1- and 2.0-Å resolution, respectively, using the same experimental approach as for rhizoxin F (Fig. S1 B and C and Table S1). We found that both compounds bound to the same site on β-tubulin as rhizoxin F (Fig. 2 A–C). Similar to rhizoxin F, one part of the macrocycle of maytansine is engaged with β-tubulin residues at the binding interface, whereas the remaining parts of the ring structure project into solution, thus creating a bulky extrusion at the tip of the β-tubulin subunit (Figs. 1D and 2A). In contrast, PM060184 assumes an extended, linear conformation with only the tert-butyl group being markedly solvent-exposed (Fig. 2B), a finding consistent with NMR measurements (14).
Fig. 2.
Structures of the tubulin–maytansine and tubulin–PM060184 complexes and pharmacophore model. (A) Close-up view of the tubulin–maytansine complex. Maytansine is in green stick representation. β-tubulin is displayed as gray ribbon. Key residues forming the interaction with the ligand are in stick representation and are labeled. Hydrogen bonds are highlighted as dashed black lines. (B) Detailed view of the tubulin–PM060184 complex. The ligand is displayed as violet-purple sticks. (C) Superposition of the binding sites of rhizoxin F (orange), maytansine (green), and PM060184 (violet-purple) highlighting the three common interaction points I, II, and III with β-tubulin. Hydrogen bond acceptors are highlighted as red spheres; the methyl groups forming the hydrophobic interaction are highlighted as yellow spheres. (D) Schematic drawing of the common pharmacophore for ligand binding to the maytansine site, using the same color code as in C.
Notably, mutation of the β-tubulin residue Asn100 in the fungus Aspergillus nidulans (Asn102, as defined here according to ref. 24) conveys resistance to rhizoxin, ansamitocin P3 (a maytansine variant), and PM060184 (14, 25). This observation is consistent with the key role of the side chain of this asparagine residue in ligand binding, as revealed by our structures (Figs. 1D and 2B). The acetylated nitrogen of the N-methyl alanine group in maytansine (N2′) is solvent-exposed in the tubulin–maytansine complex, suggesting that the attachment of even bulkier substituents to this atom should not interfere with the interactions of the maytansine macrocycle with the protein. This is in line with the strong tubulin effects that have been observed for the maytansine derivatives S-methyl-DM1 and S-methyl-DM4 (26), which contain N2′-acyl moieties that are significantly larger than the natural acetyl group in maytansine. These findings suggest that both derivatives bind to tubulin in a similar way as maytansine; this should also hold true for lysine-Nε-MCC-DM1, the pharmacologically active, intracellular metabolite of the ADC ado-trastuzumab emtansine (27).
Rhizoxin F, Maytansine, and PM060184 Share a Common Pharmacophore.
Closer inspection of the tubulin-binding mode of rhizoxin F, maytansine, and PM060184 revealed three shared key interaction points with the protein, which include hydrogen bonds between the carbonyl groups at positions 5b, 7a, and 1 of rhizoxin F, maytansine, and PM060184, respectively, and residues Asn102 and Lys105 of β-tubulin; hydrogen bonds between the hydroxyl/carbonyl oxygens at positions 13 (rhizoxin F), 1 (maytansine), and 13 (PM060184) and Val181 of β-tubulin; and hydrophobic interactions between the methyl groups at positions 8a (rhizoxin F), 6a (maytansine), and 27 (PM060184) of the ligands and a pocket shaped by residues Asn101, Asn102, Val182, Phe404, and Tyr408 of β-tubulin. These conserved tubulin interaction points constitute a common pharmacophore (Fig. 2D). Further, predominantly hydrophobic contacts are established by the side chain of rhizoxin F and the C15-C33 moiety of PM060184 that occupy adjacent pockets formed by helices H5 and H11 and by the T5 and H11-H11′ loops of β-tubulin, respectively. These interactions, in addition to those associated with the common pharmacophore, are essential for the full activity of both the ligands (28, 29).
In the following, the newly discovered drug-binding site on β-tubulin is referred to as the maytansine site.
Maytansine-Site Ligands Inhibit Longitudinal Tubulin Interactions.
Tubulin dimers experience a “curved-to-straight” conformational transition on assembly into microtubules (30). To assess possible structural changes of the maytansine-site that could be induced on tubulin assembly, we compared structures of β-tubulin in the curved and straight conformational states. Superimposition of these structures revealed that the overall conformation of the maytansine site is not significantly affected by the curved-to-straight transition (rmsd, 0.66 Å over 73 Cα atoms; Fig. S2). This analysis suggests that maytansine-site ligands can bind to both the curved and straight tubulin states.
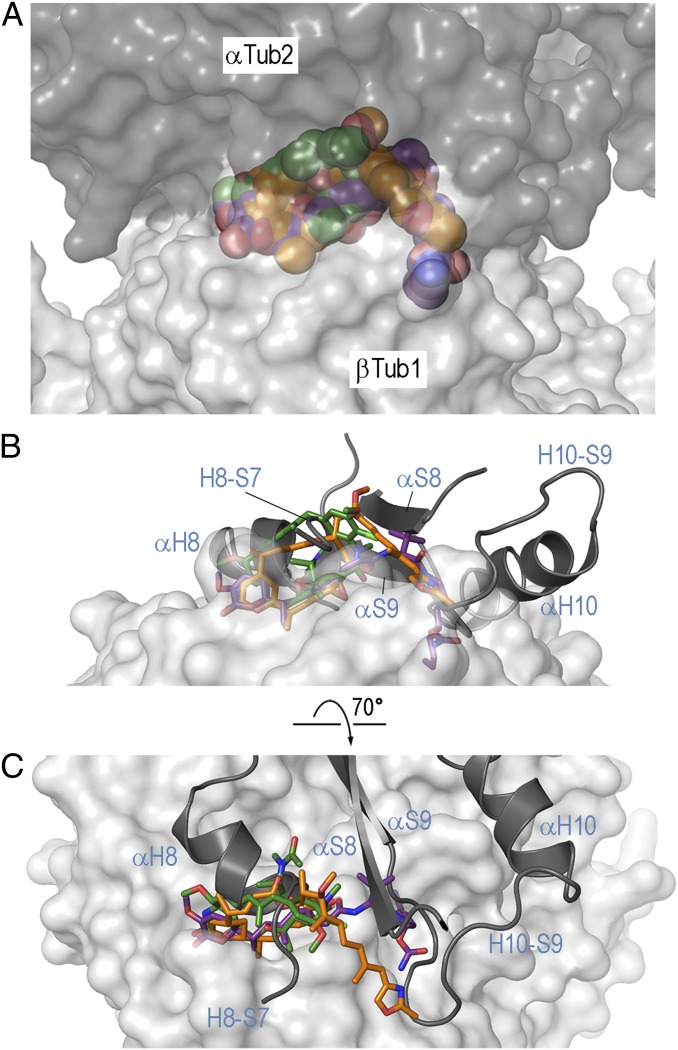
To assess the mechanism by which maytansine-site ligands may destabilize microtubules, we modeled the interactions of rhizoxin F, maytansine, and PM060184 with β-tubulin in the context of a microtubule. For this purpose, we used an atomic model of a microtubule that is based on a 3.5-Å resolution, electron crystallography structure of “straight” tubulin obtained from protofilament-based zinc sheets (24), as well as cryo-electron microscopy reconstructions of microtubules at about 8-Å resolution (31, 32). As shown in Fig. 3 A and B, binding of a ligand to the maytansine site in all three cases sterically hinders the formation of longitudinal tubulin–tubulin interactions established between the pocket that accommodates the pharmacophore, which is shaped by loops S3-H3, S5-H5, and H11-H11′ of β-tubulin, and helix H8 of α-tubulin. Additional steric clashes were observed between the side chain of rhizoxin F and the C15-C33 moiety of PM060184, as well as between the loop H10-S9 and strand S8 of α-tubulin, respectively (Fig. 3 B and C).
Fig. 3.
Binding of maytansine-site ligands in the context of a microtubule. (A) View of the tubulin–maytansine-site ligand interaction in the context of the microtubule (PDB ID no. 2XRP). The binding sites of the complexes of rhizoxin F, maytansine, and PM060184 are superimposed on the corresponding site on β-tubulin of the microtubule model. The α- and β-tubulin chains are displayed as dark and light gray surfaces, respectively. The ligands are in sphere representation, using the same color code as in Figs. 1C and 2 A and B. (B) Side view of the longitudinal tubulin–tubulin contact with superimposed maytansine-site ligands. For clarity reasons, only the secondary structure elements of α-tubulin forming the longitudinal contact are shown and labeled in light blue. β-Tubulin is in gray surface; the maytansine-site ligands are in stick representation. (C) Top view of the longitudinal tubulin–tubulin contact highlighting the prominent steric clash between the maytansine-site ligands bound to β-tubulin and the helix H8 of α-tubulin from a neighboring dimer. The same settings as in B are used.
Implications.
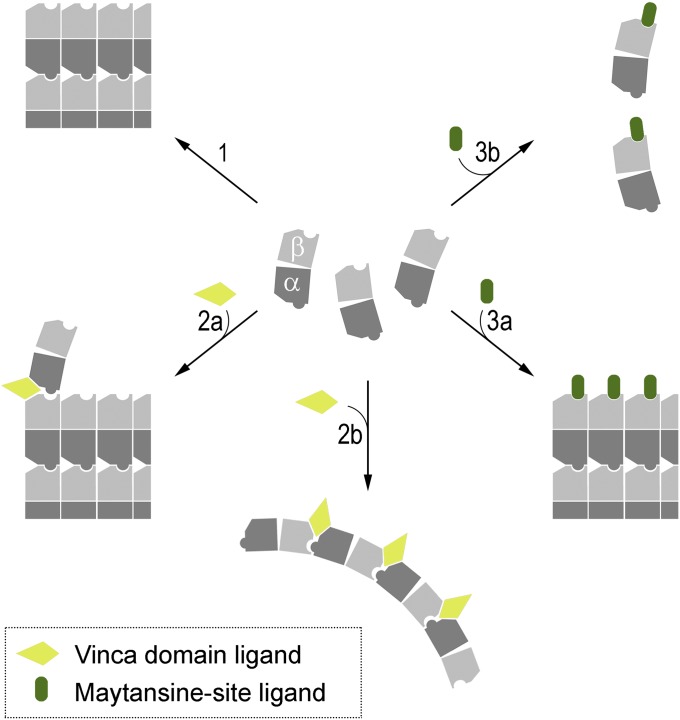
Our results establish a new ligand-binding site on β-tubulin that is targeted by clinically relevant anticancer drugs, and it is conceivable that other classes of microtubule drugs will also bind to this site. The data further suggest that maytansine-site ligands destabilize microtubules by either sequestering tubulin subunits into assembly-incompetent tubulin–drug complexes at high ligand concentrations or inhibiting the addition of tubulin subunits at the plus ends of growing microtubules by blocking longitudinal tubulin interactions at substoichiometric ligand concentrations (Fig. 4). This mechanism is distinct from the one exerted by vinca domain ligands. The latter inhibit the formation of microtubules by introducing a wedge at the longitudinal interface between two tubulin dimers and by stabilizing curved tubulin oligomers that are not compatible with the straight tubulin structure found in microtubules (21–23) (Fig. 4). The “poisoning” of microtubule plus ends by maytansine-site ligands explains the aberrant microtubule phenotypes observed in cells at subnanomolar concentrations of, for example, PM060184 (14).
Fig. 4.
Molecular mechanism of action of vinca domain- and maytansine-site ligands on tubulin and microtubules. (1) In the absence of ligands, curved αβ-tubulin heterodimers assemble into microtubules and undergo a curved-to-straight conformational transition. Formation of longitudinal contacts include the interaction between a pocket shaped by loops S3-H3, S5-H5, and H11-H11′ of the β-tubulin subunit of one dimer (cavity) and helix H8 of α-tubulin from a neighboring dimer in the microtubule lattice (knob). (2) Vinblastine binds to the vinca domain, a composite binding site that is formed by structural elements from both α- and β-tubulin monomers of two different, longitudinally aligned αβ-tubulin heterodimers. The MDA destabilizes microtubules by introducing a wedge at the interface between two tubulin dimers at the tips of microtubules, thus inhibiting the curved-to-straight conformational transition necessary to build up the microtubule lattice (2a), or by stabilizing curved, ring-like oligomers that are not compatible with the straight tubulin structure found in microtubules (2b). (3) Maytansine-site ligands bind to the site on β-tubulin that is involved in the formation of longitudinal contacts in microtubules. These types of MDAs destabilize microtubules either by binding to the plus ends of growing microtubules at substoichiometric ligand concentrations, thus inhibiting the addition of further tubulin subunits (3a), or by forming assembly incompetent tubulin–drug complexes with unassembled tubulin subunits at high ligand concentrations (3b). Note that in vitro at high Mg2+ concentrations, tubulin–PM060184 complexes can assemble into small ring-like oligomers (14).
If they do not interact with the vinca domain, why then do maytansine-site ligands interfere with vinblastine binding to tubulin dimers (10, 14)? The vinca domain is a composite binding site formed by structural elements from both α- and β-tubulin monomers of two different, consecutive αβ-tubulin heterodimers (21–23) (Fig. S3). Our data suggest that the binding of ligands to the maytansine site prevents the formation of the complete vinca domain from the two half sites, thus leading to an impairment of ligand binding; conversely, formation of the full vinca domain/vinca ligand complex prevents access to the maytansine site (Fig. 4). Conflicting hypotheses have been advanced in the literature for the mode (competitive versus noncompetitive) of mutual inhibition of tubulin binding by maytansine and vinblastine (8–10). Our model readily explains why the binding of maytansine-site and vinca-domain ligands to tubulin is mutually exclusive without having to invoke a competitive inhibition mechanism.
Although MDAs are successfully used in cancer therapy, their use is limited by substantial adverse effects. However, the incorporation of MDAs as drug cargo in ADCs has recently expanded their utility and revived strong interest in their clinical potential (4, 5). Brentuximab vedotin, which carries the MDA monomethyl auristatin E (33) and the maytansine-derived trastuzumab emtansine (12), were approved by the US Food and Drug Administration for the treatment of patients with Hodgkin lymphoma and metastatic breast cancer in 2011 and 2013, respectively. Both ADCs display excellent efficacy and are remarkably well-tolerated, thus highlighting the effect of the antibody–MDA conjugate approach. Microtubule-targeting agents are often complex, natural product-derived molecules that are highly challenging in terms of large-scale production. To the best of our knowledge, no common pharmacophore based on high-resolution structural data exists for any of the currently known drug-binding sites on tubulin. Thus, the structural information presented in this article for the maytansine site offers an opportunity for the rational design of highly potent, small-molecule MDAs that may help in the development of next-generation ADCs for cancer treatment.
Materials and Methods
Proteins and Compounds.
Bovine brain tubulin was prepared according to ref. 34. The production of the stathmin-like domain of RB3 and chicken TTL in bacteria, as well as the reconstitution of the T2R-TTL complex, is described in refs. 17, 18, and 30. The synthesis of 2,3-desepoxy rhizoxin and PM060184 has been described elsewhere (13, 19). Maytansine was obtained from the National Institutes of Health Open Chemical Repository Collection.
Crystallization, Data Collection, and Structure Solution.
Crystals of T2R-TTL were grown as described in refs. 17 and 18 and soaked overnight in reservoir solutions containing either 1 mM 2,3-desepoxy rhizoxin or 5 mM PM060184. In the case of maytansine, the crystals were soaked for 15 min in the presence of 1 mM compound in reservoir solution before flash-cooling. Crystals were fished directly from the drop and flash-cooled in a nitrogen stream at the beamline. Standard data collection at beamlines ×06DA and ×06SA at the Swiss Light Source (Paul Scherrer Institut, Villigen, Switzerland), data processing, and structure solution using the difference Fourier method were performed as described previously (17, 18). Data collection and refinement statistics are given in Table S1.
Structural Analysis and Figure Preparation.
Figures were prepared using the PyMOL Molecular Graphics System, version 1.5.0.5 (Schrödinger, LLC). Chains in the T2R-TTL complex were defined as follows: chain A, α-tubulin-1; chain B, β-tubulin-1; chain C, α-tubulin-2; chain D, β-tubulin-2; chain E, RB3; and chain F, TTL (Fig. 1B). Chains B and D were used throughout for the structural analyses and figure preparation.
Supplementary Material
Acknowledgments
We thank V. Olieric, A. Pauluhn, and M. Wang for excellent technical assistance with the collection of X-ray data at beamlines X06DA and X06SA of the Swiss Light Source (Paul Scherrer Institut, Villigen, Switzerland). We are indebted to V. de Lucas (Segovia) for providing calf brains for the tubulin purification. This work was supported by grants from the Ministerio de Economía y Competitividad (BIO2010-16351 to J.F.D. and BFU2011-23416 to J.M.A.) and the Comunidad Autónoma de Madrid (S2010/BMD-2457 to J.F.D. and S2010/BMD-2353 to J.M.A.), and by a grant from the Swiss National Science Foundation (310030B_138659 to M.O.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID code 4TUY (T2R-TTL-rhizoxin F), 4TV9 (T2R-TTL-PM060184), 4TV8 (T2R-TTLmaytansine)].
See Commentary on page 13684.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408124111/-/DCSupplemental.
References
- 1.Dumontet C, Jordan MA. Microtubule-binding agents: A dynamic field of cancer therapeutics. Nat Rev Drug Discov. 2010;9(10):790–803. doi: 10.1038/nrd3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4(4):253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 3.Canta A, Chiorazzi A, Cavaletti G. Tubulin: A target for antineoplastic drugs into the cancer cells but also in the peripheral nervous system. Curr Med Chem. 2009;16(11):1315–1324. doi: 10.2174/092986709787846488. [DOI] [PubMed] [Google Scholar]
- 4.Zolot RS, Basu S, Million RP. Antibody-drug conjugates. Nat Rev Drug Discov. 2013;12(4):259–260. doi: 10.1038/nrd3980. [DOI] [PubMed] [Google Scholar]
- 5.Teicher BA, Doroshow JH. The promise of antibody-drug conjugates. N Engl J Med. 2012;367(19):1847–1848. doi: 10.1056/NEJMe1211736. [DOI] [PubMed] [Google Scholar]
- 6.Sievers EL, Senter PD. Antibody-drug conjugates in cancer therapy. Annu Rev Med. 2013;64:15–29. doi: 10.1146/annurev-med-050311-201823. [DOI] [PubMed] [Google Scholar]
- 7.Hanauske AR, et al. The EORTC Early Clinical Trials Group Phase II clinical trials with rhizoxin in breast cancer and melanoma. Br J Cancer. 1996;73(3):397–399. doi: 10.1038/bjc.1996.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai RL, Pettit GR, Hamel E. Binding of dolastatin 10 to tubulin at a distinct site for peptide antimitotic agents near the exchangeable nucleotide and vinca alkaloid sites. J Biol Chem. 1990;265(28):17141–17149. [PubMed] [Google Scholar]
- 9.Takahashi M, et al. Rhizoxin binding to tubulin at the maytansine-binding site. Biochim Biophys Acta. 1987;926(3):215–223. doi: 10.1016/0304-4165(87)90206-6. [DOI] [PubMed] [Google Scholar]
- 10.Hamel E. Natural products which interact with tubulin in the vinca domain: Maytansine, rhizoxin, phomopsin A, dolastatins 10 and 15 and halichondrin B. Pharmacol Ther. 1992;55(1):31–51. doi: 10.1016/0163-7258(92)90028-x. [DOI] [PubMed] [Google Scholar]
- 11.Kupchan SM, et al. Maytansine, a novel antileukemic ansa macrolide from Maytenus ovatus. J Am Chem Soc. 1972;94(4):1354–1356. doi: 10.1021/ja00759a054. [DOI] [PubMed] [Google Scholar]
- 12.Verma S, et al. EMILIA Study Group Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med. 2012;367(19):1783–1791. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martín MJ, et al. Isolation and first total synthesis of PM050489 and PM060184, two new marine anticancer compounds. J Am Chem Soc. 2013;135(27):10164–10171. doi: 10.1021/ja404578u. [DOI] [PubMed] [Google Scholar]
- 14.Pera B, et al. New interfacial microtubule inhibitors of marine origin, PM050489/PM060184, with potent antitumor activity and a distinct mechanism. ACS Chem Biol. 2013;8(9):2084–2094. doi: 10.1021/cb400461j. [DOI] [PubMed] [Google Scholar]
- 15.Mandelbaum-Shavit F, Wolpert-DeFilippes MK, Johns DG. Binding of maytansine to rat brain tubulin. Biochem Biophys Res Commun. 1976;72(1):47–54. doi: 10.1016/0006-291x(76)90958-x. [DOI] [PubMed] [Google Scholar]
- 16.Batra JK, Powers LJ, Hess FD, Hamel E. Derivatives of 5,6-diphenylpyridazin-3-one: Synthetic antimitotic agents which interact with plant and mammalian tubulin at a new drug-binding site. Cancer Res. 1986;46(4 Pt 2):1889–1893. [PubMed] [Google Scholar]
- 17.Prota AE, et al. Structural basis of tubulin tyrosination by tubulin tyrosine ligase. J Cell Biol. 2013;200(3):259–270. doi: 10.1083/jcb.201211017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prota AE, et al. Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science. 2013;339(6119):587–590. doi: 10.1126/science.1230582. [DOI] [PubMed] [Google Scholar]
- 19.Neuhaus CM, Liniger M, Stieger M, Altmann KH. Total synthesis of the tubulin inhibitor WF-1360F based on macrocycle formation through ring-closing alkyne metathesis. Angew Chem Int Ed Engl. 2013;52(22):5866–5870. doi: 10.1002/anie.201300576. [DOI] [PubMed] [Google Scholar]
- 20.Kiyoto S, et al. A new antitumor complex, WF-1360, WF-1360A, B, C, D, E and F. J Antibiot (Tokyo) 1986;39(6):762–772. doi: 10.7164/antibiotics.39.762. [DOI] [PubMed] [Google Scholar]
- 21.Gigant B, et al. Structural basis for the regulation of tubulin by vinblastine. Nature. 2005;435(7041):519–522. doi: 10.1038/nature03566. [DOI] [PubMed] [Google Scholar]
- 22.Cormier A, Marchand M, Ravelli RB, Knossow M, Gigant B. Structural insight into the inhibition of tubulin by vinca domain peptide ligands. EMBO Rep. 2008;9(11):1101–1106. doi: 10.1038/embor.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranaivoson FM, Gigant B, Berritt S, Joullié M, Knossow M. Structural plasticity of tubulin assembly probed by vinca-domain ligands. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 8):927–934. doi: 10.1107/S0907444912017143. [DOI] [PubMed] [Google Scholar]
- 24.Löwe J, Li H, Downing KH, Nogales E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J Mol Biol. 2001;313(5):1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi M, Kobayashi H, Iwasaki S. Rhizoxin resistant mutants with an altered beta-tubulin gene in Aspergillus nidulans. Mol Gen Genet. 1989;220(1):53–59. doi: 10.1007/BF00260855. [DOI] [PubMed] [Google Scholar]
- 26.Lopus M, et al. Maytansine and cellular metabolites of antibody-maytansinoid conjugates strongly suppress microtubule dynamics by binding to microtubules. Mol Cancer Ther. 2010;9(10):2689–2699. doi: 10.1158/1535-7163.MCT-10-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erickson HK, et al. Antibody-maytansinoid conjugates are activated in targeted cancer cells by lysosomal degradation and linker-dependent intracellular processing. Cancer Res. 2006;66(8):4426–4433. doi: 10.1158/0008-5472.CAN-05-4489. [DOI] [PubMed] [Google Scholar]
- 28.Kato Y, et al. Studies on macrocyclic lactone antibiotics. XIII. Anti-tubulin activity and cytotoxicity of rhizoxin derivatives: Synthesis of a photoaffinity derivative. J Antibiot (Tokyo) 1991;44(1):66–75. doi: 10.7164/antibiotics.44.66. [DOI] [PubMed] [Google Scholar]
- 29.Martínez-Díez M, et al. PM060184, a new tubulin binding agent with potent antitumor activity including P-glycoprotein over-expressing tumors. Biochem Pharmacol. 2014;88(3):291–302. doi: 10.1016/j.bcp.2014.01.026. [DOI] [PubMed] [Google Scholar]
- 30.Ravelli RB, et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428(6979):198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 31.Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96(1):79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- 32.Fourniol FJ, et al. Template-free 13-protofilament microtubule-MAP assembly visualized at 8 A resolution. J Cell Biol. 2010;191(3):463–470. doi: 10.1083/jcb.201007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senter PD, Sievers EL. The discovery and development of brentuximab vedotin for use in relapsed Hodgkin lymphoma and systemic anaplastic large cell lymphoma. Nat Biotechnol. 2012;30(7):631–637. doi: 10.1038/nbt.2289. [DOI] [PubMed] [Google Scholar]
- 34.Andreu JM. Large scale purification of brain tubulin with the modified Weisenberg procedure. Methods Mol Med. 2007;137:17–28. doi: 10.1007/978-1-59745-442-1_2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.