Significance
Human noroviruses are the predominant cause of acute gastroenteritis worldwide, but they remain noncultivatable. A tractable system is needed to understand the host restriction to cultivation. We established a reverse genetics system driven by a mammalian elongation factor-1α promoter without helper virus. This system supports genome replication, particle formation, and particles containing a GFP-marked genomic RNA. RNA from these particles is infectious. The system also produces infectious murine norovirus, confirming its broad applicability to other noroviruses.
Keywords: reporter-tagged norovirus, helper-virus–free reverse genetic system
Abstract
Human norovirus (HuNoV) is the leading cause of gastroenteritis worldwide. HuNoV replication studies have been hampered by the inability to grow the virus in cultured cells. The HuNoV genome is a positive-sense single-stranded RNA (ssRNA) molecule with three open reading frames (ORFs). We established a reverse genetics system driven by a mammalian promoter that functions without helper virus. The complete genome of the HuNoV genogroup II.3 U201 strain was cloned downstream of an elongation factor-1α (EF-1α) mammalian promoter. Cells transfected with plasmid containing the full-length genome (pHuNoVU201F) expressed the ORF1 polyprotein, which was cleaved by the viral protease to produce the mature nonstructural viral proteins, and the capsid proteins. Progeny virus produced from the transfected cells contained the complete NoV genomic RNA (VP1, VP2, and VPg) and exhibited the same density in isopycnic cesium chloride gradients as native infectious NoV particles from a patient’s stool. This system also was applied to drive murine NoV RNA replication and produced infectious progeny virions. A GFP reporter construct containing the GFP gene in ORF1 produced complete virions that contain VPg-linked RNA. RNA from virions containing the encapsidated GFP-genomic RNA was successfully transfected back into cells producing fluorescent puncta, indicating that the encapsidated RNA is replication-competent. The EF-1α mammalian promoter expression system provides the first reverse genetics system, to our knowledge, generalizable for human and animal NoVs that does not require a helper virus. Establishing a complete reverse genetics system expressed from cDNA for HuNoVs now allows the manipulation of the viral genome and production of reporter virions.
Human noroviruses (HuNoVs) belong to the genus Norovirus of the family Caliciviridae and are the predominant cause of epidemic and sporadic cases of acute gastroenteritis worldwide (1, 2). HuNoVs are spread through contaminated water or food, such as oysters, shellfish, or ice, and by person-to-person transmission (3, 4). Although HuNoVs were identified more than 40 y ago, our understanding of the replication cycle and mechanisms of pathogenicity is limited, because these viruses remain noncultivatable in vitro, a robust small animal model to study viral infection is not available, and reports of successful passage of HuNoVs in a 3D cell culture system have not been reproduced (5–7). Recently, a murine model for HuNoV infection was described that involves intraperitoneal inoculation of immunocompromised mice (8); its generalizability and robustness for studying individual HuNoVs and many aspects of HuNoV biology remain to be established. Gnotobiotic pigs can support replication of a HuNoV genogroup II (GII) strain with the occurrence of mild diarrhea, fecal virus shedding, and immunofluorescent (IF) detection of both structural and nonstructural proteins in enterocytes (9). Previous systems to express the HuNoV genome from cloned DNA using T7/vaccinia systems showed that mammalian cells can produce progeny virus (10, 11), but these systems are not sufficiently efficient to be widely used to propagate HuNoVs in vitro. The factors responsible for the block(s) of viral replication using standard cell culture systems remain unknown.
The HuNoV genome is a positive-sense ssRNA of ∼7.6 kb that is organized in three ORFs: ORF1 encodes a nonstructural polyprotein, and ORF2 and ORF3 encode the major and minor capsid proteins VP1 and VP2, respectively. Because of the lack of an in vitro system to propagate HuNoV, features of their life cycle have been inferred from studies using other animal caliciviruses and murine NoV (MNV) that can be cultivated in mammalian cell cultures (12). A 3′ coterminal polyadenylated subgenomic RNA is produced within infected cells. Both genomic and subgenomic RNAs have the same nucleotide sequence motif at their 5′ ends, and they are believed for HuNoVs and shown for MNV to be covalently linked to the nonstructural protein VPg at the 5′ ends (10, 13). During MNV infection of cells, nonstructural proteins are expressed from genomic RNA and form an RNA replication complex that generates new genomic RNA molecules as well as subgenomic RNAs encoding VP1, VP2, and the unique protein called VF1 (14). After expression of the structural proteins from subgenomic RNA molecules, the capsid is assembled, and viral RNA is encapsidated before progeny release. Previous reverse genetics systems for HuNoV used helper vaccinia MVA/T7 virus-based systems. Although helper virus-free systems have been developed for MNV (15, 16), no such system is available for HuNoVs. To overcome these problems, we established a reverse genetics system driven by a mammalian elongation factor-1α (EF-1α) promoter without helper virus and then modified this system to package a reporter gene (GFP) into ORF1.
Results
ORF1 Polyprotein Is Cleaved by the ORF1-Encoded Protease.
After unsuccessful attempts to obtain HuNoV genome expression using the CMV promoter, we cloned the HuNoV GII.3 U201 genome into an expression cassette under the control of the promoter regulating the human EF-1α (Fig. S1A). This vector, named pHuNoVU201F, contains two exons and an intron from the EF-1α gene that include transcription binding sites for both Sp-1 and AP-1 for efficient transcription of the insert (17). In addition, pHuNoVU201F contains an hepatitis delta virus ribozyme that cleaves the RNA after the poly(A) sequence to produce the 3′ end of the RNA. After transcription, mRNA is produced, includes a 5′ cap and the poly(A) sequence, and is used for the translation of the HuNoV polyprotein. Based on this parental vector, other constructs were produced (SI Materials and Methods and Fig. S1 B–F).
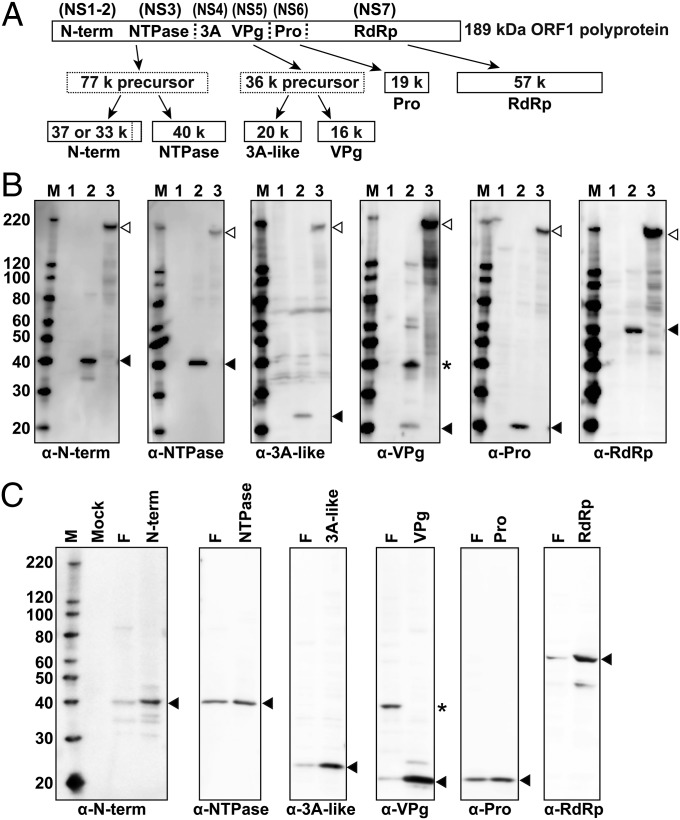
ORF1 of the HuNoV genome encodes a polyprotein that is cleaved by the viral protease into the mature proteins (Fig. 1A indicates the ORF1 proteins using their names based on function and nonstructural protein numeric designations) (18–20). To assess ORF1 synthesis and polyprotein cleavage, Western blot analysis was performed at 24 h posttransfection (hpt) (Fig. 1B) using protein-specific antibodies on lysates of COS7 cells mock-transfected or transfected with the full-length construct (pHuNoVU201F) or the protease KO mutant construct pHuNoVU201FproM, which produces a nonfunctional protease (Fig. S1C). No or few background bands were detected in mock-transfected cells (Fig. 1B, lane 1). In cells expressing the complete genome (Fig. 1B, lane 2), each protein-specific antibody detected the mature cleavage products (Fig. 1B, black arrowheads), with a larger possible precursor protein detected clearly only in the case of VPg (Fig. 1B, asterisk). Cells transfected with the protease KO construct (Fig. 1B, lane 3) primarily expressed a 189-kDa polyprotein (Fig. 1B, white arrowheads) detected with each protein-specific antibody as well as several minor bands differentially detected by a subset of the specific sera. These results indicate that a functional U201 RNA is transcribed from the EF-1α promoter that is not spliced internally within the viral genome, and this RNA expresses a complete and functional ORF1 polyprotein. Identification of each mature product was confirmed by expression of each individual protein (Fig. 1C). The lack of detection of most of the precursor proteins likely reflects the late time point examined. The structural proteins VP1 and VP2 encoded by ORF2 and ORF3, respectively, were not detected by Western blot in cells at 24 hpt. The same results were seen in HEK293T, Huh7, and Caco2 cells.
Fig. 1.
Detection of U201 protein expression in COS7 cells 24 hpt with different plasmid constructs. (A) Schematic figure of polyprotein cleavage and maturation. Functional names of the nonstructural proteins are inside the polyprotein box, and the text in parentheses shows the numeric nonstructural protein nomenclature as proposed by Sosnovtsev et al. (19). Dotted vertical lines represent estimated cleavage sites. Solid line boxes show the mature forms of each nonstructural protein. Dotted line boxes represent precursor proteins with molecular mass predicted from amino acid composition. (B) Proteins were detected by Western blot with protein-specific purified IgG (as indicated under each Western blot panel). Lane M contains the molecular mass makers MagicMark II, lane 1 is from mock-transfected COS7 cells, and lanes 2 and 3 are from pHuNoVU201F- and protease-mutant pHuNoVU201FproM-transfected COS7 cells, respectively. The black arrowheads show the mature protein size determined from the amino acid sequences using Genetyx software. The white arrowheads show the uncleaved polyprotein only observed in cells expressing the protease mutant construct (lane 3). *3A-like protein-VPg precursor protein. (C) Blots comparing nonstructural proteins expressed from full-length pHuNoVU201F (lane F) and individual expression constructs (pHuNoVU201-Nterm, -NTP, -3A-like, -VPg, -protease, and -RdRp) that produced N-term, NTPase, 3A-like protein, VPg, protease, and RdRp, respectively. M shows molecular mass markers. Black arrowheads show the mature proteins. *3A-like protein-VPg precursor protein.
Cellular Localization of the NoV Nonstructural Proteins Expressed from the Full-Length pHuNoVU201F Construct.
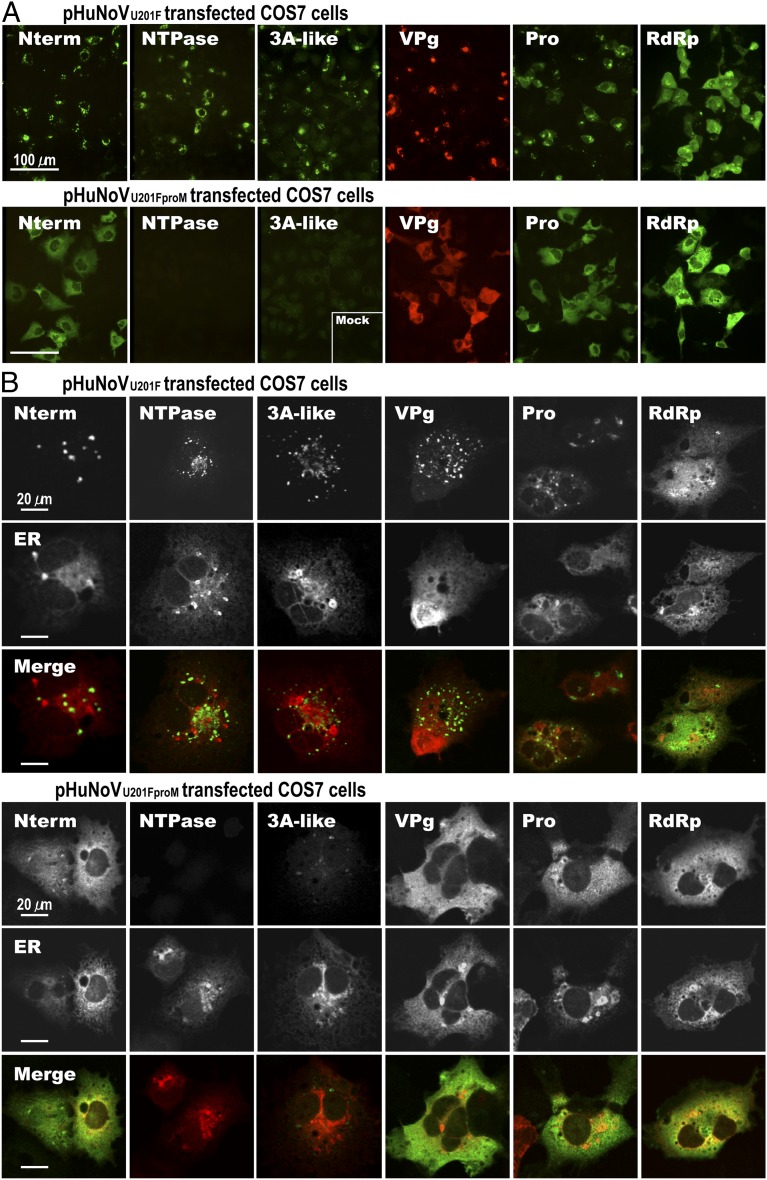
The localization of the nonstructural proteins expressed in cells transfected with the pHuNoVU201F or pHuNoVU201FproM plasmid constructs was visualized by confocal microscopy in fixed cells stained using each nonstructural protein-specific antibody; these two constructs generate the cleaved ORF1 mature proteins or the ORF1 polyprotein, respectively. Each of the detected proteins showed cytoplasmic staining in a subset of cells, with RdRp exhibiting a more diffuse distribution, whereas others showed a more punctate perinuclear staining (N-term, NTPase, 3A-like, VPg, and protease) (Fig. 2A, Upper). Together with the Western blot data, these results confirm that pHuNoVU201F expressed the complete ORF1 protein (from the N terminus to C terminus). Cells transfected with the protease KO plasmid (pHuNoVU201FproM) showed a different IF staining pattern (Fig. 2A, Lower) compared with those from pHuNoVU201F-transfected cells. The antibodies to the N-terminal protein, VPg, protease, and RdRp detected broadly diffuse cytoplasmic signals. The NTPase and 3A-like proteins were not detected in cells expressing the protease mutant by IF (Fig. 2A), although they were detected within the polyprotein by Western blot. Some background signal was also detected using the 3A-like antiserum in cells transfected with an empty vector, pKS435gateA3 (Fig. 2A, mock in 3A-like panel). These differences suggest that the uncleaved ORF1 polyprotein was present in the cells but that epitopes on the NTPase and 3A-like proteins are not accessible for binding to their respective antibodies. These results indicate that only the cleaved ORF1 proteins show a functional localization. Additional staining with an endoplasmic reticulum (ER) marker showed that the uncleaved ORF1 polyprotein was retained in the ER (Fig. 2B).
Fig. 2.
Protein expression in COS7 cells transfected with different plasmid constructs detected by IF microscopy. A, Upper represents pHuNoVU201F-transfected COS7 cells; A, Lower shows the protease mutant pHuNoVU201FproM-transfected COS7 cells at 24 hpt. IF staining for nonstructural proteins was performed using purified IgG against each protein as indicated. Inset in the 3A-like panel shows background signal in empty vector pKS435gateA3-transfected cells. (Scale bars: 100 μm.) B, Upper and B, Lower represent pHuNoVU201F- and pHuNoVU201FproM-transfected COS7 cells, respectively. IF staining for nonstructural proteins (green) and fluorescence for ER (red) are shown in rows 1 and 2, respectively. Individual channels are shown in grayscale. The merged image of each nonstructural protein and ER is shown in row 3. (Scale bars: 20 μm.)
To determine whether protease provided in trans could rescue ORF1 polyprotein cleavage, cells were transfected with the protease mutant (pHuNoVU201FproM) plasmid, and 24 h later, a wild type (WT) or mutant protease-expressing plasmid was transfected into the cells. After another 24 h, protein production was examined by IF (Fig. S2) and Western blot (Fig. S3). Protein localization was similar in the cells cotransfected with the WT protease as observed in pHuNoVU201F-transfected cells (Fig. 2A, Upper and Fig. S2). This observation indicates that the WT protease added in trans was able to cleave the polyprotein and led to redistribution of the cleaved mature products. Sequential cotransfections with mutant protease plasmids did not lead to redistribution of N-term, VPg, or protease or detection of NTPase and 3A-like proteins (Fig. S2, Lower). Western blot analysis confirmed that the protease was made when pHuNoVU201F, pHuNoVU201-pro, and pHuNoVU201-proM were transfected into cells (Fig. S3, Left). The protease made from pHuNoVU201F and pHuNoVU201-pro was functional when it was coexpressed with mutant pHuNoVU201-proM (Fig. S3, Right, lane 3), but the protease remained nonfunctional when pHuNoVU201-proM was coexpressed with the full-length pHuNoVU201F-proM construct (Fig. S3, each lane 4).
Expressed Viral Proteins Produce Negative-Sense Genomic RNA, Genomic RNA, and Subgenomic RNA.
To analyze virus genome replication, we performed Northern blot analysis to detect newly synthesized HuNoV genomic RNA, subgenomic RNA, and negative-strand RNA. We transfected COS7 cells with pHuNoVU201F and pHuNoVU201-ORF2,3 as a subgenomic-sized control. In addition, COS7 cells were transfected with pHuNoVU201F-ORF1-IRES-GFP (Fig. S1C) to monitor transfection rate and as a polymerase KO construct, because it contains a stop codon after the GFP. Total RNA was extracted from the cells at 24 and 48 hpt, and 1 μg RNA was used for Northern blot analysis. Bands corresponding to genomic RNA and subgenomic RNA were identified by analysis of in vitro transcribed RNA on a control blot (Fig. S4). A negative-sense RNA probe (spanning 7,343–5,370 nt) was used to detect positive-strand HuNoV RNA. A doublet band (Fig. S4, black arrowheads) that migrated slightly slower than the 7.6-kb genomic RNA control band was detected from cells expressing pHuNoVU201F at 24 and 48 hpt (Fig. S4A, lane 1). RNA isolated from cells transfected with the pHuNoVU201F-ORF1-IRES-GFP construct (RdRp KO) migrated slightly above the upper doublet band (Fig. S4A, white arrowhead). To characterize these RNA molecules, 5′ RACE and 3′ RACE were performed on these RNA bands after excision from the agarose gel. Two different 5′-end sequences were detected from the doublet RNA band at 7.6 kb in pHuNoVU201F-transfected cells. One RNA included 107 nt corresponding to the EF-1α exon sequence included in the pKS435gateA3 vector before the 5′ end of the genome of U201 5′-GUGAAUGAAGAUG; the other sequence was identical to the native U201 genome 5′ end, providing additional evidence that authentic replication occurred. The 3′-end sequences of both the upper and lower 7.6-kb RNA bands were identical to the U201 genome sequence, including a poly(A) tail, and did not include any other sequences. The larger-sized mRNA expressed from the pHuNoVU201F-ORF1-IRES-GFP plasmid also contained the 107-nt EF-1α exon sequence at the 5′ end and an extra 39 nt that correspond to the internal ribosome entry site (IRES-GFP) sequences inserted in the ORF1.
At 48 hpt, a subgenomic-sized band was detected in cells expressing pHuNoVU201F (Fig. S4A, 48 hpt lane 1). This band migrated slightly above the 2.6-kb in vitro transcribed subgenomic RNA control and was weaker in intensity than the genomic-sized band. This subgenomic band also migrated slightly slower than the RNA from the ORF2, ORF3 RNA construct (Fig. S4A, 48 hpt lane 2), possibly because of the presence of VPg. The subgenomic-sized band was excised, 5′ RACE and 3′ RACE were performed, and the nucleotide sequences were compared with those from viral RNA molecules obtained from the original U201 virus-containing positive stool sample. The 5′ end of the subgenomic-sized band generated from pHuNoVU201F showed 5′-GUGAAUGAAGAUG, which was identical to the sequence from the original U201 RNA molecule. This finding is the first demonstration, to our knowledge, of a newly generated subgenomic RNA sequence in a helper virus-free HuNoV GII reverse genetics system, and it confirms results reported with the prototype GI HuNoV after replication using the MVA/T7 vaccinia virus system (10). Negative-strand RNA was only detected in pHuNoVU201F-transfected cells (Fig. S4B, lane 1) with a size that was slightly larger than the genome size using a positive-sense RNA probe, and the band intensity of the negative-strand RNA increased from 24 to 48 hpt. Several extra degraded RNA or nonspecific bands were observed in the pHuNoVU201F-transfected cells at 48 hpt (Fig. S4B, 48 hpt lane 2) and cells transfected with the pHuNoVU201F-ORF1-IRES-GFP RdRp KO construct (Fig. S4B, 48 hpt lane 3).
Subgenomic RNA Produces Structural Proteins VP1 and VP2 in pHuNoVU201F-Transfected Cells.
Because subgenomic RNA was detected in the pHuNoVU201F-transfected COS7 cells (Fig. S4A), we next determined whether the structural proteins VP1 and VP2 were expressed from the subgenomic RNA. The major structural protein VP1 and minor structural protein VP2 were detected by IF using protein-specific antibodies in pHuNoVU201F-transfected COS7 cells (Fig. S4 D and E). To confirm that ORF1 protein expression led to structural protein expression, we also costained for VPg using anti-VPg mAb. VP1 was detected in pHuNoVU201F-transfected cells as a green signal, whereas VPg was detected in the same cells as red granular-like signals (Fig. S4D, white arrowheads). VP2 (Fig. S4E, green) was detected in a subset of cells also expressing VPg (Fig. S4E, red). These data indicate VP1 and VP2 were produced in pHuNoVU201F-transfected cells, and expression of these proteins was only observed in cells that expressed the ORF1 protein VPg. The capsid proteins were not able to be detected by Western blot using either rabbit or guinea pig polyclonal antibody to U201 VP1, VP2, or VLP.
To confirm VP1 expression from transcribed subgenomic RNA in cells, we evaluated GFP expression using time-lapse imaging of COS7 cells (Fig. S5A, Upper) transfected with the pHuNoVU201F-ORF2GFP (Fig. S1D). GFP was first visualized by 12 hpt, and visualization remained high through 48 hpt. Cytopathic effect (CPE) was observed within cells containing the GFP signal. In contrast, cells transfected with the RdRp KO mutant pHuNoVU201FΔ4607G-ORF2GFP failed to show any GFP signal (Fig. S5A, Upper). Additionally, we confirmed VP1 synthesis by using a full-length construct containing the highly sensitive reporter Renilla luciferase (Rluc) inserted into ORF2 (Figs. S1D and S5B). Time-dependent detection of Rluc expression in pHuNoVU201F-ORF2Rluc–transfected COS7 cells was detected 12 hpt followed by a sharp increase that peaked at 24 hpt with a maximum of 3.9 × 105 arbitrary units (A.U.). By 48 hpt, the Rluc activity sharply decreased to less than 1.0 × 105 A.U. In contrast, cells expressing the RdRp KO construct pHuNoVU201FΔ4607G-ORF2Rluc produced less than 105 A.U. These data provided clear independent confirmation that our plasmid-based reverse genetics system drives HuNoV genome replication and produces VP1 protein from subgenomic RNA.
Progeny HuNoVs Were Produced from pHuNoVU201F-Transfected Cells.
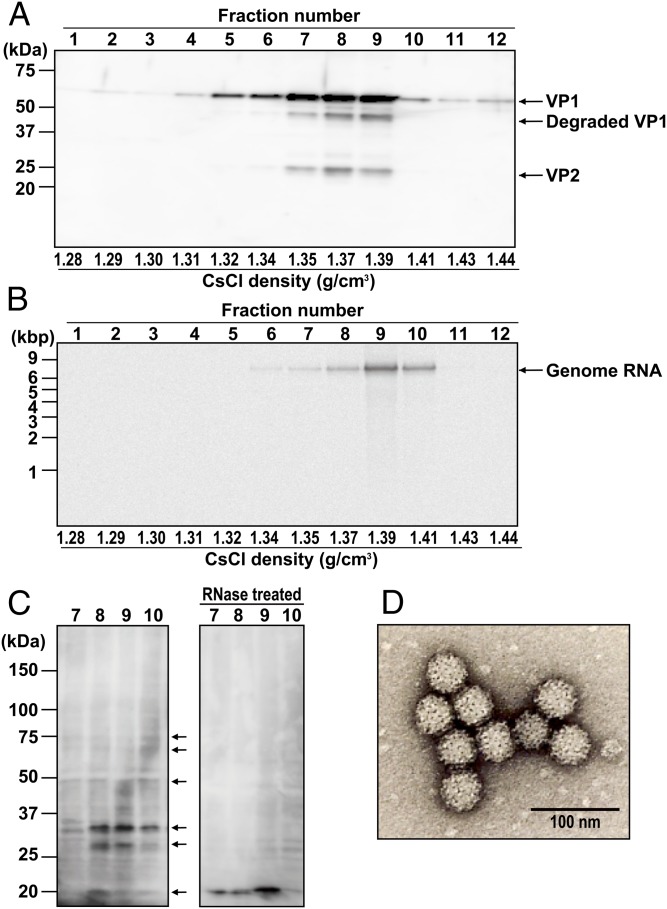
The pHuNoVU201F-transfected cells expressed nonstructural proteins and structural proteins, and they also generated genomic- and subgenomic-sized RNAs. To determine whether progeny virus was produced, large-scale cultures (10 T255 culture flasks) were transfected and processed as described in Materials and Methods. Cesium chloride (CsCl) gradients were fractionated into 12 fractions (450 μL each that were further subdivided into 50- or 100-μL aliquots for additional characterization). Fractions 1–12 were evaluated for density and the presence of the viral capsid proteins VP1 and VP2 by Western blot using rabbit anti-U201 VLP antisera that can detect VP1 and VP2 (Fig. 3A) (11). VP1 was detected in fractions 2–12, with a peak band intensity in fraction 9. VP2 was also detected in fractions 7–9. A separate aliquot of each fraction was treated with nuclease to remove any exogenous RNA and plasmid DNA. Encapsidated HuNoV genomic RNA was extracted and subsequently detected by Northern blot analysis (Fig. 3B). A 7.6-kb band was observed in fractions 6–10. The strongest band was in fraction 9, which had a density of 1.39 g/cm3 (the approximate density of native infectious HuNoV virions). No subgenomic RNA signal was detected in any fraction. We also analyzed each untreated fraction by Western blot using the U201 VPg mAb (Fig. 3C). Unexpectedly, multiple bands of VPg were observed in samples from fractions 7–10, with predominant bands of 20, 27, and 32 kDa and minor bands of 50, 65, and 75 kDa. These results suggested that VPg is attached or strongly associated with the genomic RNA, consistent with it being a multifunctional genome-linked protein. To address this possibility, aliquots from fractions 7–10 were treated with RNase, and the proteins were analyzed by Western blot. The multiple bands disappeared completely, and a single band of 20 kDa was detected (Fig. 3C, RNase treated). This band was the same size as the mature VPg detected from expression of the full-length genome (Fig. 1B) and the individually expressed VPg protein from pHuNoVU201-VPg (Fig. 1C). These results indicate that different conformations of VPg may be associated with genomic RNA and/or covalently linked with genomic RNA and packaged into virions. The multiple bands in the nonnuclease-digested samples are possibly an indication of limited hydrolysis of the VPg-linked RNA during sample preparation for SDS/PAGE. Alternatively, they may be caused by the RNA adopting multiple conformations during separation by SDS/PAGE.
Fig. 3.
Detection of HuNoV progeny particles in the supernatant of transfected COS7 cells. (A) Detection of HuNoV structural proteins VP1 and VP2 in individual fractions of a CsCl gradient by Western blot using rabbit anti-U201 VLP serum. Molecular mass markers are shown on the left side of the blot. (B) HuNoV RNA was detected with Northern blotting using a 33P-labeled antisense RNA probe. RNA molecular mass markers are shown on the left side of the blot. For A and B, the numbers above and below the blots represent the fraction number and CsCl density, respectively. (C, Left) Detection of VPg by Western blot in fractions 7–10 using VPg mAb. The arrows show multiple VPg bands. C, Right shows detection of VPg in a Western blot of RNase-treated fractions 7–10. Molecular mass markers are shown on the left side of the blot. (D) Progeny HuNoV U201 virions in fraction 9 visualized by EM after negative staining. (Magnification: 50,000×; scale bar: 100 nm.)
To confirm that HuNoV progeny virus was present in fractions 8–10, we analyzed each fraction by transmission EM. Ten to fifty native HuNoV-like virions per grid were found in fraction 9 (Fig. 3D), and a few particles were observed in fractions 8 and 10. The observed 40-nm virions exhibited the characteristic NoV structure.
Taken together, these results indicate that this reverse genetics system produces authentic progeny HuNoV particles that contain VPg as a genome-associated protein, genomic RNA, VP1, and VP2 and a structure and density similar to HuNoV virions detected in stool samples.
GFP Reporter Construct Can Produce Progeny Virus.
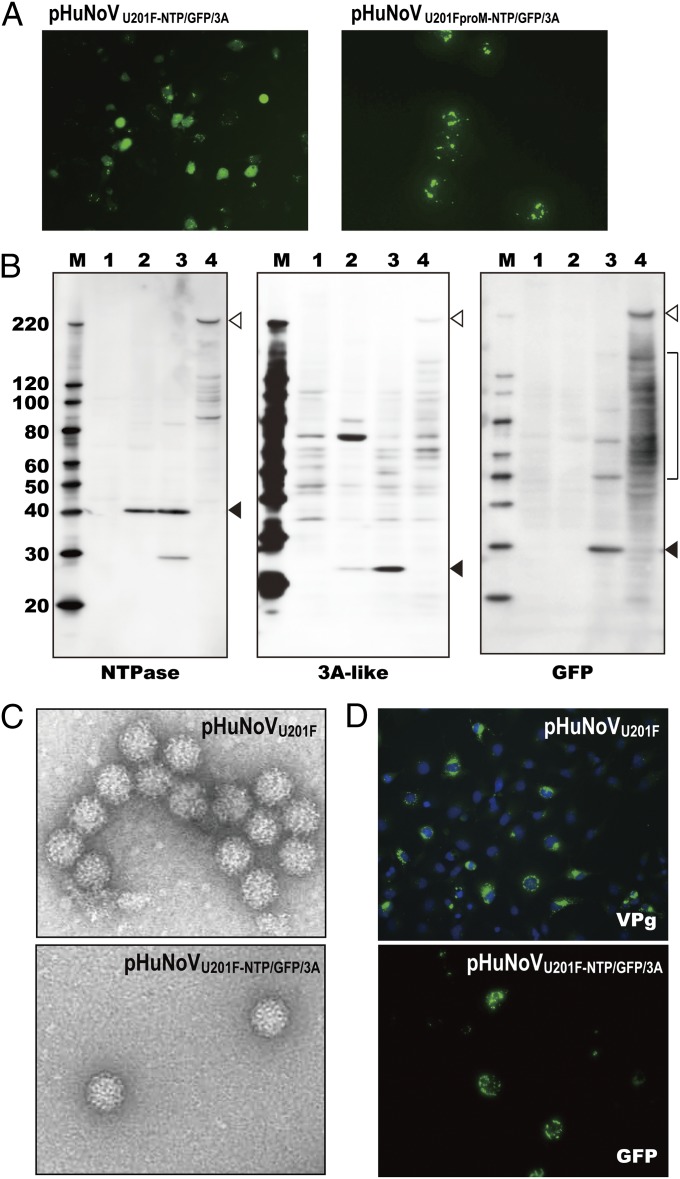
Next, we sought to produce a reporter-tagged HuNoV by inserting the GFP reporter gene in the U201 genome. The GFP gene was cloned into ORF1 between the NTPase and 3A-like proteins. A U201 protease cleavage motif QG was placed at the N terminus of GFP, and three additional amino acid residues from the native sequence of cleavage site FELQG were added at the C-terminal end of GFP (construct named pHuNoVU201F-NTP/GFP/3A) (Fig. S1E) (21). Cells transfected with the pHuNoVU201F-NTP/GFP/3A construct expressed a strong GFP signal, indicating that ORF1 translation was efficient (Fig. 4A). In contrast, GFP was not cleaved from a protease mutant pHuNoVU201FproM-NTP/GFP/3A construct (Fig. 4B, lane 4), and the GFP exhibited a punctate localization (Fig. 4A). We confirmed and compared ORF1 cleavage using Western blot with NTPase-, 3A-like protein-, and GFP-specific antibodies. GFP was efficiently cleaved from the ORF1 polyprotein produced from the pHuNoVU201F-NTP/GFP/3A along with the NTPase and 3A-like proteins (Fig. 4B, lane 3). The cleaved and mature NTPase and 3A-like protein bands were similar in size to those seen in cells expressing pHuNoVU201F when NTPase and 3A-like antibodies were used for detection (Fig. 4B, lane 2). Additional 47-, 63-, 87-, 124-, and 216-kDa bands were detected with GFP mAb. These bands likely corresponded to GFP-3A, GFP-3A-VPg, NTPase-GFP-3A, N-term-NTP-GFP-3A, and uncut polyprotein (Fig. S6). These results suggested that pHuNoVU201F-NTP/GFP/3A had the possibility to produce progeny virus containing a reporter gene.
Fig. 4.
Analyses of GFP reporter constructs pHuNoVU201F-NTP/GFP/3A and pHuNoVU201FproM-NTP/GFP/3A. (A) Images of GFP expression from (Left) pHuNoVU201F-NTP/GFP/3A–transfected and (Right) pHuNoVU201FproM-NTP/GFP/3A–transfected COS7 cells at 24 hpt. (B) Analysis of proteolytic cleavage of polyprotein translated from pHuNoVU201F-NTP/GFP/3A. M represents molecular mass markers. Lane 1 shows mock-transfected cells, lane 2 shows pHuNoVU201F-transfected cells, lane 3 shows pHuNoVU201F-NTP/GFP/3A–transfected cells, and lane 4 shows pHuNoVU201FproM-NTP/GFP/3A–transfected cells. Black arrowheads show the mature protein. White arrowheads represent uncut polyprotein and brackets show intermediate cleaved precursor proteins. Antibodies used to detect the proteins are shown below the blots. (C) Progeny HuNoV virions purified from pHuNoVU201F- and pHuNoVU201F-NTP/GFP/3A–transfected cultures. (D, Upper) VPg was detected by IF in pHuNoVU201F-transfected COS7 cells that were fixed at 24 hpt and stained with the VPg mAb and Alexa Fluor 488-labeled anti-mouse IgG. Nuclei were counterstained with DAPI. (D, Lower) GFP signal was detected 24 hpt after transfection of RNA isolated from progeny virus from supernatants of pHuNoVU201F-NTP/GFP/3A–transfected cells.
Progeny virus production from the pHuNoVU201F-NTP/GFP/3A plasmid was evaluated using the same methods and scale as for the parental pHuNoVU201F. Fraction 9 included progeny virus with the same morphology by EM as that produced from pHuNoVU201F; however, the virion production level was lower (up to 50-fold less) than pHuNoVU201F (Fig. 4C and Table 1).
Table 1.
Progeny NoV released in different cell types
| U201F | U201F-NTP/GFP/3A | TCH04-577 | Saga1 | NV68 | MNV S7 | |
| COS7 | 8.0 × 104 | 1.1 × 104 | 8.0 × 101 | 1.3 × 103 | 6.0 × 101 | 2.3 × 103 |
| 293T | 1.4 × 104 | 2.8 × 102 | 2.4 × 102 | 2.8 × 102 | 2.6 × 102 | 1.5 × 104 |
| Huh7 | 2.4 × 102 | ND | 3.3 × 102 | NT | NT | NT |
| Caco2 | 1.3 × 101 | ND | NT | NT | NT | NT |
Numbers indicate genome copies per 106 cells determined from triplicate experiments by semiquantitative long-distance RT-PCR. ND, not detected; NT, not tested.
Progeny HuNoV Particles Contain Infectious RNA.
The pHuNoVU201F system produced progeny viruses that should be infectious. However, infectivity could not be tested directly, because we still lack a small animal model and a susceptible cell culture system that supports HuNoV replication. An alternative is to determine whether the RNA encapsidated in the progeny virus produced from pHuNoVU201F and pHuNoVU201F-NTP/GFP/3A is infectious. Genomic RNA extracted from these particles was transfected into COS7 cells according to our previously reported protocol (22), and viral protein expression was monitored (Fig. 4D). Granular VPg protein was detected by IF in cells transfected with RNA extracted from particles produced from the pHuNoVU201F plasmid. This observation showed that nonstructural proteins were expressed from the transfected RNA. When we transfected RNA extracted from the particles produced from the pHuNoVU201F-NTP/GFP/3A plasmid, expression of the encoded GFP was detected. Taken together, the reporter RNA encapsidated into the NoV particle produced by the pHuNoVU201F system was active when expressed by itself. Therefore, these particles are likely to be infectious.
This Helper-Free Reverse Genetics System Is Generalizable for Other NoVs.
To test its generalizability, the EF-1α mammalian promoter system initially optimized for the GII.3 U201 strain was applied to make constructs expressing other strains of HuNoV, including a GII.P4-GII.3 chimeric virus TCH04-577 strain [nomenclature as proposed by Kroneman et al. (23)], a GII.4 Saga1 strain, and the GI.1 NV68 strain (Fig. S1F). To determine the efficiency of progeny virus release, nuclease-resistant and encapsidaded RNA in culture supernatants from 106 cells was evaluated by semiquantitative, long-distance RT-PCR (Fig. S7). COS7 cells transfected with pHuNoVU201F and pHuNoVU201F-NTP/GFP/3A produced detectable genomic RNA, whereas no RNA was amplified from cells transfected with the negative-control pHuNoVU201FΔ4607G construct or mock-transfected cells. The expression vectors for other GII strains (TCH04-577 and Saga1) produced little viral RNA in COS7 cells (Fig. S7B). The copy numbers of recovered viruses, determined by comparison of band intensities with known copy numbers of plasmid standards (Fig. S7A), are shown in Table 1. The highest yields of progeny virus (8.0 × 104 and 1.1 × 104 copies) were from 106 COS7 cells transfected with pHuNoVU201F and pHuNoVU201F-NTP/GFP/3A, respectively. Production of the TCH04-577, Saga1, and NV68 strains was 10- to 1,000-fold lower. Yields were also examined and compared after transfection of the constructs into human cell lines HEK293T, Huh7, and Caco2. RT-PCR products from HEK293T culture supernatants were detected from all plasmids, except the negative-control pHuNoVU201FΔ4607G- or mock-transfected cells (Fig. S7C). The production level of each strain varied in HEK293T cells from 2.4 × 102 to 1.4 × 104 copies/106 cells, whereas yields were lower in Huh7 and Caco2 cells (Table 1).
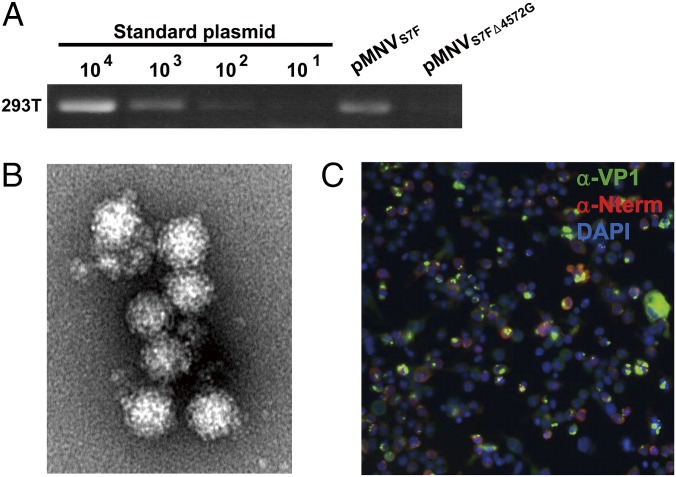
Finally, we evaluated this system using the MNV S7 strain (24–26), which is cultivable in murine macrophage cells. The pMNVS7F plasmid, which contains the full genome of MNV S7 strain (Fig. S1F), was transfected into COS7 and HEK293T cells, and 2.3 × 103 and 1.5 × 104 copies of progeny virus were released from 106 cells of COS7 and HEK293T, respectively (Table 1). Yields of infectious MNV were 3.4 × 102 and 2.0 × 103 tissue culture infectious dose-50% (TCID50) from COS7 and 293T cells, respectively. Transfection of a protease KO mutant of MNV, pMNVS7FΔ4572G (Fig. S1F), failed to produce detectable viral RNA in HEK293T cells (Fig. 5A). Progeny virus released from HEK293T cells was collected and purified from the supernatant of 10 T225 culture flasks by CsCl ultracentrifugation. The peak fraction (1.36 g/cm3) contained MNV particles as observed by negative-staining EM (Fig. 5B). The infectivity of the virus released into the culture supernatant at 48 hpt from the pMNVS7F-transfected HEK293T cells cultured in a six-well plate was tested by inoculation onto the murine macrophage line RAW264.7 cells. Expressions of the capsid protein VP1 and nonstructural N-term protein were detected in RAW264.7 cells at 48 h postinfection by IF microscopy (Fig. 5C), similar to results seen using pol II-driven systems for MNV (16). Taken together, these results show that simple single-plasmid transfection into HEK293T cells allows for the efficient and facile recovery of recombinant HuNoV strains and MNV. The recovery of infectious MNV indicates that this system has the capacity to produce infectious virus.
Fig. 5.
Recovery of infectious progeny virus from pMNVS7F-transfected HEK293T cells. (A) The pMNVS7F and pMNVS7FΔ4572G (frame shift in RdRp for KO) constructs were transfected into HEK293T cells. The yield of progeny virus released into culture supernatant at 48 hpt was determined by nested RT-PCR; 1 mL culture supernatant at 48 hpt was treated with RNase and DNase for 1 h at 37 °C. Progeny virus was immunoprecipitated with 1 μg anti-MNV rabbit serum and 5 μg protein A magnetic beads. RNA extracted from captured progeny virus was determined by long-distance nested RT-PCR with Tx30SXN/MNV F1 primers for the first step and MNV-S/MNV-R2 primers for the nested step. PCR products were subjected to agarose gel electrophoresis in the presence of 0.5 μg/mL ethidium bromide. PCR product bands were quantitated by LAS3000 (Fujifilm) with a standard curve of in vitro-transcribed complete-length MNV S7 RNA with a 10-fold dilution series (100–105 copy numbers). The pMNVS7FΔ4572G deletion mutant in the RdRp gene was used as a negative control. (B) Progeny MNV S7 virions visualized by EM after negative staining. (C) Progeny virus recovered at 48 hpt from pMNVS7F-transfected HEK293T cells were inoculated onto RAW264.7 cells. The RAW264.7 cells at 48 h postinfection were subjected to IF microscopy with anti-VP1 sera and anti–N-term sera. Blue, nucleus; green, VP1; red, N-terminal protein.
Discussion
The absence of a robust cell culture model for HuNoV infection has limited the study of the mechanisms that regulate viral replication and virus–host interactions as well as the development of effective antivirals. Previously, we reported the first experiments to establish reverse genetics systems for HuNoV GI.1 NV68 (10) and GII.3 U201 strains using recombinant vaccinia virus T7-based systems (11). These two systems produced nonstructural and structural proteins VP1 and VP2, respectively, from the subgenomic RNA, but particle production was inefficient. Recruitment of host cell translation initiation factors to the cytoplasmic replication factories produced during vaccinia virus replication (24) may have contributed to the inefficient NoV replication, structural protein translation, and particle assembly. Use of MVA/T7 to establish reverse genetics systems for calicivirus has varied and is successful with feline calicivirus (FCV) (25) but not porcine enteric calicivirus (PEC) (26) or MNV (27). A modified fowlpox virus expressing T7 RNA polymerase (FPV-T7) reverse genetics system recovers infectious MNV from cells transfected with a full-length MNV cDNA clone, but vaccinia virus inhibits MNV replication (27). The FPV-T7 system has been unable to directly recover infectious MNV from fully permissive RAW264.7 cells, presumably because of poor transfection rates and inefficient FPV-T7 infection in the RAW264.7 cells. Although reverse genetics systems with helper virus show the ability of mammalian cells to produce progeny virus, we sought to establish a simple, helper virus-free system to overcome the potential risk of inhibition of HuNoV replication by helper virus.
DNA-based systems using the pol II promoter to drive expression of viral cDNA directly or by baculovirus delivery have produced MNV (16), and a CMV promoter system recovered rabbit hemorrhagic disease virus (RHDV) (28). In vitro-transcribed RNA-based systems have recently been successful for several animal NoVs and caliciviruses, with variation in the requirement for capped or uncapped RNA [FCV (29, 30), PEC (26), MNV (15, 31), RHDV (32), and Tulane virus (33)]. Here, we report a generalizable reverse genetics system for HuNoVs that uses the EF-1α promoter and viral cDNA to produce progeny virus containing infectious RNA as well as has the ability to recover virus containing a reporter GFP gene. Infectious MNV is also recovered using this system.
These studies clarify several unanswered questions about HuNoVs. First, they show that authentic U201 RNA is produced and that the sequence of the subgenomic RNA generated by this reverse genetics system is identical to the predicted native subgenomic RNA. This finding confirms results from the MVA/T7 system that expressed the NV genome (10). Second, they show that progeny virus with a similar density to native virions (11) contains not only the predicted VP1, VP2, and genomic RNA but also, VPg (Fig. 3 A–C). Progeny HuNoV recovered from this reverse genetics system contains authentic infectious VPg-linked RNA, which was supported by the demonstration that RNA extracted from progeny virus is infectious (Fig. 4). This outcome mimics previous results showing that viral RNA extracted from the stools of infected volunteers is infectious when it contains VPg (22). An unexpected result was the detection of several distinct bands of VPg detected by Western blot that were apparently linked with RNA in the newly made, purified progeny virus (Fig. 3C). Treatment of the samples with RNase resulted in the appearance of a single VPg band. Although VPg-linked RNA is encapsidated, unfortunately, we cannot directly test the infectivity of the progeny virus, because we still lack a permissive culture system to grow HuNoVs. However, infectious NoV is made using this system, which was shown by the ability to produce MNV that infects RAW264.7 cells (Fig. 5C).
An unexplained finding is the detection of doublet bands on the Northern blot corresponding to the authentic negative genomic strand and a second larger strand (Fig. S4B, 48 hpt). We hypothesize that the larger band contains an extra 107 nt corresponding to the EF-1α exon sequence generated by the viral RdRp during negative-strand synthesis using the plasmid-generated transcript as a template, but we were not able to confirm it. Based on the previous observations that NoV RdRp activity is poly-A– and primer-independent (34) and that the authentic-sized negative- and positive-sense RNA bands are present, we suggest that the RdRp can recognize and initiate genomic RNA synthesis at the native 5′ end. A similar mechanism could be involved in the generation of subgenomic RNA.
This study made several previously unknown observations regarding HuNoV nonstructural protein processing and function. We found that six mature nonstructural proteins that comigrate with expressed recombinant proteins were detected by Western blot at 24 hpt (Fig. 1 B and C). Thus, cleavage of the precursor proteins seems to be quite efficient when the nonstructural proteins are expressed from the EF-1α promoter in COS7 cells; this finding contrasts with previous results, where intermediate precursors were detected in cells expressing the nonstructural proteins from the MVA/T7 system in HEK293T cells (11). One exception was the clear detection using only the VPg antiserum of an ∼40-kDa band by Western blot in pHuNoVU201F-transfected COS7 cells (Fig. 1B, asterisk and Fig. S3, α-VPg). This band seems to be distinct from the proposed ∼40-kDa precursor intermediate band previously detected with both VPg and 3A-like antisera in pT7U201F-transfected HEK293T cells (11). We suspect that this ∼40-kDa band is the 3A-like VPg precursor, and other faint bands at ∼60 and ∼120 kDa are additional VPg-containing precursors. It also is possible that different efficiencies of protease cleavage between different (COS7 and HEK293T) cell types explain the detection of different degrees of precursor cleavage for the U201 polyprotein.
Our studies provide information about the HuNoV 189-kDa precursor polyprotein and its cleavage products in mammalian cells. The polyprotein was only detected when constructs expressing a mutated protease were evaluated by Western blot (Fig. 1), and diffuse cytoplasmic staining of the polyprotein was detected by IF with only a subset of protein-specific antibodies (Fig. 2). The polyprotein was retained within the ER based on colocalization with an ER marker (Fig. 2B). It was susceptible to cleavage by WT protease added in trans, which resulted in a redistribution of the mature cleavage products that were all detected by protein-specific antibodies in new cellular locations (Fig. S2). The lack of detection of the polyprotein by the antibodies to NTPase and the 3A-like protein was unexpected (Fig. 2 and Fig. S2) but likely reflects masking of the epitopes in the uncleaved polyprotein structure. We also showed that the phenotype of the protease KO mutant can be negated by transfection of a plasmid that expresses a WT protease in trans (Figs. S2 and S3). This result illustrates one level of possible regulation of expression in this system. Demonstration that the mature HuNoV protease can function in trans in cells confirms results with other NoV and calicivirus systems that tested trans protease activity in bacterial or in vitro cell-free translation systems (18–20, 35–38) or cells transfected with ORF1 constructs (39). They also suggest that establishment of a cell line that expresses a mutant full-length HuNoV ORF1 construct that includes a reporter protein that would be released for detection after cleavage could lead to a useful assay to detect superinfection with an infectious HuNoV with active protease that could release the reporter.
Using this reverse genetics system, we successfully produced several reporter constructs that provide insights into HuNoV replication and HuNoV–host interactions. Two constructs were made that incorporate GFP or Rluc reporters into the capsid protein within the ORF2 gene. These constructs allowed visualization and quantitation of the kinetics of capsid protein expression that complemented detection of VP1 and VP2 protein expression by IF (Fig. S4 D and E), although these proteins were not detected by Western blotting. Although viral structural proteins and virus particles are produced, VP1 expression is likely still lower than in native infection and replication in infected humans. The expression of VP1 peaked at 24 hpt (Fig. S5B), when most of the expressed VP1 may be used for assembly of progeny virus. In addition, VP1 expression sharply decreased after 24 hpt in conjunction with the onset of CPE, which was detected in cells exhibiting the strongest GFP signal in the time-lapse analysis (Fig. S5). Cells with CPE ultimately shrank and died, and the continued reduction of VP1 expression may be because of a decrease of cells containing the HuNoV expression vector, because the produced virus particles are not transmitted to other noninfected cells in these cultures.
The observation of CPE in cells supporting HuNoV U201 replication is consistent with previous results of CPE in Huh7 cells 4–5 d posttransfection with HuNoV RNA (22). In the current studies, some cells with a weak GFP signal (from 6 to 18 hpt) were affected with CPE when transfected with the pHuNoVU201F-ORF2GFP construct. Less CPE was seen in cells transfected with the RdRp KO construct (pHuNoVU201FΔ4607-ORF2GFP), which would produce nonstructural proteins but not replicating RNA or amplification of the nonstructural proteins (Fig. S5A). The enhanced CPE seen at later times posttransfection when the capsid proteins would be expressed as well as results using constructs expressing VP1 and VP2 from a single plasmid or individual plasmids suggested that CPE may correlate with the expression of VP2 in cells. These results are consistent with the existence of replicon-bearing cell lines (40) that stably maintain the HuNoV genome without induction of CPE. The replicon cells were selected by virtue of the insertion of an antibiotic resistance gene into the VP1 capsid sequence of the HuNoV, and this system maintains an HuNoV replicon that continuously expresses nonstructural proteins but not the complete VP1 protein or any VP2 protein. Either CPE correlates with expression of VP1 and VP2 or adaptation of the replicon cell line resulted in host or viral mutations that negate induction of CPE.
Other caliciviruses induce apoptosis that correlates with the onset of CPE and is reported to involve caspase activation in FCV- (41) and MNV-infected cells (42), which may be activated by cathepsin B (43). In these systems, it is postulated that the virus may induce apoptosis to expand the window of time for virus replication, and caspase inhibitors reduce MNV replication (43). However, the best characterized noroviral or calicivirus proteins that regulate or are affected by apoptosis [the ORF1 encoded polyprotein of MNV (44), the unique VF1 protein of MNV (45), and the leader of the capsid protein of FCV (46)] are either different or not expressed from the HuNoV genome, and therefore, the mechanism of CPE induction for HuNoV remains to be determined. Several of the HuNoV nonstructural proteins could be involved. They include either or both of the two nonstructural proteins [the N-terminal protein (p48) and the 3A-like (p22) protein] that reportedly inhibit cellular protein secretion (47, 48) or the protease that cleaves the poly-(A)–binding protein and reduces cellular protein translation (49). Additional work using our reverse genetics system is required to understand the mechanism of CPE induction in HuNoV-expressing cells.
Immortalized cell lines are at least partially competent for HuNoV replication, because the RNA of Norwalk virus NV68 strain isolated from fecal samples can undergo a single round of replication when transfected into cells (22). To pursue screening and identification of a fully permissive infection system for HuNoVs in cultured human cells, we produced reporter particles with GFP inserted into ORF1 that allows detection of HuNoV protein expression in cells. Although these particles are not directly infectious in currently tested cell systems, they should facilitate additional screening of molecules needed for viral infectivity in cultured cells. Others have sought to make reporter calicivirus systems. A site in the FCV genome that produces the capsid leader protein and can tolerate GFP insertion was identified, and detection of mature protein was consistent with the progression of CPE; however, serial passage of the constructs resulted in reduced or lost foreign protein expression (50). Attempts to produce Tulane virus with GFP inserted into the N-terminal protein resulted in impaired infectivity (33). The stability of the GFP-marked progeny HuNoV remains to be assessed.
Overall, we have developed a universal system that exploits EF-1α expression of an HuNoV genome using the GII.3 U201 strain as well as other HuNoV strains. The EF-1α system produces progeny viruses, although the yield of virus differs according to the virus strain being expressed and the cell line tested. HEK293T cells are capable of progeny virus production for all strains used to date. For the U201 strain, virus yields are highest in COS7 and 293T cells (production levels measured by quantification of encapsidated genome copies were 8.0 × 104 and 1.4 × 104 copies/106 cells, respectively). Lower yields were obtained in Huh7 and Caco2 cells (2.4 × 102 and 1.3 × 101 copies/106 cells, respectively) (Table 1 and Fig. S7). Because our expression constructs contain the SV40 replication origin, the higher efficiency of progeny virus production may be affected by the presence of large T antigen in COS7 and HEK293T cells compared with Huh7 and Caco2 cells that lack T antigen. Insertion of the GFP reporter into the U201 genome decreased yields, but they were still the highest in the COS7 and HEK293 cells. The varied yields of the other HuNoV strains tested deserve comment. Expression of the TCH04-577 strain, which is a chimeric virus that contains GII.4 nonstructural proteins and the capsid proteins of a GII.3 virus, produced low yields in COS7 cells, with 10-fold higher yields in 293T cells, like the GI.1 NV68 strain. Future work will be needed to determine if transcription or translation factors of COS7 cells are better suited for expression and production for GII.3 viruses, such as the U201 genome, and are not as optimized for nonstructural protein expression of GII.4 as well GI.1 virus strains. Such work could lead to identification and understanding of strain-specific virus–host interactions.
Our success in using this system to express the MNV S7 strain (51–53) genome (Fig. 5 and Table 1) and produce infectious virus further validates this reverse genetics system. A rapid and simple single-plasmid transfection in HEK293T cells allows for the efficient and facile recovery of recombinant HuNoV strains and MNV. This system also provides the first helper-free transient replicon system, to our knowledge, for any HuNoV that can be readily modified and incorporated into screens for small-molecule inhibitors or other antivirals. The GFP-tagged progeny virus production system should be useful to find susceptible cells and cell lines. Virus recovery is highly reproducible, produces authentic genome, and will be useful for additional structure–function studies in many laboratories to allow substantial progress into the pathology and biology of virus–host interactions for HuNoV as well as will be useful to test antivirals.
Materials and Methods
Stool Sample.
An original HuNoV GII.3 U201 stool sample was collected from an outbreak of gastroenteritis in Saitama prefecture in Japan in 1998. The U201 stool sample containing 109 copies/g stool HuNoV RNA was provided by Michiyo Shinohara and Kazue Uchida (Saitama Institute of Public Health, Saitama, Japan).
Plasmid Transfection and Detection of Protein and RNA Expression.
For IF microscopy, Western blotting, and Northern blotting, cells were plated into 24-well plates at a density of 5 × 104 cells/well. After an overnight incubation at 37 °C, the cells were washed two times with DMEM containing 2% (vol/vol) FBS, and 100 ng plasmid DNA construct per well was transfected using TransIT LT-1 (Mirus) following the manufacturer’s instructions. The pDsRed2-ER Vector (Clontech) was cotransfected to visualize ER as described above. For progeny virus production, cells were plated into 10 T-225 culture plates containing 70% confluent cells, and the plasmid DNA construct (120 μg/flask) was transfected using TransIT LT-1 according to the manufacturer’s instruction and then cultured at 37 °C for 30 h.
Detection of Expressed HuNoV GII.3 U201 Proteins in Transfected Cells by IF Analysis.
At various hpt, cells were rinsed one time with 0.1 M PBS and fixed in 4% (wt/vol) paraformaldehyde for 30 min at room temperature. After incubation of cells in 0.5% Triton X-100 and 0.1 M PBS for 15 min at room temperature to permeabilize, cells were blocked at 37 °C for 2 h in 0.1 M PBS containing 1% BSA. Primary antibodies (1 μg/mL IgG final concentration) were added to the wells and incubated overnight at 4 °C. After washing with 0.1 M PBS, the cells were incubated for 2 h at room temperature in a 1:1,500 dilution of the corresponding secondary antibody conjugated to Alexa Fluor 594 or 488. Nuclei were stained with 300 nM DAPI for 15 min at room temperature. After the final wash step, IF was performed with the IX-70 inverted microscope, the IX-81 inverted microscope with the DSU spinning disk deconvolution system (Olympus), and the A1-Rs inverted laser-scanning microscope (Nikon).
Purification of Progeny Virus Particles.
The pooled culture supernatant and cells were harvested with nuclease-free water including 0.5% Zwittergent detergent (Calbiochem) and extracted with Vertrel XF (Miller-Stephenson). After centrifugation for 10 min at 12,400 × g, progeny virions were precipitated by a PEG-NaCl precipitation as described previously (22). The precipitated virus was pelleted for 15 min at 10,000 × g, and the pellet was suspended in 0.1 M PBS. The virus suspension was pelleted through a 40% (wt/vol) sucrose cushion for 3 h at 124,000 × g and further purified by isopycnic CsCl-gradient centrifugation in milli-Q water (0.44 g/mL) for 24 h at 150,000 × g by using a Beckman SW55 Ti rotor. After gradient fractionation, each fraction was diluted 10 times in milli-Q water, and viruses were recovered by ultracentrifugation for 3 h at 150,000 × g. The presence of virus in each fraction was analyzed by Western blotting, Northern blotting, and EM after staining with 2% (wt/vol) uranyl acetate (pH 4). Isolation of RNA from each fraction and produced virus was performed using the QIAamp Viral RNA Mini Kit (Qiagen) following the manufacturer’s instructions.
Observation of Reporter Expression in Live Cells.
To observe reporter gene expression in live cells, 70% confluent cells were cultured in 35-mm glass-bottomed dishes. The plasmid construct pHuNoVU201F-ORF2GFP or pHuNoVU201F∆4607G-ORF2GFP that contained the GFP gene as a reporter was transfected into the cells in the same way as described above. Live cell images were acquired at 48 hpt and reconstructed with the LCV110 Live Cell Imager (Olympus).
Detection and Transfection of Encapsidated RNA Extracted from Progeny Virus Particles.
At 30 hpt, the culture supernatants and cells were harvested, and progeny virus particles were purified in the same way as described above. Purified progeny virus particles was pretreated with 0.01 M MgCl2, 10 mM Tris (pH 8.0), and 10 units DNase and RNase A for 30 min at 37 °C. As a control, 500 ng in vitro-transcribed HuNoV U201 RNA and pHuNoVU201F plasmid were also treated in the same manner as the control described above. For extraction of encapsidated NoV RNA, purified progeny virions were treated with 20 μg/mL RNase A for 30 min at 37 °C. HuNoV RNA was extracted using the QIAamp Viral RNA Mini Kit (Qiagen) and used to transfect the cells. The GFP signals in RNA-transfected cells were observed with the Olympus inverted-system microscopes described above.
Infection of MNV to RAW264.7 Cells.
HEK293T cells cultured in a six-well plate with 2 mL culture media were transfected with pMNVS7F and pMNVS7FΔ4572G using Lipofectamine 2000 (Invitrogen). Culture supernatants were collected at 48 hpt and stored at −80 °C until use. Supernatant (500 μL) was inoculated onto RAW264.7 cells cultured in six-well plates. Cells and supernatants at 48 h postinfection were subjected to the following analysis. MNV infectivity was determined by TCID50 assays in RAW264.7 cells (27). Briefly, 10 wells of RAW264.7 cells seeded in a 96-well plate were inoculated with serially diluted culture supernatants (50 μL) of transfected 293T cells. After 7 d, cytopathic effect was observed by light microscopy.
Supplementary Material
Acknowledgments
We thank Dr. Shinji Makino for providing the Huh7 cells, Dr. Yukinobu Tohya for critical reading of the manuscript and providing the MNV-S7 strain, and Dr. Michiyo Kataoka for performing the EM observation. This work was funded by National Institutes of Health Grants P01AI57788, N01AI25465, and P30DK56338; Agriculture and Food Research Initiative Competitive Grant 2011-68003-30395; grants from the Ministry of Health, Labor, and Welfare of Japan; and Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research KAKENHI. This project was supported by the Integrated Microscopy Core at Baylor College of Medicine with funding from National Institutes of Health Grants HD007495, DK56338, and CA125123, the Dan L. Duncan Cancer Center, and the John S. Dunn Gulf Coast Consortium for Chemical Genomics.
Footnotes
Conflict of interest statement: R.L.A. and M.K.E. have received research support and served as consultants to Takeda Vaccines, Inc.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415096111/-/DCSupplemental.
References
- 1.Ramani S, Atmar RL, Estes MK. Epidemiology of human noroviruses and updates on vaccine development. Curr Opin Gastroenterol. 2014;30(1):25–33. doi: 10.1097/MOG.0000000000000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall AJ, et al. Norovirus disease in the United States. Emerg Infect Dis. 2013;19(8):1198–1205. doi: 10.3201/eid1908.130465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel MM, Hall AJ, Vinjé J, Parashar UD. Noroviruses: A comprehensive review. J Clin Virol. 2009;44(1):1–8. doi: 10.1016/j.jcv.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. N Engl J Med. 2009;361(18):1776–1785. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herbst-Kralovetz MM, et al. Lack of norovirus replication and histo-blood group antigen expression in 3-dimensional intestinal epithelial cells. Emerg Infect Dis. 2013;19(3):431–438. doi: 10.3201/eid1903.121029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papafragkou E, Hewitt J, Park GW, Greening G, Vinjé J. Challenges of culturing human norovirus in three-dimensional organoid intestinal cell culture models. PLoS ONE. 2013;8(6):e63485. doi: 10.1371/journal.pone.0063485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takanashi S, et al. Failure of propagation of human norovirus in intestinal epithelial cells with microvilli grown in three-dimensional cultures. Arch Virol. 2014;159(2):257–266. doi: 10.1007/s00705-013-1806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taube S, et al. A mouse model for human norovirus. MBio. 2013;4(4):e00450-13. doi: 10.1128/mBio.00450-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheetham S, et al. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J Virol. 2006;80(21):10372–10381. doi: 10.1128/JVI.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asanaka M, et al. Replication and packaging of Norwalk virus RNA in cultured mammalian cells. Proc Natl Acad Sci USA. 2005;102(29):10327–10332. doi: 10.1073/pnas.0408529102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katayama K, Hansman GS, Oka T, Ogawa S, Takeda N. Investigation of norovirus replication in a human cell line. Arch Virol. 2006;151(7):1291–1308. doi: 10.1007/s00705-005-0720-9. [DOI] [PubMed] [Google Scholar]
- 12.Green K. Caliciviridae: The noroviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 6th Ed. Vol 1. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 582–608. [Google Scholar]
- 13.Chaudhry Y, et al. Caliciviruses differ in their functional requirements for eIF4F components. J Biol Chem. 2006;281(35):25315–25325. doi: 10.1074/jbc.M602230200. [DOI] [PubMed] [Google Scholar]
- 14.Daughenbaugh KF, Wobus CE, Hardy ME. VPg of murine norovirus binds translation initiation factors in infected cells. Virol J. 2006;3:33. doi: 10.1186/1743-422X-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arias A, Ureña L, Thorne L, Yunus MA, Goodfellow I. Reverse genetics mediated recovery of infectious murine norovirus. J Vis Exp. 2012;64(2012):4145. doi: 10.3791/4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward VK, et al. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc Natl Acad Sci USA. 2007;104(26):11050–11055. doi: 10.1073/pnas.0700336104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uetsuki T, Naito A, Nagata S, Kaziro Y. Isolation and characterization of the human chromosomal gene for polypeptide chain elongation factor-1 alpha. J Biol Chem. 1989;264(10):5791–5798. [PubMed] [Google Scholar]
- 18.Liu B, Clarke IN, Lambden PR. Polyprotein processing in Southampton virus: Identification of 3C-like protease cleavage sites by in vitro mutagenesis. J Virol. 1996;70(4):2605–2610. doi: 10.1128/jvi.70.4.2605-2610.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sosnovtsev SV, et al. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J Virol. 2006;80(16):7816–7831. doi: 10.1128/JVI.00532-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belliot G, et al. In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J Virol. 2003;77(20):10957–10974. doi: 10.1128/JVI.77.20.10957-10974.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oka T, et al. Structural and biological constraints on diversity of regions immediately upstream of cleavage sites in calicivirus precursor proteins. Virology. 2009;394(1):119–129. doi: 10.1016/j.virol.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 22.Guix S, et al. Norwalk virus RNA is infectious in mammalian cells. J Virol. 2007;81(22):12238–12248. doi: 10.1128/JVI.01489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroneman A, et al. Proposal for a unified norovirus nomenclature and genotyping. Arch Virol. 2013;158(10):2059–2068. doi: 10.1007/s00705-013-1708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katsafanas GC, Moss B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe. 2007;2(4):221–228. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sosnovtsev SV, Garfield M, Green KY. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J Virol. 2002;76(14):7060–7072. doi: 10.1128/JVI.76.14.7060-7072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang KO, et al. Reverse genetics system for porcine enteric calicivirus, a prototype sapovirus in the Caliciviridae. J Virol. 2005;79(3):1409–1416. doi: 10.1128/JVI.79.3.1409-1416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhry Y, Skinner MA, Goodfellow IG. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J Gen Virol. 2007;88(Pt 8):2091–2100. doi: 10.1099/vir.0.82940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu G, et al. A DNA-launched reverse genetics system for rabbit hemorrhagic disease virus reveals that the VP2 protein is not essential for virus infectivity. J Gen Virol. 2008;89(Pt 12):3080–3085. doi: 10.1099/vir.0.2008/003525-0. [DOI] [PubMed] [Google Scholar]
- 29.Sosnovtsev S, Green KY. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VpG for infectivity. Virology. 1995;210(2):383–390. doi: 10.1006/viro.1995.1354. [DOI] [PubMed] [Google Scholar]
- 30.Thumfart JO, Meyers G. Feline calicivirus: Recovery of wild-type and recombinant viruses after transfection of cRNA or cDNA constructs. J Virol. 2002;76(12):6398–6407. doi: 10.1128/JVI.76.12.6398-6407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yunus MA, Chung LM, Chaudhry Y, Bailey D, Goodfellow I. Development of an optimized RNA-based murine norovirus reverse genetics system. J Virol Methods. 2010;169(1):112–118. doi: 10.1016/j.jviromet.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu G, et al. Recovery of infectious rabbit hemorrhagic disease virus from rabbits after direct inoculation with in vitro-transcribed RNA. J Virol. 2006;80(13):6597–6602. doi: 10.1128/JVI.02078-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei C, Farkas T, Sestak K, Jiang X. Recovery of infectious virus by transfection of in vitro-generated RNA from tulane calicivirus cDNA. J Virol. 2008;82(22):11429–11436. doi: 10.1128/JVI.00696-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukushi S, et al. Poly(A)- and primer-independent RNA polymerase of Norovirus. J Virol. 2004;78(8):3889–3896. doi: 10.1128/JVI.78.8.3889-3896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu BL, Viljoen GJ, Clarke IN, Lambden PR. Identification of further proteolytic cleavage sites in the Southampton calicivirus polyprotein by expression of the viral protease in E. coli. J Gen Virol. 1999;80(Pt 2):291–296. doi: 10.1099/0022-1317-80-2-291. [DOI] [PubMed] [Google Scholar]
- 36.Blakeney SJ, Cahill A, Reilly PA. Processing of Norwalk virus nonstructural proteins by a 3C-like cysteine proteinase. Virology. 2003;308(2):216–224. doi: 10.1016/s0042-6822(03)00004-7. [DOI] [PubMed] [Google Scholar]
- 37.Scheffler U, Rudolph W, Gebhardt J, Rohayem J. Differential cleavage of the norovirus polyprotein precursor by two active forms of the viral protease. J Gen Virol. 2007;88(Pt 7):2013–2018. doi: 10.1099/vir.0.82797-0. [DOI] [PubMed] [Google Scholar]
- 38.Wei C, Meller J, Jiang X. Substrate specificity of Tulane virus protease. Virology. 2013;436(1):24–32. doi: 10.1016/j.virol.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seah EL, Marshall JA, Wright PJ. Trans activity of the norovirus Camberwell proteinase and cleavage of the N-terminal protein encoded by ORF1. J Virol. 2003;77(12):7150–7155. doi: 10.1128/JVI.77.12.7150-7155.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang KO, Sosnovtsev SV, Belliot G, King AD, Green KY. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology. 2006;353(2):463–473. doi: 10.1016/j.virol.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Sosnovtsev SV, Prikhod’ko EA, Belliot G, Cohen JI, Green KY. Feline calicivirus replication induces apoptosis in cultured cells. Virus Res. 2003;94(1):1–10. doi: 10.1016/s0168-1702(03)00115-1. [DOI] [PubMed] [Google Scholar]
- 42.Bok K, Prikhodko VG, Green KY, Sosnovtsev SV. Apoptosis in murine norovirus-infected RAW264.7 cells is associated with downregulation of survivin. J Virol. 2009;83(8):3647–3656. doi: 10.1128/JVI.02028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furman LM, et al. Cysteine protease activation and apoptosis in Murine norovirus infection. Virol J. 2009;6:139. doi: 10.1186/1743-422X-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herod MR, et al. Expression of the murine norovirus (MNV) ORF1 polyprotein is sufficient to induce apoptosis in a virus-free cell model. PLoS ONE. 2014;9(3):e90679. doi: 10.1371/journal.pone.0090679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McFadden N, et al. Norovirus regulation of the innate immune response and apoptosis occurs via the product of the alternative open reading frame 4. PLoS Pathog. 2011;7(12):e1002413. doi: 10.1371/journal.ppat.1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abente EJ, et al. The feline calicivirus leader of the capsid protein is associated with cytopathic effect. J Virol. 2013;87(6):3003–3017. doi: 10.1128/JVI.02480-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ettayebi K, Hardy ME. Norwalk virus nonstructural protein p48 forms a complex with the SNARE regulator VAP-A and prevents cell surface expression of vesicular stomatitis virus G protein. J Virol. 2003;77(21):11790–11797. doi: 10.1128/JVI.77.21.11790-11797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharp TM, Guix S, Katayama K, Crawford SE, Estes MK. Inhibition of cellular protein secretion by norwalk virus nonstructural protein p22 requires a mimic of an endoplasmic reticulum export signal. PLoS ONE. 2010;5(10):e13130. doi: 10.1371/journal.pone.0013130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuyumcu-Martinez M, et al. Calicivirus 3C-like proteinase inhibits cellular translation by cleavage of poly(A)-binding protein. J Virol. 2004;78(15):8172–8182. doi: 10.1128/JVI.78.15.8172-8182.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abente EJ, Sosnovtsev SV, Bok K, Green KY. Visualization of feline calicivirus replication in real-time with recombinant viruses engineered to express fluorescent reporter proteins. Virology. 2010;400(1):18–31. doi: 10.1016/j.virol.2009.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitagawa Y, et al. Indirect ELISA and indirect immunofluorescent antibody assay for detecting the antibody against murine norovirus S7 in mice. Exp Anim. 2010;59(1):47–55. doi: 10.1538/expanim.59.47. [DOI] [PubMed] [Google Scholar]
- 52.Kitajima M, et al. Development of a broadly reactive nested reverse transcription-PCR assay to detect murine noroviruses, and investigation of the prevalence of murine noroviruses in laboratory mice in Japan. Microbiol Immunol. 2009;53(9):531–534. doi: 10.1111/j.1348-0421.2009.00152.x. [DOI] [PubMed] [Google Scholar]
- 53.Kitajima M, et al. Development and application of a broadly reactive real-time reverse transcription-PCR assay for detection of murine noroviruses. J Virol Methods. 2010;169(2):269–273. doi: 10.1016/j.jviromet.2010.07.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.