Abstract
The influence of mono-ubiquitylation of histone H2B (H2Bub) on transcription via nucleosome reassembly has been widely documented. Recently, it has also been shown that H2Bub promotes recovery from replication stress; however, the underling molecular mechanism remains unclear. Here, we show that H2B ubiquitylation coordinates activation of the intra-S replication checkpoint and chromatin re-assembly, in order to limit fork progression and DNA damage in the presence of replication stress. In particular, we show that the absence of H2Bub affects replication dynamics (enhanced fork progression and reduced origin firing), leading to γH2A accumulation and increased hydroxyurea sensitivity. Further genetic analysis indicates a role for H2Bub in transducing Rad53 phosphorylation. Concomitantly, we found that a change in replication dynamics is not due to a change in dNTP level, but is mediated by reduced Rad53 activation and destabilization of the RecQ helicase Sgs1 at the fork. Furthermore, we demonstrate that H2Bub facilitates the dissociation of the histone chaperone Asf1 from Rad53, and nucleosome reassembly behind the fork is compromised in cells lacking H2Bub. Taken together, these results indicate that the regulation of H2B ubiquitylation is a key event in the maintenance of genome stability, through coordination of intra-S checkpoint activation, chromatin assembly and replication fork progression.
Author Summary
Eukaryotic DNA is organized into nucleosomes, which are the fundamental repeating units of chromatin. Coordination of chromatin structure is required for efficient and accurate DNA replication. Aberrant DNA replication results in mutations and chromosome rearrangements that may be associated with human disorders. Therefore, cellular surveillance mechanisms have evolved to counteract potential threats to DNA replication. These mechanisms include checkpoints and specialized enzymatic activities that prevent the replication and segregation of defective DNA molecules. We employed a genome-wide approach to investigate how chromatin structure affects DNA replication under stress. We report that coordination of chromatin assembly and checkpoint activity by a histone modification, H2B ubiquitylation (H2Bub), is critical for the cell response to HU-induced replication stress. In cells with a mutation that abolishes H2Bub, replication progression is enhanced, and the forks are more susceptible to damage by environmental insults. The replication proteins on replicating DNA are akin to a train on the tracks, and movement of this train is carefully controlled. Our data indicate that H2Bub helps organize DNA in the nuclei during DNA replication; this process plays a similar role to the brakes on a train, serving to slow down replication, and maintaining stable progression of replication under environmental stress.
Introduction
Recent evidence suggests that histone modifications can affect DNA replication, under both normal or stressed conditions, through effects on nucleosome dynamics and protein recruitment [1]–[3]. One such modification is acetylation of nascent histone H3 at lysine 56 (H3K56Ac), which is regulated by the Asf1 histone chaperone and the Rtt109 acetyltransferase during the cell cycle [4], [5]. Regulation of this modification is important for DNA replication, as failure to deacetylate H3K56Ac results in impaired S phase progression [6], sensitivity to replication stress [7], and spontaneous DNA damage [6]. H3K56Ac appears to facilitate nucleosome reassembly on daughter strands during S phase [8]. Acetylation of the N terminal lysines of histone H3 by Gcn5 also contributes to nucleosome assembly during DNA replication [9]. These findings suggest that replication-coupled nucleosome assembly may impact on both fork progression and the stability of stalled forks [1], [3]. A second histone, H2B, is mono-ubiquitylated at lysine 123 (K123, K120 in human) by the E2 enzyme Rad6 and the E3 enzyme Bre1 in Saccharomyces cerevisiae [10]–[13]. Mono-ubiquitylation of H2B (H2Bub) is best characterized in terms of its effects on transcriptional regulation in budding yeast [14], [15], which are mediated through downstream methylation of lysines 4 and 79 of H3 [16]–[19]. In addition, H2Bub has been demonstrated to affect transcription independently of its regulation of H3 methylation [20], [21]. H2Bub enhances passage of RNA Polymerase II during transcription elongation by mediating nucleosome reassembly in both yeast and human [20], [22], [23]. Furthermore, H2Bub may also affect transcription and DNA repair through influencing chromatin structure [24], [25]. It has been suggested that H2Bub mediates homologous recombination repair at DNA double-strand break (DSB) sites through relaxing chromatin structure in human cells [26], [27]. H2Bub has also been shown to maintain replication fork stability by promoting replication-associated nucleosome formation in budding yeast, independently of its role in regulating H3K4 and K79 methylation [28].
During S phase, replication fork progression can be impaired by low dNTP pools or by DNA damage. Under these conditions, a sensor-response system activates the DNA replication (intra-S phase) checkpoint, which prevents fork collapse while controlling origin firing [29], [30]. The mechanism by which the intra-S checkpoint is activated is still not yet fully understood. It is hypothesized that decoupling between polymerase and helicase leads to single strand DNA accumulation and activation of the kinases Mec1/ATR and their downstream effector, Rad53 [30], [31]. Once a stalled fork has been stabilized by activation of the intra-S checkpoint, the damaged fork can resume DNA synthesis. The RecQ helicase Sgs1 is recruited to the stalled fork, where it facilitates its re-initiation through a mechanism involving the recombination repair pathway [32]. Sgs1 also facilitates the phosphorylation of Rad53 (possibly through direct physical interaction), and this process is redundant with the DNA damage checkpoint proteins Rad24 and Esc2 [33].
Activation of the intra-S phase checkpoint affects DNA synthesis by altering both the rate of replication fork progression and the rate of DNA replication initiation events [34]. For instance, a recent report suggests that Mec1 promotes chromatin accessibility at or ahead of replication forks via a mechanism independent of its checkpoint role [35]. The authors argue that a chromatin regulatory process may serve as a means of restricting fork progression, in order to control and stabilize fork progression under replication stress. However, the mechanisms through which chromatin structure regulates replication progression are still poorly understood.
In the current study, we used BrdU IP-chip to examine genome-wide DNA synthesis incorporation in wild type and H2Bub-deficient cells in the presence of hydroxyurea (HU). We demonstrate that newly-synthesized DNA in cells lacking H2Bub displays a broader distribution and enrichment at origin-distal regions; these findings suggest faster replication fork progression in the mutant. Surprisingly, this phenomenon is independent of DNA damage-induced dNTPs, and is accompanied by delayed Rad53 activation and defective chromatin assembly. All of these effects contribute to replication fork instability and reduced cell viability under replication stress. Our data indicate that H2Bub is one of the limiting factors that regulate replication fork progression, and maintain fork stability in the presence of HU-induced stress.
Results
H2B mono-ubiquitylation regulates fork progression in HU
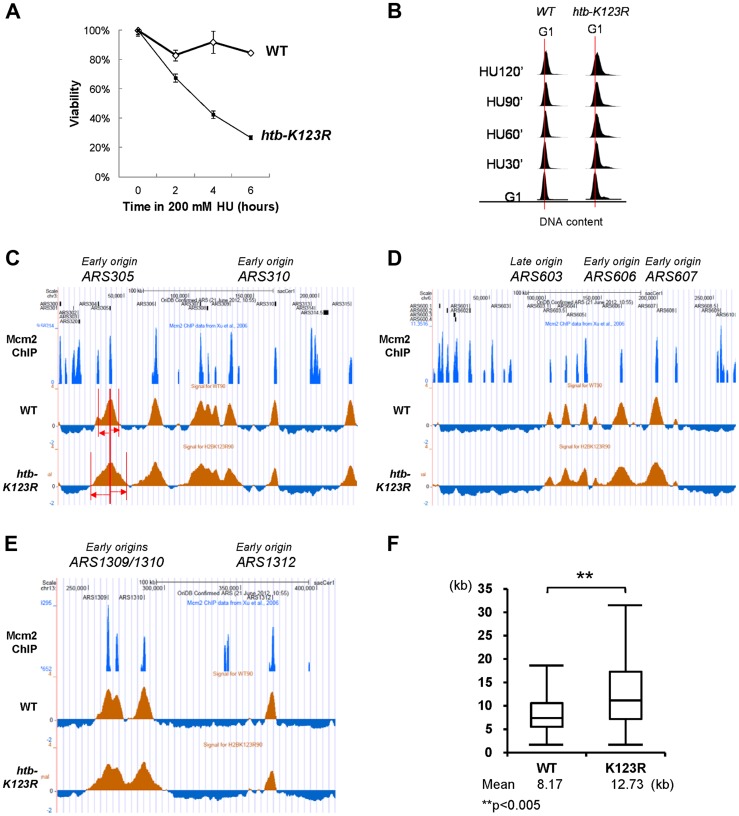
The presence of 200 mM HU increased lethality in mutant cells lacking H2Bub (htb-K123R mutant) (Fig. 1A), confirming the previously-hypothesized role of H2Bub in maintaining fork stability [28]. High doses of HU, an inhibitor of the ribonucleotide reductase, leads to a strong decrease in dNTP pools, that in turn leads to a decrease in replication speed and intra-S checkpoint activation [36], [37]. In order to investigate the mechanisms by which H2Bub sustains cell viability during replication stress, we examined genome-wide origin firing and replication fork progression under HU in wild type and in htb-K123R cells. This was achieved by performing BrdU immunoprecipitation followed by hybridization on a high density oligonucleotide array. Wild-type (WT) or H2Bub-deficient mutant (htb-K123R) cells were pre-synchronized in G1 with α-factor (Fig. 1B) and then released into fresh media containing HU and BrdU. Under such conditions, BrdU is incorporated at active origins (such as ARS305 and ARS607), and BrdU track length correlates with the replication fork progression. Positions of ARS elements were identified by Mcm2 occupancy [38]; therefore, this assay can be used to monitor origin usage and replication fork progression on a genomic scale [37].
Figure 1. H2Bub regulates fork stalling in HU.
(A) The response of WT (CFK1204) and htb-K123R (CFK1231) cells to acute doses of HU. Log-phase cells were treated with 0.2M HU for the indicated times, and dilutions were subsequently spread onto YPD plates. The plates were incubated at 30°C for 2–3 days and viability was estimated based on colony forming units (CFU). Viability was normalized to 0 min of HU treatment, which was set as 100%. (B) Flow cytometry was used to analyze the cell cycle progression of WT (CFK1204) and htb-K123R (CFK1231) cells in the presence of 0.2M HU for 120 minutes after release from α-factor-induced G1 arrest. DNA content is visualized by propidium iodide incorporation. (C–E) Replication profiles of replication origins: (C) ARS305 and ARS310, (D) ARS603, ARS606, and ARS607, and (E) ARS1309/1310 and ARS1312, in WT (CFK1419) and htb-K123R (CFK1421) cells. Cells were synchronized in G1 with α-factor, and then released into media containing 0.2M HU and 200 µg/ml BrdU for 90 minutes. After DNA extraction and fragmentation, BrdU-labeled DNA was immunoprecipitated and hybridized on high-resolution oligonucleotide tiling arrays. Orange histogram bars (BrdU) on the y axis represent the average signal ratio on a log2 scale of loci along the reported regions. Positions of ARS elements are identified by Mcm2 occupancy [72]. (F) The graph depicts the distribution of BrdU track lengths in WT (CFK1419) and htb-K123R (CFK1421) cells. Box and whiskers indicate the minimum, maximum, and 25–75 percentiles, respectively. Mean BrdU tracks lengths are indicated in kb. Asterisks indicate the P-value of the statistical test (Mann–Whitney rank sum t-test, ** P-value<0.005).
An unexpected finding was that the BrdU track lengths at most origins were significantly longer in htb-K123R cells (average 12.73 kb) than in WT cells (average 8.17 kb; Fig. 1C–F and Fig. S1), indicating extended progression of replication forks. This finding was corroborated by the observation that DNA content in the mutant was greater than in WT, as evidenced by FACS (Fig. 1B). In addition, despite the semi-quantitative aspect of this technique, BrdU incorporation peaks were clearly reduced at the majority of firing origins in the mutant. This may be indicative of a decrease in origin firing. Taken together, these results suggest that replication fork stalling is reduced in the htb-K123R mutant during HU-induced stress, and this may lead to fork destabilization.
H2Bub-mediated fork stalling is independent of Dun1-mediated dNTP regulation
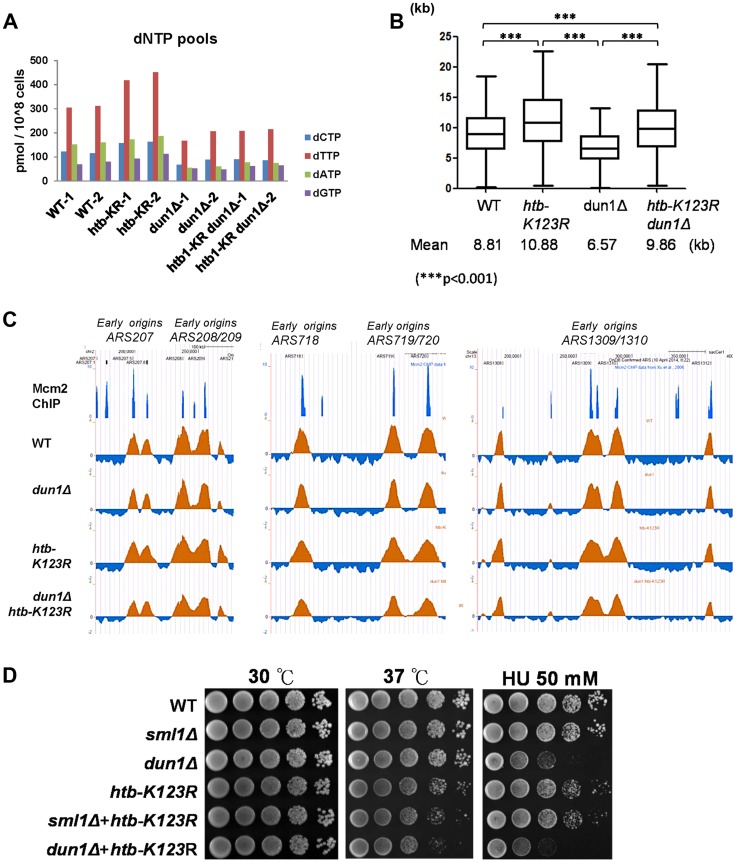
It was previously reported that yeast cells with persistently-enlarged dNTP pools are prone to DNA damage [39], and exhibit enhanced fork progression [37], [40] under replication stress. Thus, it is possible that the enhanced HU sensitivity and increased fork progression in htb-K123R cells may be a consequence of enlarged dNTP pools; this in turn may be a direct consequence of (i) increased transcription of ribonucleotide reductase (RNR) genes or (ii) spontaneous DNA damage, or otherwise via an indirect mechanism that stimulates ribonucleotide production. To test this hypothesis, we directly examined the size of dNTP pools in htb-K123R cells. We observed that the dNTP concentration in htb-K123R cells is ∼40% greater than that of WT cells (shown for four biological replicates in Fig. S2).
The cellular concentration of dNTP pools is regulated by the Rad53-Dun1 pathway during both normal and perturbed cell cycles, through multiple mechanisms [41], [42]. Deletion of DUN1 stabilizes the RNR inhibitor Sml1 and decreases the size of the cellular dNTP pool, while deletion of SML1 increases pool size [43]. To further investigate whether the effect of H2Bub on fork stalling during replication stress is dependent on the concentration of dNTP pools, we deleted the DUN1 gene from WT and htb-K123R cells. Notably, the dNTP pools of both dun1Δ and dun1Δ htb-K123R were ∼50% the size of those in WT cells (Fig. 2A). Since deletion of DUN1 suppressed the increase of dNTP in H2Bub mutants, we can conclude that the increase of dNTP level in H2Bub mutants is mediated by Dun1.We next performed BrdU-IP chip experiments to determine BrdU track length in these cells. As expected, the BrdU track length of the dun1Δ cells was significantly shorter (6.57 kb; Fig. 2B & C) than that in WT cells, probably due to the reduced concentration of dNTP [37], [39]. Surprisingly, fork progression in dun1Δ htb-K123R was significantly faster than in WT cells (9.86 kb vs. 8.81 kb; Fig. 2B & C), despite the reduced size of the dNTP pool (Fig. 2A); this finding indicates that the increase in fork progression and instability in this mutant does not arise solely from the increase in the dNTP pool. In addition, we observed that deletion of DUN1, but not SML1, increased the sensitivity of htb-K123R cells to chronic HU exposure (Fig. 2D). Therefore, we conclude that H2Bub has a role in controlling fork progression and cell survival in response to replication stress, which is independent of the Dun1-mediated regulation of ribonucleotide production.
Figure 2. H2Bub-mediated fork stalling is independent of dNTP pool size.
The size of the dNTP pools in exponentially-growing cultures of WT (CFK1419), htb-K123R (CFK1421), dun1Δ (YCL023), and dun1Δ htb-K123R (YCL025) cells in YPD media. Two independent isogenic strains of each genotype were analyzed. (B) Graph depicting the distribution of BrdU track lengths in WT (CFK1419), htb-K123R (CFK1421), dun1Δ (YCL023), and dun1Δ htb-K123R (YCL025) mutants, as shown in Fig. 1F. (C) Replication profiles of the replication origins ARS207, ARS208/209, ARS718, ARS719/720, and ARS1309/1310 in WT (CFK1419), htb-K123R (CFK1421), dun1Δ (YCL023), and dun1Δ htb-K123R (YCL025) mutants. The BrdU histogram was analyzed as described in Fig. 1C–E. (D) Temperature sensitivity and HU resistance of the indicated genotypes (WT (CFK1204), sml1Δ (CFK1481), dun1Δ (YMW069), htb-K123R (CFK1231), and htb-K123R in combination with sml1Δ (CFK1482) or dun1Δ (YMW072)). Log-phase cells were serially diluted and spotted onto YPD plates with or without HU, and incubated at 30°C or 37°C for 2–3 days.
H2B ubiquitylation sets replication dynamics and replication fork integrity under HU stress
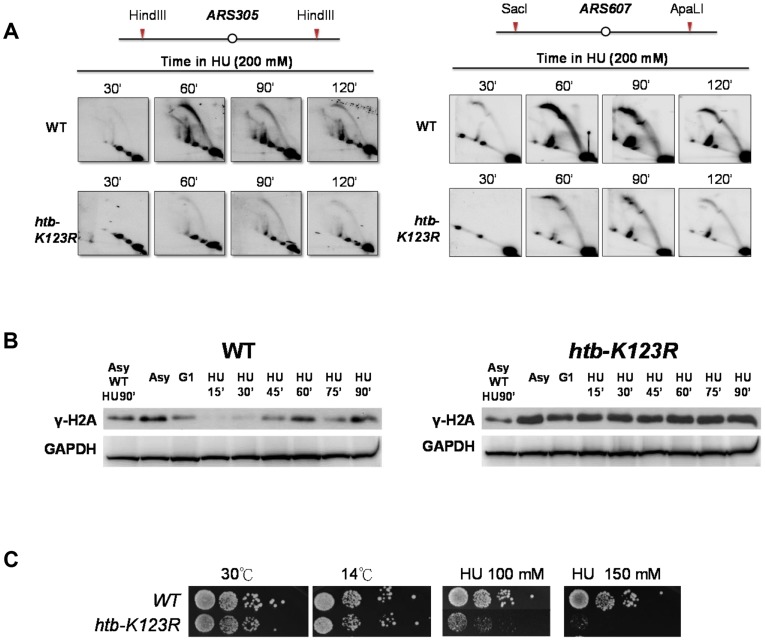
Origin firing and fork progression have been reported to be strongly co-regulated by cells in order to ensure normal completion of replication. In particular, an increase in replication speed leads to a decrease in origin firing [37]. Our BrdU immunoprecipitation and chip hybridization data are consistent with this reported tendency (Fig. 1). However, since this technique is only partially quantitative, we decided to confirm this observation using two-dimensional (2D) gel analysis (Fig. 3A). Replication intermediates migrate differently depending on their molecular weight and sterical conformation. In particular, the bubble arc reflects origin firing. Interestingly, both WT and H2Bub mutant exhibit similar replication kinetics at two early origins (ARS305 and ARS607); replication intermediates appear one hour after alpha factor release in agreement with origin firing, and start to decrease after 2 hours, reflecting fork progression outside of the restriction fragment. However, we observed a strong reduction of replication intermediates in the H2Bub mutant compared to WT, likely due to a decrease in origin efficiency.
Figure 3. H2Bub preserves replication fork stability under HU stress.
(A) Analysis of replication intermediates (RIs) at ARS305 and ARS607 in WT (CFK1204) and htb-K123R (CFK1231) mutants. Cells were synchronized at G1 phase and released into media containing 200 mM HU for 120 minutes. DNA was prepared from cells collected at the indicated times, cut with HindIII (ARS305) or SacI and ApaL1 (ARS607), and analyzed by 2D gel using the ARS305 or ARS607 probe, as described in the Materials and Methods. (B) Accumulation of damaged DNA in H2Bub-depleted cells. WT (CFK1204) and htb-K123R (CFK1231) cells were arrested in G1 and released into fresh media containing 0.2M HU for 90 minutes at 30°C. Whole cell lysates were prepared at the indicated time points, and analyzed by Western blot using antibodies against γ-H2A, a marker of DNA damage. G6PDH was used as a loading control. Asy: Asynchronized cells. (C) Cells lacking H2Bub are more sensitive to replication stress. Ten-fold serial dilutions of yeast cells (WT (CFK1204) and htb-K123R (CFK1231)) were spotted onto nonselective YPD plates under different temperatures or YPD containing 100 or 150 mM HU for a period of several days.
To further delineate the role of H2Bub in origin firing, we measured incorporation of BrdU into chromatin, using BrdU-IP combined with quantitative-PCR. This experiment was performed at 20°C to slow down DNA replication. We found that replication efficiency at ARS305 and ARS607 was much lower in mutant than in WT cells. We did not observe DNA synthesis at a telomeric region (TEL VI) in either WT or the mutant, owing to the late onset of DNA replication at telomere. These data strongly suggest that origin firing is inefficient in cells lacking H2Bub (Fig. S3).
We subsequently hypothesized that the change in replication dynamics in cells lacking H2Bub may affect the integrity of the fork, as previously observed [37]. In particular, γH2A accumulation in the H2Bub mutant confirmed the accumulation of damage in the absence of H2B ubiquitylation (Fig. 3B). This accumulation may explain the hypersensitivity to hydroxyurea that we and others [28] have observed (Fig. 3C).
The Bre1-H2Bub pathway genetically interacts with components of the intra-S phase checkpoint
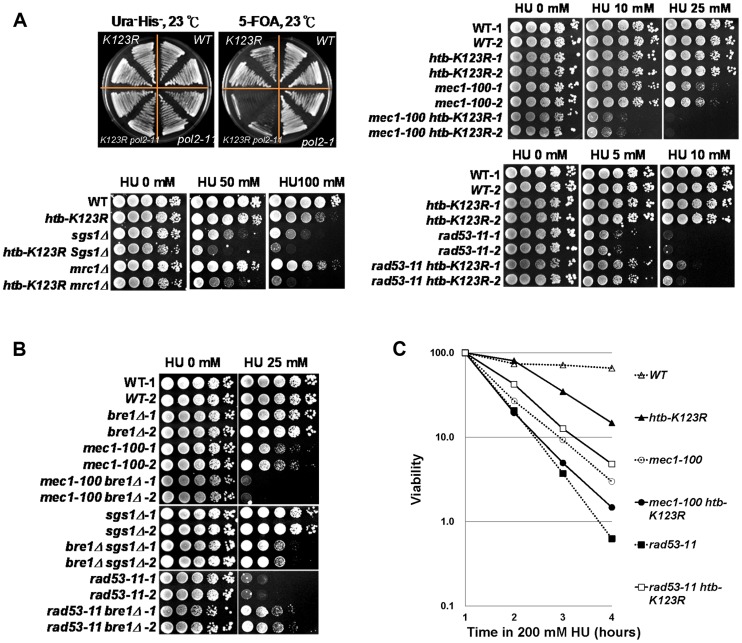
Stalled forks are detected by the intra-S phase checkpoint [30]. We reasoned that the instability of replication forks in htb-K123R cells may result from a defect in the activation of the intra-S phase checkpoint [34]. As such, we examined whether H2Bub interacts with factors that stabilize the replication fork during replication stress, by systematically examining the genetic interactions between htb-K123R and mutations in key components of this complex signaling system. Initially, we examined a hypomorphic allele of pol2-11, which encodes a mutant form of Polε that causes defects in the intra-S phase checkpoint [44]. The htb-K123R and pol2-11 double mutant exhibited synthetic growth defects at the permissive temperature (23°C) (Fig. 4A, top left panel). This interaction was confirmed to be specific, because double mutants of htb-K123R and pol1-17, pol3-14, or pri2-1 (replication defective mutants of DNA polymerase α and δ, and RNA primase, respectively [45]), exhibited subtle additive growth defects, or sensitivity to 50 mM HU at both the permissive (23°C; Fig. S4A) and non-permissive (30°C; Fig. S4B) temperature for growth.
Figure 4. The Bre1-H2Bub pathway genetically interacts with components of the intra-S-phase checkpoint.
(A) H2Bub functions in parallel with DNA polymerase (pol2-11) and intra-S-phase checkpoint cascades (Mec1, Sgs1, and Mrc1). WT and pol2-11 cells carrying HTB1 or the htb1-K123R allele on a HIS3 vector were transformed with HTB1 on a URA3 vector. The strains containing both URA3 and HIS3 (CFK2000, CFK2002, CFK2004, and CFK2006) were streaked onto 5-FOA plates to select for cells lacking H2Bub (htb1-K123R). Ten-fold serial dilutions of the indicated strains were spotted onto YPD plates in the absence or presence of different doses of HU at 30°C (WT (CFK1204, CFK2352, and CFK2414), htb-K123R (CFK1231 and CFK2416), mec1-100 (CFK2346), sgs1Δ (CFK1447), mrc1Δ (CFK1444), rad53-11 (CFK2347), and double mutants (CFK2356, CFK1453, CFK1450, and CFK2358)). (B) The H2B ubiquitin E3 ligase, Bre1, functions in parallel with intra-S-phase checkpoints under HU stress. Ten-fold serial dilutions of the indicated strains (WT (CFK2351), bre1Δ (YMW093), mec1-100 (CFK2346), mec1-100 bre1Δ (YMW095), sgs1Δ (CFK2371), bre1Δ sgs1Δ (CFK2373), rad53-11 (CFK2347), and rad53-11 bre1Δ (CFK2378)) were spotted onto YPD plates with and without HU as described in (A). (C) The response of double mutants of htb-K123R and mec1-100 or rad53-11 to acute exposure to HU. Logarithmically-growing cells were treated with 0.2M HU as described in Fig. 1A.
We next examined the effect of HU on strains containing htb-K123R and mec1-100, an intra-S phase checkpoint defective allele of Mec1/ATR [46], or deletion of MRC1 or SGS1 (Fig. 4A). Single mutants of htb-K123R and mec1-100 grew in the presence of 10 and 25 mM HU, but the double mutant was highly sensitive to these concentrations of HU (Fig. 4A, top right panel). Interestingly, similar phenotypes were observed upon combining htb-K123R with deletions of the genes encoding the checkpoint mediator protein Mrc1 or the RecQ helicase Sgs1 (Fig. 4A, bottom left panel), suggesting that H2Bub stabilizes the replication fork independently of these proteins. We then examined whether H2Bub interacts with the kinase checkpoint effector, Rad53. The rad53-11 mutant is checkpoint defective, with undetectable Rad53 activity [47]. Intriguingly, the hypersensitivity of rad53-11 to HU was partially reversed by htb-K123R (Fig. 4A, bottom-right panel). Deletion of the H2Bub-specific E3 ligase Bre1 had similar effects to htb-K123R when combined with the mec1-100, sgs1Δ, or rad53-11 mutations (Fig. 4B, S4C), suggesting that the genetic interactions between H2Bub and components required for the re-initiation of stalled forks are at the level of chromatin structure, and are linked to its chromatin modifying activities. We next examined the effect of H2Bub deficiency on the viability of mec1-100 and rad53-11 cells in the presence of HU. The absence of H2Bub exacerbated the lethality observed in mec1-100 cells in S phase, while enhancing the viability of rad53-11 cells under the same conditions (Fig. 4C). Overall, our genetic analyses suggest that H2Bub and the Mec1-dependent S-phase checkpoint function in parallel to preserve fork stability under replication stress. However, our finding that htb-K123R alleviates the rad53-11 growth defect under HU suggest that that H2Bub may function upstream of Rad53 and participate in the replication stress response.
H2Bub and Sgs1 play interdependent roles in the replication stress response
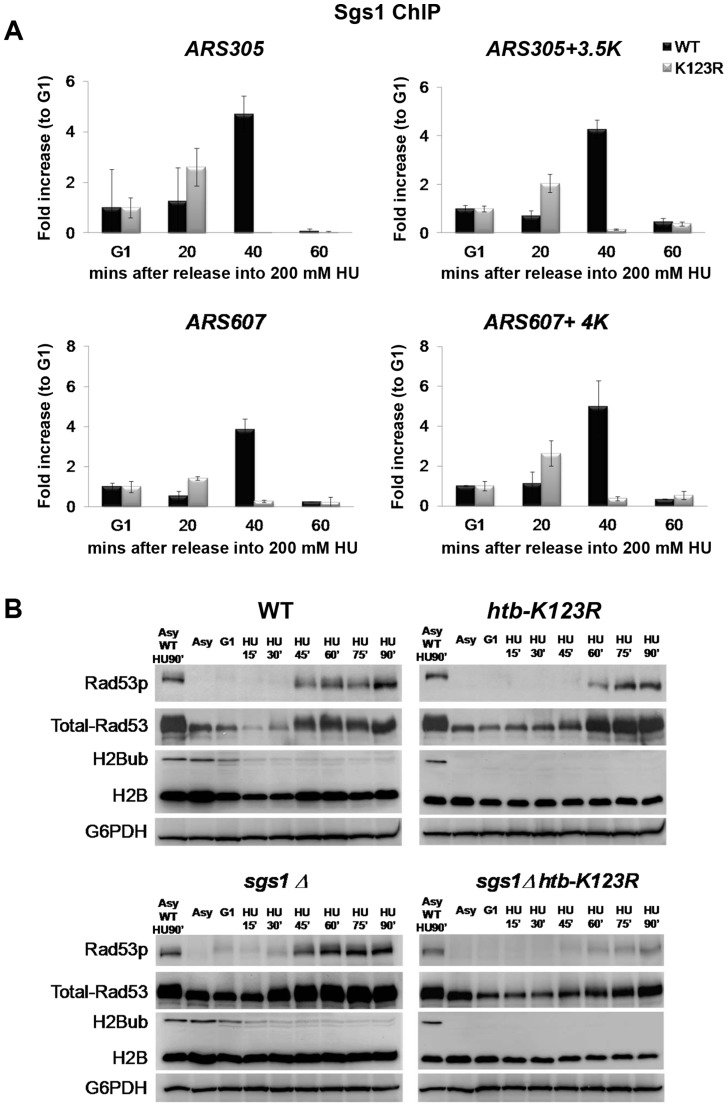
Comparing our data with earlier works [37], [48], [49] revealed several lines of evidence which suggest that H2Bub and the RecQ helicase Sgs1 have overlapping functions in maintaining fork stability under HU. First, both htb-K123R and sgs1Δ cells exhibit increased fork progression in HU (Fig. 1F; [37]). Second, the absence of either Sgs1 or H2Bub reduces the stability of stalled replication forks under HU (Fig. 1A and 3C; [48]). Third, the combination of htb-K123R or sgs1Δ with mec1-100 causes fork collapse and failure to recover from acute exposure to HU (Fig. 4C; [49]). To better delineate the role of H2Bub in the replication stress response, we decided to investigate the interaction between H2Bub and the Sgs1 helicase further. We used ChIP to measure the recruitment of Sgs1 to the ARS305 and ARS607 early origins in WT and htb-K123R cells (Fig. 5A). While Sgs1 was initially recruited efficiently to ARS305 and ARS607 in both strains, it failed to accumulate at ARS in the mutant, suggesting the association of Sgs1 with the replication fork was unstable in the absence of H2Bub (Fig. 5A). Furthermore, HU-induced phosphorylation of Rad53 was unaffected in sgs1Δ, but delayed in htb-K123R cells (Fig 5B). Rad53 activation is facilitated by the retention of Sgs1 at stalled forks [33], and the current results suggest that H2Bub may be required for such retention, and thus Rad53 phosphorylation. Interestingly, a recent study demonstrated that RPA-coated single-stranded DNA replication intermediates (ssDNA) are reduced at initiated origins in htb-K123R cells under HU [28]. RPA is postulated to interact with Sgs1 at replication forks [50]. Thus, the reduced Sgs1 occupancy at replication forks and delayed Rad53 phosphorylation in the htb-K123R mutant may be explained by the decreased amount of ssDNA at replication forks. In addition, Rad53 phosphorylation was significantly impaired in a sgs1Δ htb-K123R double mutant (Fig. 5B). This is also indicative of a Sgs1-independent role for H2Bub in Rad53 activation. Collectively, these results point to a functional role for H2B in replication and the checkpoint response, and are consistent with the observed epistatic interaction between H2Bub and Rad53 (Fig. 4A–C).
Figure 5. H2Bub and Sgs1 play interdependent roles in Rad53 phosphorylation.
(A) Sgs1 occupancy at replication origins is unstable in htb-K123R cells exposed to HU. WT (CFK1764) or htb-K123R (CFK1765) cells were synchronized in G1 and then released into fresh YPD containing 0.2M HU for 60 minutes at 30°C. Chromatin immunoprecipitation (ChIP) was performed using antibodies against Sgs1-3×Myc. DNA was quantified by qPCR using primers adjacent to ARS305 and a region 3.5 kb distal. Sgs1 occupancy at each time point was normalized to that of G1. (B) Activation of Rad53 is impaired in the absence of both H2Bub and Sgs1. WT (CFK1204), htb-K123R (CFK1231), sgs1Δ (CFK1447), and sgs1Δ htb-K123R (CFK1453) cells were arrested in G1 and released into fresh media containing 0.2M HU for 90 minutes at 30°C. Whole cell lysates were prepared at the indicated time points, and analyzed by Western blot using antibodies against Rad53 (EL7), phospho-Rad53 (F9), H2B, and mono-ubiquitylated H2B (anti-FLAG). G6PDH was used as a loading control.
H2Bub and Sgs1 cooperatively control replication fork stalling under HU
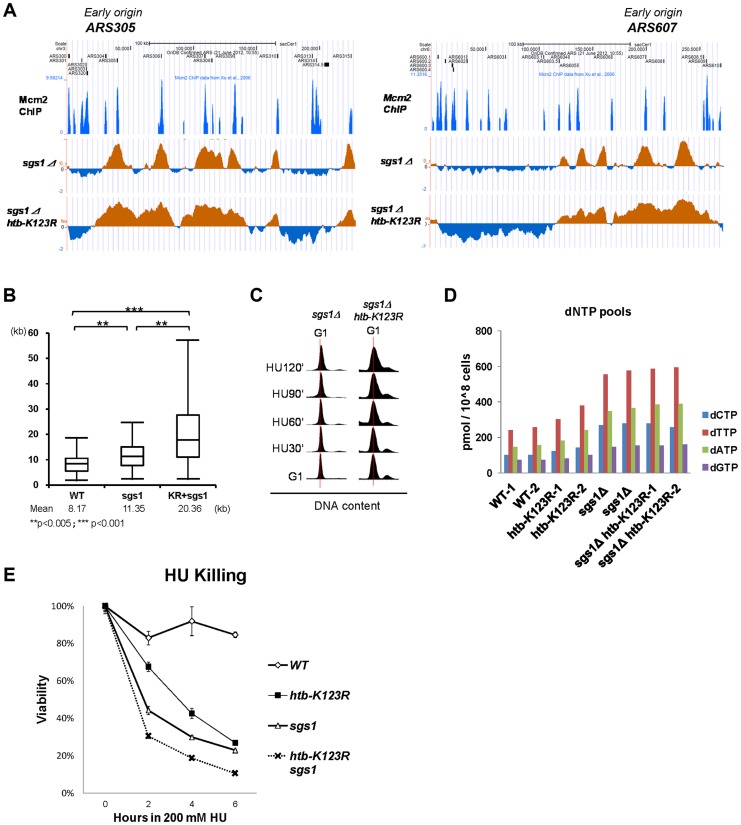
To further elucidate the interaction between Sgs1 and H2Bub, we used BrdU IP-chip to monitor fork progression in sgs1Δ and sgs1Δ htb-K123R mutant cells in the presence of HU. Consistent with a previous report [37], the average BrdU track length in sgs1Δ cells was significantly increased as compared to WT cells (11.35 kb vs. 8.17 kb, respectively; Fig. 6A and 6B), similar to the increase observed in htb-K123R cells (Fig. 1F). Remarkably, track lengths in the sgs1Δ htb-K123R double mutant (20.36 kb) were even greater, being almost 2.5-fold longer than those in WT (8.17 kb; Fig. 6A and 6B). Flow cytometry was used to confirm that the double mutant contained greater amounts of DNA in the presence of 200 mM HU (Fig. 6C). Increased fork progression in the absence of Sgs1 is believed to be a consequence of dNTP accumulation [37]. We confirmed that the dNTP concentration was increased in sgs1Δ (∼2.3 fold as compared to WT; Fig. 6D), but no additional increase was observed in the sgs1Δ htb-K123R double mutant, suggesting that elevated dNTP production does not underlie the defect in fork stalling in the double mutant.
Figure 6. H2Bub and Sgs1 cooperatively control replication fork stalling and stability under HU.
(A) Replication profiles of the early origins ARS305 and ARS607 in sgs1Δ (YCL007) and sgs1Δ htb-K123R (YCL008) mutants. The BrdU histogram was analyzed as described in Fig. 1C–E. (B) Graph depicting the distribution of BrdU track lengths in WT (CFK1419), sgs1Δ (YCL007), and sgs1Δ htb-K123R (YCL008) mutants, as shown in Fig. 1F. (C) Cell cycle progression of these mutants in the presence of 0.2M HU was analyzed by flow cytometry. (D) The size of the dNTP pools in exponentially-growing cultures of WT (CFK1419), htb-K123R (CFK1421), sgs1Δ (YCL007), and sgs1Δ htb-K123R (YCL008) cells in YPD media. Two independent isogenic strains of each genotype were analyzed. (E) Survival of WT (CFK1204), htb-K123R (CFK1231), sgs1Δ (CFK1447), and sgs1Δ htb-K123R (CFK1453) cells in response to acute doses of HU, as shown in Fig. 1A.
We have demonstrated that the replication fork becomes unstable and vulnerable to replication stress in H2Bub-deficient cells (Fig. 1A & 3B), which could be due to continuous DNA synthesis under conditions of dNTP depletion. Thus, we reasoned that the rapidly moving replication fork may become highly unstable in the absence of both H2Bub and Sgs1, a hypothesis supported by the observation that the double mutant was more sensitive to acute treatment with HU than either single mutant (Fig. 6E). Taken together, our results so far suggest that H2Bub is involved in stalling the replication fork and maintaining its stability in response to HU-induced S phase block; furthermore, this function is performed in cooperation with Rad53 kinase activity and in parallel with Mec1 and Sgs1 during S phase.
H2Bub promotes chromatin assembly in response to replication stress
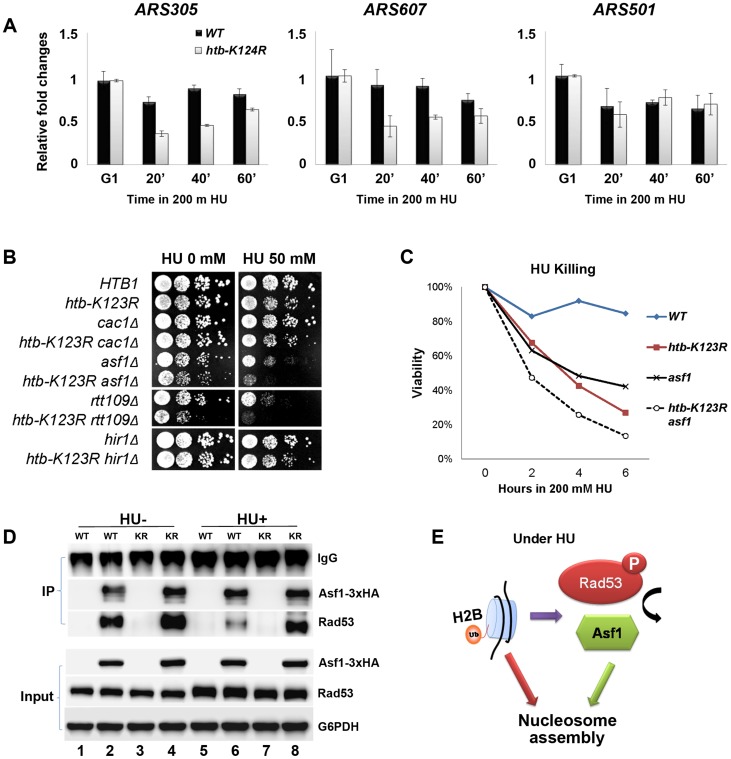
H2Bub has been shown to be required for nucleosome reassembly during RNA Polymerase II elongation [20], [23] and DNA replication [28]. We therefore hypothesized that defective fork stalling in htb-K123R cells under replication stress may be a consequence of incomplete nucleosome assembly. We first confirmed that histone assembly on newly-synthesized DNA is defective in htb-K123R under HU. In WT cells, histone H3 was associated with the early firing origins ARS305 and ARS607 at all times post-G1 release into HU. H3 occupancy at these early origins was reduced upon entry into S phase in htb-K123R cells, but occupancy at the late origin ARS501 was unaffected (Fig. 7A). These data suggest that in the absence of H2Bub, histone assembly is less efficient at firing origins.
Figure 7. H2Bub promotes chromatin assembly in response to replication stress.
(A) H2Bub is required for nucleosome assembly near replication forks under replication stress. WT (CFK1204) or htb-K123R (CFK1231) cells were arrested in G1 phase using α-factor, and were then released into 200 mM HU at 30°C for 60 minutes. At the indicated time, cells were collected and histone occupancy at two early origins (ARS305 and ARS607) and one late origin (ARS501) was determined by ChIP using antibodies against H3. IP signals at ARS sequences were normalized to IP signals at TELVI-R. The results are the mean +/− SEM of three replicates. (B) Genetic interactions between H2Bub and histone chaperones (Cac1, Asf1, and Hir1) or a histone acetyl-transferase (Rtt109). Ten-fold serial dilutions of the indicated strains (WT (CFK1204), htb-K123R (CFK1231), cac1Δ (CFK1206), cac1Δ htb-K123R (CFK1237), asf1Δ (CFK1208), asf1Δ htb-K123R (CFK1233), rtt109Δ (CFK1212), rtt109Δ htb-K123R (CFK1241), hir1Δ (CFK1202), and hir1Δ htb-K123R (CFK1235)) were spotted onto YPD plates containing HU (0 or 50 mM), and cell growth was monitored for 2–3 days. (C) The survival of asf1Δ (CFK1208) and asf1Δ htb-K123R (CFK1233) cells in response to acute treatment with HU, as described in Fig. 1A. (D) H2Bub modulates the interaction between Asf1 and Rad53 under HU stress. Asynchronous cultures of WT (YMW105) or htb-K123R (YMW104) cells were untreated (−) or treated (+) with 0.2M HU for 90 minutes. Protein extracts were prepared and incubated with pre-bound anti-HA-protein G beads to pull down Asf1-3×HA, and the immune-precipitates were resolved by SDS-PAGE, before being probed with either anti-HA or anti-Rad53 antibodies. (E) A working model depicting the role of H2Bub in nucleosome assembly under HU stress. H2Bub coordinates nucleosome assembly in response to replication stress by directly contributing to nucleosome formation and by indirectly regulating the availability of Asf1 during HU stress.
Mec1 was recently reported to increase chromatin accessibility at or ahead of replication forks, and promote fork progression in HU [35]. Thus, the mechanism promoting nucleosome assembly during DNA replication may inhibit fork progression under replication stress. We reasoned that if this were the case, deletion of genes encoding proteins involved in replication-coupled histone assembly (such as the histone chaperones CAF-1 and Asf1) should sensitize htb-K123R cells to replication stress. Asf1 has dual roles; it associates with the RCF complex and MCM helicase and facilitates nucleosome disassembly during replication [51], [52], and it assists acetylation of H3 lysine 56 (H3K56ac) by presenting newly-synthesized H3/H4 dimers to the Rtt109 acetyltransferase [4], [53]. Acetylation increases the affinity of H3 for CAF-1 [53] and promotes efficient chromatin assembly onto nascent DNA [8]. As a control, we deleted Hir1; this protein is implicated in replication-independent H3/H4 deposition [54]. We found that deletion of ASF1 or RTT109 greatly increased the HU sensitivity of htb-K123R cells, but deletion of CAC1 (the largest subunit of CAF-1) or HIR1 had no such effect (Fig. 7B). This suggests that H2Bub and Asf1-Rtt109 function synergistically to promote cell survival during replication stress. Moreover, deletion of Asf1 increased the sensitivity of htb-K123R cells to acute HU treatment (Fig. 7C), suggesting that the stability of the replication fork was decreased further.
H2Bub-mediated Rad53 activation promotes the dissociation of histone chaperone Asf1 from the Rad53 complex
Rad53 and Asf1 form a dynamic complex that dissociates in response to Rad53 phosphorylation under replication stress. Rad53 acts as a regulator of Asf1 availability and indirectly controls its chromatin assembly activity [55], [56]. Our results suggest that H2Bub may affect Rad53 phosphorylation (Fig. 5B). We hypothesized that H2Bub may contribute to nucleosome assembly by influencing the dynamic association between Asf1 and Rad53, in addition to possessing a direct role in nucleosome assembly. To test this hypothesis, we tagged the genomic ASF1 gene with a triple HA tag, thereby enabling immunoprecipitation of Asf1 with an anti-HA antibody (Fig. 7D, lanes 2, 4, 6, and 8). Rad53 co-precipitated with HA-tagged Asf1 efficiently in WT lysates (Fig. 7D, lane 2) but not with un-tagged Asf1 (Fig. 7D, lanes 1, 3, 5, and 7). Consistent with previously published results [55], [56], Rad53 association with Asf1 was reduced in the presence of HU in WT cells (Fig. 7D, lane 6). However, the association of Rad53 with Asf1 remained stable in htb-K123R cells in the presence of HU (Fig. 7D, compare lanes 6 and 8). These results suggest that H2Bub coordinates nucleosome assembly in response to replication stress by directly contributing to nucleosome formation, and by indirectly regulating the availability of Asf1, which in turn deposits histones behind the advancing replication fork (Fig. 7E).
Discussion
Here, we report that replication fork stalling is regulated by the Bre1-H2Bub pathway in the presence of HU-induced stress. We demonstrate that elimination of H2Bub enhances replication fork progression and instability in HU. Importantly, this process is independent of Dun1-mediated regulation of dNTP pools. Instead, H2Bub promotes Rad53 activation and mediates dissociation of phosphorylated Rad53 and Asf1, which may contribute to nucleosome assembly and promote cell survival in HU. These findings lead us to suggest that H2Bub plays a more direct role in fork stalling and stability under replication stress.
Chromatin state facilitates tight regulation of fork progression during replication stress
How does the Bre1-H2Bub pathway modulate the cellular response to HU-induced replication block? Interestingly, H2B ubiquitylation has been proposed to promote unwinding of the DNA chromatin complex ahead of the replication fork, and thereby stimulate fork progression in HU [28]. However, our results support an alternative role for H2Bub in restricting replication fork progression under conditions of HU stress.
We found that histone occupancy around early origins in htb-K123R cells is reduced upon S phase entry in the presence of HU (Fig. 7A). In addition, we also showed that the removal of both Asf1-Rtt109 and H2Bub synthetically increases the sensitivity to replication stress (Fig. 7B and C). Moreover, we provide evidence that H2Bub controls the availability of Asf1 during replication stress (Fig. 7D), which potentially contributes to histone deposition behind the advancing replication fork [55], [56]. Thus, we propose that enhanced chromatin assembly on nascent DNA during replication stress may facilitate replication fork stalling in response to nucleotide depletion imposed by HU (Fig. 8), akin to the brakes on a train (the replisome). Mec1 slows S phase progression by delaying late origin firing through intra-S phase checkpoint activation [30], [34], but it also promotes sustained replication fork progression at early origins [35]. The chromatin state seems to facilitate tight regulation of fork progression at early origins during replication stress.
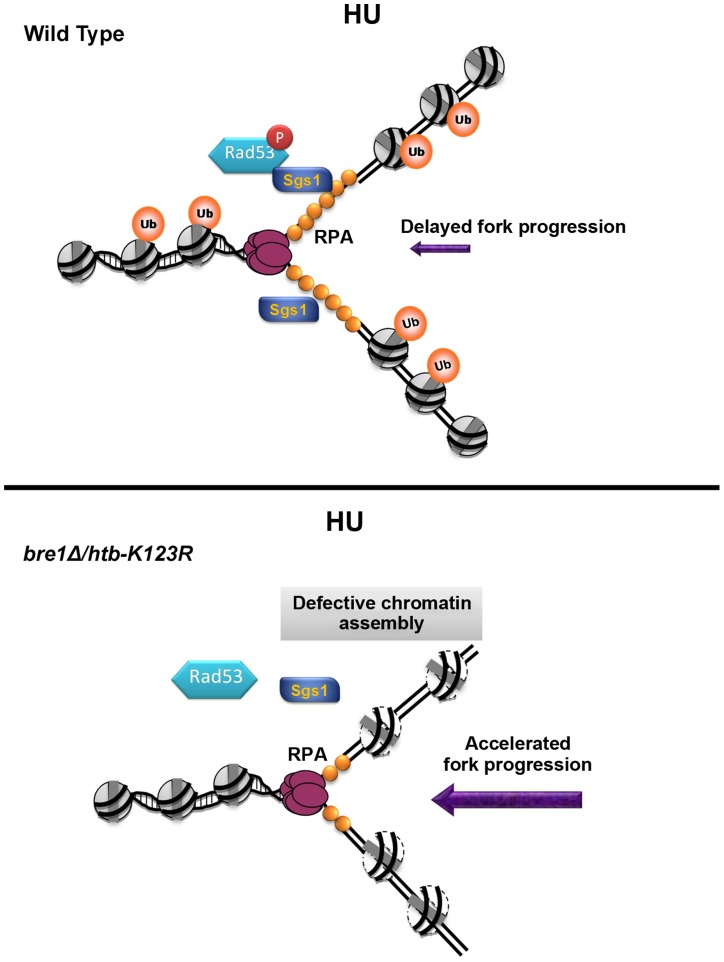
Figure 8. A model for how H2B mono-ubiquitylation facilitates fork stability under replication stress.
Upon HU-induced stress, H2Bub promotes nucleosome assembly, which assists replication fork stalling, Sgs1 recruitment, and Rad53 phosphorylation. The reassembly of chromatin on nascent DNA restricts fork progression and promotes replication fork stability and its recovery after the removal of HU. In the absence of H2Bub (bre1Δ/htb-K123R), replication fork movement is faster than even that observed under nucleotide depletion by HU, which results in shorter tracts of RPA-coated-single-stranded DNA. This in turn reduces retention of Sgs1 at the forks, and delays phosphorylation of Rad53.
We cannot exclude the possibility that continuous DNA synthesis in the htb-K123R mutant may reflect the movement of DNA polymerase through inappropriately-assembled chromatin. In this scenario, chromatin structure at or ahead of the fork would be altered in the absence of H2Bub due to defective chromatin assembly during the previous round of replication. Mec1 and Bre1-H2Bub may have antagonistic effects on chromatin dynamics ahead of the replication fork; thus in the absence of H2Bub, forks may be inclined to move faster because of Mec1-induced chromatin accessibility [35]. Although the scenario outlined above is formally possible, our molecular and genetic analyses favor a second model in which nucleosome formation on nascent DNA serves as a negative feedback mechanism to regulate the progression of the replication fork under stress. Thus, we suggest that Mec1-mediated signaling and the Bre1-H2Bub pathway synergistically interact to ensure that replisomes travel in a controlled manner, thereby maintaining fork stability under replication stress.
H2Bub is a regulator of the DNA replication stress signaling pathway
Checkpoint kinases Mec1 and Rad53 are essential for the maintenance of cell viability when replication is perturbed [30], [31]. Our genetic analyses reveal the unexpected finding that H2Bub maintains fork stability in parallel with the Mec1-mediated intra-S checkpoint, but its effect is epistatic to that of a second checkpoint kinase, Rad53. Our results support the hypothesis that Rad53 stabilizes replication forks independently of Mec1 [57], [58]. Furthermore, our findings suggest a possible mechanism for the role of H2Bub in Rad53 activation (Fig. 8 upper panel). We report that the stable association of Sgs1 with the replication fork is not only replication-dependent [48], but also H2Bub-dependent (Fig. 5A). It was previously demonstrated that Sgs1 helps recruit Rad53 to stalled forks via an interaction with RPA [50]. Intriguingly, it has been postulated that fork collapse followed by origin firing in yeast cells lacking H2Bub results in reduced levels of single-stranded DNA (ssDNA) and RPA during a G1 to HU shift [28], consistent with our observation of reduced replication intermediates and increased DNA damage in htb-K123R cells (Fig. 3). However, it is also possible that the failure to accumulate RPA in htb-K123R cells may be caused by an increase in the rate of nascent DNA synthesis, thereby reducing the accumulation of ssDNA at stalled forks; this in turn reduces Sgs1 retention and delays Rad53 phosphorylation (Fig. 8, bottom panel). The reduced activity of Rad53 may have a negative feedback effect, thereby further compromising fork stability. The absence of both H2Bub and Sgs1 therefore further disrupts Rad53 activation and fork integrity.
The mechanism by which chromatin assembly regulates fork progression and stability
In support of our model that chromatin assembly serves as a negative feedback signal to regulate the progression of replication forks, several reports in budding yeast have established that chromatin assembly at replication forks is necessary to stabilize replication forks and prevent their collapse [52], [59], [60]. A recent report in mammals demonstrated that replication fork speed is dependent on the supply of new histones and efficient nucleosome assembly during an unperturbed cell cycle [61]. Human Asf1 has been shown to associate with the MCM complex through histone H3/H4 dimers [51]. In addition, Asf1 extracted from human cells exposed to HU exhibits an enhanced ability to assemble chromatin [62]. Thus, there may be two pools of Asf1 in cells. One is coupled to replication forks, while the other is sequestered by Rad53. Replication stress triggers the release of the sequestered pool of Asf1 (which occurs at least in part through H2Bub) to promote chromatin formation (Fig. 7E) and restrict fork progression (Fig. 1C–F) under replication stress. However, defects in nucleosome assembly mediated by CAF-1 trigger DNA damage checkpoint activation and delay fork progression in human cells during an unperturbed cell cycle [63]–[65]. Our genetic analysis shows that Cac1, unlike H2Bub and Asf1, is not required by yeast cells to maintain growth under HU stress; hence chromatin assembly regulated by H2Bub and Asf1 under replication stress (Fig. 7B) likely occurs through pathways distinct from those mediated by CAF-1 [66].
In summary, we have provided evidence that H2Bub coordinates chromatin assembly and Rad53 activation during HU stress in parallel with other mechanisms that maintain fork stalling and stability during replication stress, including the intra-S phase checkpoint and the Sgs1 helicase. Our data indicate that H2Bub maintains genomic stability by creating an environment that integrates chromatin formation and checkpoint kinase activation, thereby maintaining stable replication and facilitating recovery from replication stress in concert with other components that mediate faithful DNA replication.
Materials and Methods
Yeast strains, plasmids, and phenotypic screening
Yeast strains and plasmids used in this study are shown in supplementary tables S1 and S2. All yeast cells were cultured in yeast extract peptone supplemented with 2% dextrose at 30°C. All analyses were performed during the log phase of growth. Cells were arrested in G1 by the addition of α-factor to a final concentration of 100 ng/ml (bar1Δ strain) for at least 3 hours (the exact time differed depending on the strain). Cells were released from G1 arrest by washing with sterilized H2O three times, before being re-suspended in fresh media containing hydroxyurea (HU; Sigma).
For phenotypic screening, mid-log (0.4–0.8) phase cultures were collected and counted. Ten-fold serial dilutions were spotted onto YPD plates containing different doses of HU. Plates were subsequently incubated at 30°C for several days.
Two different strain backgrounds were used in this study. With the exception of the strains used in the genetic analysis shown in Fig. 4, all strains were in the YS131 background. The YS131 parental strain is derived from W303, but both genomic copies of HTA1-HTB1 and HTA2-HTB2 are deleted, and cell viability is maintained by a plasmid-derived HTA1-HTB1 or HTA1-htb1-K123R. Earlier studies established that deletion of HTA2-HTB2 has negligible effects on mitotic growth and stress responses, and that the HTA1-HTB1 gene pair can compensate for the absence of the HTA2-HTB2 [67], [68]. Therefore, we predict that hta2-htb2Δ would not affect the htb-K123R mutation.
For the genetic analysis with the checkpoint mutants, we were conscious of an earlier report that the rad53 mutant is sensitive to histone dosage [69]. To prevent unexpected pleiotropic effects, we introduced genomic htb-K123R mutations [HTA1-htb1-K123R::NAT+ HTA2-htb2-K123R::HIS+] [70] into mec1-100 and rad53-11 for genetic analysis. We also compared the HU sensitivity of the htb-K123R mutants in both strain backgrounds to ensure that they give rise to the same replication defects, as shown in Fig. S5.
Gene replacement
For gene disruptions, the indicated gene was deleted by high efficiency transformation, using a PCR product in which the target was replaced with the KanMX gene (deletion library from SGD). The mutant alleles, pol1-17 [45], pri2-1 [45], pol2-11 [71] and pol3-14 [45], were introduced into strain CFK1204 or CFK1231 through the gene replacement technique of Scherer and Davis [72], thereby generating ts mutants. The plasmid used for gene replacement consisted of a 9-kb pol1(Ts), 3.3-kb pri2(Ts), 13-kb pol2(Ts), or 4-kb pol3(Ts) fragment cloned into the XhoI site of YlP, HpaI site of YlPA16, AgeI site of pRS306, or KpnI site of pMJ14.
BrdU-IP chip analysis
S. cerevisiae strains were designed in order to allow BrdU incorporation (TK repeats) (Tables S1 and S2). S. cerevisiae oligonucleotide microarrays were obtained from Affymetrix. BrdU-IP chip analysis was carried out as previously described [73], [74]. Briefly, cells were synchronized with α-factor and then released into fresh YPD containing 0.2M HU and 200 µg/ml BrdU for 90 minutes. The collected cells were arrested in ice-cold buffer containing 0.1% Na-azide, and genomic DNA was extracted from 2×109 cells as described in the “QIAGEN Genomic DNA Handbook”. DNA was sheared to 300 bp by sonication, denatured, and mixed with 4 µg anti-BrdU monoclonal antibody (MBL M1-11-3) as previously described [75], [76]. Antibody-bound and unbound fractions were subsequently purified, and then amplified using the WGA2 GenomePlex Complete Genome Amplification Kit. A total of 2 µg of amplified DNA was digested with DNaseI to a mean size of 100 bp; the fragments were subsequently end-labeled with biotin-N11-ddATP [77], and hybridized to the DNA chip.
Flow cytometry analysis
For DNA content analysis, approximately 1×107 cells were collected at each time point, and resuspended in 1 ml 70% ethanol (ice-cold), before being stored at −80°C for at least one night (samples were stored up to a maximum of 3 days). The cells were then washed twice with 1 ml 50 mM Tris-HCl (pH 8.0) followed by RNAase A digestion (1 mg ml−1 of RNAase A in 50 mM Tris-Cl, pH 8.0) and proteinase K digestion (16 units ml−1 in 30 mM Tris-Cl, pH 8.0). Finally, cells were stained with SYBR GREEN I buffer (in 50 mM Tris-Cl, pH 8.0) at 4°C overnight. The cell size and DNA contents of 50,000 cells were examined on a FACSCanto II (BD).
Two-dimensional (2D) electrophoresis and Southern blot
Total genomic DNA was extracted according to the protocol of the QIAGEN Genomic DNA Handbook, using genomic-tip 100/G columns. 2D gel electrophoresis was carried out as originally described by Brewer and Fangman [78]. The DNA samples were digested with HindIII or SacI/ApaL1, for ARS305 and ARS607 detection respectively, and then blotted onto a Nylon Gene Screen Plus membrane (NEN). Membranes were probed with the BamHI-NcoI 3.0 kb fragment which spans ARS305 and was purified from plasmid A6C-110 (kindly provided by C. Newlon, uMDNJ, Newark, NJ), or probed with a 3.0 kb PCR product that spans ARS607. Signals were detected using a PhosphorImager Typhoon FLA 7000 (GE Healthcare).
HU survival assay
To determine viability in response to acute doses of HU, cells were grown in culture media until they reached log phase. The cells were then arrested in G1 for 3 hours by the addition of α-factor, before being released into rich media containing 200 mM HU. Aliquots were removed from each culture at the indicated time point, plated onto YPD plates, and allowed to grow at 30°C for 2–3 days. Viability was estimated based on colony forming unit (CFU) counts, and was adjusted to that of wild-type at each time point.
Western blot
Yeast cell lysates were prepared using the TCA method [79]. Briefly, equivalent numbers of cells (1.5×108) were collected, resuspended in 200 µl TCA buffer (1.85 M NaOH and 7.4% β-mercaptoethanol), and placed on ice for 10 minutes. Following the addition of 200 µl of 20% TCA, the lysates were incubated on ice for 10 minutes. Pellets were subsequently collected, washed with 1 ml acetone, dried, and dissolved in 200 µl 0.1 M NaOH. The concentration of each sample was determined, and equal amounts were separated by SDS-PAGE, before being transferred to PVDF membranes for immunoblotting. The following antibodies were used: anti-GAPDH (Sigma), anti-Flag (Sigma) and anti-phospho-Rad53 (produced and characterized by A. Pellicioli and the IFOM antibody facility, and kindly provided by Dr. Foiani [80]). Secondary antibodies conjugated to horseradish peroxidase were detected using enhanced chemiluminescence (Amersham Biosciences).
Determination of dNTP pools
The dNTP pools were analyzed as described by a recent study [81]. At a density from 0.4 to 0.8×107 cells/ml, ∼3.7×108 cells were collected onto a 0.8 µm nitrocellulose filter (Millipore AB, Solna, Sweden). The filters were immersed in 700 ml of ice-cold extraction solution (12% w/v trichloroacetic acid, 15 mM MgCl2) in Eppendorf tubes. The following steps were carried out at 4°C. The tubes were vortexed for 30 s, incubated for 15 min, and vortexed again for 30 s. The filters were removed, and the solutions were centrifuged at 20,000× g for 1 min. After centrifugation, 700 ml of supernatant was added to 800 ml of ice-cold Freon–trioctylamine mixture [10 ml of 99% Freon (1,1,2-trichlorotrifluoroethane; Aldrich, Sigma-Aldrich Sweden AB, Stockholm, Sweden)], and 2.8 ml of>99% trioctylamine (Fluka, Sigma-Aldrich Sweden AB, Stockholm, Sweden). The samples were vortexed and centrifuged for 1 min at 20,000× g. The aqueous phase was collected and added to 700 ml of an ice-cold Freon–trioctylamine mixture. Aliquots (475 and 47.5 ml) of the resulting aqueous phase were collected. The 475 ml aliquots were pH adjusted with 1M NH4HCO3 (pH 8.9), loaded onto boronate columns [Affi-Gel 601 (Bio-Rad)], and eluted with 50 mM NH4HCO3, pH 8.9, 15 mM MgCl2 to separate dNTPs and NTPs. The eluates with purified dNTPs were adjusted to pH 3.4 with 6M HCl, and separated on a Partisphere SAX-5 HPLC column (125 mm×4.6 mm, 5 µm, Hichrom, UK) using the Hitachi HPLC EZChrom system. Nucleotides were isocratically eluted using 0.36M ammonium phosphate buffer (pH 3.4, 2.5% v/v acetonitrile). The 47.5 ml aliquots were adjusted to pH 3.4 and used to quantify NTPs by HPLC in the same way as dNTPs. The nucleotides were quantified by measuring the peak heights and comparing them to a standard curve.
Chromatin immunoprecipitation (ChIP)
Yeast strains were grown to an OD600 of 0.4–0.8, and fixed with 1% formaldehyde at room temperature (RT) for 20 min. Fixation was stopped by the addition of glycine to a final concentration of 125 mM for 5 min, and the cells were then collected and washed twice with ice-cold TBS (100 mM Tris at pH 7.5, 0.9% NaCl). Cell pellets were stored at −80°C or resuspended immediately in 500 µl of FA lysis buffer (50 mM HEPES, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% sodium deoxycholate, 0.1% SDS) supplemented with fresh protease inhibitor cocktail (Sigma), and lysed by vortexing with glass beads for 30 min at 4°C. Cell lysates were sonicated in a cooling water bath four times for 10 min each using a SONICATOR 3000 (MISONIX), with each cycle consisting of 30 sec sonication on and 30 sec off. The average size of the resulting DNA fragments was between 200 and 500 base pairs. Following centrifugation at 13.5K for 30 min at 4°C, the solubilized chromatin was collected and adjusted to 500 µl with FA lysis buffer. Twenty microliters were removed for use as input chromatin.
For immunoprecipitation, 10 OD equivalents of solubilized chromatin were incubated overnight at 4°C, together with 20 µl of protein G dynabeads (Invitrogen) that had been pre-bound with anti-H3 or anti-Myc (Sgs1-13Myc). Immunoprecipitates were collected by a step-wise washing protocol, consisting of 1.5 ml FA-lysis buffer, 1.5 ml WASH I (FA lysis buffer+0.5 M NaCl), 1.5 ml WASH II (10 mM Tris-Cl, pH 7.5, 1 mM EDTA, 0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate), and 1.5 ml TE (pH 8.0) for 5 min each at room temperature. The immuno-complexes were eluted by adding 0.25 ml Elution buffer (50 mM Tris-Cl, pH 7.5, 10 mM EDTA, 1% SDS), and incubated first at 65°C for 20 minutes, and then at room temperature for 10 minutes with vortexing. DNA was purified using Qiaquick PCR purification spin-columns (Qiagen), and used as template for quantitative-PCR. All the primers used is listed in Table S3. The primers used in the histone H3 ChIP experiment were designed to amplify DNA fragments present at nucleosomes, as depicted in Figure S6.
Co-immunoprecipitation (Co-IP)
For immunoprecipitations [56], log phase WT or htb-K123R cells untreated (−) or treated (+) with 0.2M HU were collected, resuspended in buffer containing 50 mM Tris7.5, 150 mM NaCl, 5 mM EDTA, 0.5% Triton X-100, and proteinase inhibitors, and broken open by bead beating. A total of 5 mg of protein extract was diluted in 1 ml of the same buffer, and incubated with pre-bound anti-HA-protein G beads at 4°C for 2.5 hours, and then rotated at 4°C overnight. Beads were then washed with 1 ml buffer four times. SDS-loading dye was added, and samples were boiled and resolved on SDS-PAGE.
Statistical analysis
Results are expressed as the mean ± SEM from the number of experiments indicated in the figure legends. Student's t-test was used to analyze statistical significance.
Supporting Information
(A–D) Replication profiles in WT (CFK1419) vs. htb-K123R (CFK1421) cells. Cells were synchronized in G1 with α-factor, and then released into media containing 0.2M HU and 200 µg/ml BrdU for 90 minutes. After DNA extraction and fragmentation, BrdU-labeled DNA was immunoprecipitated and hybridized on high-resolution tiling arrays. Orange (BrdU-IP) histogram bars on the y axis show the average signal ratio on a log2 scale of loci along the reported regions on (A) chromosome II, (B) chromosome V, (C) chromosome IX, and (D) chromosome XVI. The positions of potential ARS elements are identified by Mcm2 loading.
(TIFF)
The size of each dNTP pool in exponentially-growing WT (CFK1419) and htb-K123R (CFK1421) cells. Four independent isogenic strains of each genotype were analyzed as described in the Materials and Methods.
(TIFF)
H2Bub is required for efficient origin firing. htb1-K123R mutants exhibit reduced BrdU incorporation during S phase. Cells were arrested at G1 using α-factor at 23°C, and released synchronously into S phase at 20°C in YPD supplemented with BrdU. Samples were collected at the indicated times and genomic DNA was then extracted. Monoclonal BrdU antibody was used to immunoprecipitate BrdU-incorporated DNA. DNA synthesis at replication origins (ARS305 and ARS607) or telomere was detected by quantitative-PCR. Cell cycle progression was monitored by FACS at 20°C under BrdU incorporation conditions.
(TIFF)
(A) The growth of htb-K123R and DNA polymerase ts double mutants are not affected by HU at the permissive temperature (23°C). Ten-fold serial dilutions of the indicated strains (WT (CFK1204), htb-K123R (CFK1231), pol1-17 (CFK1984), pol1-17 htb-K123R (CFK1986), pri2-1 (CFK1988), pri2-1 htb-K123R (CFK1990), pol3-14 (CFK1992) and pol3-14 htb-K123R (CFK1994)) were spotted onto YPD containing different doses of HU (0–50 mM) at 23°C for several days. Growth at the restrictive temperature (37°C) is presented as a control for ts mutants. (B) The genetic interaction between H2Bub and DNA pol1, pol3, or primase. Ten-fold serial dilutions of the indicated strains were spotted onto YPD, and growth was monitored at 33°C or 30°C, or under conditions of replication stress (50 mM HU) at 30°C. (C) The histone H2B ubiquitin E3 ligase Bre1 functions in parallel with the RecQ helicase Sgs1 under replication stress. Ten-fold serial dilutions of the indicated strains (WT (CFK1204), bre1Δ (CFK1443), sgs1Δ (CFK2371) and bre1Δ sgs1Δ (CFK2373)) were spotted onto YPD containing different doses of HU (0–100 mM) at 30°C.
(TIFF)
The growth of WT and htb-K123R cells of two different backgrounds under conditions of replication stress at 30°C. Ten-fold serial dilutions of the indicated strains (WT (CFK1204), htb-K123R (CFK1231), WT (CFK2414), and htb-K123R (CFK2416)) were spotted onto YPD containing different doses of HU (0–150 mM) for 2 days. Genotypes of the strains used: CFK1024: W303 hta1-htb1Δ hta2-htb2Δ <pZS145-HTA1-Flag-HTB1 CEN HIS3> CFK1031: W303 hta1-htb1Δ hta2-htb2Δ <pZS146-HTA1-Flag-htb1-K123R CEN HIS3> CFK2414: W303 CFK2416: W303 HTA1-htb1-K123R::NAT+ HTA2-htb2-K123R::HIS+.
(TIFF)
(A) A schematic description of the nucleosome position surrounding ARS305, and the primers used in Fig. 7A to amplify ARS305 for histone chromatin immunoprecipitation. ARS305 (nuc.) (39,349–39,455) primer sequence: (F): att tca gag cct tct ttg gag, (R): atg aaa ctg gac ata ttt gag gaa. (B) A schematic description of the nucleosome position surrounding ARS607, and the primers used in Fig. 7A to amplify ARS607 for histone chromatin immunoprecipitation. ARS607 (nuc.) (199,539–199,630) primer sequence: (F): aca cat tat tcg gca cag tag, (R): tcg cag tcc ata gaa gga g. (C) A schematic description of the nucleosome position surrounding ARS501, and the primers used in Fig. 7A to amplify ARS501 for histone chromatin immunoprecipitation. ARS501 (nuc.) (549,785–549,858) primer sequence: (F): ctcct catca tcatc cc, (R): cgtac actag cccgt tg. Image created using the following software available at the Penn State Genome Cartography Project http://atlas.bx.psu.edu/cj/nucl_retrieval.html [82]
(TIFF)
Yeast strains used in this study.
(PDF)
Plasmids used in this study.
(PDF)
Primers used in this study.
(PDF)
Acknowledgments
We thank O. Aparicio, J. Campbell, S. Gasser, and J. Haber for providing strains and plasmids. We thank M. Foiani T.-S. Hsieh and Y.-C. Lo for constructive comments and suggestions.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. Experimental data are available on Gene Expression Omnibus database with accession number GSE61030 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE61030.
Funding Statement
This study was supported by a joint research project (NSC 100-2923-B-001 -001 -MY3) funded by National Science Council (NSC), Taiwan and French National Research Agency (ANR). CFK was supported by Academia Sinica, Taiwan. LM and AC were supported by the Knut and Alice Wallenberg Foundation, the Swedish Cancer Society. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ransom M, Dennehey BK, Tyler JK (2010) Chaperoning histones during DNA replication and repair. Cell 140: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Soria G, Polo SE, Almouzni G (2012) Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell 46: 722–734. [DOI] [PubMed] [Google Scholar]
- 3. Papamichos-Chronakis M, Peterson CL (2013) Chromatin and the genome integrity network. Nat Rev Genet 14: 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, et al. (2007) Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315: 653–655. [DOI] [PubMed] [Google Scholar]
- 5. Adkins MW, Carson JJ, English CM, Ramey CJ, Tyler JK (2007) The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J Biol Chem 282: 1334–1340. [DOI] [PubMed] [Google Scholar]
- 6. Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, et al. (2008) Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell 30: 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, et al. (2006) The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol 16: 1280–1289. [DOI] [PubMed] [Google Scholar]
- 8. Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, et al. (2008) Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134: 244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burgess RJ, Zhou H, Han J, Zhang Z (2010) A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell 37: 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Robzyk K, Recht J, Osley MA (2000) Rad6-dependent ubiquitination of histone H2B in yeast. Science 287: 501–504. [DOI] [PubMed] [Google Scholar]
- 11. Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, et al. (2003) A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell 11: 261–266. [DOI] [PubMed] [Google Scholar]
- 12. Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, et al. (2003) Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell 11: 267–274. [DOI] [PubMed] [Google Scholar]
- 13. Song YH, Ahn SH (2009) A Bre1-associated protein, large 1 (Lge1), promotes H2B Ubiquitylation during the early stages of transcription elongation. J Biol Chem 285: 2361–7 doi: 10.1074/jbc.M109.039255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, et al. (2003) Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17: 2648–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, et al. (2004) Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev 18: 184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, et al. (2002) Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418: 498. [DOI] [PubMed] [Google Scholar]
- 17. Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, et al. (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277: 28368–28371. [DOI] [PubMed] [Google Scholar]
- 18. Ng HH, Xu RM, Zhang Y, Struhl K (2002) Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem 277: 34655–34657. [DOI] [PubMed] [Google Scholar]
- 19. Sun ZW, Allis CD (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108. [DOI] [PubMed] [Google Scholar]
- 20. Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA (2008) H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell 31: 57–66. [DOI] [PubMed] [Google Scholar]
- 21. Margaritis T, Oreal V, Brabers N, Maestroni L, Vitaliano-Prunier A, et al. (2012) Two distinct repressive mechanisms for histone 3 lysine 4 methylation through promoting 3′-end antisense transcription. PLoS Genet 8: e1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pavri R, Zhu B, Li G, Trojer P, Mandal S, et al. (2006) Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125: 703–717. [DOI] [PubMed] [Google Scholar]
- 23. Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF (2011) Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev 25: 2254–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emre NC, Berger SL (2004) Histone H2B ubiquitylation and deubiquitylation in genomic regulation. Cold Spring Harb Symp Quant Biol 69: 289–299. [DOI] [PubMed] [Google Scholar]
- 25. Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, et al. (2011) Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol 7: 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, et al. (2011) Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell 41: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, et al. (2011) Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell 41: 515–528. [DOI] [PubMed] [Google Scholar]
- 28. Trujillo KM, Osley MA (2012) A Role for H2B Ubiquitylation in DNA Replication. Mol Cell 48: 734–46 doi: 10.1016/j.molcel.2012.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Friedel AM, Pike BL, Gasser SM (2009) ATR/Mec1: coordinating fork stability and repair. Curr Opin Cell Biol 21: 237–244. [DOI] [PubMed] [Google Scholar]
- 30. Branzei D, Foiani M (2009) The checkpoint response to replication stress. DNA Repair (Amst) 8: 1038–1046. [DOI] [PubMed] [Google Scholar]
- 31. Zegerman P, Diffley JF (2009) DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst) 8: 1077–1088. [DOI] [PubMed] [Google Scholar]
- 32. Ashton TM, Hickson ID (2010) Yeast as a model system to study RecQ helicase function. DNA Repair (Amst) 9: 303–314. [DOI] [PubMed] [Google Scholar]
- 33. Bjergbaek L, Cobb JA, Tsai-Pflugfelder M, Gasser SM (2005) Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J 24: 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Branzei D, Foiani M (2010) Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11: 208–219. [DOI] [PubMed] [Google Scholar]
- 35. Rodriguez J, Tsukiyama T (2013) ATR-like kinase Mec1 facilitates both chromatin accessibility at DNA replication forks and replication fork progression during replication stress. Genes Dev 27: 74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alvino GM, Collingwood D, Murphy JM, Delrow J, Brewer BJ, et al. (2007) Replication in hydroxyurea: it's a matter of time. Mol Cell Biol 27: 6396–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Poli J, Tsaponina O, Crabbe L, Keszthelyi A, Pantesco V, et al. (2012) dNTP pools determine fork progression and origin usage under replication stress. EMBO J 31: 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu W, Aparicio JG, Aparicio OM, Tavare S (2006) Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics 7: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chabes A, Stillman B (2007) Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 104: 1183–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Davidson MB, Katou Y, Keszthelyi A, Sing TL, Xia T, et al. (2012) Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype. EMBO J 31: 895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Georgieva B, Zhao X, Rothstein R (2000) Damage response and dNTP regulation: the interaction between ribonucleotide reductase and its inhibitor, Sml1. Cold Spring Harb Symp Quant Biol 65: 343–346. [DOI] [PubMed] [Google Scholar]
- 42. Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R (2001) The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J 20: 3544–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao X, Rothstein R (2002) The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci U S A 99: 3746–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Holmes AM, Haber JE (1999) Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell 96: 415–424. [DOI] [PubMed] [Google Scholar]
- 46. Paciotti V, Clerici M, Scotti M, Lucchini G, Longhese MP (2001) Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol Cell Biol 21: 3913–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weinert TA, Kiser GL, Hartwell LH (1994) Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev 8: 652–665. [DOI] [PubMed] [Google Scholar]
- 48. Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM (2003) DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J 22: 4325–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, et al. (2005) Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev 19: 3055–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hegnauer AM, Hustedt N, Shimada K, Pike BL, Vogel M, et al. (2012) An N-terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks. EMBO J 31: 3768–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Groth A, Corpet A, Cook AJ, Roche D, Bartek J, et al. (2007) Regulation of replication fork progression through histone supply and demand. Science 318: 1928–1931. [DOI] [PubMed] [Google Scholar]
- 52. Franco AA, Lam WM, Burgers PM, Kaufman PD (2005) Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev 19: 1365–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Driscoll R, Hudson A, Jackson SP (2007) Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315: 649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Green EM, Antczak AJ, Bailey AO, Franco AA, Wu KJ, et al. (2005) Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol 15: 2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Emili A, Schieltz DM, Yates JR 3rd, Hartwell LH (2001) Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol Cell 7: 13–20. [DOI] [PubMed] [Google Scholar]
- 56. Hu F, Alcasabas AA, Elledge SJ (2001) Asf1 links Rad53 to control of chromatin assembly. Genes Dev 15: 1061–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tercero JA, Diffley JF (2001) Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557. [DOI] [PubMed] [Google Scholar]
- 58. Segurado M, Diffley JF (2008) Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev 22: 1816–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Clemente-Ruiz M, Prado F (2009) Chromatin assembly controls replication fork stability. EMBO Rep 10: 790–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Clemente-Ruiz M, Gonzalez-Prieto R, Prado F (2011) Histone H3K56 acetylation, CAF1, and Rtt106 coordinate nucleosome assembly and stability of advancing replication forks. PLoS Genet 7: e1002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mejlvang J, Feng Y, Alabert C, Neelsen KJ, Jasencakova Z, et al. (2014) New histone supply regulates replication fork speed and PCNA unloading. J Cell Biol 204: 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, et al. (2005) Human Asf1 regulates the flow of S phase histones during replicational stress. Mol Cell 17: 301–311. [DOI] [PubMed] [Google Scholar]
- 63. Hoek M, Stillman B (2003) Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc Natl Acad Sci U S A 100: 12183–12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nabatiyan A, Krude T (2004) Silencing of chromatin assembly factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis. Mol Cell Biol 24: 2853–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, et al. (2003) Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol Cell 11: 341–351. [DOI] [PubMed] [Google Scholar]
- 66. Kats ES, Albuquerque CP, Zhou H, Kolodner RD (2006) Checkpoint functions are required for normal S-phase progression in Saccharomyces cerevisiae RCAF- and CAF-I-defective mutants. Proc Natl Acad Sci U S A 103: 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rykowski MC, Wallis JW, Choe J, Grunstein M (1981) Histone H2B subtypes are dispensable during the yeast cell cycle. Cell 25: 477–487. [DOI] [PubMed] [Google Scholar]
- 68. Norris D, Osley MA (1987) The two gene pairs encoding H2A and H2B play different roles in the Saccharomyces cerevisiae life cycle. Mol Cell Biol 7: 3473–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gunjan A, Verreault A (2003) A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115: 537–549. [DOI] [PubMed] [Google Scholar]
- 70. Hwang WW, Madhani HD (2009) Nonredundant requirement for multiple histone modifications for the early anaphase release of the mitotic exit regulator Cdc14 from nucleolar chromatin. PLoS Genet 5: e1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Budd ME, Campbell JL (1993) DNA polymerases delta and epsilon are required for chromosomal replication in Saccharomyces cerevisiae. Mol Cell Biol 13: 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Scherer S, Davis RW (1979) Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A 76: 4951–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fachinetti D, Bermejo R, Cocito A, Minardi S, Katou Y, et al. (2010) Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol Cell 39: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, et al. (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083. [DOI] [PubMed] [Google Scholar]
- 75. Scaffidi P, Misteli T (2008) Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol 10: 452–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dahl KN, Scaffidi P, Islam MF, Yodh AG, Wilson KL, et al. (2006) Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A 103: 10271–10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schubeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, et al. (2002) Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat Genet 32: 438–442. [DOI] [PubMed] [Google Scholar]
- 78. Wuhr M, Chen Y, Dumont S, Groen AC, Needleman DJ, et al. (2008) Evidence for an upper limit to mitotic spindle length. Curr Biol 18: 1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, et al. (1999) Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J 18: 6561–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bermejo R, Doksani Y, Capra T, Katou YM, Tanaka H, et al. (2007) Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev 21: 1921–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kumar D, Viberg J, Nilsson AK, Chabes A (2010) Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res 38: 3975–3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jiang C, Pugh BF (2009) A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol 10: R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A–D) Replication profiles in WT (CFK1419) vs. htb-K123R (CFK1421) cells. Cells were synchronized in G1 with α-factor, and then released into media containing 0.2M HU and 200 µg/ml BrdU for 90 minutes. After DNA extraction and fragmentation, BrdU-labeled DNA was immunoprecipitated and hybridized on high-resolution tiling arrays. Orange (BrdU-IP) histogram bars on the y axis show the average signal ratio on a log2 scale of loci along the reported regions on (A) chromosome II, (B) chromosome V, (C) chromosome IX, and (D) chromosome XVI. The positions of potential ARS elements are identified by Mcm2 loading.
(TIFF)
The size of each dNTP pool in exponentially-growing WT (CFK1419) and htb-K123R (CFK1421) cells. Four independent isogenic strains of each genotype were analyzed as described in the Materials and Methods.
(TIFF)
H2Bub is required for efficient origin firing. htb1-K123R mutants exhibit reduced BrdU incorporation during S phase. Cells were arrested at G1 using α-factor at 23°C, and released synchronously into S phase at 20°C in YPD supplemented with BrdU. Samples were collected at the indicated times and genomic DNA was then extracted. Monoclonal BrdU antibody was used to immunoprecipitate BrdU-incorporated DNA. DNA synthesis at replication origins (ARS305 and ARS607) or telomere was detected by quantitative-PCR. Cell cycle progression was monitored by FACS at 20°C under BrdU incorporation conditions.
(TIFF)
(A) The growth of htb-K123R and DNA polymerase ts double mutants are not affected by HU at the permissive temperature (23°C). Ten-fold serial dilutions of the indicated strains (WT (CFK1204), htb-K123R (CFK1231), pol1-17 (CFK1984), pol1-17 htb-K123R (CFK1986), pri2-1 (CFK1988), pri2-1 htb-K123R (CFK1990), pol3-14 (CFK1992) and pol3-14 htb-K123R (CFK1994)) were spotted onto YPD containing different doses of HU (0–50 mM) at 23°C for several days. Growth at the restrictive temperature (37°C) is presented as a control for ts mutants. (B) The genetic interaction between H2Bub and DNA pol1, pol3, or primase. Ten-fold serial dilutions of the indicated strains were spotted onto YPD, and growth was monitored at 33°C or 30°C, or under conditions of replication stress (50 mM HU) at 30°C. (C) The histone H2B ubiquitin E3 ligase Bre1 functions in parallel with the RecQ helicase Sgs1 under replication stress. Ten-fold serial dilutions of the indicated strains (WT (CFK1204), bre1Δ (CFK1443), sgs1Δ (CFK2371) and bre1Δ sgs1Δ (CFK2373)) were spotted onto YPD containing different doses of HU (0–100 mM) at 30°C.
(TIFF)
The growth of WT and htb-K123R cells of two different backgrounds under conditions of replication stress at 30°C. Ten-fold serial dilutions of the indicated strains (WT (CFK1204), htb-K123R (CFK1231), WT (CFK2414), and htb-K123R (CFK2416)) were spotted onto YPD containing different doses of HU (0–150 mM) for 2 days. Genotypes of the strains used: CFK1024: W303 hta1-htb1Δ hta2-htb2Δ <pZS145-HTA1-Flag-HTB1 CEN HIS3> CFK1031: W303 hta1-htb1Δ hta2-htb2Δ <pZS146-HTA1-Flag-htb1-K123R CEN HIS3> CFK2414: W303 CFK2416: W303 HTA1-htb1-K123R::NAT+ HTA2-htb2-K123R::HIS+.
(TIFF)
(A) A schematic description of the nucleosome position surrounding ARS305, and the primers used in Fig. 7A to amplify ARS305 for histone chromatin immunoprecipitation. ARS305 (nuc.) (39,349–39,455) primer sequence: (F): att tca gag cct tct ttg gag, (R): atg aaa ctg gac ata ttt gag gaa. (B) A schematic description of the nucleosome position surrounding ARS607, and the primers used in Fig. 7A to amplify ARS607 for histone chromatin immunoprecipitation. ARS607 (nuc.) (199,539–199,630) primer sequence: (F): aca cat tat tcg gca cag tag, (R): tcg cag tcc ata gaa gga g. (C) A schematic description of the nucleosome position surrounding ARS501, and the primers used in Fig. 7A to amplify ARS501 for histone chromatin immunoprecipitation. ARS501 (nuc.) (549,785–549,858) primer sequence: (F): ctcct catca tcatc cc, (R): cgtac actag cccgt tg. Image created using the following software available at the Penn State Genome Cartography Project http://atlas.bx.psu.edu/cj/nucl_retrieval.html [82]
(TIFF)
Yeast strains used in this study.
(PDF)
Plasmids used in this study.
(PDF)
Primers used in this study.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. Experimental data are available on Gene Expression Omnibus database with accession number GSE61030 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE61030.