Abstract
Background
Sleeping sickness caused by Trypanosoma brucei (T.b.) gambiense constitutes a serious health problem in sub-Sahara Africa. In some foci, alarmingly high relapse rates were observed in patients treated with melarsoprol, which used to be the first line treatment for patients in the neurological disease stage. Particularly problematic was the situation in Mbuji-Mayi, East Kasai Province in the Democratic Republic of the Congo with a 57% relapse rate compared to a 5% relapse rate in Masi-Manimba, Bandundu Province. The present study aimed at investigating the mechanisms underlying the high relapse rate in Mbuji-Mayi using an extended collection of recently isolated T.b. gambiense strains from Mbuji-Mayi and from Masi-Manimba.
Methodology/Principal Findings
Forty five T.b. gambiense strains were used. Forty one were isolated from patients that were cured or relapsed after melarsoprol treatment in Mbuji-Mayi. In vivo drug sensitivity tests provide evidence of reduced melarsoprol sensitivity in these strains. This reduced melarsoprol sensitivity was not attributable to mutations in TbAT1. However, in all these strains, irrespective of the patient treatment outcome, the two aquaglyceroporin (AQP) 2 and 3 genes are replaced by chimeric AQP2/3 genes that may be associated with resistance to pentamidine and melarsoprol. The 4 T.b. gambiense strains isolated in Masi-Manimba contain both wild-type AQP2 and a different chimeric AQP2/3. These findings suggest that the reduced in vivo melarsoprol sensitivity of the Mbuji-Mayi strains and the high relapse rates in that sleeping sickness focus are caused by mutations in the AQP2/AQP3 locus and not by mutations in TbAT1.
Conclusions/Significance
We conclude that mutations in the TbAQP2/3 locus of the local T.b. gambiense strains may explain the high melarsoprol relapse rates in the Mbuji-Mayi focus but other factors must also be involved in the treatment outcome of individual patients.
Author Summary
Sleeping sickness, or human African trypanosomosis, constitutes a serious health problem in sub-Sahara Africa. Treatment is a key factor in the control of this disease, not only to save the lives of the individual patients but also to stop transmission. As for other infectious diseases, drug resistance forms a constant threat for sleeping sickness control. Understanding the mechanisms underlying drug resistance is of uppermost importance for evidence-based adaptation of treatment protocols as well as for the development of new drugs. In this study we investigated the phenotype and genotype of more than 40 recently isolated strains of Trypanosoma brucei gambiense, the parasite that causes sleeping sickness in West and Central Africa. By comparing the parasites from cured and from relapsing patients in two separated sleeping sickness foci, we can explain the difference in treatment failure rates between these two sleeping sickness foci in D.R. Congo. We also provide evidence that treatment outcome in the individual patient is not exclusively defined by the drug resistance genotype of the infecting parasite but that other factors must be involved.
Introduction
Human African trypanosomosis (HAT) is a parasitic disease transmitted by tsetse flies (Glossina sp) and caused by Trypanosoma brucei (T.b.) gambiense and T.b. rhodesiense. This disease constitutes a serious public health problem in sub-Saharan Africa, particularly in central African countries like the Democratic Republic of the Congo (DRC), Central African Republic (CAR), the Republic of the Congo and the Republic of South Sudan [1]. The disease evolves from an early stage with trypanosomes invading blood, lymph and peripheral tissues towards the late or neurological stage with invasion of the brain. Chemotherapy of HAT relies on five drugs (eflornithine, melarsoprol, nifurtimox, pentamidine and suramin). Early-stage HAT is treated with pentamidine (T.b. gambiense) or suramin (T.b. rhodesiense). For treatment of the neurological stage, drugs that are able to pass the blood-brain-barrier, such as melarsoprol, nifurtimox or eflornithine, are necessary [2]. Until recently, melarsoprol was the first line treatment of late stage gambiense and rhodesiense HAT but for gambiense HAT, the nifurtimox-eflornithine combination therapy (NECT) is now recommended by the World Health Organization as first line treatment [3]. This recommendation follows the observation that NECT is as effective as melarsoprol monotherapy with less severe side effects and that NECT is able to cure patients that experienced a relapse after treatment with melarosprol monotherapy [4]. Traditionally, 5–10% of gambiense HAT patients treated with melarsoprol could not be cured but in the last decade up to >50% relapse rates were reported in Angola, Central African Republic, Democratic Republic of the Congo, Republic of South Sudan and Uganda [5]–[11]. Particularly problematic was the situation in the HAT focus of Mbuji-Mayi, East Kasai Province in DRC, where a 57% failure rate was observed in patients treated with the 10 days abridged melarsoprol regimen [8]. Various studies have been conducted to explain these unexpectedly high relapse rates, considering either the parasite or the human host being responsible for this phenomenon. Although known mechanisms may be involved, such as mutations of the P2 adenosine transporters and of aquaglyceroporin transporters in the trypanosome membrane, the explanation for treatment failure is probably more complex, including parameters of the parasite (reduced drug sensitivity and higher tissue tropism), the drug (content of active principle and correct administration) and the host including individual differences in pharmacokinetics, co-infections and disease stage [9], [12]–[24].
To investigate the mechanism underlying the high failure rates in the Mbuji-Mayi focus, Pyana and co-workers undertook the large scale isolation of the trypanosome from HAT patients in that focus [25]. Thus, we established a collection of 85 T.b. gambiense type I strains from cured and relapsed patients, some of which from the same patient before treatment and after relapse. Among these strains, 41 were adapted to Mus musculus in order to test their sensitivity to melarsoprol in an in vivo mouse infection model and to analyse some genetic features that may be related to reduced sensitivity to melarsoprol. Recently, some of them were shown to be resistant to pentamidine and melarsoprol by in vitro drug sensitivity testing and to carry a TbAQP2/3 chimera that was supposed to lead to a reduced uptake of pentamidine in the trypanosome flagellar pocket [20]. In this study, we aimed at 1° investigating the in vivo melarsoprol sensitivity phenotype of all 41 mouse adapted T.b. gambiense strains isolated in the Mbuji-Mayi focus, 2° investigating some of their genotypic characteristics and comparing them with 4 strains isolated from a sleeping focus with low relapse rates in Masi-Manimba, and 3° relating their phenotype and genotype with treatment outcome of the patients from whom they were isolated.
Materials and Methods
Ethics statement
The study in mice was approved by the Veterinary Ethics Committee of the Institute of Tropical Medicine, Antwerp, Belgium (protocol PAR-022) and adhere to the European Commission Recommendation on guidelines for the accommodation and care of animals used for experimental and other scientific purposes (18 June 2007, 2007/526/EG) and the Belgian National law on the protection of animals under experiment.
The parasite strains included in this study belong to the cryobank of the World Health Collaboration Center for Research and Training on Human African Trypanosomiasis Diagnostics at the Institute of Tropical Medicine in Antwerp, Belgium. Their isolation and use for research purposes was approved by the Ethical Committee of the Institute of Tropical Medicine (04441472) and of the Ministry of Health of DRC [25]. The strains are anonymized by using international codes and alias names.
Trypanosome strains
A list of the T.b. gambiense strains from the Mbuji-Mayi focus in East Kasai Province, DRC, is given in Table 1. The isolation history of these strains is described elsewhere [25]. The alias name of each strain indicates whether it was isolated before treatment (BT) or after treatment (AT). Eleven strains were isolated from patients that were cured after melarsoprol treatment. Thirty strains were isolated from patients that relapsed after treatment. Among these 30 strains, twenty belong to “couples”, i.e. isolated from the same patients, before treatment and after relapse. The melarsoprol sensitive and resistant strains included as reference in the in vivo drug sensitivity experiment, T.b. brucei 427 wild type and T.b. brucei 427 AT1/P2 KO (P2 adenosine transporter knock out) were received from the Swiss Tropical and Public Health Institute. For the genotype analysis, 7 extra T.b. gambiense type I strains were added (Table 1). Four of these strains were isolated in 2011 from cured patients in the Masi-Manimba focus, Bandundu Province, RDC where high relapse rates after melarsoprol treatment were never observed. Three strains are “old” isolates. LiTat 1.3 is a cloned population of the Eliane strain, isolated in Côte d'Ivoire, and previously shown to be sensitive to melarsoprol and pentamidine in vitro, while MBA and KEMLO are two Congolese strains with unknown drug sensitivity profile [26]. All the strains were kept as 250 µl cryostabilates in liquid nitrogen.
Table 1. List of T.b. gambiense strains used in this study.
| HAT focus, country | International code or name | Alias name or clone | Treatment outcome | Couple |
| Mbuji-Mayi, DRC | MHOM/CD/INRB/2008/56 | 15BT | cure | |
| MHOM/CD/INRB/2008/46 | 19BT | cure | ||
| MHOM/CD/INRB/2008/65 | 29BT | cure | ||
| MHOM/CD/INRB/2006/13 | 40BT | relapse | 1 | |
| MHOM/CD/INRB/2006/07 | 40AT | relapse | 1 | |
| MHOM/CD/INRB/2006/01 | 45BT | cure | ||
| MHOM/CD/INRB/2008/52 | 48BT | relapse | ||
| MHOM/CD/INRB/2007/28 | 57AT | relapse | ||
| MHOM/CD/INRB/2008/62 | 85BT | cure | ||
| MHOM/CD/INRB/2006/09 | 93AT | relapse | ||
| MHOM/CD/INRB/2008/63 | 95BT | cure | ||
| MHOM/CD/INRB/2008/42 | 99BT | cure | ||
| MHOM/CD/INRB/2008/49 | 104BT | relapse | 2 | |
| MHOM/CD/INRB/2008/53A | 104AT | relapse | 2 | |
| MHOM/CD/INRB/2008/50 | 105BT | relapse | ||
| MHOM/CD/INRB/2007/25B | 108AT | relapse | 3 | |
| MHOM/CD/INRB/2007/27 | 108BT | relapse | 3 | |
| MHOM/CD/INRB/2008/60 | 113AT | relapse | 4 | |
| MHOM/CD/INRB/2008/45 | 113BT | relapse | 4 | |
| MHOM/CD/INRB/2006/11A | 116AT | relapse | ||
| MHOM/CD/STI/2006/02 | 130BT | relapse | ||
| MHOM/CD/INRB/2008/64 | 141BT | cure | ||
| MHOM/CD/INRB/2005/02A | 146BT | relapse | 5 | |
| MHOM/CD/INRB/2006/05 | 146AT | relapse | 5 | |
| MHOM/CD/INRB/2007/26A | 147AT | relapse | ||
| MHOM/CD/INRB/2005/01B | 148BT | relapse | 6 | |
| MHOM/CD/INRB/2006/14 | 148AT | relapse | 6 | |
| MHOM/CD/INRB/2006/06A | 163AT | relapse | ||
| MHOM/CD/INRB/2008/47 | 167BT | relapse | 7 | |
| MHOM/CD/INRB/2008/43 | 167AT | relapse | 7 | |
| MHOM/CD/INRB/2008/59 | 174BT | relapse | 8 | |
| MHOM/CD/INRB/2007/29 | 174AT | relapse | 8 | |
| MHOM/CD/INRB/2008/37A | 186BT | cure | ||
| MHOM/CD/INRB/2006/12A | 223AT | relapse | ||
| MHOM/CD/INRB/2006/21B | 340AT | relapse | ||
| MHOM/CD/INRB/2007/22B | 346BT | relapse | 9 | |
| MHOM/CD/INRB/2007/24B | 346AT | relapse | 9 | |
| MHOM/CD/INRB/2006/23A | 348BT | cure | ||
| MHOM/CD/INRB/2006/16 | 349BT | relapse | 10 | |
| MHOM/CD/INRB/2006/19 | 349AT | relapse | 10 | |
| MHOM/CD/INRB/2007/34 | 378BT | cure | ||
| Masi-Manimba, DRC | MHOM/CD/INRB/2011/01 | MM01 | cure | |
| MHOM/CD/INRB/2011/03 | MM03 | cure | ||
| MHOM/CD/INRB/2011/05 | MM05 | cure | ||
| MHOM/CD/INRB/2011/06 | MM06 | cure | ||
| Kinshasa, DRC | MAN/ZR/74/ITMAP1811 | MBA | unknown | |
| Bwamanda, DRC | MAN/ZR/74/ITMAP1821 | KEMLO | unknown | |
| Daloa, Côte d'Ivoire | ELIANE | LiTat 1.3 | unknown |
In alias name: AT = after treatment, BT = before treatment. Treatment outcome: outcome of patient treated with melarsoprol (in Mbuji-Mayi) or with nifurtimox-eflornithine combination therapy (Masi-Manimba). Couple = number of the couple of two strains isolated from the same patient.
Expansion of parasite populations
Each stabilate was thawed in a water bath at 37°C and immediately, 250 µl of phosphate buffered saline glucose (PSG, 7.5 g/l Na2 HPO4 2H2O, 0.34 g/l NaH2 PO4 H2O, 2.12 g/l NaCl, 10 g/l D-glucose, pH 8) were added. This mixture was kept on ice until inoculated intraperitoneally (IP) into two 1–2 months old female OF-1 mice (Charles River, Belgium). Two days before infection, these mice had been immunosuppressed by IP injection with 200 mg/kg body weight (BW) of cyclophosphamide diluted in water (Endoxan, Baxter, Lessing, Belgium). Parasitaemia was monitored three times a week on a fresh preparation of 5 µl of tail blood according to the matching method of Herbert and Lumsden (1976). If needed, immunosuppression was repeated after 5 days, until the parasitaemia reached 107.5/ml or more and the trypanosome population was large enough to inoculate 12 mice for the in vivo melarsoprol sensitivity experiment. For the inoculation of the mice in the in vivo drug sensitivity experiments (see below), 30 to 40 µl of infected tail blood was diluted in about 3 ml of PSG, trypanosomes were counted in a Uriglass cell counting chamber (Menarini Diagnostics) and the suspension was further diluted in PSG to obtain a concentration of 250 trypanosomes/µl.
In vivo melarsoprol sensitivity experiments
For the treatment of mice, melarsoprol (Aventis, 5 ml vials of 180 mg in propylene glycol, lot nr. 725) was freshly diluted in 50% polyethylene glycol (PEG400, Sigma Aldrich, Belgium) to concentrations of 1 mg/ml.
For each experiment, twelve female OF-1 mice (1–2 months old, 20–30 g body weight (BW)) were immunosuppressed as described above, two days before inoculation and subsequently at days 3, 11, 25 and 85 post-infection (DPI). Each mouse was inoculated IP with 200 µl PSG containing 5×104 trypanosomes. Parasitaemia was monitored as described above from day 4 post-infection onwards. As soon as trypanosomes were detected in all mice (between 4 and 6 DPI), one group of 6 mice received IP injections of melarsoprol at 10 or 12 mg/kg BW during 4 consecutive days. On day 5–10 post infection, the mice of the control group were euthanised with an IP injection of sodium pentobarbital at 350 mg/kg BW (Nembutal, CEVA Santé Animale, Brussels, Belgium) and blood was taken by heart puncture on heparin to prepare sediments of pure trypanosomes for DNA extraction and genetic analysis (see below). The mice of the melarsoprol treated group were checked for the presence of trypanosomes in tail blood two times a week during the first two weeks and subsequently once a week for maximum 100 days. As soon as a trypanosome was detected in at least one mouse of this group, the experiment was stopped and the strain was considered “resistant”. Relapsing mice were sacrificed and blood was collected on heparin to separate the trypanosomes from the blood via DEAE chromatography for further genotypic analysis. On days 90 to 100 postinfection, all mice that remained trypanosome negative were sacrificed and blood was collected on heparin where after it was run over two mini Anion Exchange Centrifugation Technique columns to detect subpatent parasitaemia [27].
Trypanosome purification and genomic DNA extraction
Infected blood from mice was passed over a DEAE cellulose column (1∶6 blood∶gel ratio) to separate the trypanosomes from the blood [28]. The trypanosomes eluting from the column were washed three times with 5 ml ice-cold PSG by centrifugation. After the last centrifugation, the supernatant PSG was discarded and the trypanosome sediment was frozen at −80°C until DNA extraction. After thawing and addition of 200 µl of phosphate buffered saline, pH 8, genomic DNA was extracted from the trypanosome sediment with the Maxwell 16 Tissue DNA Purification kit on the Maxwell 16 robot (Promega, Madison, WI, USA) and DNA was stored at −20°C. DNA concentrations were measured with the Nanodrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, USA) and adjusted to 10 ng/µl if appropriate.
Genotyping
mlSfaNI PCR-RFLP for TbAT1
The protocol to amplify a 677-bp fragment of the TbAT1 gene and to digest it with SfaNI was based on Mäser et al. [17]. A 20 µl reaction volume contained 1× PCR buffer (Qiagen, Venlo, The Netherlands), 1 mM MgCl2 (Qiagen), 200 µM of each dNTP (Eurogentec, Seraing, Belgium), 0.8 µM of Sfa-s (5′-CGCCGCACTCATCGCCCCGTT-3)′ and Sfa-as (5′-CCACCGCGGTGAGACGTGTA-3′) (Biolegio, Nijmegen, The Netherlands), 0.4 U Hotstart Taq plus polymerase (Qiagen), 0.1 mg/ml acetylated BSA (Promega) and 2 µl target DNA. Amplification condition were as follows: initial denaturation at 94°C for 5 min, 30 cycles 1 min at 94°C, 1 min at 65°C and 2 min at 72°C and final extension at 72°C for 10 min. Five µl of the PCR products were digested 16 hours at 37°C in a 10 µl reaction mixture containing 1× NEB 3.1 buffer with 2 U of SfaNI restriction enzyme (New England Biolabs, Hitchin, UK), followed by 20 minutes incubation at 65°C. Differential banding was analysed using 10 µl on a 2% agarose gel run for 30 min at 135 V and stained with ethidium bromide.
PCR for full-length TbAT1
The protocol to amplify the full length TbAT1 gene and its flanking regions was slightly modified from Graf et al. [20]. A 20 µl reaction volume contained 1× Coral Load Buffer, 1 mM MgCl2 (Qiagen), 200 µM of each dNTP (Eurogentec), 0.8 µM of TbAT1_F (5′-GAAATCCCCGTCTTTTCTCAC-3)′ and TbAT1_R_Tbg (5′-ATGTGCTGACCCATTTTCCTT-3)′ primers (Biolegio), 0.4 U Hotstart Taq plus polymerase (Qiagen), 0.1 mg/ml acetylated BSA (Promega) and 2 µl target DNA. Amplification conditions were: initial denaturation at 95°C for 5 min, 24 cycles of 1 min at 95°C followed by 1 min at 56°C and 2 min at 72°C, final extension at 72°C for 5 min. Ten µl of the PCR product were electrophorised in a 2% agarose gel for 30 min at 135 V and stained with ethidium bromide for detection of the amplicons. For selected strains, the amplicons were cleaned up and concentrated using a PCR clean-up kit (Qiagen) and sent out for bidirectional direct sequencing at the VIB Genetic Sequencing Facility (Antwerp, Belgium) using the described PCR primers and additional internal primers to cover TbAT1.
Locus specific PCR for aquaglyceroporin genes
The protocols to amplify either the AQP2 locus or the combined AQP2 and AQP3 locus, including the AQP2/3 chimera were slightly modified from Graf et al. [20]. To amplify the wild-type APQ2 locus, a 20 µl reaction volume contained 1× Coral Load Buffer (Qiagen), 1 mM MgCl2 (Qiagen), 200 µM of each dNTP (Eurogentec), 0.8 µM of AQP2/3_F 5′-AAGAAGGCTGAAACTCCACTTG-3′ and AQP2_R 5′-CTTCGGGAGAAACAAAACCTC -3′ primers (Biolegio), 0.4 U Hotstart Taq plus polymerase (Qiagen), 0.1 mg/ml acetylated BSA (Promega) and 2 µL target DNA. Amplification conditions were: initial denaturation at 95°C for 5 min, 24 cycles of 1 min at 95°C followed by 1 min at 60°C and 2 min at 72°C, final extension at 72°C for 10 min. Ten µl of the PCR product were electrophorised in a 2% agarose gel for 30 min at 135 V and stained with ethidium bromide for detection of the amplicons. To amplify the combined APQ2 and APQ3 locus, a 20 µl reaction volume contained 1× Coral Load Buffer (Qiagen), 1 mM MgCl2 (Qiagen), 200 µM of each dNTP (Eurogentec), 0.8 µM of AQP2/3_F 5′-AAGAAGGCTGAAACTCCACTTG-3′ and AQP2/3_R 5′-TGCACTCAAAAACAGGAAAAGA-3′ primers (Biolegio), 0.4 U Hotstart Taq plus polymerase (Qiagen), 0.1 mg/ml acetylated BSA (Promega) and 2 µL target DNA. Amplification conditions were: initial denaturation at 95°C for 5 min, 24 cycles of 1 min at 95°C followed by 1 min at 60°C and 4 min at 72°C, final extension at 72°C for 10 min. Ten µl of the PCR product were electrophorised in a 0.8% agarose gel for 30 min at 135 V and stained with ethidium bromide for detection of the amplicons. For selected strains, amplicons were cleaned up and concentrated using a PCR clean-up kit (Qiagen) and sent out for bidirectional direct sequencing at the VIB Genetic Sequencing Facility (Antwerp, Belgium) using the described PCR primers and additional internal primers to cover TbAQP2.
Cloning of aquaglyceroporin variants
The full-length ORFs of the AQP2 and AQP2/3 genes were amplified from genomic DNA using a proofreading polymerase (Phusion High-Fidelity DNA Polymerase, Thermo Scientific, Waltham, MA, USA). All cloning primers (Biolegio) contained a 5′ extension of 15 nucleotides from the trypanosomal expression vector pHD309, as required for the In-Fusion Cloning reaction (Clontech, Takara Bio, Otsu, Japan) [29]. The primer sets consisted of a forward primer (IF HindIII 5′-AACTGCAACG AAGCTT ATGCAGAGCCAACCAGACA-3′) and an AQP2 specific reverse primer (IF BamHI AQP2 5′-TAAATGGGCA GGATCC TTAGTGTGGAAGAAAATATTTGTACAG-3′) for amplification of T.b. gambiense MM01, LiTaR1 and MBA or an AQP2/3 specific reverse primer (IF BamHI AQP2/3 5′-TAAATGGGCA GGATCC TTAGTGTGGCACAAAATATTTGTACA-3′) for amplification of T.b. gambiense 348BT. Plasmids were purified from several transformed E. coli and sent out for bidirectional direct sequencing at the VIB Genetic Sequencing Facility (Antwerp, Belgium).
PCR-RFLP for AQP2/3 chimeric alleles from Mbuji-Mayi
A 278-bp fragment was amplified from the predicted AQP2/3 chimeras from Mbuji-Mayi in a 20 µl reaction volume that contained 1× PCR buffer (Qiagen), 1 mM MgCl2 (Qiagen), 200 µM of each dNTP (Eurogentec), 0.4 µM of AQP2-RFLP-F (5′-GAACTCATTTCCACCGCAGT-3)′ and AQP2-RFLP-R (5′-AGTCCAAAGATACCTCCAAACA-3′) (Biolegio), 0.4 U Hotstart Taq plus polymerase (Qiagen), 0.1 mg/ml acetylated BSA (Promega) and 2 µl target DNA. Amplification conditions were as follows, initial denaturation at 94°C for 5 min, 24 cycles of 94°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec and final annealing at 72°C for 5 min. Five µl of the PCR products or 1 µg of plasmid DNA, containing a cloned AQP2/3 variant, were digested for 15 min at 37°C in a 15 µl reaction mixture containing 1 µl FastDigest Green buffer with 1 µl of either AvaI or SduI FastDigest restriction enzymes (Thermo Scientific), followed by 20 minutes inactivation at 80°C. Differential banding was analysed using electrophoresis of 10 µl on a 3% small fragment agarose gel run for 30 min at 135 V and stained with ethidium bromide.
Results
Phenotype
In a preparatory experiment, mice were infected with strain 348BT, isolated from a patient who had been cured, and were treated with melarsoprol at 1, 2, 3, 4, 5, 8 and 10 mg/kg BW. The minimum melarsoprol dosage needed to cure the mice from the infection appeared to be 10 mg/kg BW. This dosage was then used to treat mice infected with all the 41 T.b. gambiense strains and with the two T.b. brucei strains. Some mice infected with T.b. gambiense strains 15BT, 163AT and 346AT experienced a relapse that was detectable only about three months after the infection and after immunosuppression with cyclophosphamide (Table 2). All mice infected with the other T.b. gambiense strains and with both T.b. brucei strains (including the AT1/P2 KO strain) got cured by melarsoprol at 10 mg/kg/BW, as defined by the absence of detectable relapse up to 100 days after infection. To confirm the apparent melarsoprol resistance of 15BT, 163AT and 346AT, the experiment was repeated with these strains and with melarsoprol treatment at 10 and 12 mg/kg BW. All mice infected with 15BT remained without detectable parasitaemia after treatment. Among the mice infected with 163AT, one mouse relapsed at DPI 28 after treatment with 10 mg/kg BW and one mouse relapsed at DPI 31 after treatment with 12 mg/kg BW melarsoprol. All mice infected with 346AT and treated with 10 and 12 mg/kg BW melarsoprol, relapsed at DPI 20 (Table 2).
Table 2. Phenotype of melarsoprol resistant strains.
| 10 mg/kg BW | 10 mg/kg BW (rep) | 12 mg/kg BW | ||||
| Alias name | numbers relapsed | DPI | numbers relapsed | DPI | numbers relapsed | DPI |
| 15BT | 2 | 85* | 0 | na | 0 | na |
| 163AT | 3 | 88* | 1 | 28 | 1 | 31 |
| 346AT | 1 | 90* | 6 | 20 | 6 | 20* |
Number of relapsing mice (out of 6 infected) and day post-infection that relapses were observed after treatment with melarsoprol at different dosages and repetitions. DPI: days post-infection, BW: body weight, rep: repetition, na: not applicable,
*: relapsing population used for AQP2/3 RFLP analysis.
Genotype
TbAT1 P2 adenosine transporter gene
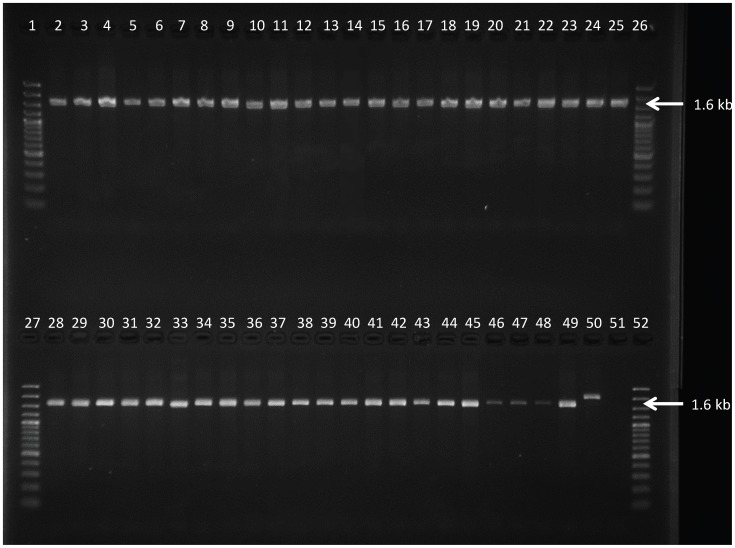
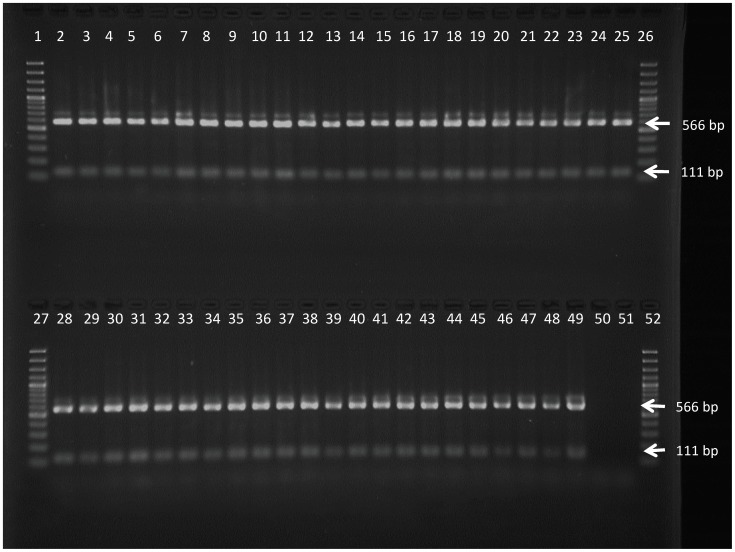
PCR with the Sfa primers yielded the expected 677 bp amplicon in all T.b. gambiense and in the T.b. brucei 427 WT strain but not in the T.b. brucei 427 AT1/P2 KO strain. Restriction enzyme digestion of the PCR amplicons with SfaNI showed the wild type pattern, i.e. one fragment of 566 bp and one of 111 bp, in all the T.b. gambiense and in the T.b. brucei 427 WT strains (Figure 1). With all the 45 T.b. gambiense strains and with the T.b. brucei 427 WT, the TbAT1 PCR generated an amplicon of about 1600 bp, indicating the presence of the P2 adenosine transporter gene (Figure 2). Only the T.b. brucei 427 AT1/P2 KO strain yielded a fragment of about 2000 bp indicative of the replacement of P2 adenosine transporter gene by antibiotic resistance genes in the Tbat1 null mutant. For selected strains the entire TbAT1 gene was sequenced (Table 3). Twelve strains from Mbuji-Mayi and all four strains from Masi-Manimba had a TbAT1 sequence identical to the wild-type TbAT1 sequence of T.b. gambiense 930 (KF564940). However, two old Congolese T.b. gambiense strains, MBA and KEMLO, contained a not yet described trinucleotide deletion in TbAT1 Δ516–518. Aquaglyceroporin transporters
Figure 1. Restriction digest profile generated with SfaNI PCR-RFLP on DNA of the T.b. gambiense strains as listed in Table 1 and of the two T.b. brucei control strains.
Lanes 1, 26, 27 and 52 = GeneRuler 100 bp Plus DNA Ladder (Fermentas), lanes 2 to 44 = T.b. gambiense strains isolated from Mbuji-Mayi, lanes 45–48 = T.b. gambiense strains isolated from Masi-Manimba, lane 49 = T.b. brucei 427 WT, lane 50 = T.b. brucei 427 AT1/P2 KO, lane 51 = negative PCR control.
Figure 2. Amplicons generated with TbAT1-PCR on DNA of the T.b. gambiense strains as listed in Table 1 and of the two T.b. brucei control strains.
Lanes 1, 26, 27 and 52 = GeneRuler 100 bp Plus DNA Ladder (Fermentas), lanes 2 to 44 = T.b. gambiense strains isolated from Mbuji-Mayi, lanes 45 to 48 = T.b. gambiense strains isolated from Masi-Manimba, lane 49 = T.b. brucei 427 WT, lane 50 = T.b. brucei 427 AT1/P2 KO, lane 51 = negative PCR control.
Table 3. Genotype, strains and accession numbers for TbAT1.
| Genotype | Strains | Accession number |
| TbAT1 | 15BT | KM282018 |
| 45BT | KM282019 | |
| 108BT | KM282020 | |
| 108AT | KM282021 | |
| 130BT | KM282022 | |
| 146AT | KM282023 | |
| 163AT | KM282024 | |
| 174BT | KM282025 | |
| 174AT | KM282026 | |
| 346AT | KM282027 | |
| 348BT | KM282028 | |
| 378BT | KM282029 | |
| MM01 | KM282030 | |
| MM03 | KM282031 | |
| MM05 | KM282032 | |
| MM06 | KM282033 | |
| TbAT1 Δ516–518 | MBA | KM282016 |
| KEMLO | KM282017 |
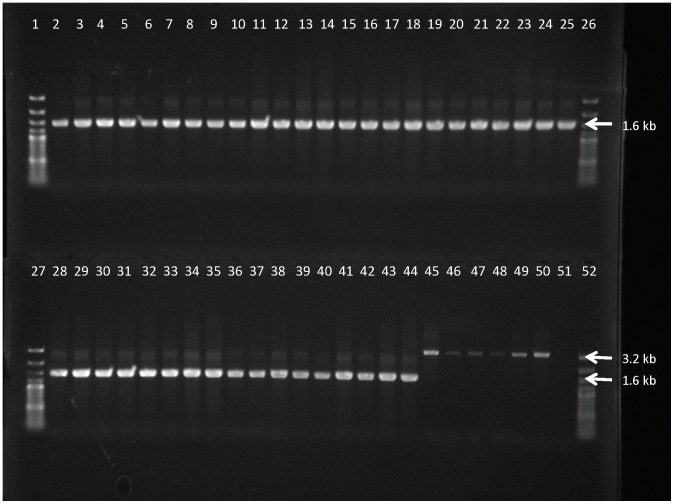
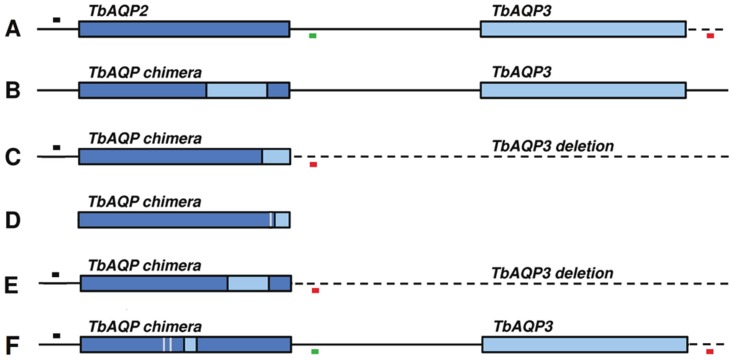
With all the 41 T.b. gambiense strains from Mbuji-Mayi and in both MBA and KEMLO, no amplification was seen when the wild type AQP2 locus was amplified with an AQP2 locus specific PCR. In contrast AQP2/3-PCR generated one single amplicon of about 1500 bp (Figure 3) previously shown to represent the chimeric AQP2/3 gene and the deletion of the AQP3 gene [20]. With the 4 T.b. gambiense strains from Masi-Manimba and the T.b. brucei 427 WT as well as the T.b. brucei 427 AT1/P2 KO, one single amplicon of about 1500 bp was found with the AQP2 locus specific PCR, while in AQP2/3-PCR a band of 3200 bp was generated, indicating the presence of both the AQP2 and the AQP3 genes. Direct sequencing of the AQP2/3 amplicons of twelve of the Mbuji-Mayi strains showed that the band was indeed the previously described AQP2/3 chimera (Table 4), containing the first 813 bp from AQP2 and the last 126 bp from AQP3, abbreviated here as AQP2/3 (814) to indicate that the switch from AQP2 to AQP3 is first detected at nucleotide 814 (Figure 4 – C). Amplicons from MBA and KEMLO, revealed a yet unknown AQP2/3 chimera, containing the first 677 bp from AQP2, followed by a stretch of 202 bp of AQP3 and ending with the last 60 bp from AQP2, abbreviated here as AQP2/3 (678–880) to indicate that the switch from AQP2 to AQP3 is first detected at nucleotide 678 and the switch back to AQP2 is first detected at nucleotide 880 (Figure 4 – E). Direct sequencing results from the circa 1500 bp AQP2 amplicons from 4 strains from Masi-Manimba consistently showed a total of 18 heterozygous single nucleotide polymorphisms in comparison to the wild-type AQP2 sequence of T.b. gambiense STIB 930 (KF564925). After cloning of the full length AQP2 sequence from one strain of Masi-Manimba, MM01, we obtained either the wild-type sequence or another novel chimera that shares the first 616 bp from AQP2, with 2 point mutations at position T548C and G573A, followed by a stretch of 41 bp identical to AQP3 and ending with the last 282 bp from AQP2, abbreviated here as AQP2/3 (617–658) (Figure 4 – F). Cloning AQP2 sequences from LiTat 1.3, the West-African T.b. gambiense strain sensitive to melarsoprol and pentamidine, revealed only wild-type sequences (Figure 4 – A), while cloned AQP2/3 sequences from the old T.b. gambiense MBA strain did not reveal any differences with results already obtained by direct sequencing (Figure 4 – E). Surprisingly, cloning of the AQP2/3 gene from T.b. gambiense 348 BT revealed next to the chimera found by direct sequencing yet another AQP2/3 chimera that shared the first 879 bp from AQP2, with one point mutation at T869C and only the last 60 bp from AQP3, abbreviated as AQP2/3(880) (Figure 4 – D). A partial alignment of the obtained sequences of the AQP2, AQP3 and AQP2/3 variants is given in figure S1. When T.b. gambiense 348 BT was amplified with a cloning primer specific for AQP2, only a very faint amplification pattern was seen, probably attributable to primer mismatch on AQP2/3 (figure S2).
Figure 3. Amplicons generated with AQP2/3-PCR on DNA of the T.b. gambiense strains as listed in Table 1 and of the two T.b. brucei control strains.
Lanes 1, 26, 27 and 52 = GeneRuler 100 bp Plus DNA Ladder (Fermentas), lanes 2 to 44 = T.b. gambiense strains isolated from Mbuji-Mayi, lanes 45–48 = T.b. gambiense strains isolated from Masi-Manimba, lane 49 = T.b. brucei 427 WT, lane 50 = T.b. brucei 427 AT1/P2 KO, lane 51 = negative PCR control.
Table 4. Genotype, strains and accession numbers for TbAQP2 and TbAQP2/3.
| Genotype | Strains | Accession number |
| AQP2 | LiTat 1.3 | KM282048 |
| MM01* | KM282049 | |
| AQP2/3 (814) | 15BT | KM282036 |
| 45BT | KM282037 | |
| 108BT | KM282038 | |
| 108AT | KM282039 | |
| 130BT | KM282040 | |
| 146AT | KM282041 | |
| 163AT | KM282042 | |
| 174BT | KM282043 | |
| 174AT | KM282044 | |
| 346AT | KM282045 | |
| 348BT | KM282046 | |
| 378BT | KM282047 | |
| AQP2/3 (880) | 348BT | KM282050 |
| AQP2/3 (678–880) | MBA | KM282034 |
| KEMLO | KM282035 | |
| AQP2/3 (617–658) | MM01* | KM282051 |
*direct sequencing results suggested 18 heterozygous single nucleotide polymorphisms in the AQP2 coding sequence of T.b. gambiense MM01, MM03, MM05, and MM06.
Figure 4. Schematic view of the AQP2/3 variants identified in this study (adapted from Graf et al [20]).
Sequence of AQP3 was not verified. Positions of primer: black box = AQP2/3_F, green box = AQP2_R, red box = AQP2/3_R. A) Reference locus of AQP2 and AQP3, with wild-type AQP2 found in the melarsoprol and pentamidine sensitive strain T.b. gambiense LiTat 1.3 and in all strains from Masi-Manimba. B) Chimera of AQP2 and AQP3 occurring in a melarsoprol and pentamidine resistant T.b. brucei strain as described by Baker et al. [19]. C) Chimera of AQP2 and AQP3 plus loss of AQP3 in all T.b. gambiense strains from Mbuji-Mayi as described in this article and by Graf et al. [20]. D) New chimera of AQP2 and AQP3, possibly outside the known locus, found in all T.b. gambiense strains from Mbuji-Mayi. E) New chimera of AQP2 and AQP3 plus loss of AQP3 found in two old Congolese T.b. gambiense strains, MBA and KEMLO. F) New chimera of AQP2 and AQP3, without loss of AQP3, found in all four T.b. gambiense strains isolated in Masi-Manimba.
RFLP to discriminate Mbuji-Mayi AQP2/3 variants
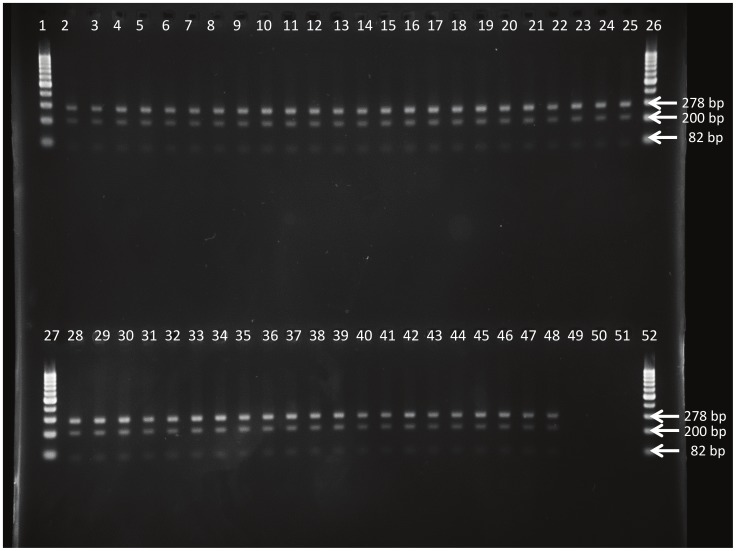
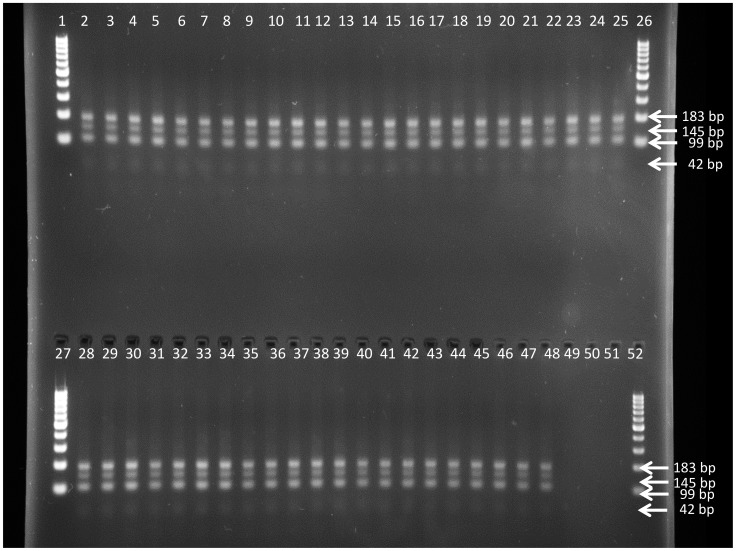
Since T.b. gambiense 348 BT was isolated from a cured patient and seemed to contain both AQP2/3 (814) and AQP2/3 (880), we hypothesised that only AQP2/3 (814) would be present in strains isolated from relapsing patients. For this reason, we amplified a part of the AQP2/3 gene using a PCR specific to the Mbuji-Mayi strains and used RFLP with either AvaI or SduI restriction enzymes to discriminate between both AQP2/3 variants. RFLP with AvaI does not cut the AQP2/3 (814) chimera, but generates bands of 200 bp and 82 bp in the AQP2/3 (880) chimera. RFLP with SduI cuts AQP2/3 (814) chimera in bands of 99 bp and 183 bp, while AQP2/3 (880) chimera is cut in bands of 99 bp, 145 bp and 42 bp. PCR-RFLP on plasmids containing only a single AQP2/3 variant could unambiguously differentiate between both AQP2/3 (814) and AQP2/3 (880) (figure S3). However, PCR-RFLP, with either AvaI (Figure 5) or SduI (Figure 6), on all T.b. gambiense strains from Mbuji-Mayi, including four strains isolated from relapses from mice, indicated the presence of both variants of the AQP2/3 chimera, suggesting that strains that cause a relapse in either mice and humans are not genetically characterised by a loss of AQP2/3 (880).
Figure 5. Restriction digest profile generated with AvaI PCR-RFLP on DNA of the T.b. gambiense strains as listed in Table 1, including the four strains isolated from relapsed mice.
Lanes 1, 26, 27 and 52 = GeneRuler 100 bp Plus DNA Ladder (Fermentas), lanes 2 to 44 = T.b. gambiense strains isolated from Mbuji-Mayi, lane 45 = T.b. gambiense 15BT relapse 10 mg/kg BW, lane 46 = T.b. gambiense 163AT relapse 10 mg/kg BW, lane 47 = T.b. gambiense 346AT relapse 10 mg/kg BW, lane 48 = T.b. gambiense 346AT relapse 12 mg/kg BW, lane 49 = T.b. gambiense MBA, lane 50 = T.b. gambiense MM01, lane 51 = negative PCR control.
Figure 6. Restriction digest profile generated with SduI PCR-RFLP on DNA of the T.b. gambiense strains as listed in Table 1, including the four strains isolated from relapsed mice.
Lanes 1, 26, 27 and 52 = GeneRuler 100 bp Plus DNA Ladder (Fermentas), lanes 2 to 44 = T.b. gambiense strains isolated from Mbuji-Mayi, lane 45 = T.b. gambiense 15BT relapse 10 mg/kg BW, lane 46 = T.b. gambiense 163AT relapse 10 mg/kg BW, lane 47 = T.b. gambiense 346AT relapse 10 mg/kg BW, lane 48 = T.b. gambiense 346AT relapse 12 mg/kg BW, lane 49 = T.b. gambiense MBA, lane 50 = T.b. gambiense MM01, lane 51 = negative PCR control.
Discussion
This study was undertaken to investigate the mechanisms underlying the high relapse rates observed in second stage gambiense HAT patients treated with melarsoprol in Mbuji-Mayi, DRC. The in vivo melarsoprol sensitivity experiment showed that a minimum dose of 10 mg/kg BW melarsoprol was needed to cure mice infected with trypanosomes that were isolated from a cured patient. This is 4 times higher than the dose needed to cure mice infected with T.b. gambiense strains isolated from Ibba in South Sudan, another HAT focus known for high melarsoprol relapse rates [9]. On the other hand, Kibona and co-workers considered 3 out of 35 tested T.b. rhodesiense strains as resistant when mice relapsed after treatment with 5 mg/kg BW melarsoprol [30]. Among the 41 strains from Mbuji-Mayi, 2 induced infections that could not be cured with 12 mg/kg BW melarsoprol. Both were isolated from patients after treatment with melarsoprol.
When setting up the in vivo drug sensitivity experiment, we were confronted with the lack of a standardised protocol, especially for T.b. gambiense. Most studies have been dealing with T.b. brucei or T.b. rhodesiense, both behaving quite virulent in laboratory mice. Although melarsoprol is a drug that can cure the chronic phase of trypanosomosis, we opted for an acute phase in vivo model for several reasons: i. the concentration of melarsoprol that reaches the central nervous system is only a minor fraction of what reaches the plasma and is more prone to uncontrolled variations among individual outbred animals, ii. the acute phase model is expected to correspond better with the standard in vitro model, iii. since T.b. gambiense can cause subclinical or even silent infections, assessing treatment outcome in a chronic model via examination of blood and organs, including the brain, is unreliable [31]. The protocol we used here is mainly based on the study that Maina et al. carried out on recent isolates of T.b. gambiense from Sudan [9]. We also immunosuppressed the mice before inoculation with 5×104 trypanosomes to guarantee that all mice would become infected. Some major differences however are to be noted. During the preparatory experiments, we noted that melarsoprol precipitates immediately when diluted in water, the usual diluent in other in vitro and in vivo studies [7], [9], [30]. In our final protocol, we diluted melarsoprol in polyethylene glycol to keep it in solution facilitating correct dosage when treating the mice. In contrast to the custom 60 days after treatment follow-up, we monitored the mice for up to 100 days after treatment and we immunosuppressed them on day 85 after treatment. In addition, at the end of the follow-up period, we sacrificed the mice and passed all the blood on DEAE cellulose columns instead of checking only a few drops of tail blood with the microhaematocrit technique. This allowed us to observe relapses at days 85–90 after treatment that otherwise would have been missed. Still, some treated mice showed paralysis but without any detectable trypanosome in the blood, suggesting that the real relapse rate was higher than what we actually can report based on trypanosome detection only. A weakness in our study is the absence of well documented T.b. gambiense melarsoprol resistant control strain. In the absence of such a strain, we had to rely on the T.b. brucei 427 AT1/P2 KO strain and its corresponding T.b. brucei 427 wild-type strain. Both strains appeared to be sensitive to melarsoprol at 10 mg/kg BW which is consistent with what has been observed in previous studies on a Tbat1 null mutant [14]. On the other hand, for 5 strains included in our in vivo experiment, it was shown in vitro that they were 2–4 times less sensitive for melarsoprol than the reference sensitive T.b. gambiense strain STIB 930 [20]. Basing our treatment dose for the in vivo study solely on the dose required to cure a stabilate isolated from a cured patient proved to be a limitation for further interpretation of the in vivo results. Our initial hypothesis, including the rationale for the isolation of couples, was that strains originating from cured patients would be more sensitive to melarsoprol than strains from relapsed patients. It was clearly not expected that all strains would carry drug resistance markers. The different in vivo melarsoprol sensitivity phenotypes observed in our experiment do not correspond with the low variability observed within the two studied genetic markers associated with melarsoprol resistance. Indeed, all strains from Mbuji-Mayi, irrespective of their isolation from cured or relapsing patients, carry the wild type TbAT1 allele. In addition, in all strains from Mbuji-Mayi, the TbAQP2 and TbAQP3 are replaced by chimeric TbAQP2/3 variants, of which one has been reported previously to correlate with in vitro pentamidine and melarsoprol resistance by Graf and co-workers [20]. The latter study included 5 strains from the Mbuji-Mayi collection (40 AT, 45 BT, 130 BT, 349 BT, and 349 AT). According to Graf and co-workers, these 5 strains contained the TbAQP2/3 chimera and the wild type TbAT1, what is confirmed in our study, and showed decreased sensitivity for pentamidine and for melarsoprol in vitro. In our study we found a second variant of a chimeric AQP2/3 gene in these strains, which was only observed after cloning and not by direct sequencing, possibly indicating the presence of such variant outside the known AQP2 locus. Surprisingly, the strains from Masi-Manimba were heterozygous for the AQP2 locus. One allele contained the wild-type sequence, but the second allele contained a yet undescribed TbAQP2/3 chimera. Munday and co-workers recently described that the presence of a functional wild-type AQP2 sequence renders strains sensitive to melarsoprol and pentamidine [24]. However, the effect on pentamidine and melarsoprol uptake of the newly described AQP2/3 chimeras is unknown. All variants were cloned in a trypanosomal expression vector for future evaluation and are available upon request. That this probable drug resistant genotype is found in all strains from Mbuji-Mayi and not in the strains from Masi-Manimba could be sufficient to explain the difference in melarsoprol relapse rates observed in East-Kasai (high) and in Bandundu (low). Within this context, it is interesting to note that we observed yet another TbAQP2/3 chimera genotype in two “old” T.b. gambiense type I strains isolated in 1974 in Kinshasa and in Nord Equateur Province. Their AQP2/3 variant is similar, but shorter, than the AQP2/3 mutation described by Baker and co-workers and is therefore possibly capable of reducing melarsoprol and pentamidine uptake [19]. However, in contrast to the locus described by Baker and co-workers, AQP3 seems not preserved in these strains. Both “old” isolates also contain a new variant of TbAT1 with unknown effect on drug uptake. Due to the fact that both AQP2/3 and TbAT1 genes are different, these “old” isolates are probably not closely related to the contemporary strains circulating in Masi-Manimba and Mbuji-Mayi. Appearance of resistance to arsenicals, including melarsoprol, and to pentamidine in several HAT foci in DRC (former Congo Belge and Zaire) has already been described decades ago and was considered a result of mass treatment and chemoprophylaxis [32], [33]. The finding of the pentamidine/melarsoprol resistant genotype in the “old” T.b. gambiense strains may be the basis for molecular studies into the appearance and spread of pentamidine/melarsoprol resistant T.b. gambiense strains. In the present study, we didn't carry out microsatellite analysis to verify the similarity between strains from the Mbuji-Mayi focus, in particular from strains isolated from the same patient before treatment and after relapse. However, results from mobile genetic element PCR (MGE-PCR) as described by Simo et al suggest only very small differences between all strains from Mbuji-Mayi (figure S4) [34]. Thus, most probably, all strains isolated in Mbuji-Mayi are probably the clonal progeny of one strain acquiring the AQP2/3 chimeras. This is the first large scale in vivo drug sensitivity study on T.b. gambiense strains that were isolated within a short period of time, in one single HAT focus, from cured as well as from relapsing patients and for 10 cases from the same patient before and after treatment. As such, the fact that the homogeneity observed within the TbAQ2/3 and the TbAT1 loci does not correspond with the heterogeneity in treatment outcome of the patients and of the mice, suggests that other factors, such as virulence and tissue tropism of the parasite or genotype and phenotype of the individual patient, may influence the treatment outcome. For example, in several independent studies it was found that high cell count in the cerebrospinal fluid (>100 cell/µl) is arisk factor for relapse [8], [23], [35]–[37]. This may indicate that patients in advanced second stage of the disease are less responsive for the standard melarsoprol treatment schedule. It is even not excluded that such patients are also less responsive for other drugs or therapeutic regimes such as NECT, the current first line treatment of second stage gambiense HAT. Therefore, further investigations into treatment failure in HAT and into alternative drugs or treatment regimes should not only focus on differential genotypes of the parasites but also on differential virulence and tissue tropism.
In conclusion, this study confirms that the high melarsoprol relapse rates observed in the Mbuji-Mayi focus can be explained by mutations in the TbAQP2/3 locus of the trypanosomes that circulate in that focus. However, other factors will also influence the treatment outcome of individual patients.
Supporting Information
Partial alignment of AQP2, AQP3 and AQP2/3 variants with GenBank accession numbers.
(TIF)
Amplification of T.b. gambiense DNA with a cloning primer specific for AQP2. Lanes 1 and 14: GeneRuler 100 bp Plus DNA Ladder (Fermentas), lanes 2–10: T.b. gambiense strains isolated from Mbuji-Mayi, lane 11 = T.b. gambiense LiTat 1.3, lane 12 = T.b. gambiense MM01, lane 13: negative PCR control.
(TIF)
RFLP with either AvaI or SduI restriction enzymes to discriminate between both AQP2/3 variants. RFLP with AvaI (panel A) does not cut the AQP2/3(814) chimera (lane 1), but generates bands of 200 bp and 82 bp in the AQP2/3(880) chimera (lane 2). RFLP with SduI (panel B) cuts AQP2/3(814) chimera in bands of 99 bp and 183 bp (lane 4), while AQP2/3(880) chimera is cut in bands of 99 bp, 145 bp and 42 bp (lane 5). Lanes 3 and 6: GeneRuler 100 bp Plus DNA Ladder (Fermentas).
(TIF)
Amplicons generated with mobile genetic element PCR (MGE-PCR) on DNA of diverse T. brucei strains. Lanes 1, 26, 27 and 51 = GeneRuler 100 bp Plus DNA Ladder (Fermentas), lanes 2 to 43 = T.b. gambiense strains isolated from Mbuji-Mayi, lanes 44 = T.b. gambiense MBA, lane 45: T.b. gambiense AnTat 22.1, lane 46 = T.b. gambiense AnTat 9.1, lane 47: T.b. gambiense LiTat 1.3, lane 48 = T.b. rhodesiense AnTat 12.1, lane 49: T.b. rhodesiense AnTat 25.1, lane 50 = T.b. gambiense type II Abba.
(TIF)
Acknowledgments
We are grateful to Nicolas Bebronne for the assistance with the cryopreservation of the trypanosome strains and with the in vivo experiments. We thank Reto Brun and Pascal Mäser from the Swiss Tropical and Public Health Institute for providing the T.b. brucei wild type and the T.b. brucei 427 AT1/2 KO strains. The pHD309 vector was a kind gift from George Cross from the Rockefeller University. Melarsoprol was kindly provided by Pere Simarro from the World Health Organization. We acknowledge Fabrice Graf from the Swiss Tropical and Public Health Institute for advise on aquaglyceroporin specific PCR and sequencing. We thank Harry de Koning from the University of Glasgow for his suggestion on the classification of the aquaglyceroporin chimera. We would like to thank “Les Amis de l'Institut Pasteur de Bruxelles” a.s.b.l. for their support.
Funding Statement
This study was funded by the Belgian Development Cooperation (DGCD) under the Framework Agreement 3 (FA3) with the Institute of Tropical Medicine (ITM), Antwerp (Belgium). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Simarro PP, Diarra A, Ruiz Postigo JA, Franco JR, Jannin JG (2011) The human African trypanosomiasis control and surveillance programme of the World Health Organization 2000–2009: the way forward. PLoS Negl Trop Dis 5: e1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bacchi CJ (2009) Chemotherapy of human African trypanosomiasis. Interdiscip Perspect Infect Dis 2009: 195040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simarro PP, Franco J, Diarra A, Postigo JA, Jannin J (2012) Update on field use of the available drugs for the chemotherapy of human African trypanosomiasis. Parasitology 139: 842–846. [DOI] [PubMed] [Google Scholar]
- 4. Priotto G, Kasparian S, Mutombo W, Ngouama D, Ghorashian S, et al. (2009) Nifurtimox-eflornithine combination therapy for second-stage African Trypanosoma brucei gambiense trypanosomiasis: a multicentre, randomised, phase III, non-inferiority trial. Lancet 374: 56–64. [DOI] [PubMed] [Google Scholar]
- 5. Stanghellini A, Josenando T (2001) The situation of sleeping sickness in Angola: a calamity. Trop Med Int Health 6: 330–334. [DOI] [PubMed] [Google Scholar]
- 6. Legros D, Fournier C, Gastellu Etchegorry M, Maiso F, Szumilin E (1999) Echecs thérapeutiques du mélarsoprol parmi des patients traités au stade tardif de trypanosomose humaine africaine à T. b. gambiense en Ouganda. Bull Soc Pathol Exot Fil 92: 171–172. [PubMed] [Google Scholar]
- 7. Brun R, Schumacher R, Schmid C, Kunz C, Burri C (2001) The phenomenon of treatment failures in human African trypanosomiasis. Trop Med Int Health 6: 906–914. [DOI] [PubMed] [Google Scholar]
- 8. Mumba Ngoyi D, Lejon V, Pyana P, Boelaert M, Ilunga M, et al. (2010) How to shorten patient follow-up after treatment for Trypanosoma brucei gambiense sleeping sickness? J Infect Dis 201: 453–463. [DOI] [PubMed] [Google Scholar]
- 9. Maina N, Maina KJ, Mäser P, Brun R (2007) Genotypic and phenotypic characterization of Trypanosoma brucei gambiense isolates from Ibba, South Sudan, an area of high melarsoprol treatment failure. Acta Trop 104: 84–90. [DOI] [PubMed] [Google Scholar]
- 10. Robays J, Nyamowala G, Sese C, Kande Betu-Ku-Mesu V, Lutumba P, et al. (2008) High failure rates of melarsoprol for sleeping sickness, Democratic Republic of Congo. Em Inf Dis 14: 966–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Matovu E, Enyaru JCK, Legros D, Schmid C, Seebeck T, et al. (2001) Melarsoprol refractory T. b. gambiense from Omugo, north-western Uganda. Trop Med Int Health 6: 407–411. [DOI] [PubMed] [Google Scholar]
- 12. Burri C, Keiser J (2001) Pharmacokinetic investigations in patients from northern Angola refractory to melarsoprol treatment. Trop Med Int Health 6: 412–420. [DOI] [PubMed] [Google Scholar]
- 13. Nerima B, Matovu E, Lubega GW, Enyaru JC (2007) Detection of mutant P2 adenosine transporter (TbAT1) gene in Trypanosoma brucei gambiense isolates from northwest Uganda using allele-specific polymerase chain reaction. Trop Med Int Health 12: 1361–1368. [DOI] [PubMed] [Google Scholar]
- 14. Matovu E, Stewart ML, Geiser F, Brun R, Mäser P, et al. (2003) Mechanisms of arsenical and diamidine uptake and resistance in Trypanosoma brucei . Eukaryot Cell 2: 1003–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matovu E, Geiser F, Schneider V, Mäser P, Enyaru JCK, et al. (2001) Genetic variants of the TbAT1 adenosine transporter from African trypanosomes in relapse infections following melarsoprol therapy. Mol Biochem Parasitol 117: 73–81. [DOI] [PubMed] [Google Scholar]
- 16. Claes F, Vodnala SK, Van Reet N, Boucher N, Lunden-Miguel H, et al. (2009) Bioluminescent imaging of Trypanosoma brucei shows preferential testis dissemination which may hamper drug efficacy in sleeping sickness patients. PLoS Negl Trop Dis 3: e486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mäser P, Sütterlin C, Kralli A, Kaminsky R (1999) A nucleoside transporter from Trypanosoma brucei involved in drug resistance. Science 285: 242–244. [DOI] [PubMed] [Google Scholar]
- 18. Bridges DJ, Gould MK, Nerima B, Mäser P, Burchmore RJS, et al. (2007) Loss of the high-affinity pentamidine transporter is responsible for high levels of cross-resistance between arsenical and diamine drugs in African trypanosomes. Mol Pharmacol 71: 1098–1106. [DOI] [PubMed] [Google Scholar]
- 19. Baker N, Glover L, Munday JC, Aguinaga Andrés D, Barrett MP, et al. (2012) Aquaglyceroporin 2 controls susceptibility to melarsoprol and pentamidine in African trypanosomes. Proc Natl Acad Sci USA 109: 10996–11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Graf FE, Ludin P, Wenzler T, Kaiser M, Brun R, et al. (2013) Aquaporin 2 mutations in Trypanosoma brucei gambiense field isolates concur with decreased susceptibility to pentamidine and melarsoprol. PLoS Negl Trop Dis 7: e2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pépin J, Ethier L, Kazadi C, Milord F, Ryder R (1992) The impact of human immunodeficiency virus infection on the epidemiology and treatment of Trypanosoma brucei gambiense sleeping sickness in Nioki, Zaire. Am J Trop Med Hyg 47: 133–140. [DOI] [PubMed] [Google Scholar]
- 22. Pépin J, Mpia B, Iloasebe M (2002) Trypanosoma brucei gambiense African trypanosomiasis: differences between men and women in severity of disease and response to treatment. Trans R Soc Trop Med Hyg 96: 421–426. [DOI] [PubMed] [Google Scholar]
- 23. Legros D, Evans S, Maiso F, Enyaru JCK, Mbulamberi D (1999) Risk factors for treatment failure after melarsoprol for Trypanosoma brucei gambiense trypanosomiasis in Uganda. Trans R Soc Trop Med Hyg 93: 439–442. [DOI] [PubMed] [Google Scholar]
- 24. Munday JC, Eze AA, Baker N, Glover L, Clucas C, et al. (2014) Trypanosoma brucei aquaglyceroporin 2 is a high-affinity transporter for pentamidine and melaminophenyl arsenic drugs and the main genetic determinant of resistance to these drugs. J Antimicrob Chemother 69: 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pyana PP, Ngay Lukusa I, Mumba Ngoyi D, Van Reet N, Kaiser M, et al. (2011) Isolation of Trypanosoma brucei gambiense from cured and relapsed sleeping sickness patients and adaptation to laboratory mice. PLoS Negl Trop Dis 5: e1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Van Reet N, Van de Vyver H, Pyana PP, Van der Linden AM, Büscher P (2014) A panel of Trypanosoma brucei strains tagged with blue and red shifted luciferases for bioluminescent imaging in murine infection models. PLoS Negl Trop Dis In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Büscher P, Mumba Ngoyi D, Kaboré J, Lejon V, Robays J, et al. (2009) Improved models of mini anion exchange centrifugation technique (mAECT) and modified single centrifugation (MSC) for sleeping sickness diagnosis and staging. PLoS Negl Trop Dis 3: e471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lanham SM, Godfrey DG (1970) Isolation of salivarian trypanosomes from man and other mammals using DEAE-cellulose. Exp Parasitol 28: 521–534. [DOI] [PubMed] [Google Scholar]
- 29. Wirtz E, Hartmann C, Clayton C (1994) Gene expression mediated by bacteriophage T3 and T7 RNA polymerase in transgenic trypanosomes. Nucleic Acids Res 22: 3887–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kibona SN, Matemba L, Kaboya JS, Lubega GW (2006) Drug-resistance of Trypanosoma b. rhodesiense isolates from Tanzania. Trop Med Int Health 11: 144–155. [DOI] [PubMed] [Google Scholar]
- 31. Giroud C, Ottones F, Coustou V, Dacheux D, Biteau N, et al. (2009) Murine models for Trypanosoma brucei gambiense disease progression-from silent to chronic infections and early brain tropism. PLoS Negl Trop Dis 3: e509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burke J (1971) Historique de la lutte contre la maladie du sommeil au Congo. Ann Soc Belg Méd Trop 51: 465–477. [PubMed] [Google Scholar]
- 33. Ollivier G, Legros D (2001) Trypanosomiase humaine africaine: historique de la thérapeutique et ses échecs. Trop Med Int Health 6: 11–855. [DOI] [PubMed] [Google Scholar]
- 34. Simo G, Herder S, Nijokou F, Asonganyi T, Tilley A, et al. (2005) Trypanosoma brucei s.l.: Characterisation of stocks from Central Africa by PCR analysis of mobile genetic elements. Exp Parasitol 110: 353–362. [DOI] [PubMed] [Google Scholar]
- 35. Lejon V, Roger M, Mumba Ngoyi D, Menten J, Robays J, et al. (2008) Novel markers for treatment outcome in late stage T.b. gambiense trypanosomiasis. Clin Infect Dis 47: 15–22. [DOI] [PubMed] [Google Scholar]
- 36. Schmid C, Nkunku S, Merolle A, Vounatsou P, Burri C (2004) Efficacy of 10-day melarsoprol schedule 2 years after treatment for late-stage gambiense sleeping sickness. Lancet 364: 789–790. [DOI] [PubMed] [Google Scholar]
- 37. Balasegaram M, Harris S, Checchi F, Hamel C, Karunakara U (2006) Treatment outcomes and risk factors for relapse in patients with early-stage human African trypanosomiasis (HAT) in the Republic of the Congo. Bull World Health Organ 84: 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Partial alignment of AQP2, AQP3 and AQP2/3 variants with GenBank accession numbers.
(TIF)
Amplification of T.b. gambiense DNA with a cloning primer specific for AQP2. Lanes 1 and 14: GeneRuler 100 bp Plus DNA Ladder (Fermentas), lanes 2–10: T.b. gambiense strains isolated from Mbuji-Mayi, lane 11 = T.b. gambiense LiTat 1.3, lane 12 = T.b. gambiense MM01, lane 13: negative PCR control.
(TIF)
RFLP with either AvaI or SduI restriction enzymes to discriminate between both AQP2/3 variants. RFLP with AvaI (panel A) does not cut the AQP2/3(814) chimera (lane 1), but generates bands of 200 bp and 82 bp in the AQP2/3(880) chimera (lane 2). RFLP with SduI (panel B) cuts AQP2/3(814) chimera in bands of 99 bp and 183 bp (lane 4), while AQP2/3(880) chimera is cut in bands of 99 bp, 145 bp and 42 bp (lane 5). Lanes 3 and 6: GeneRuler 100 bp Plus DNA Ladder (Fermentas).
(TIF)
Amplicons generated with mobile genetic element PCR (MGE-PCR) on DNA of diverse T. brucei strains. Lanes 1, 26, 27 and 51 = GeneRuler 100 bp Plus DNA Ladder (Fermentas), lanes 2 to 43 = T.b. gambiense strains isolated from Mbuji-Mayi, lanes 44 = T.b. gambiense MBA, lane 45: T.b. gambiense AnTat 22.1, lane 46 = T.b. gambiense AnTat 9.1, lane 47: T.b. gambiense LiTat 1.3, lane 48 = T.b. rhodesiense AnTat 12.1, lane 49: T.b. rhodesiense AnTat 25.1, lane 50 = T.b. gambiense type II Abba.
(TIF)