Abstract
The Yersinia pestis chromosome contains a large variety and number of insert sequences that have resulted in frequent chromosome rearrangement events. To identify the chromosomal rearrangement features of Y. pestis strains from five typical plague foci in China and study spontaneous DNA rearrangements potentially stabilized in certain lineages of Y. pestis genomes, we examined the linking mode of locally collinear blocks (LCBs) in 30 Y. pestis strains by a polymerase chain reaction-based method. Our results suggest most strains have relatively stable chromosomal arrangement patterns, and these rearrangement characteristics also have a very close relationship with the geographical origin. In addition, some LCB linking modes are only present in specific strains. We conclude Y. pestis chromosome rearrangement patterns may reflect the genetic features of specific geographical areas and can be applied to distinguish Y. pestis isolates; furthermore, most of the rearrangement events are stable in certain lineages of Y. pestis genomes.
Introduction
Plague is a zoonotic disease primarily spread among wild rodents and small animals inhabiting natural plague foci around the world.1 Yersinia pestis, the etiological agent of plague, is transmitted between hosts by fleas, but sometimes is transmitted by air during pneumonic plague pandemics. Humans are likely to be affected by the bite of an infected flea or by contacting an infected host. Throughout human history, plague pandemic waves have led to hundreds of thousands of human deaths.2,3
The bacterial genus Yersinia includes three pathogenic species: Yersinia enterocolitica, Yersinia pseudotuberculosis, and Yersinia pestis. The first two are enteric pathogens that cause easily recoverable gastrointestinal diseases in humans and are transmitted by the fecal–oral route, whereas Y. pestis is a blood-borne pathogen of mammals and usually results in often fatal systemic diseases.4 Previous studies have indicated that Y. pestis is a clone recently derived from Y. pseudotuberculosis.5 Horizontal gene acquisition, massive gene loss, and genome rearrangement events have all played important roles in the evolution of Y. pestis from its progenitor.5,6 Thus far, closed completed genomes of 12 Y. pestis strains and four Y. pseudotuberculosis strains have been available and the sequences analyzed by several groups. Those studies revealed that the number of insertion sequence (IS) elements in the Y. pestis genome is unusually large compared with its ancestral Y. pseudotuberculosis genome. It has been suggested that IS-mediated genomic recombination often leads to genome rearrangement events such as translocation, inversion, and inverted translocation, which may have frequently occurred on the chromosome of Y. pestis.7 Based on their guanine-cytosine base composition bias results, Parkhill and others4 reported three rearrangement events (one translocation and two inversions) in the chromosome of the Y. pestis CO92 strain, an Orientalis biovar, and then confirmed these rearrangements by using a polymerase chain reaction (PCR) method. Moreover, they suggested that such rearrangements might occur during the bacteria culture process. Deng and others8 divided the genomes of Y. pestis strains CO92 and KIM (a Mediaevalis biovar) into 27 conserved segments and, in comparison with CO92, detected three multiple inversion regions on the chromosome of KIM. Large-scale genome rearrangements of other completely sequenced Y. pestis strains have also been described and analyzed.9–11 Taken together, these findings suggest the genome of Y. pestis is dynamic and it exhibits a high degree of fluidity. In this study, we designed a simple but practical PCR amplification method for investigating the chromosomal rearrangement features of some representative Y. pestis strains in China.
Materials And Methods
Bacterial strains.
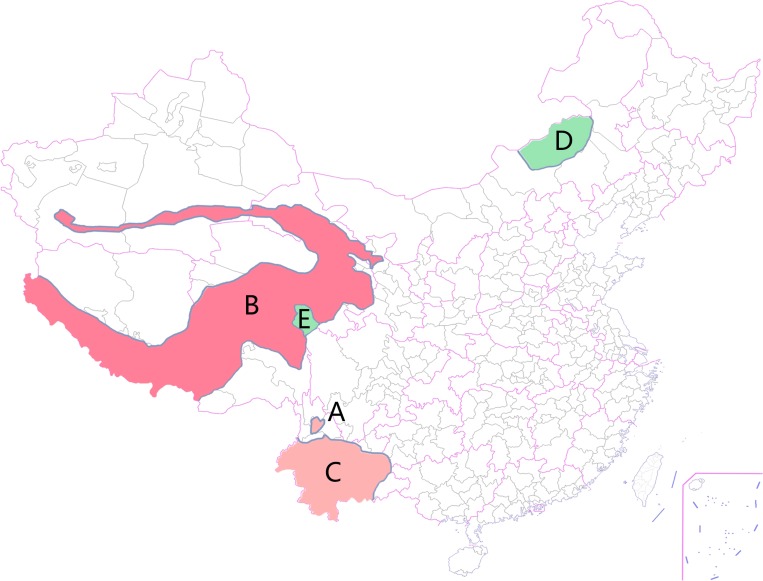
We selected 30 Y. pestis strains from five plague foci in China (Table 1), and there were six strains chosen to test in each plague focus. Plague epidemic of animals is quite severe in Focus B, and cases of human infection have been reported almost every year since 1999. In Focus A five human cases with two dead were observed in the year 2005. Plague Focus C is the place where the third world plague pandemic started, whereas human cases are not reported in the last decade. Focus D and Focus E are the only two known natural plague foci with the main reservoir as Microtus in China, and Y. pestis strains from the two places have never infected human beings. The distribution of these plague foci is shown in Figure 1. All strains used here are from a collection maintained by the National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention.
Table 1.
Yersinia pestis strains used for screening in this study
| No. | Strain name | Geographical origin | Isolated host | Isolated year | Biovar |
|---|---|---|---|---|---|
| 1 | 331 | Yunnan | Eothenomys miletus | 1954 | Antiqua |
| 2 | 84017 | Yunnan | Neopsylla specialis | 1977 | Antiqua |
| 3 | D182038 | Yunnan | Apodemus chevrieri | 1982 | Antiqua |
| 4 | 2083 | Yunnan | Rattus nitidus | 1994 | Antiqua |
| 5 | D106004 | Yunnan | Apodemus chevrieri | 2006 | Antiqua |
| 6 | Z13 | Yunnan | Neopsylla specialis | 2006 | Antiqua |
| 7 | Z176003 | Northern Tibet | Himalayan marmot | 1976 | Antiqua |
| 8 | 33001 | Northern Tibet | Himalayan marmot | 1978 | Antiqua |
| 9 | 19029 | Qinghai | Himalayan marmot | 1992 | Antiqua |
| 10 | 315006 | Southern Tibet | Himalayan marmot | 1998 | Antiqua |
| 11 | 373001 | Southern Tibet | Himalayan marmot | 1994 | Antiqua |
| 12 | 34003 | Southern Tibet | patient | 1966 | Antiqua |
| 13 | 540 | Yunnan | Rattus flavipectus | 1982 | Orientalis |
| 14 | 86022 | Yunnan | Rattus flavipectus | 1990 | Orientalis |
| 15 | 80069 | Yunnan | Rattus flavipectus | 1955 | Orientalis |
| 16 | 1804 | Yunnan | Rattus flavipectus | 1991 | Orientalis |
| 17 | 2202 | Yunnan | Suneus murinus | 1995 | Orientalis |
| 18 | 2381 | Yunnan | Norway rat | 1997 | Orientalis |
| 19 | 91001 | Inner Mongolia | Microtus brandti | 1970 | Medievalis |
| 20 | b1 | Inner Mongolia | Meriones unguiculatus | 1970 | Medievalis |
| 21 | b3 | Inner Mongolia | Microtus brandti | 1970 | Medievalis |
| 22 | b12 | Inner Mongolia | Microtus brandti | 1976 | Medievalis |
| 23 | b15 | Inner Mongolia | Microtus brandti | 1987 | Medievalis |
| 24 | b19 | Inner Mongolia | Microtus brandti | 1989 | Medievalis |
| 25 | N010001 | Sichuan | Microtus fuscus | 1997 | Medievalis |
| 26 | N010008 | Sichuan | Microtus fuscus | 1997 | Medievalis |
| 27 | N010031 | Sichuan | Microtus fuscus | 2000 | Medievalis |
| 28 | 18011 | Qinghai | Microtus fuscus | 2001 | Medievalis |
| 29 | 18015 | Qinghai | Microtus fuscus | 2001 | Medievalis |
| 30 | 18016 | Qinghai | Microtus fuscus | 2001 | Medievalis |
Figure 1.
Distribution characteristics of five natural plague foci in China in this study. (A) Represents Plague Focus A, major hosts are Apodemus chevrieri and Eothenomys miletus, humans are infected by Yersinia pestis occasionally, and the area is filled with pink; (B) represents Plague Focus B, major host is Himalayan marmot, plague epidemic among animals is widespread and often affects human beings, and is marked in red; (C) represents Plague Focus C, major host is Rattus flavipectus, human bubonic plague epidemic had happened historically, and the area is marked in pink; (D) represents Plague Focus D with Microtus brandti as the major host; (E) represents Plague Focus E, and major host is Microtus fuscus. Both D and E are filled with green because cases of human infection have never been observed in the two areas.
Selection of chromosomal rearrangement sites.
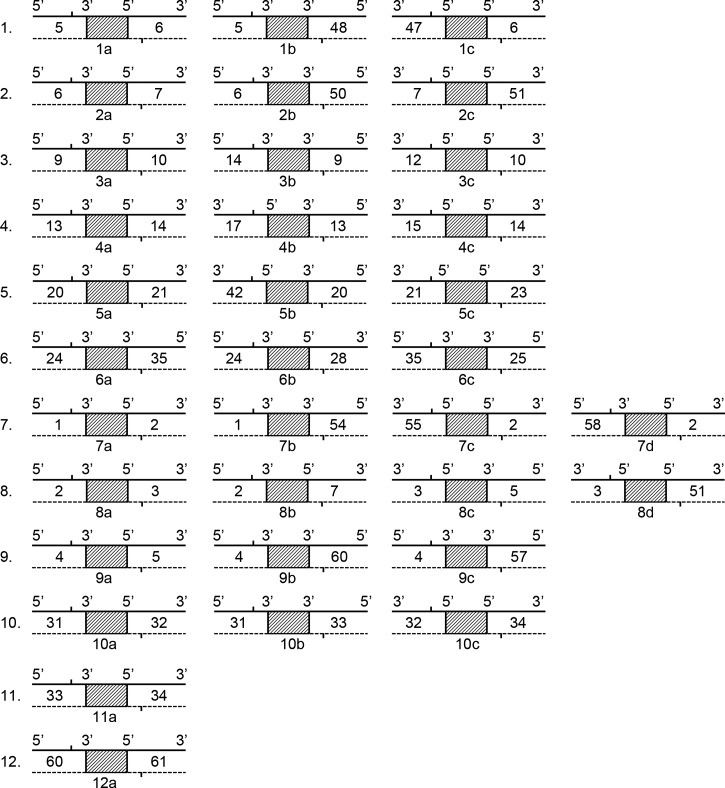
In previous studies, we divided the chromosome of Y. pestis into 61 large DNA segments (numbered according to the CO92 strain's chromosome order), based on the Coding Sequences similarity in a comparison of eight chromosomes of completely sequenced Y. pestis strains.12 Those DNA segments were very closely related to the locally collinear blocks (LCBs) reported by Darling and others.13 In this study, we used the LCB term to represent the large DNA segments. The gene content of, and structure within, each LCB is well conserved and stable, but different LCBs are relatively independent and mobile, which results in a variety of different LCB arrangement patterns in the chromosomes of different Y. pestis strains. The regions joining two neighboring LCBs, so-called breakpoint regions, are composed of IS elements and/or rRNA sequences and are responsible for the rearrangements among LCBs. Based on the arrangement patterns of LCBs in the completely sequenced Y. pestis strains, strain 91001 had the same LCB arrangement patterns as strain D182038, strain D106004 and strain Z176003 in six rearrangeable sites termed Site 7 to Site 12, but different in the other six rearrangeable sites termed Site 1 to Site 6. The LCB linkage patterns of all 12 sites were identical among strains D182038, D106004, and Z176003. Thereafter, we chose the previous 12 rearrangeable sites and identified 34 possible LCB linkage patterns (Figure 2).
Figure 2.
Twelve possible chromosome rearrangement sites and their related locally collinear block (LCB) linking modes. The solid line represents the coding strand, whereas the dotted line represents the complementary strand. The number between the solid line and the dotted line is the LCB number. The shaded rectangle represents a breakpoint region joining two LCBs. The vertical line indicates the approximate position of the primer. The bottom number and alphabet character indicates the name of the specific primer.
PCR amplification.
Using CO92 chromosome sequence as the reference sequence, we designed appropriate primers by using Primer Premier 5.0 software. The primers all located near the LCB boundaries and within the LCBs. Details on the primers are summarized in Table 2. The Y. pestis genomic DNA was extracted by DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) and diluted to 2 ng/μL for use as template DNA. Reaction mixtures for PCR amplifications were prepared in a final volume of 25 μL containing 12.5 μL 2× TransTaq-T PCR SuperMix, 10 pM forward primer, 10 pM reverse primer, 2 ng template DNA, and 9.5 μL filtered sterile water. The MyCycler thermal cycler (Bio-Rad, Hercules, CA) was programmed to a sequence of 95°C for 5 min for initial denaturation, followed by 30 cycles consisting of 95°C for 1 min, 62°C for 1 min, and 72°C for different periods (ranging from 2 min to 16 min according to the expected product's length). Final extension was performed at 72°C for 20 min.
Table 2.
Primers used for amplification in this study
| Primer name | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| 1a | TGGTAGGGCAAGCAGCAC | GGGTGGCTCGGGTTATCA |
| 1b | CAGATTATGCCGATGGTTTAG | CCTGCTCAGATTACGTCTACC |
| 1c | GCTGGCGATGGTTGGGCGGCTATT | TCGGTGGGTGGCTCGGGTTA |
| 2a | TTGCCAGAGGCGGTTTGTG | AAGAAAGAGGCTAACGCAGAGG |
| 2b | ATCTGGGAAGGCTCAGGCAAT | CGTAGTGGGCTTTGTGCAGTTT |
| 2c | TTTCGGCTTGGACCTGTTC | AGACCATCCTATTCATCAAGAG |
| 3a | GGCAGCCTATGCGTTGGGTAT | CCTGACAGCGTTTATCTTCCAC |
| 3b | GCACAGCACGGTAACCTTTC | GGATCATTCGGAACTCGCAAC |
| 3c | GTGAGCCACGAGTATTACCGAAAC | TTACGGATAGCACTAACCAACTG |
| 4a | GTATGCCACAAGAGGTCACG | GCTGGAGCACTCTGGTGATGTC |
| 4b | GCCCTCTATCCGTTCCCTG | GGTCACGCTCAAGCCGATG |
| 4c | AGTGCTTTATTGAGCCGTTC | GCGGGAAGATGAGTTGCTGTT |
| 5a | TCCGCTGCTGAAGGCTTAGATAC | GGTGGCAAATCAGGGAGAAGGTAT |
| 5b | TCACGAGTTCTGAATATGAGGAG | GCAACCGATACGCTGACCATT |
| 5c | TCGATCCCGACGATATGCTG | TGAAACCGTTGGTGCTCGTC |
| 6a | CAGGCGAGTAATCAAGCAGAG | GCAAGCCGCATCCAGAAGT |
| 6b | GGCGAGTAATCAAGCAGAG | CTTTCACGAAGTCCTTATTTACC |
| 6c | CGCTCTATTGTCATCCCATCAGTTG | GATTACTACGGTCCCATAGGTTC |
| 7a | TTTTCAGCCAGTAAGTAGGA | GGCAAGTGATAAGCCAATA |
| 7b | CTGCGAGAATACGCTGAC | CTGGAGTGGCGAGTTAGA |
| 7c | CTGAGTAGAGCACGGCGGATTG | AGCCCGAACAGATTACGGAGTT |
| 7d | GGCAAGTGATAAGCCAATA | CTCCCATAGAGCGACAATA |
| 8a | AGTGCCGTGAGCGTGGTGTA | CGTTTGCGGTGCCGTTATCT |
| 8b | TTTTAGCGATGAGAATGACA | TATGAGAACGAAAGAAAGAGG |
| 8c | TGGCTTGCGTAACATTTC | CGATTGGGTTTAGCAGATT |
| 8d | TGGCTTGCGTAACATTTC | GGGAGGCTAAGTCTTGGTG |
| 9a | TGAGTTGAGTTATGCGATTT | AGCGAGCGATTATTGAGA |
| 9b | GCAGTGAAACGGCATTAGAGGAG | GGCTGAGTTGAGTTATGCGATTTGT |
| 9c | TTTTGACGGTCTGGATAAT | TTTGATGGCTGACTTGC |
| 10a | TAAAGCCGCCTGCTGTTCG | ACCCACGCTACCCACTGAT |
| 10b | CGGCGTTAGATTCTCACAT | ACAGGTCGTTATTGGTGGC |
| 10c | CTGATGGTGGGTGATACGC | TGCTGGCTTGGTTAGATGA |
| 11a | GTAGCCGTTACCCGACAG | AAGAACCGTGACCGAAGG |
| 12a | AACAGATTACCTTGGCGGATTT | TGTGGCGACTGCGATTGA |
Identification of PCR products.
The PCR-amplified products were assayed by using 1% agarose gels. For each positive PCR product, DNA was extracted from the agarose gel and sent to Beijing Genomics Institute (Beijing, China) for sequencing. The obtained sequencing data were compared with known Y. pestis genome sequences using the basic local alignment search tool (BLAST).14
Results
If one pair of primers could amplify a single, bright band, and that band was confirmed as a breakpoint region connecting two neighboring LCBs (determined by sequencing the PCR products), we then reasoned that the two LCBs are linked together, that is, the corresponding arrangement is present in the tested strain. The LCB linkages for Y. pestis strains 91001, D182038, D106004, and Z176003 known according to their chromosomal sequences from Genbank.7,11 In addition, the PCR and sequencing data showed the same LCB linkage pattern in the DNA sample of the previous strains except 91001.
When we amplified one strain's DNA sample using a set of primers, it was supposed that if we got a positive amplification result using a primer pair named a, the results of other pairs of primers should then be negative. Most of the PCR results were in line with the expectations. Notably, we observed a single band using primer pairs 1a, 1b, and 1c, respectively, from PCR amplification of strain 33001 and strain N010031(Figure 3). The PCR products of primer pairs 1a, 1b, and 1c of strain N010031 were of the expected size as shown in Figure 3, however, the PCR product of primer pair 1b of strain 33001 was less than the expected size. After sequencing the unique PCR product, we found that a 5s–23s–16s rRNA gene cluster lacked in comparison with the expected PCR product. These results suggest that two kinds of LCB linkage patterns, which should be incompatible with each other exist in the same DNA sample.
Figure 3.
Illustration of gel imaging for particular polymerase chain reaction (PCR) products. M1 and M2 show the DNA molecular marker. Lanes 1–3 represent amplifications of strain D182038 using primer set 1a, 1b, and 1c. Lanes 4–6 represent amplifications of strain 91001 using primer set 1a, 1b, and 1c. Lanes 7–9 represent amplifications of strain 33001 using primer set 1a, 1b, and 1c. Lanes 10–12 represent amplifications of strain N010031 using primer set 1a, 1b, and 1c. Lanes 13–15 represent amplifications of strain N010008 using primer set 1a, 1b, and 1c. Lane 16 shows the blank control.
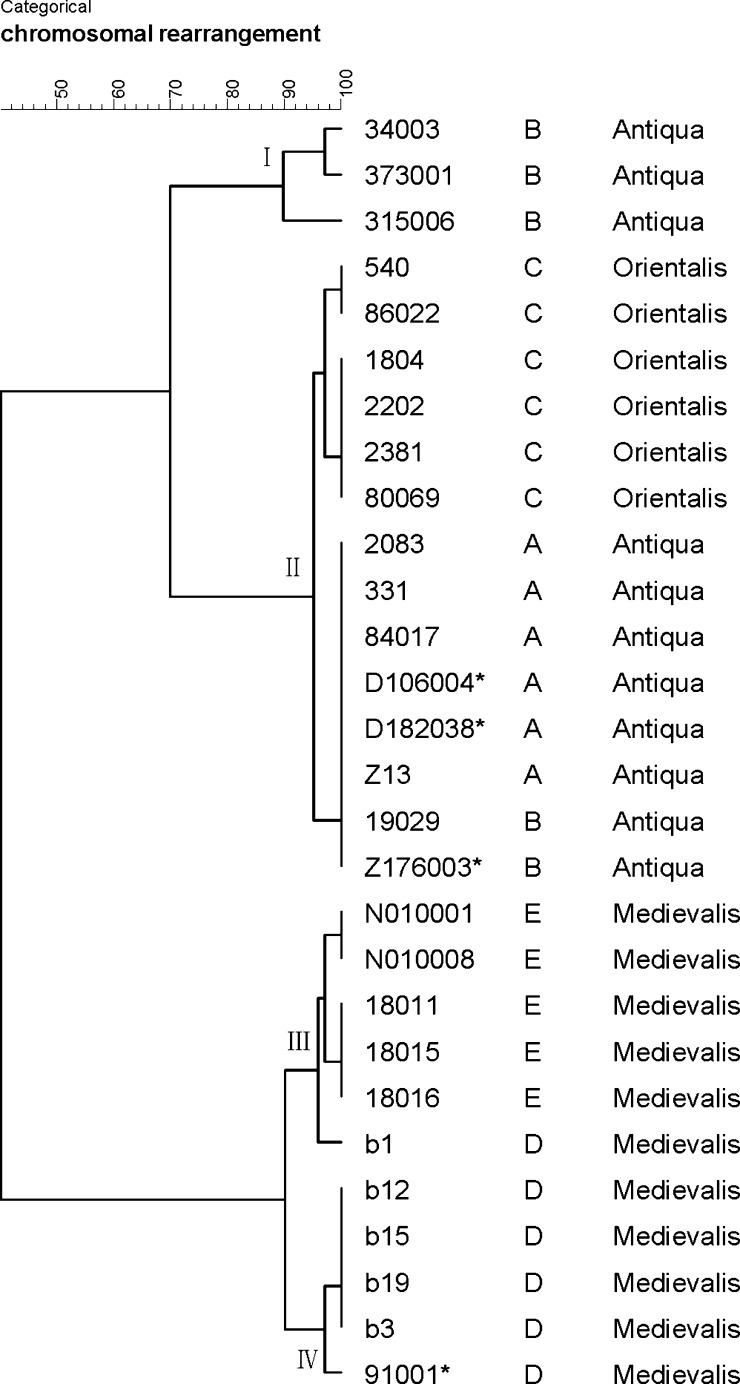
Allowing for the abnormal PCR results, strains 33001 and N010031 were kept away from further analysis. On the basis of all PCR results of the other 28 tested strains (Supplemental Table 1), a dendrogram was then finally generated by Bionumerics Software (Figure 4). According to the dendrogram, we eventually identified 11 groups among 28 strains (Table 3), corresponding to 11 LCBs linking modes. The dendrogram illustrated that Y. pestis strains isolated from plague Focus D and Focus E were closely related phylogenetically, though the distance between two foci was very far. In contrast, strains from the other three plague foci were clustered together. Strains from Focus B owned the most various LCBs linking modes, and two of six strains had the same LCBs linking mode as the six strains from Plague Focus A. Plague Focus C is mainly located in Yunnan Province of China as is Focus A, but strains isolated from two places belong to biovar Orientalis and biovar Antiqua separately. Therefore, strains of Focus C were identified as two LCB-groups that are different from isolates of Focus A. We also found that the amplification result of strains N010001 and N010008 (isolated in Sichuan Province) by primer pair 7c was positive, but negative in strains 18011, 18015, and 18016 (isolated in Qinghai Province), and other PCR results were identical among these five strains.
Figure 4.
Phylogenetic relationships among 28 Yersinia pestis strains inferred from the polymerase chain reaction (PCR) results using BioNumerics v5.10 software.
Table 3.
Locally collinear blocks (LCBs) linkage modes of 28 tested strains excluding strain 33001 and N010031
| Strain name | LCBs linking mode | Grouping in this study | Natural plague focus in China | Focus designation in this study |
|---|---|---|---|---|
| 331 | 1a-2a-3a-4a-5a-6a-7a-8a-9a-10a-11a-12a | IIB | Apodemus chevrieri-Eothenomys miletus Plague Focus of the highland of Northwestern Yunnan Province | A |
| 84017 | ||||
| D182038 | ||||
| 2083 | ||||
| D106004 | ||||
| Z13 | ||||
| Z176003 | 1a-2a-3a-4a-5a-6a-7a-8a-9a-10a-11a-12a | IIB | Marmota himalayana Plague Focus of the Qinghai-Tibet Plateau | B |
| 19029 | ||||
| 315006 | 1a-2a-3a-4a-5a-6a-7b-7c-8b-8c-9b-10a-11a | IB | ||
| 373001 | 1a-2b-3a-4a-5a-7b-7c-8b-8c-9b-10a-11a | IA2 | ||
| 34003 | 1a-2b-3a-4a-5a-7b-8b-8c-9b-10a-11a | IA1 | ||
| 540 | 1a-2a-3a-4a-5a-7a-8a-9a-10a-11a-12a | IIA1 | Rattus flavipectus Plague Focus of the Yunnan-Guangdong-Fujian provinces | C |
| 86022 | ||||
| 80069 | 1a-2a-3a-4a-5a-8a-9a-10a-11a-12a | IIA2 | ||
| 1804 | ||||
| 2202 | ||||
| 2381 | ||||
| 91001 | 1b-1c-2b-2c-3b-3c-4b-4c-5b-5c-6b-6c-8a-9b-10a-11a | IVB | Microtus brandti Plague Focus of the Xilin Gol Grassland | D |
| b1 | 1b-1c-2b-2c-3c-4b-5b-5c-6b-6c-7b-8a-9b-10a-11a | IIIB | ||
| b3 | 1b-1c-2b-2c-3b-3c-4b-4c-5b-5c-6b-6c-7b-8a-9b-10a-11a | IVA | ||
| b12 | ||||
| b15 | ||||
| b19 | ||||
| N010001 | 1b-1c-2b-2c-3c-4b-5c-6b-6c-7b-7c-8a-9b-10a-11a | IIIA1 | Microtus fuscus Plague Focus of the Qinghai-Tibet Plateau | E |
| N010008 | ||||
| 18011 | 1b-1c-2b-2c-3c-4b-5c-6b-6c-7b-8a-9b-10a-11a | IIIA2 | ||
| 18015 | ||||
| 18016 |
Discussion
Breakpoint regions between two neighboring LCBs are composed of IS100, IS1541, IS285, IS1661, or rRNA gene clusters. In this study, we designed primers localized near the boundaries and within the LCBs; thus, we could amplify the entire breakpoint region between two LCBs. After sequencing and alignment analysis, we determined the actual linkage situation between two LCBs in the tested strains. Using this relatively simple PCR-based method, we determined the chromosomal arrangement patterns of 30 Y. pestis strains in China.
The results show that strains from Focus B have diverse LCBs linkage patterns. The DFR (different region) analysis also obtained multiple genomovars in the strains of Focus B.15 The higher genomic polymorphism may be relevant to the complicated composition of ecosystem inside this Focus. Such rearrangement events alter the genetic features of Y. pestis strains being able to adapt to different ecological niches. Isolates from Focus A and Focus C formed two independent populations, but they all show a closer genetic relationship with the strains of Qinghai Province and Northern Tibet inside the Plague Focus B. Focus E is adjacent to Focus B and far apart from Focus D, but strains of Focus E possess the same phenotypic characteristics, genomovar and MLVA-type as the strains of Focus D. They also have a very similar LCBs linking mode in this study, therefore, we infer that strains from both Focus D and E are evolving from a common old ancestor of Y. pestis. Previously, MLVA and DFR could not differentiate strains of Foci D and E, although the presented method in this study can easily distinguish strains of Focus D from strains of Focus E. Even strains from two places (Qinghai Province and Sichuan Province) in the Focus E can be well separated by the LCB linkage mode.
In regard to strains 33001 and N010031, we think the LCBs linkage patterns presented by primer pair 1a, 1b, and 1c are indeed in their chromosomes according to the PCR results and amplification products' sequences. This means that the rearrangement event occurs frequently during the course of cultivation. This may be caused by the lack of selection pressure in the culture condition and, as a result, a sub-clone with the rearrangement mutations can survive, or other unknown reasons. Therefore, the tested strains should be natural isolates because chromosomal instability increases as the cell passage is increasing.
Our results show that Y. pestis strains from different plague foci have different chromosomal rearrangement features, indicating the presence of genetic diversity within Y. pestis. It was suggested that genome rearrangement is a better way to represent vertical inheritance.16 Thus, the observed chromosomal rearrangements may help us better understand the genetic characteristics and intraspecies evolution of Y. pestis strains among plague foci in China. Detecting information of chromosomal rearrangements need the completely sequenced genome data, and this work is time-consuming and expensive. On the contrary, the PCR-based method presented here is a simple and low-cost method. However, only part of the rearrangement situations can be identified in tested strains by way of our method and the effects of genome rearrangement on virulence, gene transcription level, and pathogenicity in Y. pestis are still unknown. To improve knowledge of chromosome rearrangement in Y. pestis, more LCB linking modes in different strains need to be determined.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to Zhanyong Jiang for preparing the figures in this manuscript.
Footnotes
Financial support: This work was supported by a grant (award 201202021) from the Special Fund for Health Sector, People's Republic of China, and grant (2012ZX10004215) from the National Key Programs for Infectious Diseases of China.
Authors' addresses: Ying Liang, Fang Xie, Enmin Zhang, Zhikai Zhang, Hong Cai, Yanhua Wang, Xiaona Shen, Hongqun Zhao, Dongzheng Yu, Lianxu Xia, and Rong Hai, State Key Laboratory for Infectious Disease Prevention and Control, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China, E-mails: liangying@icdc.cn, 513596429@qq.com, zhangenmin@icdc.cn, zhangzhikai@icdc.cn, caihong@icdc.cn, wangyanhua@icdc.cn, shenxiaona@icdc.cn, zhaohongqun@icdc.cn, yudongzheng@icdc.cn, xialianxu@icdc.cn, and hairong@icdc.cn. Xinyuan Tang and Mei Wang, Qinghai Institute for Endemic Disease Control and Prevention, Xining, China, E-mails: tang19790624@126.com and wangmei19790624@126.com.
References
- 1.Perry RD, Fetherston JD. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anisimov AP, Lindler LE, Pier GB. Intraspecific diversity of Yersinia pestis. Clin Microbiol Rev. 2004;17:434–464. doi: 10.1128/CMR.17.2.434-464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler T. Plague into the 21st century. Clin Infect Dis. 2009;49:736–742. doi: 10.1086/604718. [DOI] [PubMed] [Google Scholar]
- 4.Parkhill J, Wren BW, Thomson NR, Titball RW, Holden TG, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, Baker S, Basham D, Bentley SD, Brooks K, Cerdeño-Tárraga AM, Chillingworth T, Cronin A, Davies RM, Davis P, Dougank G, Feltwell T, Hamlin N, Holroyd S, Jagels K, Karlyshev AV, Leather S, Moule S, Oyston CF, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 5.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chain SG, Carniel E, Larimer W, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, Brubaker RR, Fowler J, Hinnebusch J, Marceau M, Medigue C, Simonet M, Chenal-Francisque V, Souza B, Dacheux D, Elliott JM, Derbise A, Hauser LJ, Garcia E. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song Y, Tong Z, Wang J, Wang L, Guo Z, Han Y, Zhang J, Pei D, Zhou D, Qin H, Pang X, Han Y, Zhai J, Li M, Cui B, Qi Z, Jin L, Dai R, Chen F, Li S, Ye C, Du Z, Lin W, Wang J, Yu J, Yang H, Wang J, Huang P, Yang R. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 2004;11:179–197. doi: 10.1093/dnares/11.3.179. [DOI] [PubMed] [Google Scholar]
- 8.Deng W, Burland V, Plunkett G III, Boutin A, Mayhew GF, Liss P, Perna NT, Rose DJ, Mau B, Zhou S, Schwartz DC, Fetherston JD, Lindler LE, Brubaker RR, Plano GV, Straley SC, McDonough KA, Nilles ML, Matson JS, Blattner FR, Perry RD. Genome sequence of Yersinia pestis KIM. J Bacteriol. 2002;184:4601–4611. doi: 10.1128/JB.184.16.4601-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chain SG, Hu P, Malfatti SA, Radnedge L, Larimer F, Vergez LM, Worsham P, Chu MC, Andersen GL. Complete genome sequence of Yersinia pestis strains Antiqua and Nepal516: evidence of gene reduction in an emerging pathogen. J Bacteriol. 2006;188:4453–4463. doi: 10.1128/JB.00124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eppinger M, Worsham PL, Nikolich MP, Riley DR, Sebastian Y, Mou S, Achtman M, Lindler LE, Ravel J. Genome sequence of the deep-rooted Yersinia pestis strain Angola reveals new insights into the evolution and pangenome of the plague bacterium. J Bacteriol. 2010;192:1685–1699. doi: 10.1128/JB.01518-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen X, Wang Q, Xia L, Zhu X, Zhang Z, Liang Y, Cai H, Zhang E, Wei J, Chen C, Song Z, Zhang H, Yu D, Hai R. Complete genome sequences of Yersinia pestis from natural foci in China. J Bacteriol. 2010;192:3551–3552. doi: 10.1128/JB.00340-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang Y, Hou X, Wang Y, Cui Z, Zhang Z, Zhu X, Xia L, Shen X, Cai H, Wang J, Xu D, Zhang E, Zhang H, Wei J, He J, Song Z, Yu X, Yu D, Hai R. Genome rearrangements of completely sequenced strains of Yersinia pestis. J Clin Microbiol. 2010;48:1619–1623. doi: 10.1128/JCM.01473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darling AE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Dai E, Cui Y, Li M, Zhang Y, Wu M, Zhou D, Guo Z, Dai X, Cui B, Qi Z, Wang Z, Wang H, Dong X, Song Z, Zhai J, Song Y, Yang R. Different region analysis for genotyping Yersinia pestis isolates from China. PLoS ONE. 2008;3:e2166. doi: 10.1371/journal.pone.0002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darling AE, Miklos I, Ragan MA. Dynamics of genome rearrangement in bacterial populations. PLoS Genet. 2008;4:e1000128. doi: 10.1371/journal.pgen.1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.