Significance
Cholesterol is the major sterol in blood and in excess causes cardiovascular disease. In addition to cholesterol, numerous other sterols of unknown function and pathogenicity circulate in the bloodstream. Here, we use chemical methods to screen for over 60 different sterols and sterol derivatives in the sera of 3,230 clinically well-characterized individuals. Twenty-seven sterols and two sterol derivatives (vitamin D2 and D3) were routinely detected in vastly different amounts in a majority of individuals. Genes, ethnicity, gender, age, clinical phenotype, and anatomy were identified as significant sources of interindividual variation in these lipid metabolites.
Keywords: human genetics, genotype–phenotype correlation
Abstract
An unknown fraction of the genome participates in the metabolism of sterols and vitamin D, two classes of lipids with diverse physiological and pathophysiological roles. Here, we used mass spectrometry to measure the abundance of >60 sterol and vitamin D derivatives in 3,230 serum samples from a well-phenotyped patient population. Twenty-nine of these lipids were detected in a majority of samples at levels that varied over thousands of fold in different individuals. Pairwise correlations between sterol and vitamin D levels revealed evidence for shared metabolic pathways, additional substrates for known enzymes, and transcriptional regulatory networks. Serum levels of multiple sterols and vitamin D metabolites varied significantly by sex, ethnicity, and age. A genome-wide association study identified 16 loci that were associated with levels of 19 sterols and 25-hydroxylated derivatives of vitamin D (P < 10−7). Resequencing, expression analysis, and biochemical experiments focused on one such locus (CYP39A1), revealed multiple loss-of-function alleles with additive effects on serum levels of the oxysterol, 24S-hydroxycholesterol, a substrate of the encoded enzyme. Body mass index, serum lipid levels, and hematocrit were strong phenotypic correlates of interindividual variation in multiple sterols and vitamin D metabolites. We conclude that correlating population-based analytical measurements with genotype and phenotype provides productive insight into human intermediary metabolism.
Lipids are an important component of serum and there play essential roles in energy metabolism, signaling, and transport. A recent survey revealed an unexpectedly large complexity in the human serum lipidome, which was found to be composed of hundreds of different molecular species in each major lipid class (1). For example, cholesterol and other sterols were detected in concentrations ranging from milligrams per milliliter to nanograms per milliliter, and over 200 different triglyceride species were found. A pooled plasma sample derived from multiple individuals was analyzed in this study; thus, whether the observed complexity reflected functional diversity in the roles played by different lipids, was environmentally driven or was genetically determined could not be ascertained.
To address these issues with respect to sterols and vitamin D metabolites (secosteroids), we developed analytical methods to measure more than 60 different types of these lipids in small volumes (<200 µL) of human serum (2). An initial analysis of 200 human serum samples showed that 22 of the >60 compounds were routinely detected and began to define the ranges and distributions of these analytes in the population (2).
The steady-state concentration of a given lipid is determined by rates of formation and degradation, the kinetics of movement into and out of cells and tissues, the levels of lipoproteins that transport sterols through the bloodstream, their availability in the diet, and the physiological state of the individual. For some sterols, the individual contributions of these variables can be determined from known metabolic pathways, clinical measurements of lipoprotein levels, and nutritional and health information. In other cases, where biosynthetic, catabolic, and transport pathways are unknown, genome-wide association studies can be used to identify loci that are linked to serum sterol levels (3, 4). Subsequent genetic and functional studies then define the role of the product specified by the identified gene (5, 6).
In the current study, we used the analytical methods of McDonald et al. (2) to measure serum sterols in 3,230 individuals from a clinically well-defined cohort, the Dallas Heart Study (DHS) (7). In this large patient population, 27 sterols and vitamin D derivatives were consistently detected and each showed marked interindividual variation in their serum levels. Through further studies, we identified genetic, anatomic, and clinical phenotypes that were associated with many of these lipids.
Results
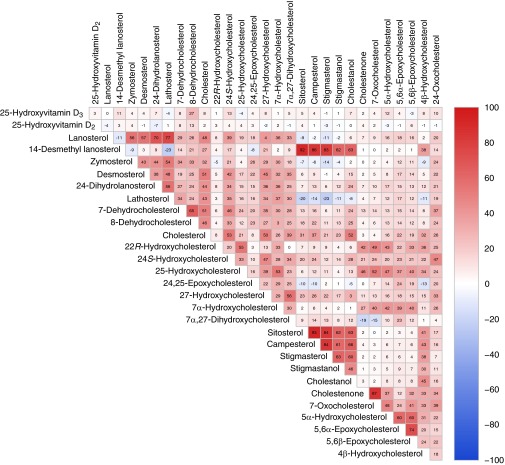
The data of Table 1 show the 27 sterols and vitamin D metabolites that were detected in the DHS (n = 3,230) together with their mean and median concentrations. Interindividual variation in the levels of these analytes ranged from as little as 31-fold (24S-hydroxycholesterol) to as large as 7,760-fold (24-dihydrolanosterol). Fig. 1A shows the raw data generated for one analyte, 24S-hydroxycholesterol. The mean concentration of this oxysterol in the population was 60 ng/mL, and the range was 10–314 ng/mL. Levels of 24S-hydroxycholesterol correlated with those of cholesterol (Fig. 1B, r = 0.53). This association accounted for 31% of the observed variance in interindividual 24S-hydroxycholesterol levels. Most analytes showed log-normal distributions as exemplified by 24S-hydroxycholesterol and the overlaid best-fit curves shown in Fig. 1 C and D. Comparisons between all possible pairs of analytes revealed additional significant correlations between many sterols (Fig. 2). A majority of correlations were positive, and in general these were stronger than the smaller number of negative correlations identified.
Table 1.
Serum sterol and vitamin D levels in 3,230 subjects
| Metabolite | LIPID MAPS ID | Mean, ng/mL | Median, ng/mL | Range, ng/mL |
| 25-Hydroxyvitamin D3 | LMST03020246 | 43 | 39 | 5–160 |
| 25-Hydroxyvitamin D2 | LMST03010030 | 5 | 2 | 0.1–232 |
| Lanosterol | LMST01010017 | 145 | 125 | 13–4,667 |
| 14-Desmethyl lanosterol | LMST01010176 | 552 | 486 | 62–2,671 |
| Zymosterol | LMST01010066 | 41 | 30 | 2–843 |
| Desmosterol | LMST01010016 | 923 | 838 | 22–18,810 |
| 24-Dihydrolanosterol | LMST01010087 | 34 | 17 | 2–15,333 |
| Lathosterol | LMST01010089 | 2,016 | 1,842 | 1–13,850 |
| 7-Dehydrocholesterol | LMST01010069 | 642 | 529 | 7–21,143 |
| 8-Dehydrocholesterol | LMST01010242 | 765 | 667 | 142–11,762 |
| 22R-Hydroxycholesterol | LMST01010086 | 1 | 1 | 0.1–16 |
| 24S-Hydroxycholesterol | LMST01010019 | 60 | 57 | 10–314 |
| 25-Hydroxycholesterol | LMST01010018 | 10 | 8 | 1–56 |
| 24,25-Epoxycholesterol | LMST01010012 | 2 | 1 | 0.1–56 |
| 27-Hydroxycholesterol | LMST01010057 | 158 | 150 | 25–990 |
| 7α-Hydroxycholesterol | LMST01010013 | 114 | 90 | 12–2,762 |
| 7α,27-Dihydroxycholesterol | LMST04030081 | 10 | 10 | 2–90 |
| Sitosterol | LMST01040129 | 2,460 | 2,148 | 308–19,476 |
| Campesterol | LMST01030097 | 3,596 | 3,191 | 141–34,143 |
| Stigmasterol | LMST01040123 | 131 | 116 | 3–10,000 |
| Stigmastanol | LMST01040128 | 23 | 19 | 1–1,138 |
| Cholestanol | LMST01010077 | 2,883 | 2,676 | 431–63,333 |
| Cholestenone | LMST01010015 | 110 | 53 | 4–1,457 |
| 7-Oxocholesterol | LMST01010049 | 55 | 39 | 8–375 |
| 5α-Hydroxycholesterol | LMST01010275 | 37 | 34 | 2–370 |
| 5,6α-Epoxycholesterol | LMST01010011 | 99 | 90 | 9–388 |
| 5,6β-Epoxycholesterol | LMST01010010 | 277 | 253 | 36–1,410 |
| 4β-Hydroxycholesterol | LMST01010014 | 39 | 36 | 9–500 |
| 24-Oxocholesterol | LMST01010133 | 6 | 5 | 0.4–116 |
The vitamin D derivatives and sterols routinely detected in the study population are listed together with their mean and median concentrations and ranges. Information regarding the structure and function of each analyte may be obtained by searching for the individual LIPID MAPS ID number at www.lipidmaps.org/data/structure. Information regarding quality assessment and control in the measurement process is available in ref. 2.
Fig. 1.
Distribution of serum 24S-hydroxycholesterol levels in 3,230 DHS participants. (A) Raw data showing 24S-hydroxycholesterol levels in individual subjects. (B) Correlation between serum cholesterol and 24S-hydroxycholesterol levels in individuals. (C) Distribution of 24S-hydroxycholesterol levels after normalization to cholesterol levels. The red line shows the probability density function for the best-fitting log-normal distribution. (D) Normalized distribution following log transformation of 24S-hydroxycholesterol/cholesterol levels. The red line shows the probability density function for the best-fitting normal distribution.
Fig. 2.
Pairwise correlations between serum levels of analyte. Values are Spearman’s rank correlation coefficients (r) × 100 between the indicated pairs of analytes; r values for individual comparisons are depicted using a bipolar color progression as indicated by the scale on the right of the figure. Values greater than ±4 are statistically different from 0 at a 5% significance level.
Additional factors contributing to interindividual variation in sterol and vitamin D levels were identified by regression analyses. As shown in Fig. 3, sex, ethnicity, and age explained a large fraction of the variation in multiple lipids. For example, three intermediates of bile acid biosynthesis, 27-hydroxycholesterol, 7α,27-dihydroxycholesterol, and 7α-hydroxycholesterol were significantly lower in females than in males, confirming an earlier study (8). An intermediate in the Bloch pathway of cholesterol synthesis, desmosterol, showed similar sexual dimorphism. For these and other lipid species (Fig. 3 and Fig. S1), sex explained as much as 24.7% of interindividual variability (R2).
Fig. 3.

Effects of sex, ethnicity, and age on analytes levels. Box and whisker plots depict median values for the indicated analyte (thick black bars), first-third quartile (interquartile range, box), 5th and 95th percentiles (thin horizontal lines), and outliers beyond this range (x). In sex comparisons: F, female; M, male. In ethnicity comparisons: AA, African American; EA, European American; HIS, Hispanic. R2 values indicate the percentage of variance explained by each of the indicated covariates. P values indicate significance of the observed relationship.
Ethnicity was a significant determinant of variability for a different set of analytes. As noted (9), 25-hydroxyvitamin D3 levels were highest in individuals of European-American descent, intermediate in Hispanics, and lowest in African Americans (Fig. 3). Levels of 8-dehydrocholesterol showed a similar trend across these ethnic groups, whereas IgG showed an opposite trend (i.e., were highest in African Americans). As with sex, ethnicity explained as much as 25% of interindividual variability in these traits (Fig. 3 and Fig. S2).
Age was a weaker but significant determinant of variability, with increasing age correlated with elevated levels of some sterols such as 7-dehydrocholesterol and decreased levels of others such as 24S-hydroxycholesterol (10) and lathosterol (Fig. 3 and Fig. S3). Additional principal-component analyses between analytes showed that, overall, sex was the largest determinant of variation between individuals and that ethnicity was the second largest determinant (Fig. S4).
The observed effects of ancestry on interindividual variation in sterol and secosteroids suggested that genetic factors were an additional effector of analyte levels (11). To identify DNA sequence variations that contributed to variation, the exomes of the 3,230 individuals in the DHS cohort were genotyped for ∼240,000 single-nucleotide polymorphisms (SNPs) by chip-based oligonucleotide hybridization (Illumina HumanExome BeadChip). A majority of SNPs present on the genotyping chip were nonsynonymous sequence variations. Each genetic variant was tested for association with individual analytes as described in Materials and Methods. Variants in 16 different loci located on 10 different chromosomes were associated with one or more of 19 sterols and vitamin D metabolites at exome-wide significance (P = 10−74 to 10−7) (Fig. 4 and Table S1). The levels of some sterols were influenced by variants at several genomic loci (e.g., ABCG5/ABCG8, HSD3B7, and cholestanol), whereas other sterols and secosteroids were associated with a single locus (e.g., EPHX2 and 24,25-epoxycholesterol, and GC and 25-hydroxyvitamin D3).
Fig. 4.

Chromosomal locations of genes significantly linked to individual lipid levels. Schematics of human chromosomes stained with Giemsa are shown together with the locations of genes significantly linked (P ≤ 10−7) to individual sterol and vitamin D metabolite levels, which are color-coded at the bottom of the figure.
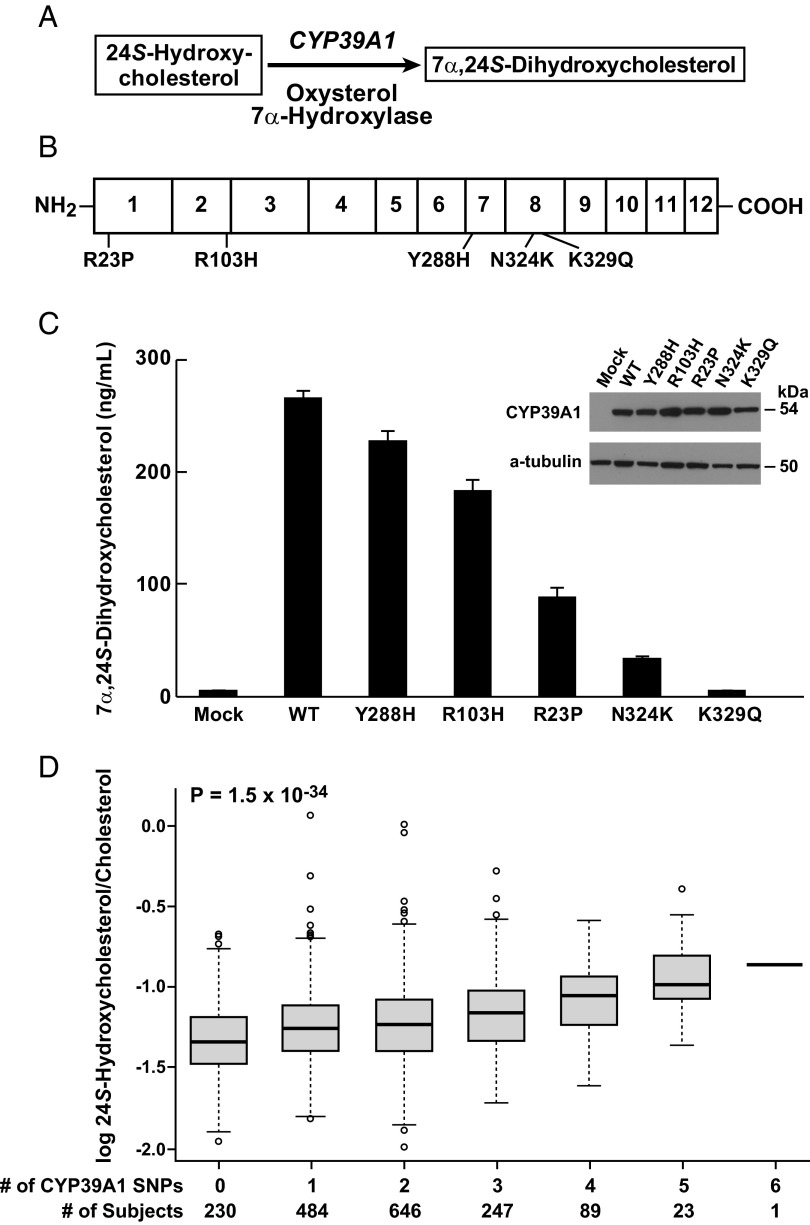
The rs2277119 variant in CYP39A1, which specifies an oxysterol 7α-hydroxylase, was associated with elevated levels of 24S-hydroxycholesterol (P = 10−74) and is a G-to-A transition that alters codon 103 in the gene from arginine to histidine (R103H). Given the reaction catalyzed by the encoded P450 (Fig. 5A), the association of this variant with higher serum levels of the oxysterol implied that R103H was a hypomorphic allele of CYP39A1.
Fig. 5.
Expression analysis of genetic variants linked to high serum 24S-hydroxycholesterol levels. (A) Biochemical reaction catalyzed by the CYP39A1 oxysterol 7α-hydroxylase. (B) Schematic of the 500-aa CYP39A1 protein showing the sequences and locations of the five variants (rs12192544, R23P; rs2277119, R103H; rs17856332, Y288H; rs7761731, N324K; rs41273654, K329Q) identified in individuals with high levels of 24S-hydroxycholesterol. (C) Expression analysis of normal and variant enzymes. Plasmids expressing normal (N) and the indicated variant CYP39A1 enzymes were transfected into cultured HEK 293 cells and assayed for oxysterol 7α-hydroxylase activity. Inset shows levels of CYP39A1 protein in transfected cells as determined by immunoblotting; α-tubulin served as a loading control. (D) Cumulative effects of multiple SNPs on serum levels of 24S-hydroxycholesterol. Log-normalized serum levels of the oxysterol (y axis) are indicated by whisker plots showing median values (thick black bars), first-third quartile (interquartile range, box), 5th and 95th percentiles (thin horizontal lines), and outliers beyond this range (open circles), and are plotted versus the total number of CYP39A1 SNPs present in an individual (x axis). The number of subjects who inherited a given number of SNPs is indicated at the bottom of the plot.
To determine whether there were other CYP39A1 variants associated with serum 24S-hydroxycholesterol levels, the 12 exons of this gene were sequenced in the 30 DHS individuals with the highest normalized 24S-hydroxycholesterol levels (Fig. 1D). This resequencing revealed four additional nonsynonymous sequence variants: R23P, T288H, N324K, and K329Q (Fig. 5B and Table S2). The biochemical effects of these alterations and the R103H variant were determined in transfection experiments. Fig. 5C shows averaged results from three separate experiments in which enzyme activity encoded by each variant was assayed in triplicate dishes. Relative to the normal enzyme, all sequence variants reduced enzyme activity from ∼15% (rs17856332; Y288H) to 100% (rs41273654; K329Q), but did not have an obvious effect on CYP39A1 mRNA or protein expression as judged by real-time PCR or immunoblotting (Fig. 5C, Inset), indicating that these amino acid changes directly reduced enzyme activity.
To determine the association of these variants with serum 24S-hydroxycholesterol levels, the DHS population was genotyped for the four CYP39A1 alleles identified by resequencing. Three of the five alleles were independently associated with an increase in serum 24S-hydroxycholesterol levels in the combined DHS cohort (African-American, European-American, and Hispanic participants), with two variants (rs2277119; R103H; and rs7761731; N324K) making significantly larger contributions than the third (Table S2). The variants appeared to act additively, with no evidence of a statistical interaction detected between alleles (P for pairwise interactions, >0.05). As indicated in Fig. 5D, individuals with one or more variant CYP39A1 alleles had progressively higher serum 24S-hydroxycholesterol levels compared with those with no variant alleles (P for trend, 1.5 × 10−34). Together, the CYP39A1 alleles explained ∼10.8% of the interindividual variation observed for serum 24S-hydroxycholesterol levels.
Earlier studies in a small number of subjects with cognitive impairment revealed a modestly significant association (P = 0.03) between gray matter volume and serum 24S-hydroxycholesterol levels (12), and based on indirect measurements, a similar relationship was detected between the size of the brain and the capacity of the liver to metabolize the oxysterol (13). These findings suggested that 24S-hydroxycholesterol was synthesized in gray matter neurons, a hypothesis subsequently confirmed by histochemical studies (14). To determine whether brain anatomy influenced serum 24S-hydroxycholesterol levels, MRI was used to measure total brain, gray matter, and white matter volumes in 2,109 DHS participants. These analyses were adjusted for age, race, sex, and cholesterol levels. Male and female values were separated to control for the known sexual dimorphism in brain size (15). Total brain volume was modestly but significantly correlated with oxysterol levels in men and women (r = 0.21 in men, r = 0.16 in women; P = 5.5 × 10−5 and 2.5 × 10−4, respectively). Gray matter volume in both females and males was more strongly correlated with serum 24S-hydroxycholesterol levels, whereas the association with white matter volume was weaker than that for gray matter but statistically significant (Fig. S5). Gray matter volumes explained 1.75% of variance in serum 24S-hydroxycholesterol levels after adjustment for white matter volume. White matter was not significantly associated with this trait after accounting for gray matter volume. Total brain volume explained the same amount of variance (1.75%) as did gray matter volume. Thus, gray matter volume is directly related to serum 24S-hydroxycholesterol levels, whereas white matter is only indirectly associated through correlation to gray matter volume.
Additional phenotypic determinants of interindividual variation in other sterols and vitamin D metabolites were identified by correlating further clinical measurements in each participant with individual serum lipid levels. Using an arbitrary cutoff value of greater than or equal to ±10 to simplify presentation of these data, positive and negative correlations were found between many clinical parameters and different classes of lipids (Fig. 6). Strong positive correlations existed within lipid classes, such as those between sterol synthesis intermediates and total serum cholesterol and lipoprotein levels. Comparisons between classes revealed shared patterns of positive and negative correlations as exemplified by those between multiple clinical parameters and 14-desmethyl lanosterol, 4β-hydroxycholesterol, and five plant sterols (Fig. 6).
Fig. 6.
Pairwise correlations between serum levels of analytes and clinical phenotypes. Values are partial correlation coefficients × 100 after adjustment for age, sex, and ethnicity between the indicated pairs of analytes and clinical parameters; r values for individual comparisons are depicted using a bipolar color progression as indicated by the scale below the figure. Only traits for which at least one r value was greater than or equal to ±10 are shown.
Discussion
In the current study, we used mass spectrometry to quantify vitamin D metabolite and sterol levels in sera from 3,230 unselected subjects and then correlated interindividual variation in these lipids with genotype and phenotype. Screening for >60 molecular species identified 29 that were consistently present at widely varying levels in a majority of individuals. Variation in specific lipids correlated with disparities in serum cholesterol levels, ethnicity, sex, age, genetic variation, anatomy, and clinical phenotypes.
For some analytes, such as 24S-hydroxycholesterol, the observed correlations were consistent with known metabolic pathways (Fig. 5). This oxysterol is synthesized in the brain by a neuronal enzyme (cholesterol 24-hydroxylase, CYP46A1) and thereafter secreted into the circulation where it associates with circulating lipoproteins. Synthesis is required for normal brain function (16, 17), and once synthesized, 24S-hydroxycholesterol is a potential ligand for the liver X receptor and a substrate for CYP39A1 and hepatic bile acid synthesis (18–20).
Correlations between individual levels of different lipids revealed shared metabolic pathways (Fig. 2). Most correlations were positive, and these were usually stronger than the smaller number of negative correlations detected. Common origins may explain many positive correlations such as those between most sterols and cholesterol, reflecting cotransport in serum lipoprotein particles (21), and those between the plant sterols sitosterol, campesterol, stigmasterol, and stigmastanol, reflecting a shared dietary origin and their absorption and excretion via ABCG5/ABCG8 (22). Unexpectedly, serum levels of a cholesterol biosynthetic intermediate, 14-desmethyl lanosterol, and the ring-structure oxysterol 4β-hydroxycholesterol also correlated significantly (r > 0.38) with plant sterols (Fig. 2). These findings suggested serum 14-desmethyl lanosterol and 4β-hydroxycholesterol may derive from the diet and/or that these sterols are ABCG5/ABCG8 substrates. A unique origin for these two sterols was also suggested by the negative or weak positive correlations between 14-desmethyl lanosterol and other intermediates in the cholesterol biosynthetic pathways such as lathosterol and lanosterol, and by weaker correlations between 4β-hydroxycholesterol and other ring-structure oxysterols such as 7α-hydroxycholesterol (Fig. 2).
A common origin related to formation by spontaneous oxidation may explain the positive associations between cholestenone, 7-oxocholesterol, 5α-hydroxycholesterol, and the 5,6-epoxycholesterols (Fig. 2) (23), as enzymatic pathways for the formation of these sterols have not been defined. Similarly, positive correlations between these sterols and 22R-hydroxycholesterol and 25-hydroxycholesterol suggest that some amount of these two oxysterols reflects formation by spontaneous as opposed to enzymatic oxidation (24).
Precursor–product relationships explained several positive correlations, such as that between 27-hydroxycholesterol and 7α,27-dihydroxycholesterol, which are sequential intermediates in the alternate pathway of bile acid synthesis (25), and that between 7-dehydrocholesterol and 8-dehydrocholesterol. Similarly, sterol intermediates that are unique to the Bloch pathway of cholesterol biosynthesis (lanosterol, zymosterol, and desmosterol) were positively correlated as were intermediates in the Kandutsch–Russell pathway (lanosterol, 24,25-dihydrolanosterol, and lathosterol). These compounds are indices of whole-body cholesterol synthesis (26), and levels of pathway-specific sterols may thus indicate relative outputs from Bloch versus Kandutsch–Russell pathways; however, positive associations were also detected between intermediates unique to each cholesterol biosynthetic pathway (e.g., between zymosterol and lathosterol). These associations may reflect crossover of intermediates between pathways (27), or regulation of pathway genes by sterol regulatory element binding protein (SREBP) transcription factors (28).
Exome-wide association studies revealed significant associations between levels of sterols and variants in genes encoding enzymes or proteins that are known to synthesize, metabolize, or transport the analytes to which they were linked (Fig. 4 and Table S1). For example, a variant in CYP27A1 (rs114768494), which specifies sterol 27-hydroxylase (29), was significantly associated (P = 6.9 × 10−20) with decreased serum 27-hydroxycholesterol levels. Multiple variants in CYP39A1, which encodes an oxysterol 7α-hydroxylase (20), were strongly associated with increased levels of the oxysterol 24S-hydroxycholesterol (Fig. 5). A variant (rs751141) of EPHX2, which encodes a soluble epoxide hydrolase (30), was robustly associated (P = 7.5 × 10−39) with elevated serum levels of 24,25-epoxycholesterol. Higher levels of an intermediate in the classic pathway of bile acid synthesis, 7α-hydroxycholesterol, and those of an intermediate in the alternate pathway, 7α,27-dihydroxycholesterol, were associated (P = 1.4 × 10−21 and P = 1.7 × 10−40, respectively) with the same variant (rs34212827) of HSD3B7, which encodes an enzyme that catalyzes an essential step in both pathways (31). Based on these findings, other strong genetic associations shown in Fig. 4 may identify substrates of enzymes specified by variant alleles, including 8-dehydrocholesterol and SDR42E1, which encodes a short chain dehydrogenase/reductase, and 7-dehydrocholesterol and 24-oxocholesterol with CYP39A1.
With respect to transport, elevated levels of multiple plant sterols were strongly associated with variants in ABCG5/ABCG8 (Fig. 4 and Table S1), confirming prior studies indicating the encoded heterodimeric protein transports this class of sterols across hepatocyte and enterocyte membranes and that mutations in these genes underlie the genetic disease sitosterolemia in which plant sterols accumulate to pathologic levels (4, 22). Levels of 14-desmethyl lanosterol were associated with these same ABCG5/ABCG8 variants, which confirmed the correlation between plant sterols and 14-desmethyl lanosterol (Fig. 2), and suggested that the latter sterol was an ABCG5/ABCG8 substrate. As in earlier studies (32), levels of 25-hydroxyvitamin D3 were significantly associated with the serum vitamin D transport gene (GC) on chromosome 4.
Shared transport mechanisms may also underlie the strong positive correlations between multiple clinical phenotypes and interindividual variation in different sterols (Fig. 6). Most serum sterols are associated with circulating lipoprotein particles (LDL, very low-density lipoprotein, HDL) leading to strong correlations between these lipids and cholesterol and triglyceride levels (8, 21, 22). Similarly, the positive correlations between some sterols and hematocrit most likely represent the association and transport of these analytes within reticulocyte membranes. Sterols are spontaneously transferred from donor membranes to red blood cells (33), and in the mouse this movement may account for a substantial portion of reverse cholesterol transport (34, 35), the movement of cholesterol and presumably other sterols from peripheral tissues to the liver. This pathway may underlie the correlation (r = 0.4; Fig. 6) between 27-hydroxycholesterol and hematocrit in that a majority of this oxysterol in serum is formed in the lung (36), a tissue in which reticulocyte–cell membrane interactions are frequent, and is thereafter converted to bile acids in liver. Plant sterol, 4β-hydroxycholesterol, and 14-desmethylanosterol levels did not correlate with hematocrit, suggesting that a shared origin, physiochemistry, or biology excludes these sterols from association with circulating cells.
The experimental approaches taken here identify numerous relationships between serum levels of vitamin D metabolites and sterols, genes, and clinical phenotypes, and in the case of the oxysterol 24S-hydroxycholesterol and CYP39A1, identify biochemical and anatomical bases for the observed relationship. A key aspect of this study is the availability of a well-phenotyped population cohort that allows useful information to be derived from static measurements of serum analytes. A limitation is that this is an observational/cross-sectional study, and therefore we cannot draw causal conclusions, only detect associations between clinical phenotypes and the analytes measured. Nevertheless, additional studies in this population may allow the definition of mechanisms underlying observed correlations and to determine whether analyte levels are therapeutically informative.
Materials and Methods
Materials and methods are described at length in SI Materials and Methods. This description includes patient population, analytical chemistry, statistical analyses, genotyping, and biochemical and molecular biology assays, together with references. Analytical standards were from Avanti Polar Lipids (Alabaster, AL).
Supplementary Material
Acknowledgments
We thank Jonathan Cohen, Helen Hobbs, and Jay Horton for critically reading the manuscript. This research was supported by grants awarded to D.W.R. from the National Institutes of Health (5U54GM069338 and 2P01HL20948) and the Clayton Foundation for Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413561111/-/DCSupplemental.
References
- 1.Quehenberger O, et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010;51(11):3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDonald JG, Smith DD, Stiles AR, Russell DW. A comprehensive method for extraction and quantitative analysis of sterols and secosteroids from human plasma. J Lipid Res. 2012;53(7):1399–1409. doi: 10.1194/jlr.D022285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia CK, et al. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science. 2001;292(5520):1394–1398. doi: 10.1126/science.1060458. [DOI] [PubMed] [Google Scholar]
- 4.Berge KE, et al. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 2000;290(5497):1771–1775. doi: 10.1126/science.290.5497.1771. [DOI] [PubMed] [Google Scholar]
- 5.He G, et al. ARH is a modular adaptor protein that interacts with the LDL receptor, clathrin, and AP-2. J Biol Chem. 2002;277(46):44044–44049. doi: 10.1074/jbc.M208539200. [DOI] [PubMed] [Google Scholar]
- 6.Yu L, et al. Overexpression of ABCG5 and ABCG8 promotes biliary cholesterol secretion and reduces fractional absorption of dietary cholesterol. J Clin Invest. 2002;110(5):671–680. doi: 10.1172/JCI16001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Victor RG, et al. Dallas Heart Study Investigators The Dallas Heart Study: A population-based probability sample for the multidisciplinary study of ethnic differences in cardiovascular health. Am J Cardiol. 2004;93(12):1473–1480. doi: 10.1016/j.amjcard.2004.02.058. [DOI] [PubMed] [Google Scholar]
- 8.Burkard I, von Eckardstein A, Waeber G, Vollenweider P, Rentsch KM. Lipoprotein distribution and biological variation of 24S- and 27-hydroxycholesterol in healthy volunteers. Atherosclerosis. 2007;194(1):71–78. doi: 10.1016/j.atherosclerosis.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 9.Looker AC, Dawson-Hughes B, Calvo MS, Gunter EW, Sahyoun NR. Serum 25-hydroxyvitamin D status of adolescents and adults in two seasonal subpopulations from NHANES III. Bone. 2002;30(5):771–777. doi: 10.1016/s8756-3282(02)00692-0. [DOI] [PubMed] [Google Scholar]
- 10.Lütjohann D, et al. Cholesterol homeostasis in human brain: Evidence for an age-dependent flux of 24S-hydroxycholesterol from the brain into the circulation. Proc Natl Acad Sci USA. 1996;93(18):9799–9804. doi: 10.1073/pnas.93.18.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romeo S, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40(12):1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solomon A, et al. Plasma levels of 24S-hydroxycholesterol reflect brain volumes in patients without objective cognitive impairment but not in those with Alzheimer’s disease. Neurosci Lett. 2009;462(1):89–93. doi: 10.1016/j.neulet.2009.06.073. [DOI] [PubMed] [Google Scholar]
- 13.Bretillon L, et al. Plasma levels of 24S-hydroxycholesterol reflect the balance between cerebral production and hepatic metabolism and are inversely related to body surface. J Lipid Res. 2000;41(5):840–845. [PubMed] [Google Scholar]
- 14.Ramirez DMO, Andersson S, Russell DW. Neuronal expression and subcellular localization of cholesterol 24-hydroxylase in the mouse brain. J Comp Neurol. 2008;507(5):1676–1693. doi: 10.1002/cne.21605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gur RC, et al. Gender differences in age effect on brain atrophy measured by magnetic resonance imaging. Proc Natl Acad Sci USA. 1991;88(7):2845–2849. doi: 10.1073/pnas.88.7.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotti TJ, Ramirez DM, Pfeiffer BE, Huber KM, Russell DW. Brain cholesterol turnover required for geranylgeraniol production and learning in mice. Proc Natl Acad Sci USA. 2006;103(10):3869–3874. doi: 10.1073/pnas.0600316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maioli S, et al. Is it possible to improve memory function by upregulation of the cholesterol 24S-hydroxylase (CYP46A1) in the brain? PLoS One. 2013;8(7):e68534. doi: 10.1371/journal.pone.0068534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXR α. Nature. 1996;383(6602):728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 19.Bjorkhem I, et al. From brain to bile. Evidence that conjugation and ω-hydroxylation are important for elimination of 24S-hydroxycholesterol (cerebrosterol) in humans. J Biol Chem. 2001;276(40):37004–37010. doi: 10.1074/jbc.M103828200. [DOI] [PubMed] [Google Scholar]
- 20.Li-Hawkins J, Lund EG, Bronson AD, Russell DW. Expression cloning of an oxysterol 7α-hydroxylase selective for 24-hydroxycholesterol. J Biol Chem. 2000;275(22):16543–16549. doi: 10.1074/jbc.M001810200. [DOI] [PubMed] [Google Scholar]
- 21.Babiker A, Diczfalusy U. Transport of side-chain oxidized oxysterols in the human circulation. Biochim Biophys Acta. 1998;1392(2-3):333–339. doi: 10.1016/s0005-2760(98)00047-2. [DOI] [PubMed] [Google Scholar]
- 22.Berge KE, et al. Heritability of plasma noncholesterol sterols and relationship to DNA sequence polymorphism in ABCG5 and ABCG8. J Lipid Res. 2002;43(3):486–494. [PubMed] [Google Scholar]
- 23.Breuer O, Björkhem I. Simultaneous quantification of several cholesterol autoxidation and monohydroxylation products by isotope-dilution mass spectrometry. Steroids. 1990;55(4):185–192. doi: 10.1016/0039-128x(90)90109-o. [DOI] [PubMed] [Google Scholar]
- 24.Dzeletovic S, Breuer O, Lund E, Diczfalusy U. Determination of cholesterol oxidation products in human plasma by isotope dilution-mass spectrometry. Anal Biochem. 1995;225(1):73–80. doi: 10.1006/abio.1995.1110. [DOI] [PubMed] [Google Scholar]
- 25.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 26.Björkhem I, et al. Correlation between serum levels of some cholesterol precursors and activity of HMG-CoA reductase in human liver. J Lipid Res. 1987;28(10):1137–1143. [PubMed] [Google Scholar]
- 27.Ačimovič J, Rozman D. Steroidal triterpenes of cholesterol synthesis. Molecules. 2013;18(4):4002–4017. doi: 10.3390/molecules18044002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horton JD, Goldstein JL, Brown MS. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109(9):1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cali JJ, Russell DW. Characterization of human sterol 27-hydroxylase. A mitochondrial cytochrome P-450 that catalyzes multiple oxidation reaction in bile acid biosynthesis. J Biol Chem. 1991;266(12):7774–7778. [PubMed] [Google Scholar]
- 30.Newman JW, Morisseau C, Hammock BD. Epoxide hydrolases: Their roles and interactions with lipid metabolism. Prog Lipid Res. 2005;44(1):1–51. doi: 10.1016/j.plipres.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Schwarz M, et al. The bile acid synthetic gene 3β-hydroxy-Δ5-C27-steroid oxidoreductase is mutated in progressive intrahepatic cholestasis. J Clin Invest. 2000;106:1175–1184. doi: 10.1172/JCI10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGrath JJ, Saha S, Burne TH, Eyles DW. A systematic review of the association between common single nucleotide polymorphisms and 25-hydroxyvitamin D concentrations. J Steroid Biochem Mol Biol. 2010;121(1-2):471–477. doi: 10.1016/j.jsbmb.2010.03.073. [DOI] [PubMed] [Google Scholar]
- 33.Steck TL, Kezdy FJ, Lange Y. An activation-collision mechanism for cholesterol transfer between membranes. J Biol Chem. 1988;263(26):13023–13031. [PubMed] [Google Scholar]
- 34.Xie C, Turley SD, Dietschy JM. ABCA1 plays no role in the centripetal movement of cholesterol from peripheral tissues to the liver and intestine in the mouse. J Lipid Res. 2009;50(7):1316–1329. doi: 10.1194/jlr.M900024-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hung KT, Berisha SZ, Ritchey BM, Santore J, Smith JD. Red blood cells play a role in reverse cholesterol transport. Arterioscler Thromb Vasc Biol. 2012;32(6):1460–1465. doi: 10.1161/ATVBAHA.112.248971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Babiker A, et al. Elimination of cholesterol as cholestenoic acid in human lung by sterol 27-hydroxylase: Evidence that most of this steroid in the circulation is of pulmonary origin. J Lipid Res. 1999;40(8):1417–1425. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.