Significance
Methylation of the N6 position of selected internal adenines (m6A) in mRNAs and noncoding RNAs is widespread in eukaryotes, and the YTH domain in a collection of proteins recognizes this modification. We report the crystal structure of the splicing factor YT521-B homology (YTH) domain of Zygosaccharomyces rouxii methylated RNA-binding protein 1 in complex with a heptaribonucleotide with an m6A residue in the center. The m6A modification is recognized by an aromatic cage, and there are also interactions with other regions of the RNA. Mutations in the RNA binding site can abolish the formation of the complex. Overall, our structural and biochemical studies have defined the molecular basis for how the YTH domain functions as a reader of methylated adenines.
Abstract
Methylation of the N6 position of selected internal adenines (m6A) in mRNAs and noncoding RNAs is widespread in eukaryotes, and the YTH domain in a collection of proteins recognizes this modification. We report the crystal structure of the splicing factor YT521-B homology (YTH) domain of Zygosaccharomyces rouxii MRB1 in complex with a heptaribonucleotide with an m6A residue in the center. The m6A modification is recognized by an aromatic cage, being sandwiched between a Trp and Tyr residue and with the methyl group pointed toward another Trp residue. Mutations of YTH domain residues in the RNA binding site can abolish the formation of the complex, confirming the structural observations. These residues are conserved in the human YTH proteins that also bind m6A RNA, suggesting a conserved mode of recognition. Overall, our structural and biochemical studies have defined the molecular basis for how the YTH domain functions as a reader of methylated adenines.
The methylation of the N6 position of selected internal adenines (m6A) modification is widespread in eukaryotic mRNAs and noncoding RNAs (1–3), reviewed in refs. 4–6. Recent studies have linked this modification to the regulation of alternative splicing (2), RNA processing, and mRNA degradation (7). Although the exact cellular functions of this modification are still not completely understood, m6A has been linked to the regulation of the circadian clock (8) and m6A levels are highest during yeast meiosis (1). The m6A methyl group can be removed by the dioxygenases FTO (9) and ALKBH5 (10, 11), suggesting that m6A is a reversible modification on the RNA.
A consensus sequence G(m6A)C has been identified for this modification based on transcriptome-wide mapping (1, 2), which is consistent with that identified from earlier biochemical studies (4–6). Several YTH domain (12) family members, YTHDF1, YTHDF2, and YTHDF3 in humans (2, 7) and methylated RNA-binding protein 1 (MRB1) in yeast (1), have been shown to bind RNAs with m6A modification, and the binding consensus for the YTH domain of YTHDF2 is also G(m6A)C (7), consistent with that found by transcriptome-wide mapping.
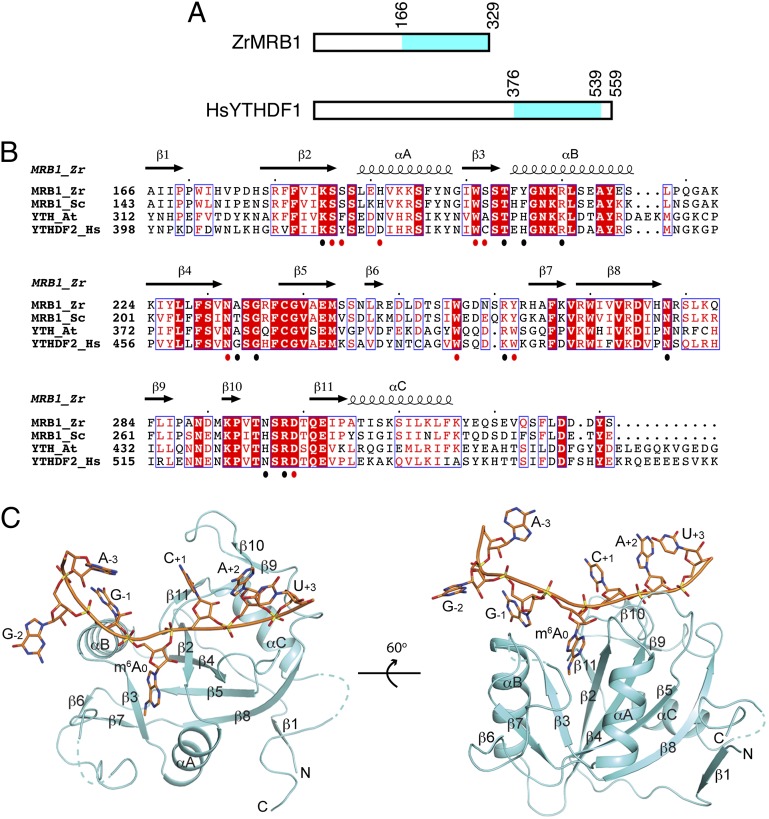
The YTH domain contains ∼160 residues and is found in yeast, plants, and animals (Fig. S1) (12, 13). The domain is located at the C-terminal end of yeast MRB1 and human YTHDF1-3 (Fig. 1A), and its sequence is well conserved among these proteins (Fig. 1B and Fig. S1). The N-terminal regions of these proteins are poorly conserved, although that of YTHDF2 mediates its function in regulating mRNA localization and degradation (7). Yeast MRB1 regulates phosphate metabolism by destabilizing the mRNA of a transcription factor of the pathway and, hence, it is also known as Pho92 (14), although direct evidence of MRB1 regulating m6A-containing mRNAs in yeast cells is lacking.
Fig. 1.
Crystal structure of the YTH domain of Z. rouxii MRB1 in complex with an m6A RNA. (A) Domain organization of ZrMRB1 and human YTHDF1. The YTH domain is shown in cyan. (B) Sequence alignment of the YTH domains of Z. rouxii (Zr) and S. cerevisiae (Sc) MRB1, A. thaliana (At) YTH protein, and human (Hs) YTHDF2. The secondary structure elements in the structure of the ZrMRB1 YTH domain are indicated. Residues that contact the m6A residue are indicated with the red dots below the alignment, and those that contact the rest of the RNA are indicated with the black dots. Produced with ESPript (25). (C) Two views of the structure of the YTH domain of ZrMRB1 (in cyan) in complex with a 7-mer m6A RNA (in orange). The secondary structure elements in the YTH domain are labeled. The structure figures were produced with PyMOL (www.pymol.org).
The structures of the YTH domains of two related proteins, human YTH domain containing protein 1 [YTHDC1; Protein Data Bank (PDB) ID code 2YUD] and YTHDC2 (2YU6), have been reported. Human YTHDC1 has 29% sequence identity to human YTHDF1 for the YTH domain (Fig. S1). YTHDC1 binds a degenerate unmethylated RNA sequence (13), which does not have similarity to the G(m6A)C consensus. The interaction between its YTH domain and an unmethylated RNA was studied by chemical-shift perturbation (13), but the structure of a complex is not available. The molecular mechanism for how the YTH domain recognizes the m6A modification is not known.
Results and Discussion
We have determined the crystal structure at 2.7 Å resolution of the YTH domain of Z. rouxii MRB1 (ZrMRB1), a close homolog of Saccharomyces cerevisiae MRB1 (Fig. 1B), in complex with a 7-mer oligoribonucleotide with the sequence A–3G–2G–1(m6A)0C+1A+2U+3. The atomic model has good agreement with the X-ray diffraction data and the expected geometric parameters (Table S1). Roughly 96.5% of the residues are in the favored region of the Ramachandran plot, 3.5% in the allowed region, and no residues in the outlier region.
There are six copies of the YTH–RNA complex in the asymmetric unit. Interestingly, the six RNA molecules form three unusual, parallel dimers through extensive base-stacking, but no base-pairing, interactions (Fig. S2). The three dimers then associate into a hexamer, primarily through base stacking of the G–2 nucleotides of neighboring molecules. The m6A base is not involved in the formation of this dimer or hexamer. This hexameric assembly of the RNA molecules is, in turn, flanked by three YTH domains on each face (Fig. S2). The YTH–RNA complex has 1:1 stoichiometry in solution based on our gel filtration data and, therefore, the 6:6 complex is likely formed during crystallization. Nonetheless, the unusual assembly mechanism of this dimer and hexamer could have relevance for RNA structures in general.
The structure of the YTH domain of ZrMRB1 has a central, six-stranded β-sheet (Fig. 1C), although the strand at each edge of the sheet makes only two hydrogen bonds with the neighboring strand. There are also two smaller β-sheets, one with three strands and the other with two. Three helices cover some of the surfaces of these β-sheets. The overall structures of the six YTH domains in the crystal asymmetric unit are similar, with rms distance of ∼0.3 Å among equivalent Cα atoms of any pair of the YTH domains (Fig. S3). Variations in the conformations of several side chains on the surface of the structure are observed. There are also conformational differences for several side chains in the RNA binding site (see below). The overall structures of the six RNA molecules are also similar to each other except for the last nucleotide (Fig. S3), which has weaker electron density in some of the molecules. Two different conformations are observed for the 5′ phosphate group of G–2, and, in fact, several of them assume both conformations.
The closest structural homologs of ZrMRB1 are the YTH domains of human YTHDC1 and YTHDC2, with rms distance of ∼2 Å and sequence identity of ∼32% among structurally equivalent residues (Fig. S4). Other structural homologs, as identified in an earlier study based on YTHDC1 (13), include the DUF55 domain of human thymocyte nuclear protein 1 (15) and the EVE domain that is found in a collection of prokaryotic proteins (16) such as Pyrococcus horikoshii protein PH1033 (17), Agrobacterium tumefaciens Atu2648 (16), and a Leishmania major protein (18) (Fig. S4), with Z scores between 9 and 13 from the program DaliLite (19) and sequence identities between 10 and 18%. However, the functions of most of these proteins are not known.
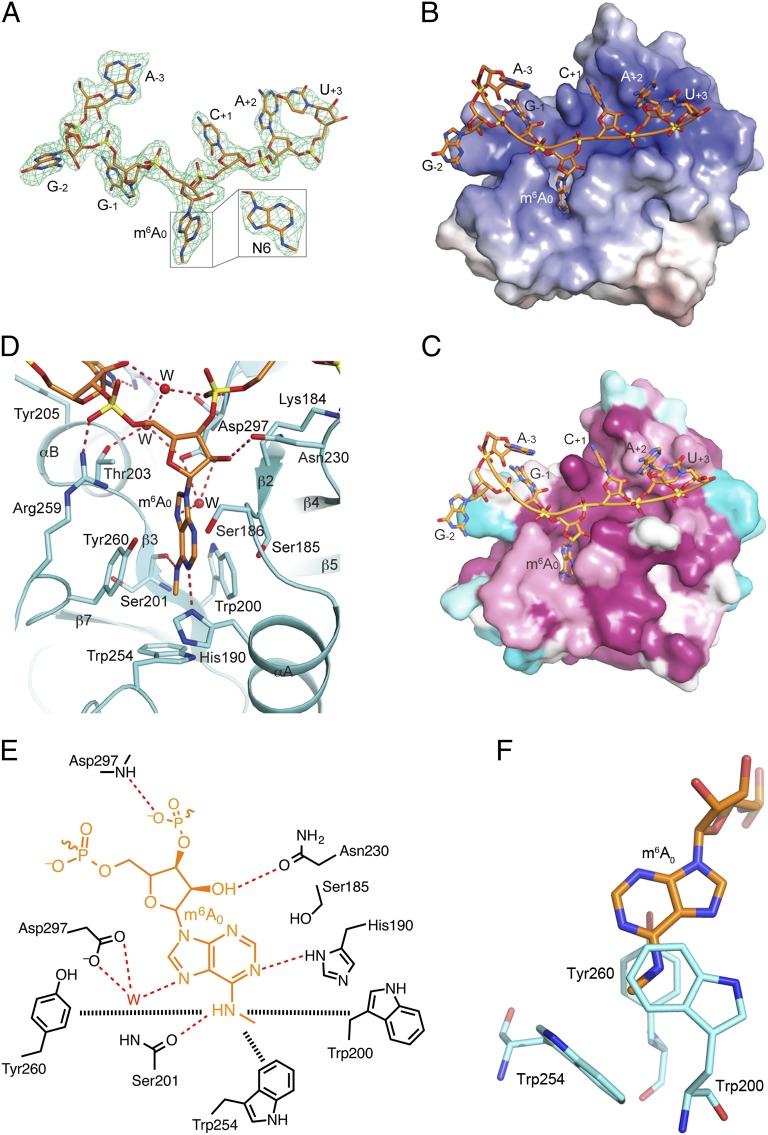
Clear electron density is observed for most of the m6A heptanucleotide (Fig. 2A). The RNA is positioned across the top of the central β-sheet of the YTH domain (Fig. 1C), and the m6A base is inserted into a deep pocket in the structure (Fig. 2B), providing the anchoring contacts with the protein. Residues in the RNA binding site are highly conserved among YTHDF1-3 and the MRB1 proteins, especially those involved in m6A binding (Figs. 1B and 2C). The 6-methylamino group is recognized by an aromatic cage, being sandwiched between the side chains of Trp200 and Tyr260, and the methyl group is pointed toward the side chain of Trp254 (Fig. 2 D and E). Interestingly, the Trp200 and Tyr260 side chains flank the methylated N6 rather than the adenine base (Fig. 2F), indicating that this aromatic cage is organized to recognize the methylated base. The remaining hydrogen atom on the N6 amino group is hydrogen bonded to the main-chain carbonyl oxygen of Ser201, thereby precluding the binding of doubly methylated adenine. The N1 nitrogen atom in the adenine ring is hydrogen bonded to the side chain of His190, and the N7 atom interacts with the side chain of Asp297 through a well-ordered water molecule, which is present in all six copies of the complex in the asymmetric unit. Ser185 is positioned against one face of the adenine base, and the Ser186 side chain is near the N3 atom of the base. Overall, the structure indicates that this YTH domain produces a well-defined pocket that recognizes the m6A base.
Fig. 2.
Recognition of the m6A RNA by the YTH domain. (A) Omit Fo–Fc electron density for the RNA at 2.7 Å resolution, contoured at 2.5σ. (B) Electrostatic surface of the ZrMRB1 YTH domain in the region of m6A RNA binding. The RNA is located in a positively charged surface patch (blue) in the protein. (C) Sequence conservation of residues in the RNA binding site, generated based on an alignment of 50 sequences by the program ConSurf (26). Purple, conserved residues; cyan, variable residues. (D) Detailed interactions between the m6A nucleotide and the YTH domain. Water molecules are shown as red spheres. Hydrogen-bonding interactions are indicated by dashed lines in red. (E) Schematic drawing of the interactions between the m6A nucleotide and the YTH domain. The interactions between the m6A modification and the aromatic cage are indicated by the dashed lines in black. (F) Trp200 and Tyr260 flank the methylated N6 rather than the adenine base.
Interestingly, the aromatic cage observed here for the m6A modification has similarity to that seen in chromo and tudor domains for recognizing methylated lysine and arginine residues (20, 21), despite the fact that the YTH domain shares no similarity in backbone fold with these other proteins. By manually superposing the methyl-lysine and its aromatic cage in the structures of polycomb chromo domain (22), 53BP tudor domain (23), and JMJD2A tudor domain (24) with the pocket in the YTH domain, the three aromatic residues in each cage are placed at roughly the same position (Fig. S5). This structural similarity demonstrates the remarkable conservation in the recognition of a methylated amino group.
However, this aromatic cage is not well conserved in the other structural homologs of the YTH domain (Fig. S4). Although the EVE domain in PH1033 appears to have a complete aromatic cage, Tyr260 is replaced by a Leu residue in YTHDC1 and YTHDC2, and only Trp200 is conserved in most of the other homologs. Therefore, these proteins may not be able to recognize m6A with good affinity. This is supported by our mutagenesis studies (see below) and the fact that YTHDC1 failed to complement the function of MRB1 in yeast, whereas YTHDF2 was able to complement (14).
The overall conformations of the three residues in the aromatic cage of MRB1 are similar to the residues in YTHDC1 and YTHDC2 (Fig. S4), which are in the absence of any bound RNA. This structural similarity suggests that the aromatic cage in MRB1 may be preformed and has a similar conformation in the absence of m6A binding. Our attempts at crystallizing free MRB1 YTH domain have not been successful.
The 2′-OH of the m6A ribose has hydrogen-bonding interactions with Asn230 (Fig. 2D). The 5′ phosphate group of the m6A residue is located ∼6 Å from the N-terminal end of helix αB and may have some favorable interactions with the dipole of this helix. In one of the six complexes in the crystal, the side chain of Arg259 has ionic interactions with this phosphate group. In the other five complexes, this side chain or the entire residue is disordered (Fig. S3).
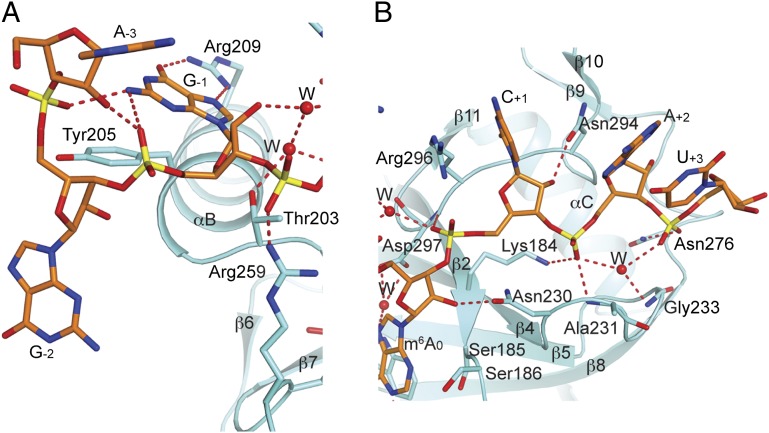
The base of the G–1 nucleotide is recognized by bidentate hydrogen-bonding interactions with the side chain of Arg209, consistent with the consensus for a G at this position (Fig. 3A). The base is also π-stacked with the side chain of Tyr205. It is in the syn conformation, and the 2-amino group of guanine has hydrogen-bonding interactions with 5′ phosphate groups of the –1 and –2 residues. However, nucleotides A–3 and G–2 have little direct contacts with the YTH domain. This conformation might be due to the formation of the RNA dimer (Fig. S2), and these two nucleotides might contact the protein in a 1:1 complex. The A–3 base is also in the syn conformation.
Fig. 3.
Interactions between the other nucleotides of the m6A RNA and the YTH domain. (A) Detailed interactions between the 5′-end of the RNA and the YTH domain. (B) Detailed interactions between the 3′-end of the RNA and the YTH domain.
Following the m6A residue, the base of residue C+1 has π-stacking interaction with the side chain of Arg296, although it does not appear to be recognized specifically by the YTH domain (Fig. 3B). In addition, the base has weak electron density in several of the RNA molecules, and the guanidinium group of Arg296 assumes different conformations in the other YTH domains, possibly because it is also stacked with A–3 of the other monomer of the RNA dimer (Fig. S3). The bases of the +2 and +3 nucleotides are projected away from the YTH domain and do not have direct contacts with the protein (Fig. 1C), but their phosphodiester backbone has favorable electrostatic interactions with the positively charged protein surface (Fig. 2B). Specifically, the 5′ phosphate group of A+2 interacts with the side chain of Lys184 and the main chain amide of Ala231, and the 5′ phosphate groups of both A+2 and U+3 interact with the main chain amide of Gly233 through a water molecule (Fig. 3B).
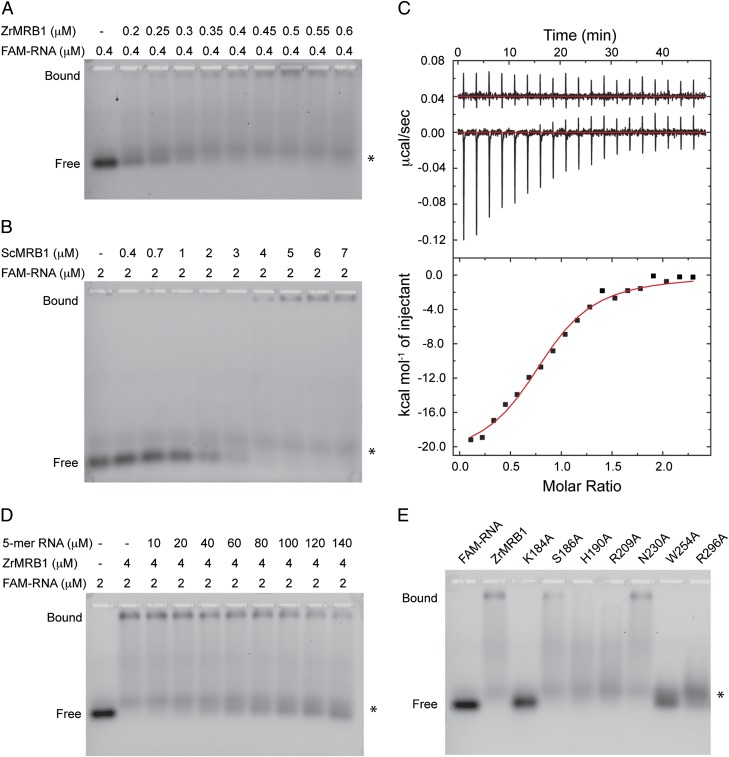
To assess the structural observations, we characterized the interactions between the MRB1 YTH domain and the RNA by electrophoretic mobility shift assay (EMSA) and isothermal titration calorimetry (ITC). For EMSA experiments, the 7-mer RNA with a 5′ FAM fluorophore was used as the probe, at 0.4 or 2 μM concentration. Our structure showed that the 5′-end of the RNA has few interactions with the YTH domain and, therefore, the introduction of the FAM label is unlikely to affect binding substantially. The experiments show that the YTH domains of ZrMRB1 (Fig. 4A), S. cerevisiae MRB1 (ScMRB1; Fig. 4B), and Kluyveromyces lactis MRB1 (KlMRB1; Fig. S6) have strong affinity for the RNA. At 0.4 μM concentration for the ZrMRB1 YTH domain, almost all of the RNA (at 0.4 μM) is shifted to the complex, indicating that the Kd of the complex is likely below 0.4 μM. There was a minor contaminating species in the labeled RNA sample (Fig. 4 A and B), but it did not interact with the proteins and was unlikely to have affected the outcome of the experiments.
Fig. 4.
Characterization of the interactions between the YTH domain and the m6A RNA. (A) A titration experiment assessing the affinity between the ZrMRB1 YTH domain and the 7-mer m6A RNA with a 5′ FAM fluorophore. The asterisk indicates a minor contaminating species in the RNA sample that does not interact with the YTH domain. (B) A titration experiment for the ScMRB1 YTH domain. (C) ITC data for the interaction between ZrMRB1 YTH domain and the labeled 7-mer RNA. Inset shows the heat release from the titration into buffer alone. (D) A competition binding assay testing the affinity of an unlabeled 5-mer m6A RNA for the ZrMRB1 YTH domain. (E) Effects of YTH domain mutations in the RNA interface on the formation of the complex. The FAM-labeled RNA is at 2 μM concentration, and the protein samples are at 4 μM concentration.
We carried out ITC experiments with the ZrMRB1 YTH domain to obtain a more quantitative measurement of the affinity (Fig. 4C). The Kd of the complex was determined to be 0.20 μM, consistent with our EMSA data. The enthalpy and entropy changes for the formation of the complex were –21.5 kcal/mol and –41.4 cal/mol⋅K, respectively. The molar ratio of the complex was 0.84, possibly reflecting some errors in the concentrations of the protein and the RNA.
We also tested the binding affinity of a 5-mer RNA, missing one nucleotide from each end of the 7-mer RNA. The 5-mer RNA competed weakly with the labeled 7-mer RNA for binding to the ZrMRB1 YTH domain, and could not completely disrupt the 7-mer RNA complex even at 120 μM concentration (Fig. 4D), indicating that the Kd of this complex could be ∼60 μM. This is consistent with the structural observations that the 3′-end of the 7-mer RNA has favorable interactions with the YTH domain (Figs. 2B and 3B). The nucleotide at the –2 position probably makes little contribution to the increased affinity of the 7-mer RNA. As a control, the unlabeled 7-mer RNA competed with the labeled 7-mer RNA for binding at roughly the same concentration (Fig. S6), also confirming that the 5′ FAM label did not substantially affect binding.
We next introduced mutations in the RNA binding site of ZrMRB1 YTH domain based on the structural information and determined their effects on complex formation. The mutants were purified by following the same protocol as the wild-type protein (Fig. S6) and produced similar profiles on a gel filtration column. The K184A (Fig. 3B), W254A (Fig. 2D), and R296A (Fig. 3B) mutations completely blocked RNA binding (Fig. 4E), whereas the S186A (Fig. 2D), H190A (Fig. 2D), and R209A (Fig. 3A) mutations substantially reduced the interaction. In contrast, the N230A (Fig. 2D) mutation had only a small effect on the binding. Overall, the mutagenesis data confirm the structural observations and demonstrate the importance of the aromatic cage and other residues in binding the methylated RNA.
We also assessed the interaction between the ZrMRB1 YTH domain and an RNA of the same sequence but without the m6A modification. The EMSA produced a smear of bands, and much higher concentrations of the protein were needed to shift most of the RNA molecule (Fig. S6), suggesting that the affinity of the YTH domain for this RNA is substantially lower. Mutations in the YTH domain that disrupt binding to the m6A RNA (Fig. 4E) also interfere with the binding to this unmethylated RNA, with the interesting exception of the W254A mutation in the aromatic cage (Fig. S6). These observations indicate that the unmethylated RNA likely assumes a similar binding mode in the YTH domain, although the aromatic cage is not crucial for this interaction. Therefore, the YTH domain in MRB1 and YTHDF1-3 may also bind unmethylated RNAs, with lower affinity, consistent with the observation that some targets of YTHDF2 do not appear to contain m6A sites (7).
ZrMRB1 residues in the RNA interface are highly conserved among MRB1 and YTHDF1-3 proteins (Fig. S1). Especially, the aromatic cage of YTHDF1-3 contains three Trp residues, with Tyr260 of ZrMRB1 replaced by a Trp residue. Therefore, our observations with ZrMRB1 should be directly relevant to how these other YTH domains bind the m6A RNA. Overall, our structural and biochemical studies have defined the molecular basis for the recognition of methylated adenines by the YTH domain.
Materials and Methods
The YTH domains of MRB1 proteins from Zygosaccharomyces rouxii, S. cerevisiae, and Kluyveromyces lactis were overexpressed in Escherichia coli and purified by nickel agarose and gel filtration chromatography. The RNAs were chemically synthesized by Dharmacon (GE Healthcare). Crystals of the YTH domain in complex with the m6A RNA were obtained by the sitting-drop vapor-diffusion method at 4 °C. The crystals belong to space group P6122, and there are six complexes in the asymmetric unit. The structure was determined by the selenomethionyl single-wavelength anomalous diffraction method. The interactions between the YTH domain and the RNA were assessed with the EMSA and ITC, using an RNA with a 5′ 6-FAM fluorophore label. Full experimental details are provided in the SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Neil Whalen and Annie Heroux for access to the X25 beamline. The in-house instrument for X-ray diffraction screening was purchased with a National Institutes of Health (NIH) Grant S10OD012018 (to L.T.). This research is supported by NIH Grant R01GM077175 (to L.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4U8T).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412742111/-/DCSupplemental.
References
- 1.Schwartz S, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155(6):1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 3.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013;29(2):108–115. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niu Y, et al. N6-methyl-adenosine (m6A) in RNA: An old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics. 2013;11(1):8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15(5):313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fustin JM, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155(4):793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 9.Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu C, et al. Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation. J Biol Chem. 2014;289(25):17299–17311. doi: 10.1074/jbc.M114.550350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoilov P, Rafalska I, Stamm S. YTH: A new domain in nuclear proteins. Trends Biochem Sci. 2002;27(10):495–497. doi: 10.1016/s0968-0004(02)02189-8. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, et al. The YTH domain is a novel RNA binding domain. J Biol Chem. 2010;285(19):14701–14710. doi: 10.1074/jbc.M110.104711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang HJ, et al. A novel protein, Pho92, has a conserved YTH domain and regulates phosphate metabolism by decreasing the mRNA stability of PHO4 in Saccharomyces cerevisiae. Biochem J. 2014;457(3):391–400. doi: 10.1042/BJ20130862. [DOI] [PubMed] [Google Scholar]
- 15.Yu F, et al. Determining the DUF55-domain structure of human thymocyte nuclear protein 1 from crystals partially twinned by tetartohedry. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 3):212–219. doi: 10.1107/S0907444908041474. [DOI] [PubMed] [Google Scholar]
- 16.Bertonati C, et al. Structural genomics reveals EVE as a new ASCH/PUA-related domain. Proteins. 2009;75(3):760–773. doi: 10.1002/prot.22287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugahara M, Asada Y, Morikawa Y, Kageyama Y, Kunishima N. Nucleant-mediated protein crystallization with the application of microporous synthetic zeolites. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 6):686–695. doi: 10.1107/S0907444908009980. [DOI] [PubMed] [Google Scholar]
- 18.Arakaki T, et al. Structure of Lmaj006129AAA, a hypothetical protein from Leishmania major. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62(Pt 3):175–179. doi: 10.1107/S1744309106005902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24(23):2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khorasanizadeh S. Recognition of methylated histones: New twists and variations. Curr Opin Struct Biol. 2011;21(6):744–749. doi: 10.1016/j.sbi.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Yap KL, Zhou MM. Structure and mechanisms of lysine methylation recognition by the chromodomain in gene transcription. Biochemistry. 2011;50(12):1966–1980. doi: 10.1021/bi101885m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischle W, et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17(15):1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127(7):1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J, Thompson JR, Botuyan MV, Mer G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat Struct Mol Biol. 2008;15(1):109–111. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gouet P, Courcelle E, Stuart DI, Métoz F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15(4):305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 26.Armon A, Graur D, Ben-Tal N. ConSurf: An algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol. 2001;307(1):447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.