Significance
Reversible and repetitive structural changes are essential for ligand-gated ion channels to mediate biological signaling. The inositol 1,4,5-trisphosphate receptor (IP3R) assembles ligand-gated ion channels that mediate calcium signaling. IP3 activates channels at a distance by reversible allosteric changes in the IP3R tetramer. Here we show a new mode of posttranslational modification that irreversibly blocks allosteric changes in the IP3R. We identified an IP3R-modifying enzyme as tissue transglutaminase that inhibits IP3R function by locking subunit configurations. This modification chronically impaired calcium signaling and autophagy regulation in living cells, and up-regulated modification was observed in Huntington disease models. To our knowledge, this is the first demonstration of transglutaminase-catalyzed posttranslational modification in ligand-gated channel allostery and provides a new framework for enzymatic regulation of allostery.
Keywords: allosteric regulation, IP3 receptor, isopeptide bond, transamidation, deamidation
Abstract
The inositol 1,4,5-trisphosphate receptor (IP3R) in the endoplasmic reticulum mediates calcium signaling that impinges on intracellular processes. IP3Rs are allosteric proteins comprising four subunits that form an ion channel activated by binding of IP3 at a distance. Defective allostery in IP3R is considered crucial to cellular dysfunction, but the specific mechanism remains unknown. Here we demonstrate that a pleiotropic enzyme transglutaminase type 2 targets the allosteric coupling domain of IP3R type 1 (IP3R1) and negatively regulates IP3R1-mediated calcium signaling and autophagy by locking the subunit configurations. The control point of this regulation is the covalent posttranslational modification of the Gln2746 residue that transglutaminase type 2 tethers to the adjacent subunit. Modification of Gln2746 and IP3R1 function was observed in Huntington disease models, suggesting a pathological role of this modification in the neurodegenerative disease. Our study reveals that cellular signaling is regulated by a new mode of posttranslational modification that chronically and enzymatically blocks allosteric changes in the ligand-gated channels that relate to disease states.
Ligand-gated ion channels function by allostery that is the regulation at a distance; the allosteric coupling of ligand binding with channel gating requires reversible changes in subunit configurations and conformations (1). Inositol 1,4,5-trisphosphate receptors (IP3Rs) are ligand-gated ion channels that release calcium ions (Ca2+) from the endoplasmic reticulum (ER) (2, 3). IP3Rs are allosteric proteins comprising four subunits that assemble a calcium channel with fourfold symmetry about an axis perpendicular to the ER membrane. The subunits of three IP3R isoforms (IP3R1, IP3R2, and IP3R3) are structurally divided into three domains: the IP3-binding domain (IBD), the regulatory domain, and the channel domain (3–6). Fitting of the IBD X-ray structures (7, 8) to a cryo-EM map (9) indicates that the IBD activates a remote Ca2+ channel by allostery (8); however, the current X-ray structure only spans 5% of each tetramer, such that the mechanism underlying allosteric coupling of the IBD to channel gating remains unknown.
The IP3R in the ER mediates intracellular calcium signaling that impinges on homeostatic control in various subsequent intracellular processes. Deletion of the genes encoding the type 1 IP3R (IP3R1) leads to perturbations in long-term potentiation/depression (3, 10, 11) and spinogenesis (12), and the human genetic disease spinocerebellar ataxia 15 is caused by haploinsufficiency of the IP3R1 gene (13–15). Dysregulation of IP3R1 is also implicated in neurodegenerative diseases including Huntington disease (HD) (16–18) and Alzheimer’s disease (AD) (19–22). IP3Rs also control fundamental cellular processes—for example, mitochondrial energy production (23, 24), autophagy regulation (24–27), ER stress (28), hepatic gluconeogenesis (29), pancreatic exocytosis (30), and macrophage inflammasomes (31). On the other hand, excessive IP3R function promotes cell death processes including apoptosis by activating mitochondrial or calpain pathways (2, 17). Considering these versatile roles of IP3Rs, appropriate IP3R structure and function are essential for living systems, and aberrant regulation of IP3R closely relates to various diseases.
Several factors such as cytosolic molecules, interacting proteins, and posttranslational modifications control the IP3-induced Ca2+ release (IICR) through allosteric sites in IP3Rs. Cytosolic Ca2+ concentrations strictly control IICR in a biphasic manner with activation at low concentrations and inhibition at higher concentrations. The critical Ca2+ sensor for activation is conserved among the three isoforms of IP3 and ryanodine receptors, and this sensor is located in the regulatory domain outside the IBD and the channel domain (32). A putative ATP regulatory region is deleted in opisthotonos mice, and IICR is also regulated by this mutation in the regulatory domain (33). Various interacting proteins, such as cytochrome c, Bcl-2-family proteins, ataxin-3, huntingtin (Htt) protein, Htt-associated protein 1A (HAP1A), and G-protein–coupled receptor kinase-interacting protein 1 (GIT1), target allosteric sites in the carboxyl-terminal tail (3–5). The regulatory domain and the carboxyl-terminal tail also undergo phosphorylation by the protein kinases A/G and B/Akt and contain the apoptotic cleavage sites for the protease caspase-3 (4, 5). These factors allosterically regulate IP3R structure and function to control cellular fates; therefore, understanding the allosteric coupling of the IBD to channel gating will elucidate the regulatory mechanism of these factors.
Transglutaminase (TG) catalyses protein cross-linking between a glutamine (Gln) residue and a lysine (Lys) residue via an Nε-(γ-glutamyl)lysine isopeptide bond (34, 35). TG type 2 (TG2) is a Ca2+-dependent enzyme with widespread distribution and is highly inducible by various stimulations such as oxidative stress, cytokines, growth factors, and retinoic acid (RA) (34, 35). TG2 is considered a significant disease-modifying factor in neurodegenerative diseases including HD, AD, and Parkinson’s diseases (PD) (34, 36–45) because TG2 might enzymatically stabilize aberrant aggregates of proteins implicated in these diseases—that is, mutant Htt, β-amyloid, and α-synuclein; however, the causal role of TG2 in Ca2+ signaling in brain pathogenesis has been unclear. Ablation of TG2 in HD mouse models is associated with increased lifespan and improved motor function (46, 47). However, TG2 knockout mice do not show impaired Htt aggregation, suggesting that TG2 may play a causal role in these disorders rather than TG2-dependent cross-links in aberrant protein aggregates (47, 48).
In this study, we discovered a new mode of chronic and irreversible allosteric regulation in IP3R1 in which covalent modification of the receptor at Gln2746 is catalyzed by TG2. We demonstrate that up-regulation of TG2 modifies IP3R1 structure and function in HD models and propose an etiologic role of this modification in the reduction of neuronal signaling and subsequent processes during the prodromal state of the neurodegenerative disease.
Results
Identification of an IP3R1-Modifying Enzyme as TG.
In exploring a new allosteric mechanism of IP3R tetramers, we developed an agarose gel electrophoresis (AGE) system that can evaluate the large quaternary structure of IP3R1 (>1 MDa) (Fig. 1 A and B). We found that the incubation of IP3R1 with brain cytosol resulted in cross-linking of the IP3R1subunits (Fig. 1C), indicating the existence of an endogenous cross-linking enzyme in the brain. We fractionated the cross-linking activity using chromatography (Fig. S1 A–C). The Ca2+ dependence, elution profile of the activity, and inhibition with cystamine led us to postulate that the cross-linking might be catalyzed by a TG, most likely TG2 (34, 49).
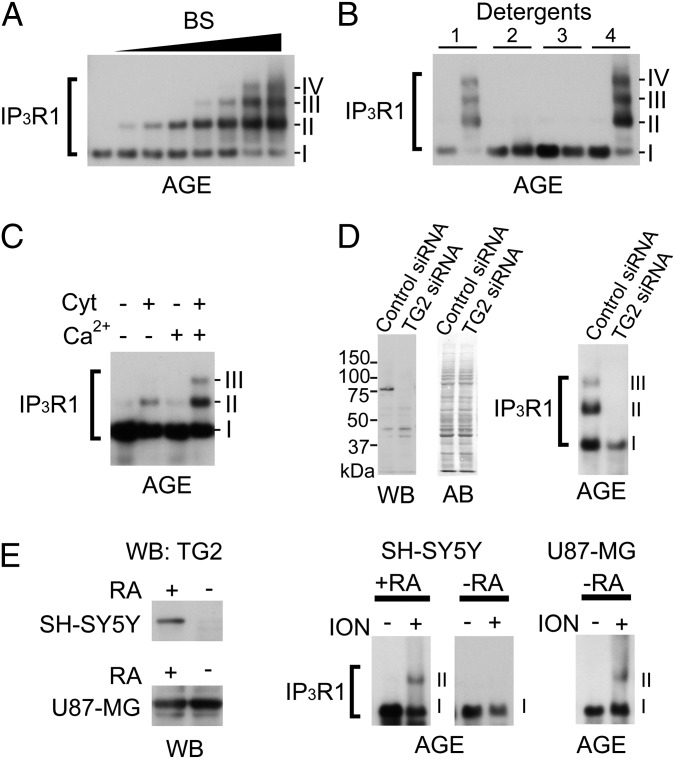
Fig. 1.
Identification of an IP3R1-modifying enzyme as TG. (A) Detection of cross-linked IP3R1 by AGE. IP3R1 subunits were cross-linked by the chemical cross-linker bis[sulfosuccinimidyl] suberate (BS). Roman numerals indicate cross-linked subunits observed by AGE; IP3R1 sizes were compared with the thyroglobulin monomer (335 kDa) and dimer (670 kDa). (B) IP3R1 (left lane) pretreated with Triton X-100 (1), Zwittergent 3–14 (2), SDS (3), or no detergent (4) was cross-linked with BS (right lane), indicating the oligomeric status (97). (C) Endogenous activity of Ca2+-dependent cross-linking between IP3R1 subunits in the brain cytosol (Cyt). (D) Cross-linking activity in HeLa cells pretreated with control siRNA or TG2 siRNA for 48 h. Western blots (WBs) with αTG2 and amido black (AB) staining show the cellular levels of TG2 and total proteins, respectively. (E) TG2 expression levels in SH-SY5Y or U87-MG cells treated with RA or DMSO. The Right panels show cross-linking of IP3R1 by 10 μM ionomycin (ION) in human SH-SY5Y or U87-MG cells, as revealed by AGE.
The direct cross-linking of IP3R1 by TG2 was confirmed using purified IP3R1 and TG2 (Fig. S1D). To examine the role of TG2 in the cross-linking of IP3R1, we silenced TG2 expression using siRNA in HeLa cells. TG2 knockdown completely abolished the in vitro cross-linking activity in HeLa cell lysates (Fig. 1D), indicating that TG2 is essential for the cross-linking of IP3R1. In contrast, induction of TG2 expression by RA enhanced the cross-linking activity in HeLa cells (Fig. S1E). Similar cross-linking was observed in living cells including human neuroblastoma (SH-SY5Y) and human glioblastoma (U87-MG) cells (Fig. 1E). These results support the conclusion that IP3R1 is an endogenous substrate of TG2 in living cells.
TG2 Inhibits IP3R-Mediated Ca2+ Release.
The AGE analysis revealed that TG2 modifies the IP3R1 structure; therefore, we examined whether TG2 regulates IP3R1 function. We measured IICR using P2/P3 vesicles and found that purified TG2 inhibited IICR in a dose-dependent manner, without altering ATP-dependent Ca2+ uptake or the total pool of Ca2+ (Fig. 2A). The inhibitory effects were not observed using boiled TG2 or N-ethylmaleimide (NEM)-treated TG2, indicating that the catalytic activity of TG2 was required for inhibition (Fig. 2A).
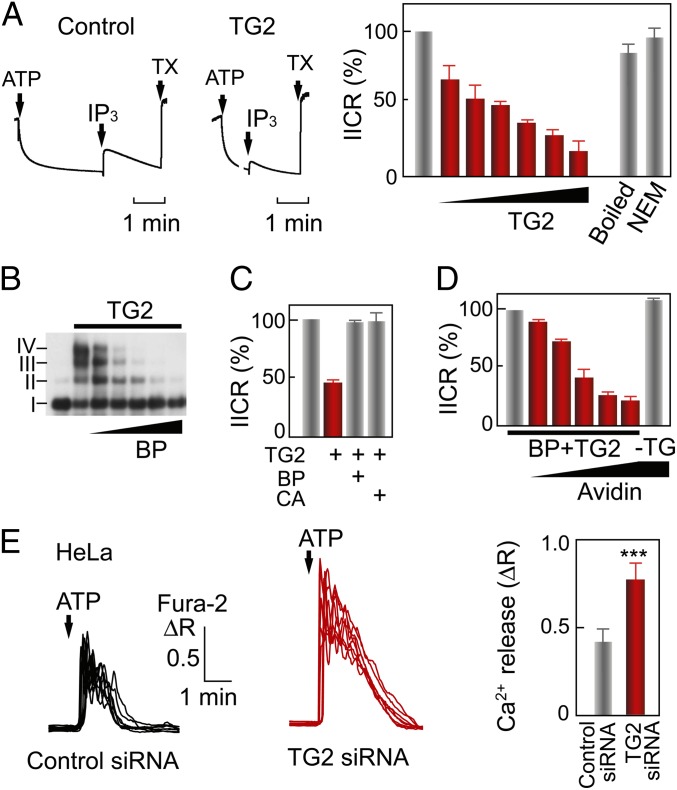
Fig. 2.
TG2 inhibits IP3R1-mediated Ca2+ release. (A) Effects of purified TG2 on IICR from P2/P3 vesicles after Ca2+ loading by adding 1 mM Mg–ATP. IICRs were evoked with 0.1 μM IP3, and the total Ca2+ pool was estimated using 10 mM Triton X-100 (TX). The bar graph summarizes the mean + SD from three independent experiments using P2/P3 vesicles pretreated with TG2 (0, 1, 2, 5, 10, 20, and 50 μg/mL), 10 μg/mL boiled TG2, or 10 μg/mL NEM–TG2. The IC50 value of TG2 is 34 nM, calculated by a nonlinear regression analysis using a four-parameter logistic function. (B) The BP was incorporated into the IP3R1 by TG2-catalyzed reactions. IP3R1 was pretreated with both TG2 and BP (1, 2, 5, 10, and 20 mM), and the oligomeric status was confirmed using AGE. (C) The IICR from TG2-pretreated P2/P3 vesicles prepared in the presence of 10 mM BP or 10 mM cystamine (CA). (D) The BP-containing (BP + TG) or BP-lacking (−TG) P2/P3 vesicles were pretreated with 10 or 0 μg/mL TG2. The IICR was measured in the presence of 0, 31, 63, 94, 125, and 156 μg/mL (slope) or 156 μg/mL (−TG) of avidin. (E) Fura-2 ratio images of living HeLa cells transfected with control (black lines) or TG2 (red lines) siRNA for 48 h were obtained under a nominally Ca2+-free condition. The bar graph represents the mean + SD of peak amplitudes (control siRNA, n = 117; TG2 siRNA, n = 119). ΔR indicates changes in the fura-2 ratio in response to 3 μM ATP. ***P = 8 × 10−18.
To elucidate the mechanism by which TG2 negatively regulates channel activity through covalent modification, we used a monoamine compound 5-biotinamido-pentylamine (BP) that contains both primary amine and biotin moieties. BP mimics a Lys residue, and TG2 can enzymaticaly incorporate BP into the Gln residue, which can be traced by horseradish peroxidase (HRP)-conjugated streptavidin. Therefore, we prepared BP-containing IP3R1 using purified TG2 and P2/P3 vesicles (Fig. 2B). The IICR assay revealed that the BP-containing IP3R1 has intact activity (Fig. 2C), indicating that the structures of both IP3-binding and Ca2+-permeating domains are not affected by the covalent incorporation of BP molecules. Removal of TG2 and BP followed by the addition of avidin proteins with a neutralized isoelectric point (NeutrAvidin) to the BP-containing IP3R1 inhibited IICR in a dose-dependent manner (Fig. 2D). These results indicate that steric hindrance near the BP-containing Gln residue is important for the allosteric inhibition of IICR.
To examine the inhibitory role of endogenous TG2 in living cells, Ca2+ imaging was performed using HeLa cells. TG2 silencing significantly enhanced ATP-evoked Ca2+ release in the Ca2+-free condition (Fig. 2E), indicating that endogenous TG2 inhibits IICR activity in living cells.
TG2 Negatively Regulates IP3R-Mediated Autophagy.
Our data indicate that TG2 inhibits IP3R1 function. To examine the physiological role of this regulation in the cellular functions of IP3R1, we studied the role of TG2 and IP3R in autophagy. IP3R1 regulates autophagy (24–27), and TG2 is also thought to regulate autophagy (50, 51); however, the interplay between TG2 and IP3R1 in autophagy regulation has not been studied. We hypothesized that TG2 affects IP3R1-mediated autophagy regulation. To examine autophagy, we used HeLa cells in which both IP3R1 and TG2 were expressed dominantly. IP3R1 knockdown increases LC3-II (lapidated microtubule-associated protein light chain 3, LC3, an index of autophagy) content without changing the amount of actin (Fig. 3A), consistent with previous reports (24–27). Furthermore, we found that TG2 knockdown had the opposite effect (Fig. 3A).
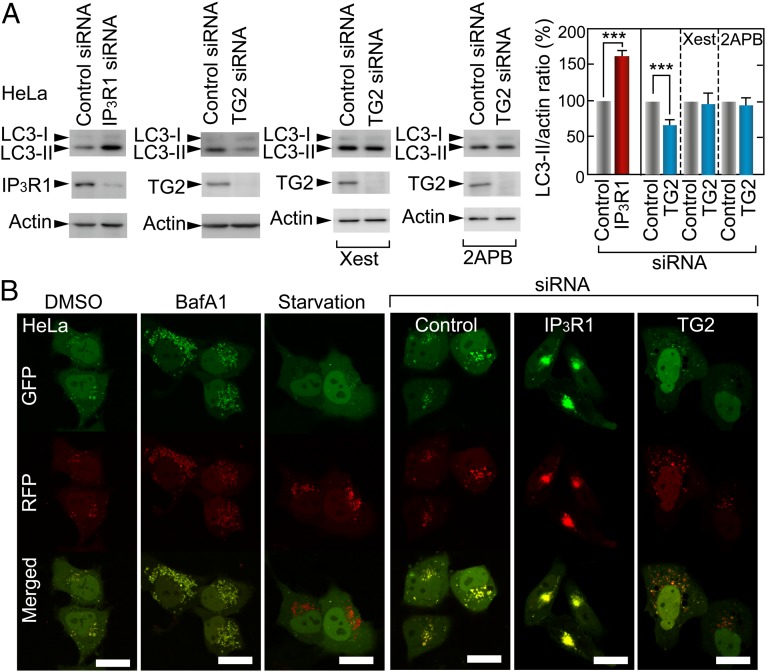
Fig. 3.
TG2 negatively regulates IP3R1-mediated autophagy processes. (A) LC3-I, LC3-II, IP3R1, TG2, and actin levels in HeLa cells were evaluated 48 h after transfection with control, IP3R, or TG2. We added 2 μM Xest or 100 μM 2-APB to HeLa cells 6 h after siRNA transfection. The bar chart represents the mean + SEM of LC3-II (LC3-II/actin ratio). ***P = 0.0011 (control versus IP3R1), ***P = 0.0031 (control versus TG2). (B) Loss of IP3R1 or TG2 affects autophagy as measured by GFP and RFP signals of an autophagosome indicator with tandem fluorophores (tfLC3) (52) in HeLa cells. We adjusted the sensitivity of microscopy for green and red signals using 10 nM BafA1- and amino-acid–deprived HeLa cells as negative and positive controls, respectively.
To clarify whether IP3R1 mediated the TG2-dependent regulation of autophagy, we examined the IP3R1 inhibitors xestospongin C (Xest) and 2-aminoethoxydiphenyl borate (2-APB). Xest and 2-APB block the TG2-dependent effect on autophagy (Fig. 3A) without affecting TG2 levels (Fig. S2A), suggesting that TG2 regulates autophagy through IP3R1. To confirm this possibility in living cells, we observed the maturation of autophagosomes using a tandem fluorescent LC3 indicator containing both red (RFP) and green (GFP) fluorescent proteins (tfLC3) that distinguishes between autophagosomes and autolysosomes by quenching GFP fluorescence at lysosomal pH (52). In control siRNA-transfected cells, GFP and RFP puncta were independently distributed, indicating the presence of both autophagosomes and autolysosomes in the cells (Fig. 3B). However, IP3R1 knockdown caused significant accumulation of autophagosomes that were colabeled with GFP and RFP fluorescence, whereas TG2 knockdown yielded predominantly RFP signals, as observed in amino acid deprivation (Fig. 3B). These results confirm that TG2 and IP3R1 have opposing effects in the control of autophagy.
To verify the opposing effects of TG2 and IP3R1 on autophagy, we monitored the dynamics of autophagosomes in living cells. Time-lapse imaging of tfLC3 revealed that the autophagosomes in control and TG2 siRNA-transfected cells actively moved inside cells, whereas the altered trafficking of autophagosomes was observed in IP3R1 siRNA-transfected cells (Fig. S2B).
To confirm these observations, we monitored the distribution of endogenous autophagosomes and lysosomes in siRNA-transfected HeLa cells. In control and TG2–siRNA-transfected cells, LC3-labeled autophagosomes and lysosomal-associated membrane protein 1 (LAMP1)-labeled lysosomes were widely distributed in the cells and stochastically overlapped with each other (Fig. S2C). However, autophagosomes accumulated near the nucleus in cells transfected with IP3R1 siRNA (Fig. S2C). These data confirmed the observations using tfLC3 indicators that autophagy was regulated by the opposing effects of TG2 and IP3R1.
The IP3R1 Gln2746 Residue Is a Potent TG2 Substrate.
To elucidate the mechanism by which TG2 inhibited IP3R1-mediated signaling, we performed limited digestion experiments to identify the site of TG2 modification in IP3R1. Calpain digestion of IP3R1 pretreated with TG2 revealed a marked decrease in the C-terminal 130-kDa (C130) and 85-kDa (C85) fragments but not the N-terminal 180-kDa (N180) or 100-kDa fragments (N100) (Fig. S3 A and B). Consistent with these observations, cross-linking by TG2 after limited digestion also resulted in a marked decrease in C130 recovery, whereas the amount of N180 was not altered by TG2 (Fig. S3C). These results indicate that cross-linking occurs between the C130 fragments but not the residual N180 fragments. To confirm this, we performed limited digestion of BP-containing IP3R1 (Fig. S3 D and E). The apparent sizes of BP-labeled bands corresponded to the C130 and C85 fragments. Further digestion confirmed that the BP-labeled bands corresponded to the C-terminal fragment. Together, these results indicated that subunit cross-linking occurred within the C130 fragments.
To precisely determine the Gln (Q) donor site for IP3R1 cross-linking, we performed mass spectrometric analyses. We pulled down endogenous substrates of TG2 from mouse brains by incorporation with BP. A band of ∼300 kDa was identified as mouse IP3R1 by in-gel digestion and mass spectrometry (MS), indicating that IP3R1 is a preferred substrate of TG2 (Fig. S4 A and B). Calculating the mass of BP, we found that the carboxyl-terminal peptide was exclusively modified among 63 Gln residues assigned by mass analysis (Fig. S4C). We determined the precise modified residue in IP3R1 using liquid chromatography–MS (LC–MS), followed by tandem MS (MS/MS). This analysis identified one transamidation site of the Gln residue at position 2746 (Gln2746) (Fig. 4A). Compared with the wild-type peptide, a clear mass shift corresponding to BP was detected in the modified peptide (Fig. 4A). The substrate specificity of TG2 was determined using the sequence motif QxPl (x, any; l, aliphatic amino acids) (53). The sequence containing Gln2746 is consistent with this motif, indicating that Gln2746, but not Gln2747, is a target of TG2. Peptides containing QxP motifs at Gln524, Gln1137, and Gln1219 in mouse IP3R1 were detected by MS, but no BP incorporation was observed at these sites. Therefore, these Gln residues are not accessible to TG2 in the native conformation of tetrameric IP3R1.
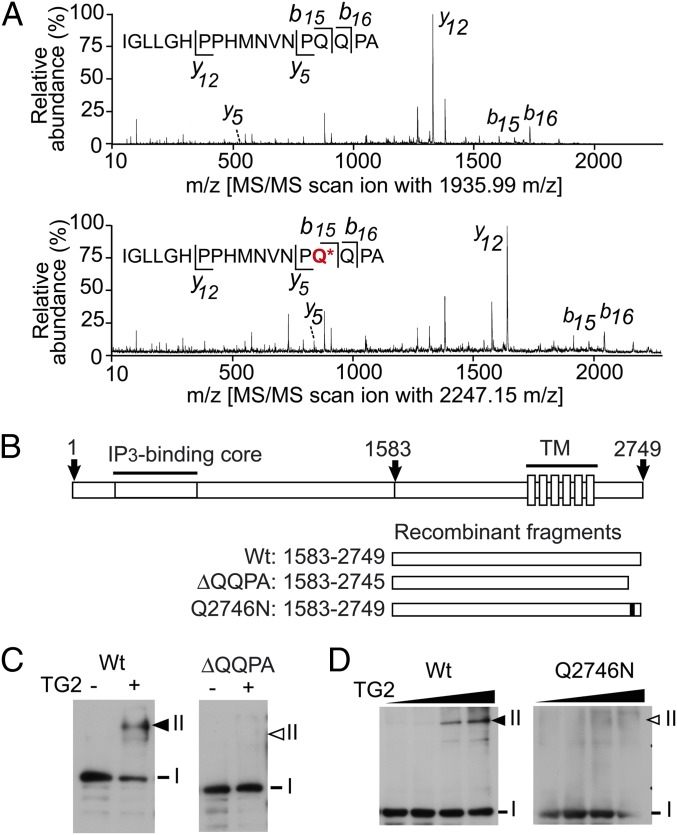
Fig. 4.
Gln2746 of IP3R1 is modified by TG2. (A) IP3R1 was modified at Gln2746 by TG2. MS/MS scanned the C-terminal peptide of IP3R1 with ion m/z 1,935.99 (Upper) and the biotinylated peptide with m/z 2,247.15 (Lower). The b and y ions are indicated on the MS/MS spectra. The asterisk denotes a modified Gln residue (see also Fig. S4). (B) A scheme shows the primary structure of IP3R1 and recombinant proteins corresponding to C-terminal 130-kDa fragments generated by calpain cleavage (Fig. S3). WT, a mutant in which four C-terminal amino acids were deleted (ΔQQPA), and a point mutant containing a Gln2746 to Asn substitution (Q2746N) are shown in the scheme. (C) The lower gels show intersubunit cross-linking of WT and ΔQQPA expressed in COS-7 cells. (D) Intersubunit cross-linking of WT and Q2746N expressed in COS-7 cells. Black and white arrows indicate the cross-linked dimer and its corresponding size, respectively.
The QxP motif at Gln1705 might be modified by TG2 modification; therefore, we examined the cross-linking of mutants. We generated deletion mutants of the C-terminal domain of mouse IP3R1 (calculated mass, 132.4 kDa, corresponding to the C130 fragment) and expressed them in COS-7 cells (Fig. 4B). We found that the recombinant wild-type (WT) channel was cross-linked by TG2, but the deletion mutant ΔQQPA (Fig. 4C) and the Q2746N mutant (Fig. 4D) were not cross-linked. These results indicate that the C-terminal QxP motif at Gln2746 was essential for subunit cross-linking. This motif is conserved among vertebrates including fish, birds, and mammals but not among invertebrates, suggesting that this regulation might be specific to vertebrates. The Gln2746-containing QxP motif is not conserved among other isoforms, but mouse IP3R2 (Gln897, Gln1221, and Gln1997) and mouse IP3R3 (Gln896, Gln1211, and Gln1948) also contain three QxP motifs. If these motifs are exposed and accessible to TG2, IP3R2/3 might also be TG2 targets.
Next, we examined whether the Gln2746-containg motif is sufficient for cross-linking by TG2. The NPQQPA peptide, but not NPNQPA, reacted with tetramethylrhodamine cadaverine (TMR-cad) to produce a cross-linked product with TG2 (Fig. S4D). We confirmed the structure of this purified product using MS and subsequently subjected it to a de–cross-linking reaction with TG2. Incubation of the product with both TG2 and CaCl2 formed a product that was consistent with TMR-cad (Fig. S4E). These data indicate that the Gln2746-containg motif is essential and sufficient for cross-linking by TG2.
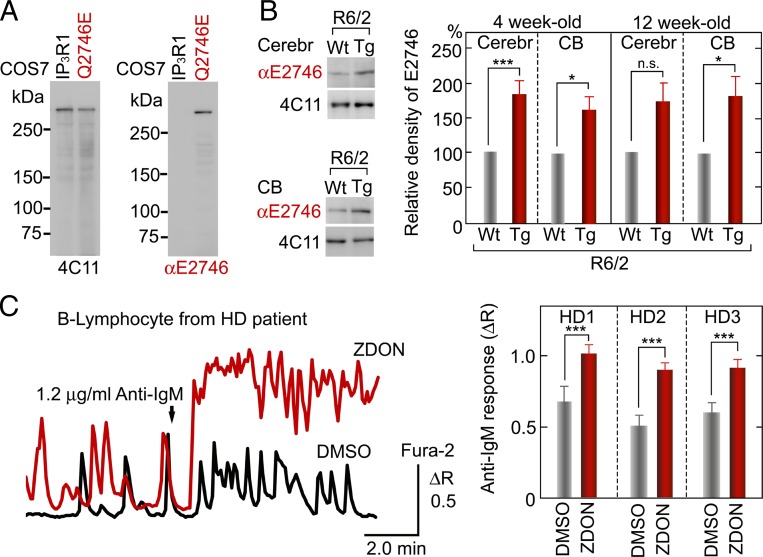
Either isopeptide hydrolysis and/or deamidation by TG2 can generate a glutamate residue (E) at the position of the intrinsic Gln residue (Q) (34). To test this possibility, we generated the antibody αE2746, which specifically recognizes deamidated IP3R1 (Fig. S5A). The site-specific αE2746 antibody detected TG2-catalyzed deamidation of tetrameric IP3R1 in P2/P3 vesicles (Fig. S5B). The αE2746 antibody revealed the existence of deamidated IP3R1 in cerebellar IP3R1 (Fig. S5C), and LC–MS analysis also confirmed the deamidated IP3R1 (Fig. S5 D and E). The specificity of the αE2746 antibody was further validated using the full-length IP3R1 with Q2746E mutation (Fig. 5A and Fig. S5F).
Fig. 5.
IP3R1 structure and function are modified in HD models. (A) The 4C11 monoclonal antibody (Left) detected both full-length IP3R1 and Q2746E mutant expressed in COS-7 cells, whereas our newly developed αE2746 antibody (Right) detected only the Q2746E mutant. (B) Gln2746 modification in endogenous IP3R1 in HD models. Blotted endogenous IP3R1 in mouse P2/P3 fractions was prepared from the cerebrum (cerebr) containing the cortex and the striatum (Upper) or the CB (Lower) of 4-wk-old WT or Tg male R6/2 mice. The loading volumes were adjusted based on the total IP3R1 content. The bar charts represent the mean + SEM of normalized Gln2746 modification in three-litter pairs of 4-wk-old and 12-wk-old R6/2 mice. Relative amounts of Gln2746 modification were calculated from the ratio of the αE2746 signal to the 4C11 signal. Four weeks, *P = 0.025, ***P = 0.0099; 12 wk, P = 0.051 (not significant, n.s), *P = 0.049. (C) Rescue of Ca2+ signaling in HD patient cells by inhibition of TG2. Black and red lines show IP3R-mediated Ca2+ release by anti-IgM (1.2 μg/mL) stimulation in B-lymphocytes obtained from an HD patient (HD1). Bar charts represent the mean + SEM of peak amplitudes of anti–IgM-induced Ca2+ release in lymphocytes from three HD patients [HD1, ***P = 0.0078 (control, n = 21; ZDON, n = 38); HD2, ***P = 7.3 × 10−5 (control, n = 34; ZDON, n = 54); HD3, ***P = 5.9 × 10−5 (control, n = 15; ZDON, n = 48)] pretreated with DMSO (control) or a site-directed TG inhibitory peptide ZDON (100 μM) for 30 min. HD1, HD2, and HD3 correspond to GM5560, GM6773, and GM4830, respectively. The peak amplitude was calculated as the difference between the mean value before stimulation and the peak value during stimulation.
Modification of IP3R1 in HD Models.
Our data indicated that the modification by TG2 impaired IP3R1-mediated cellular processes; therefore, we hypothesized that this regulation might lead to cellular dysregulation under pathological conditions. TG2 is up-regulated in the brain in HD model mice, including R6/2 and YAC128 mice (54–57), and HD patients (49, 58–61). Therefore, we tested whether IP3R was modified by TG2 in these HD models.
We investigated TG2-mediated modification of IP3R1 in an animal model of HD R6/2, which is transgenic (Tg) for exon 1 of the human Htt gene with 120 polyQ repeats, using the specific antibody that recognizes modified Gln2746 in IP3R1 (Fig. 5A). The brain P2/P3 vesicles were obtained from the cerebrum including the striatum and the cortex or the cerebellum (CB) of three litter pairs of 4-wk-old and 12-wk-old HD model mice (R6/2 Tg or WT); we observed endogenous IP3R1 using αE2746 and 4C11 that detects both modified and unmodified IP3R1. The IP3R1 expression levels were suppressed particularly in 12-wk-old HD mice (16, 28); thus, we normalized the αE2746 signal to the 4C11 signal. The relative level of the αE2746 signal was significantly higher in the brains of both 4-wk-old and 12-wk-old R6/2 Tg mice compared with WT mice (Fig. 5B).
TG2 up-regulation has been observed in the brain and lymphocytes of patients with HD (49, 58–61); therefore, we examined a role of TG on IP3R activity in B-lymphocytes from HD patients. Pretreatment with a specific TG blocker (Z-DON–Val–Pro–Leu–OMe, ZDON) (56, 62) enhanced IP3R-mediated Ca2+ release during B-cell antigen receptor stimulation by the anti-IgM antibody, which evoked IICR (63) in lymphocytes from a patient with HD (Fig. 5C). Notably, this effect was consistently observed in three individuals with HD. This indicates that in addition to the modification in the structure of IP3R in HD model mice (Fig. 5B), TG modifies the function of IP3R in B-lymphocytes from HD patients (Fig. 5C).
We observed the effects of the human Htt mutant on Ca2+ signaling in other HD models. The induced expression of the full-length human Htt including 145 polyQ in PC12 cells impaired acetylcholine (ACh)-evoked Ca2+ release under Ca2+-free conditions, whereas the enhanced ACh-evoked Ca2+ increase (Fig. S6A) was observed in the presence of extracellular Ca2+ in line with the sensitized Ca2+ increase in YAC128 neurons (64–66). To verify the inhibitory effect of the Htt mutant, we observed Ca2+ release in Neuro2a cells transiently expressing the myc-tagged full-length Htt including 23 polyQ (Htt23Q–myc) or 145 polyQ (Htt145Q–myc). The Ca2+ release upon stimulation by bradykinin (BK) in Neuro2a cells expressing with Htt145Q–myc was less than Htt23Q–myc under Ca2+-free conditions (Fig. S6B). Transient expression of Htt exon1 including 60 polyQ (eHtt60Q–EGFP) or 150 polyQ (eHtt150Q–EGFP) also impaired Ca2+ release in Neuro2a cells compared with the control (eHtt18Q–EGFP) (Fig. S6C), and the 3,5-dihydroxy phenylglycine (DHPG)-evoked Ca2+ increase in eHtt150Q–EGFP-expressing neurons was lower compared with eHtt18Q–EGFP-expressing neurons (Fig. S6D). To examine the involvement of TG2-dependent modification, we transfected Neuro2a cells coexpressing eHtt150Q–EGFP with either a Gln2746 mutant fragment (AQPA) or a Gln2746-containing fragment (QQPA). Overexpression of QQPA enhanced Ca2+ release compared with AQPA (Fig. S6D), suggesting the involvement of TG2-dependent modification. We also examined the Gln2746 modification in Neuro2a cells inducibly expressing Htt exon1 including 16 polyQ (eHtt16Q–EGFP) or 150 polyQ (eHtt150Q–EGFP) using the αE2746 antibody. Western blotting of endogenous IP3R1 (Fig. 6E) and immunostaining of the overexpressed IP3R1 C terminus (Fig. 6F) demonstrate that the αE2746-detectable modification was enhanced in the Neuro2 cells with eHtt150Q–EGFP after 48-h induction compared to those with eHtt16Q–EGFP.
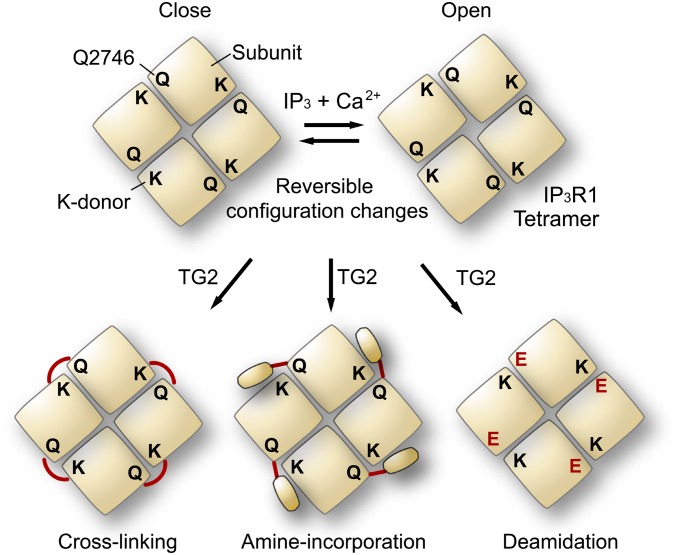
Fig. 6.
Model for TG2-mediated regulation of IP3R1. Schematic diagrams indicating the subunit arrangement of the allosteric coupling domain within the C-terminal 130-kDa region (C130) of IP3R1. The Q-donor at the C terminus is located in close proximity to the K-donor region of the neighboring subunit. We propose reversible configuration changes between the closed and open states in tetrameric IP3R1. TG-catalyzed transamidation causes covalent intersubunit cross-linking, which locks the closed configuration. A large molecule such as avidin but not a small moiety such as BP inhibits the motion by amine incorporation. Gln2746 of IP3R1 is deamidated by TG2 and converted into a glutamate residue that is resistant to TG2-mediated inhibition.
Discussion
The role of defective allostery in ER-resident calcium channels in subsequent cellular defects in neurological disorders remains unclear. The current study addresses this issue, demonstrating a novel allosteric mechanism—the covalent modification of IP3R structure and function—as well as its implications for the etiology of neurological diseases. Specifically, we discovered that TG2 in the brain regulates IP3R1 by catalyzing a posttranslational modification at the Gln side chain of the Gln2746 residue. Up-regulation of TG2 enhanced the Gln modification of IP3R1, and TG2 suppressed IP3R1-mediated Ca2+ signaling and subsequent cellular processes. Modification of Gln2746 and IP3R1 function was observed in HD models and B-lymphocytes from HD patients; these results therefore suggest a novel mechanism linking the activation of brain TG to the control of IP3R-mediated Ca2+ signaling in HD. Based on these results, we propose a general model for the progressive brain dysfunction observed in brain pathogenesis that involves aberrant IP3R signaling.
An important point in this study is the identification of TG2 as a chronic regulator of IP3R1 structure and function. This finding has broad implications for pathological neuronal function and degeneration because it links TG2 functions in cellular differentiation, cellular stress, and inflammation (34, 35) to the chronic defects in Ca2+ signaling. Numerous literatures have demonstrated that TG2 is up-regulated in various diseases including HD (49, 54, 55, 59–61), AD (37, 38, 40, 42, 43, 45), and PD (39, 44). The splicing mechanism (67, 68) and stress-dependent inductions (34, 35, 69–72) are considered to elucidate the up-regulation, and the increased Ca2+ levels observed in many diseases (4, 17–22) are also competent to trigger TG2 activation. The Ca2+-dependent activation of TG2 inhibits the IP3R1-mediated Ca2+ release, as revealed in the current study; thus, this enzymatically allosteric modulation should primarily contribute to a negative feedback mechanism to maintain homeostatic control of intracellular Ca2+ levels. However, this modification subsequently would have a chronically negative effect on Ca2+ signaling, because it is much more static than noncovalent allosteric modulators required for the control of rapid and dynamic Ca2+ signaling such as Ca2+ oscillation. Therefore, the TG2-dependent allosteric modulation should chronically set in motion cascades of defective signaling related to pathological states; up-regulation of TG2 may disrupt the IP3R1-mediated plasticity of neuronal signaling (3, 10–12) and the homeostatic control of cellular processes including mitochondria energy production (23), autophagy regulation (24–27), ER stress (28), and others (29–31) and ultimately increases cellular stress, resulting in further TG2 up-regulation proceeding a vicious cycle. Given the widespread brain expression profiles of TG2 and IP3R1, we propose that the mechanism described in this study might potentially serve as a general principle for many other diseases of the brain or other tissues in which TG2 is up-regulated by a disease-initiating agent. In this study, we assume that the disease-initiating agent is the mutant Htt; future studies are required to examine whether other disease-initiating agents such amyloid proteins in AD and α-synuclein in PD also would cause chronic TG2 activation, resulting in IP3R modification and aberrant calcium signaling, and suggesting its presumable role in brain dysfunctions including cognitive declines occurs frequently during the prodromal states of these neurodegenerative diseases (73–77).
This study revealed that the covalent modification by TG2 results in three alternative states, as shown in Fig. 6; thus, the other roles of TG2 in Ca2+ signaling are also conceivable. TG2 inhibits IICR activity by both cross-linking and steric hindrance around Gln2746 (Fig. 2), whereas functional analysis of Q2746E indicates that deamidation of IP3R1 suppresses TG2-mediated inhibition (Fig. S6G). All potential modifications are shown in the schematic diagram, but the overall activity of a single channel would depend significantly on the relative number of cross-linked isopeptide bonds, amine incorporations, and deamidation in the tetramer. Acute increases in Ca2+ levels cause subunit cross-linking in neuroblastoma and glioblastoma cells (Fig. 1E), whereas amine incorporation and deamidation were observed in resting cells and brain tissues without stimulation by increasing Ca2+ concentrations (Fig. 5B and Figs. S5 C–E and S6 H and I). The deamidation reaction by TG2 is dependent on pH (78) and substrate affinity (79); therefore, the relative abundance of these three modifications is presumably determined by the cellular state and the conformation around Gln2746. These irreversible processes would contribute to chronic changes in homeostasis of Ca2+ signaling and subsequent cellular processes, whereas the TG2-resistant states including the small amine-containing IP3R1, such as BP incorporation shown in Fig. 2C and/or the deamidated IP3R1 shown in Fig. S6G, might contribute to Ca2+-dependent cell death, including apoptosis (2, 17), due to the lack of negative feedback regulation by Ca2+. Although deletion of the C-terminal 12 amino acids has no effect on IICR (80), binding of the specific monoclonal antibody 18A10 (81) and GIT1 to this region inhibits IICR (82), similar to the BP–avidin complex in this study. Because both IgG and GIT1 form bulky architectures, these observations are consistent with our conclusion that large molecules such as avidin but not small moieties such as BP interfere with configuration changes by steric hindrance. IICR is regulated by binding of the C-terminal region near Gln2746 by various proteins governing cellular fate—for example, Bcl-XL, Bcl-2, cytochrome c, HAP1, and Htt (4, 5, 17, 18); thus, we speculate that the IP3R1 C terminus might act as a signaling hub whose structure affects cellular processes by regulating the subunit configuration by steric hindrance. Considering the direct activation of IP3R1 by the Htt mutant (18), the net activity of IP3R1 may be comprehensively determined by the Htt mutant and TG2 in HD. The impairment of Ca2+ release by eHtt150Q–EGFP was more potent than Htt145Q–myc (Fig. S6 B and C), consistent with higher TG2 activities in R6/2 than YAC128 mice expressing the full-length Htt mutant (55, 57) and the sensitized Ca2+ increase in YAC128 mice more than shortstop mice expressing Htt exon1/2 (64). The poly-Gln repeat of the Htt mutant is a favorable substrate of TG2 (41); thus, the possibility of substrate competition and steric hindrance around Gln2746 is also interesting. Thereby, we suggest that the prevalence of full-length Htt mutant and TG2 modulations on IP3R1 may determine the total net activity of Ca2+ signaling from the ER and the pathological states in HD—that is, the prodromal decline of neural signaling or the degeneration by apoptosis.
The first insight into the mechanism underlying the coupling between the IBD and the channel domain by IP3-evoked structural changes was described more than two decades ago (83). From our early EM studies of negatively stained IP3R1 using heparin-gold labeling, we proposed a long-range coupling of IBD to the channel domain by Ca2+-dependent structural changes (84–86). However, direct binding of the IBD to the channel domain was postulated, and a model was proposed in which an N-terminal suppressor domain directly binds a loop between putative transmembrane helices (87–90). Recently, highly improved cryo-EM reconstruction was reported (9), and by fitting calculations using X-ray structures (7, 8), it was concluded that direct interaction of the IBD with the channel domain is unlikely (8). However, because the X-ray structure of the IBD is limited to only 5% of each tetramer, which is equivalent to 20% of each subunit, the role of the other domains in channel gating remains unclear (91). Here, we show new evidence demonstrating that configuration changes outside the IBD are required for channel activation (Fig. 6). Our data are consistent with the conclusion that the Gln2746 in the C-terminal tail is proximal to the neighboring IP3R1 subunit, and TG2 covalently tethers Gln2746 to the adjacent subunit by the Gln–Lys isopeptide bond that locks the channel in a closed configuration (Fig. 6). Notably, our BP incorporation experiments indicate that the isopeptide bond occurs outside the IBDs without impairing the intrinsic structures required for IP3 binding and Ca2+ permeation (Fig. 2 B–D). IP3R1 function was not inhibited by transamidation of a small molecule such as BP; however, the subsequent addition of a bulky molecule such as avidin completely blocked receptor function (Fig. 2 B–D), indicating that steric hindrance near Gln2746 is crucial. We propose that an “allosteric coupling domain” in the C-terminal 130-kDa region (C130) exists between the IBD and the transmembrane domains to couple the IBD to the channel and that configuration changes in this domain are required for channel activation (Fig. 6). The EcoRI fragment-encoded short domain (ES; amino acids 2216–2749) of IP3R1 is sufficient for tetramer formation (92), but the channel in the ES tetramer is always open (93). Therefore, the allosteric coupling domain presumably keeps the channel closed, and the transition to the open configurations could revert to the default channel state that constitutively permeates Ca2+—for instance, the leaky channel assembled from ES domains.
What is the key mechanism regulating the allosteric transitions during channel activation? Two intrinsic determinants stabilizing the subunit configurations can be considered a priori—IP3 and Ca2+—because channel activation requires both IP3 and Ca2+. In fact, the recent X-ray structures of the IBD indicate that conformational changes occur upon IP3 binding (7, 8); our group and others have demonstrated Ca2+-induced structural changes (84, 85, 94, 95). Fluorescence resonance energy transfer (FRET) analysis of recombinant IP3Rs also showed reversible structural changes induced by physiological concentrations of IP3 and Ca2+ (96). Considering these observations and our current data, we propose that structural changes by IP3 and Ca2+ determine the configuration of the allosteric coupling domain. Given the Ca2+-sensor site at E2100 (32) and the deletion site in opisthotonos mice (33) are located in this domain, these allosteric sites presumably control subunit configurations within this domain. These ligands—that is, IP3, Ca2+, and ATP—might confer a predominantly open configuration by noncovalent binding, but the covalent Gln–Lys isopeptide bonds and steric hindrance around Gln2746 are more energetically stable; therefore, these modifications can robustly and chronically lock the closed subunit configuration.
In summary, we identified TG2 as an IP3R1-modifying enzyme that irreversibly controls the essential function of IP3R1 in Ca2+-signaling and subsequent cellular processes whose homeostasis is crucial for cellular fate and uncovered the allosteric mechanism by which IP3R channel activation is controlled by ligand binding. Many functional proteins undergo allosteric regulation; therefore, understanding allosteric mechanisms can elucidate the mechanisms underlying functions of these proteins and pave the way for drug design (1). This is the first demonstration, to our knowledge, of IP3R1 allosteric transitions locked by TG2-catalyzed cross-linking, and these observations might provide a new framework for TG2-dependent enzymatic regulation of ligand-gated ion channels and allosteric proteins similar to IP3R1.
Materials and Methods
Cell Culture.
SH-SY5Y, U87-MG, HeLa, and COS-7 cells were cultured in DMEM containing 10% (vol/vol) heat-inactivated FBS, 50 U/mL penicillin, and 50 μg/mL streptomycin. Human B-lymphocytes from patients with HD were obtained from the Coriell Institute (GM5560, GM6773, and GM4830) and cultured in RPMI medium 1640 containing 10% (vol/vol) heat-inactivated FBS, 50 U/mL penicillin, and 50 μg/mL streptomycin. All cells were maintained in 5% (vol/vol) CO2 at 37 °C. The differentiation medium supplemented with 20 μM RA was replaced every 48 h for the SH-SY5Y and U87-MG cells. All of the animals were ethically treated according to the guidelines of the Animal Experiments Committee of the Institute of Physical and Chemical Research (RIKEN) Brain Science Institute.
Transfection of siRNAs or Plasmids.
The siRNAs and human negative controls were purchased from B-Bridge. The nucleotide sequence of the TG2 siRNA was ggucaaugccgacguggua. IP3R1 knockdown was performed as described previously (28). The siRNA and ptfLC3 were transfected in HeLa cells using Lipofectamine 2000 (Invitrogen) and TransIT (Mirus) transfection reagents, respectively.
Preparation of P2/P3 Fractions.
Mouse brains were washed extensively with chilled PBS and were then homogenized in a homogenate buffer (pH 7.5) composed of 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes)/sodium hydroxide (NaOH), 0.32 M sucrose, 1 mM ethylene diamine tetraacetic acid (EDTA), 2 mM Tris(2-carboxyethyl)phosphine (TCEP), and protease inhibitors (0.2 mM phenylmethylsulfonyl fluoride, 10 μM leupeptin, 10 μM pepstatin A, and 10 μM E-64). The homogenates were centrifuged at 800 × g for 5 min at 4 °C, and the resultant supernatants were subjected to ultracentrifugation at 105,000 × g for 60 min at 2–4 °C as previously described (97). The precipitates were suspended in resuspension buffer ResB (pH 7.5), containing 50 mM Hepes–NaOH, 150 mM NaCl, 1 mM EDTA, 2 mM TCEP, and protease inhibitors. The supernatant obtained after ultracentrifugation was used as the cytosolic fraction. The protein concentrations in the solution were estimated, adjusted to 3 mg/mL using ResB, and the samples were then quickly frozen in liquid nitrogen for storage until use in various experiments.
Analysis of Intersubunit Cross-Linking Within IP3R1 by Electrophoresis.
We estimated the cross-linking between IP3R1 subunits using 1% AGE. The samples containing mouse cerebellar P2/P3 proteins (0.75–2.25 μg) were loaded in individual wells. Agarose was dissolved in a buffer containing 0.1% SDS using a microwave device. The agarose was heated and then gently mixed by manual rotation. The 1% agarose solution was poured into an acrylic tray (Nihon Eido) in which a 26-lane comb was placed. After 30 min at room temperature, the apparatus was filled with the electrolyte buffer, and the comb was carefully removed. Electrophoresis was performed at 100 V for 1–2 h. Immediately following electrophoresis, the agarose gels were placed in transfer buffer for 30 min. The separated proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore). IP3R1 was detected using the 4C11 monoclonal antibody as described previously (28, 97). A molecular marker was prepared by cross-linking bovine thyroglobulin with 1 mM 1,8-bis-maleimido-diethyleneglycol overnight at 4 °C.
Measurement of Ca2+ Release Activity of IP3R1.
We loaded fura-2AM (Dojindo) into HeLa cells and B-lymphocytes by incubation with 5 μM fura-2AM in balanced salt solution (pH 7.4) containing 115 mM NaCl, 5.4 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 20 mM Hepes, and 10 mM glucose for 30 min at room temperature, and performed Ca2+ imaging using inverted fluorescence microscopy with a 20× objective lens (S Fluor 20× N.A. 0.75) (Nikon) and AQUACOSMOS (Hamamatsu Photonics) to obtain F340/F380 ratio images as described previously (28, 98). For measurements of IICR, the P2/P3 membranes (3 mg/mL protein) of mouse cerebella were washed three times with a buffer (pH 7.4) containing 50 mM Hepes–KOH, 110 mM KCl, 10 mM NaCl, and 5 mM KH2PO4. The washed 10-μL P2/P3 membranes (2 mg/mL protein) of the cerebella were added to 287 μL of a buffer containing 1 mM 2-mercaptoethanol, 1 μM CaCl2, and 0.2 μM fura-2 in a cuvette. To introduce Ca2+ into P2/P3 membrane vesicles, we added 3 μL of 100 mM Mg–ATP to the cuvette with stirring. Ratiometric measurement of fura-2 was performed at 30 °C using a CAF-110 (Jasco) or an F-2500 spectrofluorometer (Hitachi).
Analysis of Autophagosome Dynamics by Fluorescence Microscopy.
We used ptfLC3 plasmids (52) expressing mRFP–GFP–LC3 to monitor autophagic processes in living cells. Cells plated in a 35-mm glass-bottomed dish were transfected with siRNA in Lipofectamine 2000 transfection reagent (Invitrogen) and then with 1 μg ptfLC3 by TransIT transfection reagent (Mirus) 6 h after the siRNA transfection. We observed GFP and RFP fluorescence signals in living cells under an FV1000D confocal microscope (Olympus) with a stage top incubator in 5% CO2 at 37 °C.
Affinity Purification of TG2 Substrates.
We incubated the reaction solution containing the P2/P3 fractions (1 mg/mL protein), 50 mM Hepes–NaOH (pH 7.5), 150 mM NaCl, 10 mM BP, 1 mM EGTA, 2 mM CaCl2, and 30 ng/mL TG for 30 min at 37 °C. The mixture was centrifuged at 20,000 × g for 30 min at 4 °C, and the pellets were washed with 50 mM Hepes–NaOH (pH 7.5). The proteins were solubilized with 1% (wt/vol) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) and mixed with avidin–agarose. After washing three times with the 1% CHAPS-containing Hepes buffer, the biotin-containing proteins bound to avidin–agarose were subjected to SDS/PAGE and were detected by Coomassie brilliant blue (CBB) staining or Western blotting analysis. The CBB-stained band was excised, digested using an in-gel digestion protocol, and analyzed by MS using MALDI-TOF MS (ABI).
Specific Antibody Against Modified IP3R1.
To examine the status of Gln2746 in mouse IP3R1, a deamidated peptide (PPHMNVNPEQPA) corresponding to amino acids 2738–2749 was synthesized and conjugated to keyhole limpet hemocyanin via a cysteine added to the N terminus of the peptide. Japanese white rabbits were immunized with these peptides. The resulting polyclonal antibody was first applied to an affinity resin immobilized with a control peptide (PPHMNVNPQQPA) via cysteine added to the amino terminus, and the flow-through was applied onto a resin immobilized with the deamidated peptides. The specific antibody against the deamidated peptide was eluted with 100 mM glycine–HCl buffer (pH 2.5). To verify the specificity of the antibodies, we prepared recombinant IP3R1, which was expressed as an EGFP–fusion protein (28). The PCR product was inserted into EGFP–IP3R1-containing pcDNA3.1 vector with EcoRI and XhoI. Mutation of the Gln residue at the amino acid position of 2746 was confirmed by sequence analysis using an ABI 3730xl automatic sequencer. To confirm the specificity of the E2746 antibody, the C-terminal fragment of IP3R1 (66 amino acids) was subcloned into the bacterial expression vector pGEX-6p (GE Healthcare) using EcoRI and SalI. Quantification of modified or native IP3R1 was performed by densitometric analysis of Western blots using Image J software (National Institute of Health).
Statistical Analysis.
All probability calculations were performed using unpaired and two-tailed Student t tests. The significance after multiple comparisons was estimated by the Bonferroni method or by Dunnett’s method. For all data, no statistics were used for predetermination of the sample size, randomization, or blinding.
Supplementary Material
Acknowledgments
We are grateful to Dr. T. Yoshimori (Osaka University) for the ptfLC3 plasmid and Dr. C. Yokoyama (RIKEN) for constructive comments on our manuscript. We thank the RIKEN Research Resources Center (RRC) for common-use equipment, M. Usui and K. Otsuki (RIKEN RRC) for assistance with MS, the RIKEN Brain Science Institute (BSI)–Olympus Collaboration Center for technical support, as well as M. Kurosawa, H. Miyazaki, and T. Yamanaka (RIKEN) for the HD model mice, cells, and plasmids. We are grateful to Y. Shimizu (Japan Science and Technology Agency, JST) for JST office management. This study was supported by JST International Cooperative Research Project–Solution Oriented Research for Science and Technology (K.M.); Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research S [25221002 (to K.M.)], Scientific Research C [24500451 (to C.H.)], and Encouragement of Scientists [24926012 (to A.T.)]; and RIKEN BSI Grant and President's Fund (to K.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409730111/-/DCSupplemental.
References
- 1.Changeux JP. 50 years of allosteric interactions: The twists and turns of the models. Nat Rev Mol Cell Biol. 2013;14(12):819–829. doi: 10.1038/nrm3695. [DOI] [PubMed] [Google Scholar]
- 2.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1(1):11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 3.Mikoshiba K. IP3 receptor/Ca2+ channel: From discovery to new signaling concepts. J Neurochem. 2007;102(5):1426–1446. doi: 10.1111/j.1471-4159.2007.04825.x. [DOI] [PubMed] [Google Scholar]
- 4.Foskett JK, White C, Cheung KH, Mak DO. Inositol trisphosphate receptor Ca2+ release channels. Physiol Rev. 2007;87(2):593–658. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patterson RL, Boehning D, Snyder SH. Inositol 1,4,5-trisphosphate receptors as signal integrators. Annu Rev Biochem. 2004;73:437–465. doi: 10.1146/annurev.biochem.73.071403.161303. [DOI] [PubMed] [Google Scholar]
- 6.Furuichi T, et al. Primary structure and functional expression of the inositol 1,4,5-trisphosphate-binding protein P400. Nature. 1989;342(6245):32–38. doi: 10.1038/342032a0. [DOI] [PubMed] [Google Scholar]
- 7.Lin CC, Baek K, Lu Z. Apo and InsP3-bound crystal structures of the ligand-binding domain of an InsP3 receptor. Nat Struct Mol Biol. 2011;18(10):1172–1174. doi: 10.1038/nsmb.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seo MD, et al. Structural and functional conservation of key domains in InsP3 and ryanodine receptors. Nature. 2012;483(7387):108–112. doi: 10.1038/nature10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ludtke SJ, et al. Flexible architecture of IP3R1 by Cryo-EM. Structure. 2011;19(8):1192–1199. doi: 10.1016/j.str.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishiyama M, Hong K, Mikoshiba K, Poo MM, Kato K. Calcium stores regulate the polarity and input specificity of synaptic modification. Nature. 2000;408(6812):584–588. doi: 10.1038/35046067. [DOI] [PubMed] [Google Scholar]
- 11.Harnett MT, Bernier BE, Ahn KC, Morikawa H. Burst-timing-dependent plasticity of NMDA receptor-mediated transmission in midbrain dopamine neurons. Neuron. 2009;62(6):826–838. doi: 10.1016/j.neuron.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugawara T, et al. Type 1 inositol trisphosphate receptor regulates cerebellar circuits by maintaining the spine morphology of purkinje cells in adult mice. J Neurosci. 2013;33(30):12186–12196. doi: 10.1523/JNEUROSCI.0545-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Gregorio E, et al. Two Italian families with ITPR1 gene deletion presenting a broader phenotype of SCA15. Cerebellum. 2010;9(1):115–123. doi: 10.1007/s12311-009-0154-0. [DOI] [PubMed] [Google Scholar]
- 14.Hara K, et al. Total deletion and a missense mutation of ITPR1 in Japanese SCA15 families. Neurology. 2008;71(8):547–551. doi: 10.1212/01.wnl.0000311277.71046.a0. [DOI] [PubMed] [Google Scholar]
- 15.Yamazaki H, et al. Functional characterization of the P1059L mutation in the inositol 1,4,5-trisphosphate receptor type 1 identified in a Japanese SCA15 family. Biochem Biophys Res Commun. 2011;410(4):754–758. doi: 10.1016/j.bbrc.2011.06.043. [DOI] [PubMed] [Google Scholar]
- 16.Luthi-Carter R, et al. Decreased expression of striatal signaling genes in a mouse model of Huntington’s disease. Hum Mol Genet. 2000;9(9):1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- 17.Bezprozvanny I. Calcium signaling and neurodegenerative diseases. Trends Mol Med. 2009;15(3):89–100. doi: 10.1016/j.molmed.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang TS, et al. Huntingtin and huntingtin-associated protein 1 influence neuronal calcium signaling mediated by inositol-(1,4,5) triphosphate receptor type 1. Neuron. 2003;39(2):227–239. doi: 10.1016/s0896-6273(03)00366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung KH, et al. Mechanism of Ca2+ disruption in Alzheimer’s disease by presenilin regulation of InsP3 receptor channel gating. Neuron. 2008;58(6):871–883. doi: 10.1016/j.neuron.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popugaeva E, Bezprozvanny I. Role of endoplasmic reticulum Ca2+ signaling in the pathogenesis of Alzheimer disease. Front Mol Neurosci. 2013;6:29. doi: 10.3389/fnmol.2013.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430(7000):631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shilling D, et al. Suppression of InsP3 receptor-mediated Ca2+ signaling alleviates mutant presenilin-linked familial Alzheimer’s disease pathogenesis. J Neurosci. 2014;34(20):6910–6923. doi: 10.1523/JNEUROSCI.5441-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White C, et al. The endoplasmic reticulum gateway to apoptosis by Bcl-X(L) modulation of the InsP3R. Nat Cell Biol. 2005;7(10):1021–1028. doi: 10.1038/ncb1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cárdenas C, et al. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell. 2010;142(2):270–283. doi: 10.1016/j.cell.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Decuypere JP, et al. Ins(1,4,5)P3 receptor-mediated Ca2+ signaling and autophagy induction are interrelated. Autophagy. 2011;7(12):1472–1489. doi: 10.4161/auto.7.12.17909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vicencio JM, et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ. 2009;16(7):1006–1017. doi: 10.1038/cdd.2009.34. [DOI] [PubMed] [Google Scholar]
- 27.Criollo A, et al. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ. 2007;14(5):1029–1039. doi: 10.1038/sj.cdd.4402099. [DOI] [PubMed] [Google Scholar]
- 28.Higo T, et al. Mechanism of ER stress-induced brain damage by IP(3) receptor. Neuron. 2010;68(5):865–878. doi: 10.1016/j.neuron.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, et al. Inositol-1,4,5-trisphosphate receptor regulates hepatic gluconeogenesis in fasting and diabetes. Nature. 2012;485(7396):128–132. doi: 10.1038/nature10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Futatsugi A, et al. IP3 receptor types 2 and 3 mediate exocrine secretion underlying energy metabolism. Science. 2005;309(5744):2232–2234. doi: 10.1126/science.1114110. [DOI] [PubMed] [Google Scholar]
- 31.Lee GS, et al. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature. 2012;492(7427):123–127. doi: 10.1038/nature11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyakawa T, et al. Ca(2+)-sensor region of IP(3) receptor controls intracellular Ca(2+) signaling. EMBO J. 2001;20(7):1674–1680. doi: 10.1093/emboj/20.7.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tu H, et al. Functional characterization of the type 1 inositol 1,4,5-trisphosphate receptor coupling domain SII(+/-) splice variants and the Opisthotonos mutant form. Biophys J. 2002;82(4):1995–2004. doi: 10.1016/S0006-3495(02)75548-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iismaa SE, Mearns BM, Lorand L, Graham RM. Transglutaminases and disease: Lessons from genetically engineered mouse models and inherited disorders. Physiol Rev. 2009;89(3):991–1023. doi: 10.1152/physrev.00044.2008. [DOI] [PubMed] [Google Scholar]
- 35.Eckert RL, et al. Transglutaminase regulation of cell function. Physiol Rev. 2014;94(2):383–417. doi: 10.1152/physrev.00019.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilhelmus MM, van Dam AM, Drukarch B. Tissue transglutaminase: A novel pharmacological target in preventing toxic protein aggregation in neurodegenerative diseases. Eur J Pharmacol. 2008;585(2-3):464–472. doi: 10.1016/j.ejphar.2008.01.059. [DOI] [PubMed] [Google Scholar]
- 37.Wang DS, et al. Cognitive performance correlates with cortical isopeptide immunoreactivity as well as Alzheimer type pathology. J Alzheimers Dis. 2008;13(1):53–66. doi: 10.3233/jad-2008-13106. [DOI] [PubMed] [Google Scholar]
- 38.Ruan Q, Johnson GV. Transglutaminase 2 in neurodegenerative disorders. Front Biosci. 2007;12:891–904. doi: 10.2741/2111. [DOI] [PubMed] [Google Scholar]
- 39.Junn E, Ronchetti RD, Quezado MM, Kim SY, Mouradian MM. Tissue transglutaminase-induced aggregation of alpha-synuclein: Implications for Lewy body formation in Parkinson’s disease and dementia with Lewy bodies. Proc Natl Acad Sci USA. 2003;100(4):2047–2052. doi: 10.1073/pnas.0438021100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selkoe DJ, Abraham C, Ihara Y. Brain transglutaminase: In vitro crosslinking of human neurofilament proteins into insoluble polymers. Proc Natl Acad Sci USA. 1982;79(19):6070–6074. doi: 10.1073/pnas.79.19.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahlem P, Green H, Djian P. Transglutaminase action imitates Huntington’s disease: Selective polymerization of Huntingtin containing expanded polyglutamine. Mol Cell. 1998;1(4):595–601. doi: 10.1016/s1097-2765(00)80059-3. [DOI] [PubMed] [Google Scholar]
- 42.Bonelli RM, Aschoff A, Niederwieser G, Heuberger C, Jirikowski G. Cerebrospinal fluid tissue transglutaminase as a biochemical marker for Alzheimer’s disease. Neurobiol Dis. 2002;11(1):106–110. doi: 10.1006/nbdi.2002.0535. [DOI] [PubMed] [Google Scholar]
- 43.Wilhelmus MM, et al. Transglutaminases and transglutaminase-catalyzed cross-links colocalize with the pathological lesions in Alzheimer’s disease brain. Brain Pathol. 2009;19(4):612–622. doi: 10.1111/j.1750-3639.2008.00197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vermes I, Steur EN, Jirikowski GF, Haanen C. Elevated concentration of cerebrospinal fluid tissue transglutaminase in Parkinson’s disease indicating apoptosis. Mov Disord. 2004;19(10):1252–1254. doi: 10.1002/mds.20197. [DOI] [PubMed] [Google Scholar]
- 45.Johnson GV, et al. Transglutaminase activity is increased in Alzheimer’s disease brain. Brain Res. 1997;751(2):323–329. doi: 10.1016/s0006-8993(96)01431-x. [DOI] [PubMed] [Google Scholar]
- 46.Mastroberardino PG, et al. ‘Tissue’ transglutaminase ablation reduces neuronal death and prolongs survival in a mouse model of Huntington’s disease. Cell Death Differ. 2002;9(9):873–880. doi: 10.1038/sj.cdd.4401093. [DOI] [PubMed] [Google Scholar]
- 47.Bailey CD, Johnson GV. Tissue transglutaminase contributes to disease progression in the R6/2 Huntington’s disease mouse model via aggregate-independent mechanisms. J Neurochem. 2005;92(1):83–92. doi: 10.1111/j.1471-4159.2004.02839.x. [DOI] [PubMed] [Google Scholar]
- 48.Chun W, Lesort M, Tucholski J, Ross CA, Johnson GV. Tissue transglutaminase does not contribute to the formation of mutant huntingtin aggregates. J Cell Biol. 2001;153(1):25–34. doi: 10.1083/jcb.153.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeitner TM, Pinto JT, Krasnikov BF, Horswill M, Cooper AJ. Transglutaminases and neurodegeneration. J Neurochem. 2009;109(Suppl 1):160–166. doi: 10.1111/j.1471-4159.2009.05843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akar U, et al. Tissue transglutaminase inhibits autophagy in pancreatic cancer cells. Mol Cancer Res. 2007;5(3):241–249. doi: 10.1158/1541-7786.MCR-06-0229. [DOI] [PubMed] [Google Scholar]
- 51.D’Eletto M, et al. Transglutaminase 2 is involved in autophagosome maturation. Autophagy. 2009;5(8):1145–1154. doi: 10.4161/auto.5.8.10040. [DOI] [PubMed] [Google Scholar]
- 52.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 53.Csosz E, et al. Substrate preference of transglutaminase 2 revealed by logistic regression analysis and intrinsic disorder examination. J Mol Biol. 2008;383(2):390–402. doi: 10.1016/j.jmb.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 54.Karpuj MV, et al. Prolonged survival and decreased abnormal movements in transgenic model of Huntington disease, with administration of the transglutaminase inhibitor cystamine. Nat Med. 2002;8(2):143–149. doi: 10.1038/nm0202-143. [DOI] [PubMed] [Google Scholar]
- 55.Van Raamsdonk JM, et al. Cystamine treatment is neuroprotective in the YAC128 mouse model of Huntington disease. J Neurochem. 2005;95(1):210–220. doi: 10.1111/j.1471-4159.2005.03357.x. [DOI] [PubMed] [Google Scholar]
- 56.McConoughey SJ, et al. Inhibition of transglutaminase 2 mitigates transcriptional dysregulation in models of Huntington disease. EMBO Mol Med. 2010;2(9):349–370. doi: 10.1002/emmm.201000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dedeoglu A, et al. Therapeutic effects of cystamine in a murine model of Huntington’s disease. J Neurosci. 2002;22(20):8942–8950. doi: 10.1523/JNEUROSCI.22-20-08942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karpuj MV, et al. Transglutaminase aggregates huntingtin into nonamyloidogenic polymers, and its enzymatic activity increases in Huntington’s disease brain nuclei. Proc Natl Acad Sci USA. 1999;96(13):7388–7393. doi: 10.1073/pnas.96.13.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krzysztoń-Russjan J, et al. A study of molecular changes relating to energy metabolism and cellular stress in people with Huntington’s disease: Looking for biomarkers. J Bioenerg Biomembr. 2013;45(1-2):71–85. doi: 10.1007/s10863-012-9479-3. [DOI] [PubMed] [Google Scholar]
- 60.Cariello L, et al. Transglutaminase activity is related to CAG repeat length in patients with Huntington’s disease. Hum Genet. 1996;98(6):633–635. doi: 10.1007/s004390050273. [DOI] [PubMed] [Google Scholar]
- 61.Munsie L, et al. Mutant huntingtin causes defective actin remodeling during stress: Defining a new role for transglutaminase 2 in neurodegenerative disease. Hum Mol Genet. 2011;20(10):1937–1951. doi: 10.1093/hmg/ddr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaertl S, et al. A profiling platform for the characterization of transglutaminase 2 (TG2) inhibitors. J Biomol Screen. 2010;15(5):478–487. doi: 10.1177/1087057110366035. [DOI] [PubMed] [Google Scholar]
- 63.Sugawara H, Kurosaki M, Takata M, Kurosaki T. Genetic evidence for involvement of type 1, type 2 and type 3 inositol 1,4,5-trisphosphate receptors in signal transduction through the B-cell antigen receptor. EMBO J. 1997;16(11):3078–3088. doi: 10.1093/emboj/16.11.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang H, et al. Full length mutant huntingtin is required for altered Ca2+ signaling and apoptosis of striatal neurons in the YAC mouse model of Huntington’s disease. Neurobiol Dis. 2008;31(1):80–88. doi: 10.1016/j.nbd.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang TS, et al. Disturbed Ca2+ signaling and apoptosis of medium spiny neurons in Huntington’s disease. Proc Natl Acad Sci USA. 2005;102(7):2602–2607. doi: 10.1073/pnas.0409402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang TS, Chen X, Liu J, Bezprozvanny I. Dopaminergic signaling and striatal neurodegeneration in Huntington’s disease. J Neurosci. 2007;27(30):7899–7910. doi: 10.1523/JNEUROSCI.1396-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Datta S, Antonyak MA, Cerione RA. GTP-binding-defective forms of tissue transglutaminase trigger cell death. Biochemistry. 2007;46(51):14819–14829. doi: 10.1021/bi701422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Citron BA, SantaCruz KS, Davies PJ, Festoff BW. Intron-exon swapping of transglutaminase mRNA and neuronal Tau aggregation in Alzheimer’s disease. J Biol Chem. 2001;276(5):3295–3301. doi: 10.1074/jbc.M004776200. [DOI] [PubMed] [Google Scholar]
- 69.Lee J, et al. Transglutaminase 2 induces nuclear factor-kappaB activation via a novel pathway in BV-2 microglia. J Biol Chem. 2004;279(51):53725–53735. doi: 10.1074/jbc.M407627200. [DOI] [PubMed] [Google Scholar]
- 70.Basso M, et al. Transglutaminase inhibition protects against oxidative stress-induced neuronal death downstream of pathological ERK activation. J Neurosci. 2012;32(19):6561–6569. doi: 10.1523/JNEUROSCI.3353-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caccamo D, Currò M, Ferlazzo N, Condello S, Ientile R. Monitoring of transglutaminase 2 under different oxidative stress conditions. Amino Acids. 2012;42(2-3):1037–1043. doi: 10.1007/s00726-011-1018-8. [DOI] [PubMed] [Google Scholar]
- 72.Yi SJ, et al. Arachidonic acid activates tissue transglutaminase and stress fiber formation via intracellular reactive oxygen species. Biochem Biophys Res Commun. 2004;325(3):819–826. doi: 10.1016/j.bbrc.2004.10.122. [DOI] [PubMed] [Google Scholar]
- 73.Sheline YI, et al. Amyloid plaques disrupt resting state default mode network connectivity in cognitively normal elderly. Biol Psychiatry. 2010;67(6):584–587. doi: 10.1016/j.biopsych.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Höhn S, et al. Behavioral and in vivo electrophysiological evidence for presymptomatic alteration of prefrontostriatal processing in the transgenic rat model for huntington disease. J Neurosci. 2011;31(24):8986–8997. doi: 10.1523/JNEUROSCI.1238-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sorg C, et al. Selective changes of resting-state networks in individuals at risk for Alzheimer’s disease. Proc Natl Acad Sci USA. 2007;104(47):18760–18765. doi: 10.1073/pnas.0708803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sepers MD, Raymond LA. Mechanisms of synaptic dysfunction and excitotoxicity in Huntington’s disease. Drug Discov Today. 2014;19(7):990–996. doi: 10.1016/j.drudis.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 77.Stout JC, et al. Neurocognitive signs in prodromal Huntington disease. Neuropsychology. 2011;25(1):1–14. doi: 10.1037/a0020937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fleckenstein B, et al. Gliadin T cell epitope selection by tissue transglutaminase in celiac disease. Role of enzyme specificity and pH influence on the transamidation versus deamidation process. J Biol Chem. 2002;277(37):34109–34116. doi: 10.1074/jbc.M204521200. [DOI] [PubMed] [Google Scholar]
- 79.Stamnaes J, Fleckenstein B, Sollid LM. The propensity for deamidation and transamidation of peptides by transglutaminase 2 is dependent on substrate affinity and reaction conditions. Biochim Biophys Acta. 2008;1784(11):1804–1811. doi: 10.1016/j.bbapap.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Uchida K, Miyauchi H, Furuichi T, Michikawa T, Mikoshiba K. Critical regions for activation gating of the inositol 1,4,5-trisphosphate receptor. J Biol Chem. 2003;278(19):16551–16560. doi: 10.1074/jbc.M300646200. [DOI] [PubMed] [Google Scholar]
- 81.Nakade S, Maeda N, Mikoshiba K. Involvement of the C-terminus of the inositol 1,4,5-trisphosphate receptor in Ca2+ release analysed using region-specific monoclonal antibodies. Biochem J. 1991;277(Pt 1):125–131. doi: 10.1042/bj2770125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang S, Hisatsune C, Matsu-Ura T, Mikoshiba K. G-protein-coupled receptor kinase-interacting proteins inhibit apoptosis by inositol 1,4,5-triphosphate receptor-mediated Ca2+ signal regulation. J Biol Chem. 2009;284(42):29158–29169. doi: 10.1074/jbc.M109.041509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mignery GA, Südhof TC. The ligand binding site and transduction mechanism in the inositol-1,4,5-triphosphate receptor. EMBO J. 1990;9(12):3893–3898. doi: 10.1002/j.1460-2075.1990.tb07609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hamada K, Miyata T, Mayanagi K, Hirota J, Mikoshiba K. Two-state conformational changes in inositol 1,4,5-trisphosphate receptor regulated by calcium. J Biol Chem. 2002;277(24):21115–21118. doi: 10.1074/jbc.C200244200. [DOI] [PubMed] [Google Scholar]
- 85.Hamada K, Terauchi A, Mikoshiba K. Three-dimensional rearrangements within inositol 1,4,5-trisphosphate receptor by calcium. J Biol Chem. 2003;278(52):52881–52889. doi: 10.1074/jbc.M309743200. [DOI] [PubMed] [Google Scholar]
- 86.Sato C, et al. Inositol 1,4,5-trisphosphate receptor contains multiple cavities and L-shaped ligand-binding domains. J Mol Biol. 2004;336(1):155–164. doi: 10.1016/j.jmb.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 87.Boehning D, Joseph SK. Direct association of ligand-binding and pore domains in homo- and heterotetrameric inositol 1,4,5-trisphosphate receptors. EMBO J. 2000;19(20):5450–5459. doi: 10.1093/emboj/19.20.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schug ZT, Joseph SK. The role of the S4-S5 linker and C-terminal tail in inositol 1,4,5-trisphosphate receptor function. J Biol Chem. 2006;281(34):24431–24440. doi: 10.1074/jbc.M604190200. [DOI] [PubMed] [Google Scholar]
- 89.Chan J, et al. Structural studies of inositol 1,4,5-trisphosphate receptor: Coupling ligand binding to channel gating. J Biol Chem. 2010;285(46):36092–36099. doi: 10.1074/jbc.M110.140160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rossi AM, et al. Synthetic partial agonists reveal key steps in IP3 receptor activation. Nat Chem Biol. 2009;5(9):631–639. doi: 10.1038/nchembio.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hamada K, Mikoshiba K. Revisiting channel allostery: A coherent mechanism in IP3 and ryanodine receptors. Sci Signal. 2012;5(225):pe24. doi: 10.1126/scisignal.2003148. [DOI] [PubMed] [Google Scholar]
- 92.Sayers LG, et al. Intracellular targeting and homotetramer formation of a truncated inositol 1,4,5-trisphosphate receptor-green fluorescent protein chimera in Xenopus laevis oocytes: Evidence for the involvement of the transmembrane spanning domain in endoplasmic reticulum targeting and homotetramer complex formation. Biochem J. 1997;323(Pt 1):273–280. doi: 10.1042/bj3230273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nakayama T, et al. The regulatory domain of the inositol 1,4,5-trisphosphate receptor is necessary to keep the channel domain closed: Possible physiological significance of specific cleavage by caspase 3. Biochem J. 2004;377(Pt 2):299–307. doi: 10.1042/BJ20030599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Anyatonwu G, Joseph SK. Surface accessibility and conformational changes in the N-terminal domain of type I inositol trisphosphate receptors: Studies using cysteine substitution mutagenesis. J Biol Chem. 2009;284(12):8093–8102. doi: 10.1074/jbc.M806932200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Anyatonwu G, et al. Calcium-dependent conformational changes in inositol trisphosphate receptors. J Biol Chem. 2010;285(32):25085–25093. doi: 10.1074/jbc.M110.123208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shinohara T, et al. Mechanistic basis of bell-shaped dependence of inositol 1,4,5-trisphosphate receptor gating on cytosolic calcium. Proc Natl Acad Sci USA. 2011;108(37):15486–15491. doi: 10.1073/pnas.1101677108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maeda N, et al. Structural and functional characterization of inositol 1,4,5-trisphosphate receptor channel from mouse cerebellum. J Biol Chem. 1991;266(2):1109–1116. [PubMed] [Google Scholar]
- 98.Nakamura K, et al. Distinct roles of M1 and M3 muscarinic acetylcholine receptors controlling oscillatory and non-oscillatory [Ca] increase. Cell Calcium. 2013;54(2):111–119. doi: 10.1016/j.ceca.2013.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.