Abstract
Hepatocellular carcinoma (HCC) is a frequent cancer with limited treatment options and poor prognosis. Tumorigenesis has been linked with macrophage-mediated chronic inflammation and diverse signaling pathways including the Epidermal Growth Factor Receptor (EGFR) pathway. The precise role of EGFR in HCC is unknown, and EGFR-inhibitors have shown disappointing clinical results. Here we discover that EGFR is expressed in liver macrophages in both human HCC and in a mouse HCC model. Mice lacking EGFR in macrophages show impaired hepatocarcinogenesis, whereas mice lacking EGFR in hepatocytes unexpectedly develop more HCC due to increased hepatocyte damage and compensatory proliferation. Mechanistically, following IL-1 stimulation, EGFR is required in liver macrophages to transcriptionally induce IL-6, which triggers hepatocyte proliferation and HCC. Importantly, the presence of EGFR-positive liver macrophages in HCC-patients is associated with poor survival. This study demonstrates a tumor-promoting mechanism for EGFR in non-tumor cells, which could lead to more effective precision medicine strategies.
INTRODUCTION
HCC is the third most common cause of cancer-related mortality worldwide1. Main risk factors for HCC include hepatitis B or C infection, alcoholic liver injury, non-alcoholic steatohepatitis, environmental carcinogens and hereditary metabolic diseases2, which can lead to chronic hepatitis or cirrhosis, conditions regarded as preneoplastic stages3. Current treatment options are limited, which may be due to the lack of biomarkers for patient stratification, since there is limited use of biopsies for HCC diagnosis, which is largely based on radiological criteria4. Therefore, a better understanding of the mechanisms driving HCC development is needed.
Persistent infections, activation of liver-resident macrophages (Kupffer cells) and recruitment of inflammatory cells can lead to chronic inflammation5-8 accompanied by many factors favoring HCC development9. The molecular link between inflammation and HCC is not completely understood. Cytokines such as IL-1 and IL-6 play a central role in liver carcinogenesis. IL-6 is produced by Kupffer cells following stimulation with IL-1, which is released by dying hepatocytes10,11. IL-6 is responsible for compensatory proliferation of damaged hepatocytes leading eventually to HCC development12. Many signaling pathways involved in HCC development, such as MyD88, JNK1/2, p38α and IKKβ, can regulate hepatic IL-6 production, although the precise mechanism is unclear. Moreover, JNK1, p38 and IKKβ have been shown to be involved in human HCC13-15 and their deletion in parenchymal versus non-parenchymal cells can differentially affect hepatocarcinogenesis in mice7,10,12,13,16-19.
EGFR overexpression, which occurs in 40-70% of human HCCs, has been linked with tumorigenesis20. Elevated expression of the EGFR ligand TGFα has been reported in preneoplastic lesions suggesting a role in early HCC21. EGFR antagonists were effective in human HCC cells and in a rat HCC model22,23. In clinical trials with unselected patients, Erlotinib has shown moderate effects in Phase II, whereas Gefitinib and Cetuximab have provided only disappointing results in advanced stage HCC patients2. Moreover, the SEARCH trial, the only Phase III study performed, was unable to show survival improvement with Erlotinib in advanced stage HCC24. Therefore, a better understanding of the mechanisms whereby EGFR signaling influences HCC progression is needed. Genetically modified mouse models represent an invaluable tool to dissect the interplay between tumor and stromal cells during HCC development and to identify important signaling pathways in the respective cell types. In this study, we employed mice lacking EGFR in different cell types of the liver to dissect the role of different cellular players and signaling pathways in HCC development.
RESULTS
HCC formation in mice lacking EGFR in all liver cells
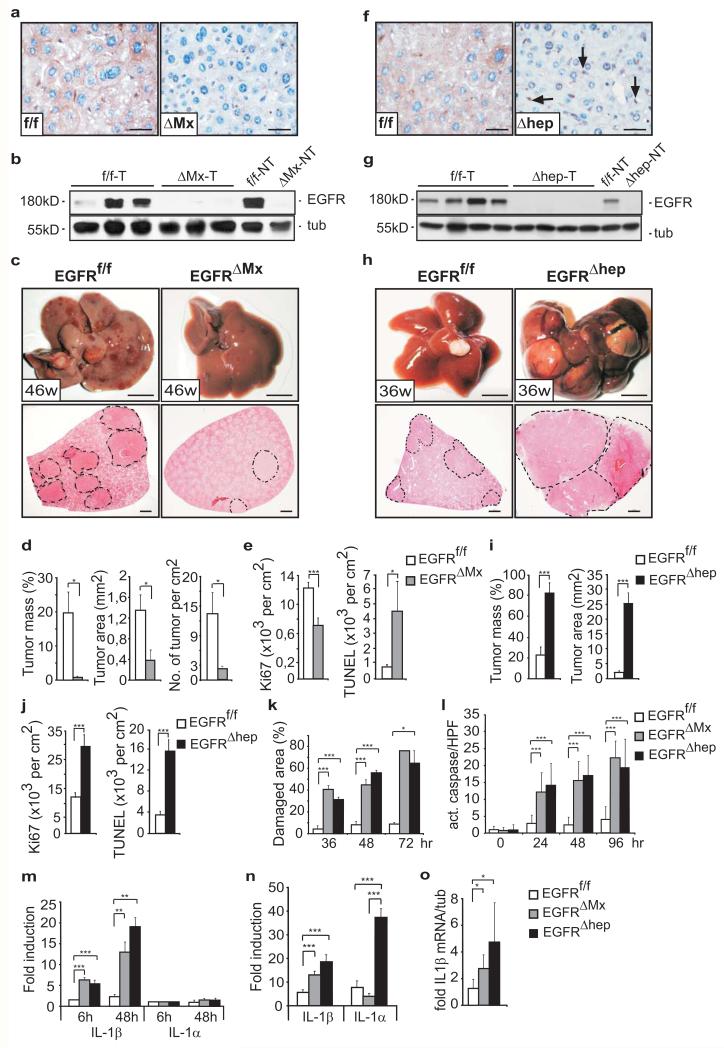
To investigate the function of EGFR during HCC formation, we employed polyinosinic-polycytidylic acid (pIpC)-inducible Mx-Cre transgenic mice (EGFRf/f;Mx-Cre=EGFRΔMx), which delete the EGFR in parenchymal and non-parenchymal liver cells as well as several other organs25. We used the diethylnitrosamine/phenobarbital (DEN/PB) protocol26 to induce HCC (Supplementary Fig.1a), which occurs similar to the human disease. DEN-damaged hepatocytes undergo apoptosis and are replaced via compensatory proliferation of surviving hepatocytes, which can give rise to HCC if mutated by DEN27. Complete EGFR deletion in EGFRΔMx tumor and non-tumor tissue was confirmed by immunohistochemistry, Southern and Western blot analysis (Fig.1a,b, Supplementary Fig.1b). By 46 weeks, EGFRf/f livers developed tumors, whereas EGFRΔMx livers showed a dramatic decrease in tumor mass, area, and number (Fig.1c,d). Analysis of EGFRΔMx mice revealed a significant decrease in proliferation and an increase in apoptosis in HCC (Fig.1e), but not in adjacent non-tumor tissue (Supplementary Fig.1c). These results suggest that EGFR in liver cells promotes HCC formation by protecting them from DEN-induced apoptosis.
Figure 1. HCC formation in mice lacking EGFR in hepatocytes or all liver cells.
(a) EGFR staining on tumor sections of EGFRf/f and EGFRΔMx livers. Scale bar: 100μm. (b) Western Blot of tumor (T) and non-tumor (NT) tissue. (c) Representative livers (top, scale bar: 1cm) and haematoxylin and eosin (H&E) stainings of liver sections (bottom, scale bar: 1mm). (d) Tumor mass (left), area (middle), and number (right) of EGFRf/f (n=8) and EGFRΔMx (n=10) mice. Two pooled independent experiments. (e) Ki67-positive (left, EGFRf/f: n=103; EGFRΔMx: n= 27 and TUNEL-positive cells (right, EGFRf/f: n=153; EGFRΔMx: n=49) in tumours. n=High Power Field (HPF). 6 mice per genotype were analyzed. (f) EGFR-staining on liver tumor sections of EGFRf/f and EGFRΔhep mice. Arrows point to EGFR-expressing non-parenchymal cells. Scale bar: 100μm. (g) Western Blot of tumor (T) and non-tumor tissue (NT). (h) Representative livers (top, scale bar: 1cm) and H&E stainings of liver sections (bottom, scale bar: 1mm). Dotted lines in c and h mark tumor nodules. (i) Tumor mass (left) and area (right) of EGFRf/f (n=12) and EGFRΔhep (n=14) mice. Two pooled independent experiments. (j) Ki67-positive (left, EGFRf/f: n=26; EGFRΔhep: n=68) and TUNEL-positive cells (right, EGFRf/f: n=19; EGFRΔhep: n=48) in tumors. n=HPF, 3 EGFRf/f and 4 EGFRΔhep mice were analyzed. (k) Damaged liver areas after DEN intoxication. 36hrs (EGFRf/f: n=5, EGFRΔhep: n=4, EGFRΔMx: n=4), 48hrs (EGFRf/f: n=4, EGFRΔhep: n=3, EGFRΔMx: n=3), and 72hrs (EGFRf/f: n=4, EGFRΔhep: n=3, EGFRΔMx: n=2). n=mice (l) Caspase-3-positive cells after DEN intoxication. 0hrs: EGFRf/f : n=23 (6 mice), EGFRΔhep: n=23 (6 mice), EGFRΔMx: n=23 (5 mice), 24hrs: EGFRf/f : n=19 (4 mice), EGFRΔhep: n=10 (2 mice), EGFRΔMx: n= 20 (4 mice), 48hrs: EGFRf/f : n=5 (1 mouse), EGFRΔhep: n=12 (2 mice), EGFRΔMx: n=15 (3 mice), 96hrs: EGFRf/f : n=32 (6 mice), EGFRΔhep: n=21 (3 mice), EGFRΔMx: n=6 (1 mouse). n=HPF (m) qRT-PCR showing IL-1β and IL-1α expression in isolated hepatocytes after DEN intoxication in vivo. (n=3 mice per genotype and time point). (n) qRT-PCR for IL-1β and IL-1α of total liver (n=6 mice per genotype) 24 hours after DEN intoxication. Two pooled independent experiments. (o) qRT-PCR for IL-1β in liver tumors of EGFRf/f (n=5), EGFRΔMx (n=3) and EGFRΔhep (n=5) mice. Data (d, e, i, j) represent mean±s.e.m. Data (k-o) represent mean±s.d. Student‘s t-test for independent samples and unequal variances was used to assess statistical significance (*p<0,05, **p<0,01, ***p<0,001). Original data are provided in Supplementary Table 1 and uncropped blots in Supplementary Fig. 6.
HCC formation in mice lacking EGFR in parenchymal cells
Given the complexity of HCC and the involvement of different cell types, we induced HCC in mice lacking EGFR specifically in hepatocytes and bile duct cells (EGFRf/f;Alfp-Cre=EGFRΔhep)25(Supplementary Fig.1a). Absence of EGFR in parenchymal cells was confirmed by immunohistochemistry and Western blot analysis in tumor and non-tumor tissue (Fig.1f,g). In EGFRΔhep tumors, EGFR was detectable in non-parenchymal cells (Fig.1f), where Alfp-Cre is not expressed, which explains the unrecombined flox allele in the Southern blot (Supplementary Fig.1b). In direct contrast to EGFRΔMx mice, EGFRΔhep mice developed significantly larger HCCs than littermate control mice (Fig.1h,i). This result was unexpected, as EGFR is believed to be tumor-promoting. Proliferation in EGFRΔhep tumors was increased, contrary to what was observed in EGFRΔMx tumors (Fig.1j). However, similar to EGFRΔMx, EGFRΔhep tumors showed increased apoptosis (Fig.1j), suggesting that EGFR protects hepatocytes against DEN-induced apoptosis.
To investigate this in more detail, we monitored liver damage at different time points after DEN injection. Serum aspartate transaminase (AST) and alanine transaminase (ALT) levels, markers of acute liver toxicity, were significantly increased in EGFRΔMx and EGFRΔhep livers compared to controls (Supplementary Fig.1e,f). Moreover, after DEN injection, damaged areas were also significantly increased (Fig.1k, Supplementary Fig.1d) and higher levels of cleaved caspase 3 were detected in Zone III of the liver lobule, where DEN is mainly metabolized28 (Fig.1l, Supplementary Fig.1g). To investigate if this hepatocyte damage was cell-autonomous, we stimulated isolated hepatocytes with DEN, and performed immunofluorescent staining for the necrotic marker high-mobility group box 1 (HMGB1)29, which revealed a strong necrotic response specifically in EGFR-deficient hepatocytes (Supplementary Fig.1h). They were also more sensitive to apoptosis after TNFα/cycloheximide treatment (Supplementary Fig.1i). Thus, apoptosis and necrosis are responsible for increased DEN-induced liver damage in EGFRΔMx and EGFRΔhep mice, revealing that EGFR fulfills an important death-protecting function in hepatocytes.
During toxic hepatic injury, pro-inflammatory cytokines such as IL-1 and IL-6 activate an inflammatory repair process11. qRT-PCR analysis of hepatocytes isolated after DEN injection revealed a significant increase in IL-1β expression in EGFRΔMx and EGFRΔhep mice, whereas IL-1α expression was low and unchanged (Fig.1m). In contrast, when IL-1α/β levels were measured in total livers after DEN treatment, we observed that IL-1α was higher than IL-1β expression (Fig.1n), which confirms previous results11. Together, these results suggest that IL-1α is mainly released by non-parenchymal cells, possibly Kupffer cells/liver macrophages, and not by damaged hepatocytes. Increased IL-1β expression was also detected in EGFRΔMxand EGFRΔhep tumors (Fig.1o). Moreover, increased IL-1β release was observed when EGFR-deficient hepatocytes were treated in vitro with DEN or TNFα (Supplementary Fig.2a,b). Thus, hepatoprotection via EGFR signaling prevents continuous hepatocyte death and IL-1β release in HCC.
In both EGFRΔMx and EGFRΔhep mice, EGFR protects cells from DEN-induced damage, but still EGFR plays opposing roles in promoting HCC formation illustrating its complex role in tumorigenesis. To exclude that timing of EGFR deletion (late gestation in EGFRΔhep mice versus 7-week old mice in EGFRΔMx mice) is responsible for the differences in HCC development between the two models, we deleted EGFR shortly after birth in EGFRf/f;Mx-Cre mice (EGFRΔMx*) (Supplementary Fig.1a, 2c). Similar to EGFRΔMx mice, also EGFRΔMx* mice developed significantly less and smaller tumors than their littermate controls (Supplementary Fig.2d,e), suggesting that the cell types, in which EGFR is deleted, account for the differences in HCC development between EGFRΔMx and EGFRΔhep mice.
EGFR expression in Kupffer cells/liver macrophages promotes HCC development
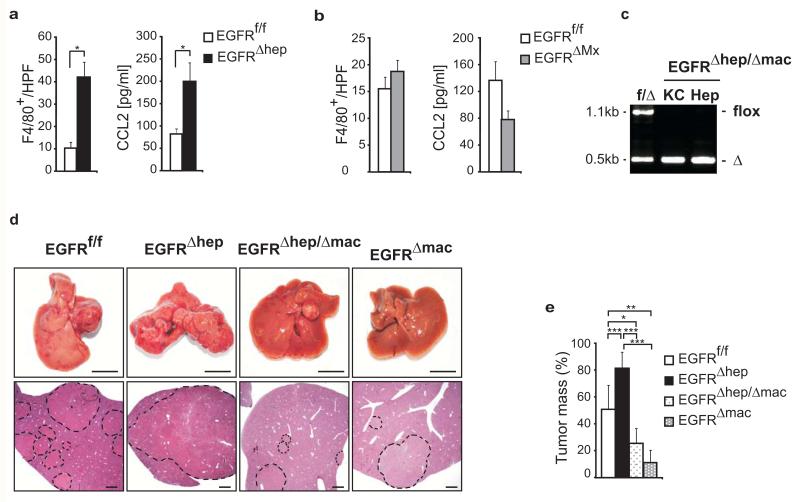
We hypothesized that the difference in HCC formation between EGFRΔhep and EGFRΔMx mice was caused by EGFR function in non-parenchymal cells. We therefore performed immunohistochemistry for non-parenchymal cell markers and observed a 4-fold increase of F4/80-positive cells, which could be Kupffer cells or infiltrating macrophages in EGFRΔhep tumors (Fig.2a,b). There was significant upregulation of serum CCL2, a chemokine known to attract F4/80-positive cells, in EGFRΔhep, but not in EGFRΔMx tumors (Fig.2a,b). To test if EGFR-expressing liver macrophages could contribute to increased HCC formation in EGFRΔhep mice, we employed LysM-Cre transgenic mice to generate EGFRΔhep/Δmac (EGFRf/f;Alfp-Cre;LysM-Cre) mice, which lack EGFR in both hepatocytes and Kupffer cells/macrophages (Fig.2c). Similar to EGFRΔMx, EGFRΔhep/Δmac mice showed significantly smaller tumors compared to control and EGFRΔhep mice (Fig.2d,e). Thus, increased HCC formation in EGFRΔhep mice is caused by increased numbers of EGFR-expressing Kupffer cells/infiltrating macrophages. Finally, to test whether EGFR-expressing macrophages are responsible for HCC formation, we induced HCC in mice lacking EGFR only in macrophages (EGFRf/f;LysM-Cre=EGFRΔmac). Similar to EGFRΔMx and EGFRΔhep/Δmac mice, EGFRΔmac mice displayed significantly smaller tumors when compared to the respective controls (Fig.2d,e). Thus, our results reveal an unexpected tumor-promoting role for EGFR in Kupffer cells/liver macrophages during HCC formation.
Figure 2. EGFR expression in Kupffer cells/liver macrophages promotes HCC development.
(a, b) Numbers of F4/80+ cells in tumors of mice (left, EGFRf/f: n=66 (4 mice), EGFRΔhep: n=54 (4 mice), EGFRf/f: n=37 (3 mice), EGFRΔMx: n=36 HPF (4 mice)) and CCL2 serum levels in HCC mice (right, EGFRf/f: n=4, EGFRΔhep: n=7, EGFRf/f: n=4 and EGFRΔMx: n=6 mice). (c) Representative PCR showing EGFR deletion in isolated hepatocytes (Hep) and Kupffer cells (KC) of control (f/Δ) and EGFRΔhep/Δmac mice. flox = not deleted (1.1kb) and Δ = deleted EGFR (0.5kb). (d) Representative livers (top, scale bar: 1cm) and H&E stainings of sections of indicated genotypes (bottom, scale bars: 1mm) 63 weeks after tumor initiation. Dotted lines mark tumor nodules. Note: Tumors of EGFRf/f mice are bigger than in Fig. 2c, d, because the tumors were analyzed 27 weeks later due to a change in the genetic background of the mice. (e) Tumor mass in livers of EGFRf/f (n=10), EGFRΔhep (n=5), EGFRΔhep/Δmac (n=5) and EGFRΔmac (n=4) mice. Data (a, b) represent mean±s.e.m. Data (e) represent mean ±s.d. Student‘s t-test for independent samples and unequal variances was used to assess statistical significance (*p<0,05, **p<0,01, ***p<0,001). Original data are provided in Supplementary Table 1 and uncropped gel in Supplementary Fig. 6.
EGFR expression is induced in activated Kupffer cells/liver macrophages under pathological conditions
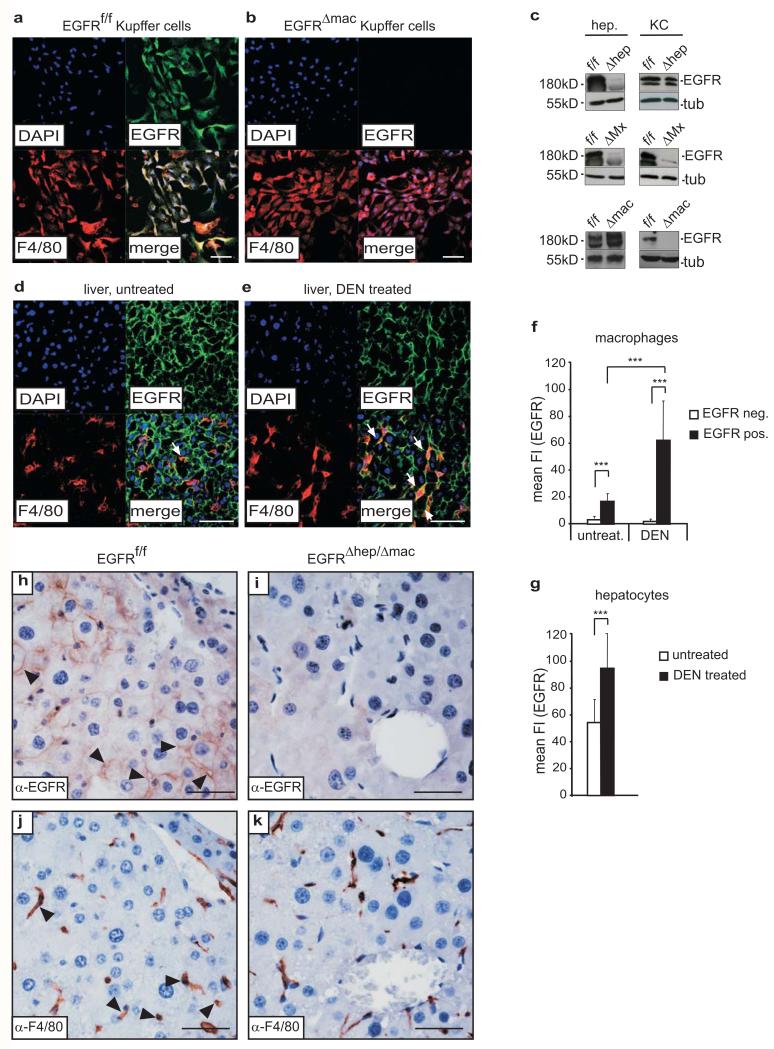
Immunofluorescent staining revealed that EGFR was expressed in Kupffer cells/macrophages isolated from EGFRf/f livers and stimulated with IL-1β in vitro (Fig.3a), a finding that could be confirmed by Western Blot (Fig.3c). As expected, no EGFR expression was detected in Kupffer cells isolated from EGFRΔmac and EGFRΔMx mice (Fig.3b,c). EGFR levels in EGFRΔhep Kupffer cells were similar to control cells, whereas EGFR was present in hepatocytes isolated from EGFRΔmac, but not from EGFRΔhep and EGFRΔMx mice (Fig.3c). Healthy and untreated EGFRf/f livers contained only very few EGFR-positive Kupffer cells (Fig.3d,f). However, 5 days after DEN injection, prominent EGFR expression was detected in F4/80-positive cells (Fig.3e,f). Interestingly, increased levels of EGFR were also present in hepatocytes following DEN treatment (Fig.3g). We next investigated whether EGFR was induced in Kupffer cells/liver macrophages of HCCs. Immunohistochemistry on serial sections of EGFRf/f and EGFRΔhep/Δmac tumors revealed co-expression of EGFR and F4/80 in tumor and adjacent tissue of EGFRf/f (Fig.3h,j), but not of EGFRΔhep/Δmac mice (Fig.3i,k). These findings suggest that EGFR expression is induced in activated Kupffer cells/liver macrophages under pathological conditions.
Figure 3. EGFR expression is induced in activated Kupffer cells/liver macrophages under pathological conditions.
(a-b) Representative immunofluorescent confocal image showing co-staining for F4/80 and EGFR in cultured Kupffer cells/liver macrophages isolated from (a) EGFRf/f and (b) EGFRΔmac livers and stimulated with IL-1β for 24 h. Cultures contained ≥ 98% Kupffer cells/liver macrophages as confirmed by F4/80 staining. Scale bar: 50μm. (c) Representative Western Blot showing EGFR expression in isolated hepatocytes and Kupffer cells of EGFRf/f, EGFRΔMx, EGFRΔhep and EGFRΔmac mice. (d-e) Representative immunofluorescent confocal images showing F4/80 and EGFR expression in liver sections of (d) untreated and (e) DEN treated (5 days) EGFRf/f mice. White arrows indicate EGFR-positive Kupffer cells. Scale bar: 50μm. (f-g) Mean fluorescence intensity (mean FI) showing EGFR expression levels (Alexa 488, green) in (f) liver macrophages (EGFRf/f untreated: EGFR negative (n=9), EGFR positive (n=10); EGFRf/f 5 days after DEN: EGFR negative (n=4), EGFR positive (n=26) and (g) hepatocytes (EGFRf/f untreated (n=12), EGFRf/f 5 days after DEN (n=13). Analysis of stainings shown in (d) and (e). Two pooled independent experiments for (f) and (g). (h-k) Representative anti-EGFR (h, i) and anti-F4/80 (j, k) staining performed on serial sections of control (h, j) and EGFRΔhep/Δmac (i, k) HCC showing EGFR expression in tumor cells and co-expression of EGFR and F4/80 in Kupffer cells/liver macrophages of EGFRf/f HCC and no EGFR expression in EGFRΔhep/Δmac tumors. Scale bar: 50μm. (a-b, d-e) Nuclei (DAPI, blue), EGFR (Alexa 488, green) and F4/80 (Alexa 594, red), merge (bottom right). Data (f-g) represent mean±s.d. Student‘s t-test for independent samples and unequal variances was used to assess statistical significance (*p<0,05, **p<0,01, ***p<0,001). Original data are provided in Supplementary Table 1 and uncropped blots in Supplementary Fig. 6.
The presence of EGFR-expressing Kupffer cells/liver macrophages in human HCC correlates with poor prognosis
We next investigated if EGFR expression in Kupffer cells/liver macrophages is relevant for human HCC and analyzed EGFR expression in 129 surgically resected HCC samples, predominantly Hepatitis B (HBV)-positive from China, in the respective adjacent non-cancerous tissue, and in 15 “normal” livers (Table 1a,b). Since chronic HBV is a less common cause of HCC in Europe, we additionally investigated EGFR expression in 108 European HCC patients, who underwent liver transplantation for HCC (Table 1a). This cohort had predominantly alcohol-, non-alcoholic steatohepatitis- and Hepatitis C (HCV)-induced liver cirrhosis (Table 1b). EGFR staining for both cohorts was examined and blindly scored by two independent pathologists using a previously published scale30.
Table 1(a). EGFR expression in hepatocytes and Kupffer cells of the Chinese (n=129 patients) and European (in brackets) (n=108 patients) cohort.
| Hepatocytes | Kupffer cells | |||||
|---|---|---|---|---|---|---|
| EGFR expression | HCC | Tissue adjacent to carcinoma | Normal liver | HCC | Tissue adjacent to carcinoma | Normal liver |
| 0 | 70/129 (69/108) |
83/129 (57/108) |
11/15 | 71/129 (71/108) |
114/129 (76/104*) |
15/15 |
| + | 35/129 (31/108) |
30/129 (40/108) |
3/15 | 33/129 (36/108) |
8/129 (27/104*) |
0/15 |
| ++ | 18/129 (6/108) |
13/129 (7/108) |
1/15 | 20/129 (1/108) |
5/129 (1/104*) |
0/15 |
| +++ | 6/129 (2/108) |
3/129 (0/108) |
0/15 | 5/129 (0/108) |
2/129 (0/104*) |
0/15 |
In 4 patients, no adjacent tissue was available
Table 1(b). Relationship between EGFR-positivity in Kupffer cells and the clinicopathological characteristics.
| Variable | EGFR expression in Kupffer cells | ||
|---|---|---|---|
|
0
China: n=71 EU: n= 49 * |
+, ++, +++ n=58 n=59** |
P
value |
|
|
Age
China: median years (range) EU: median years (range) |
49 (30-78) 57 (33-68) |
49 (21-68) 55 (28-67) |
0.8646# 0.349# |
|
Sex
China: , M:F EU: M:F |
65:6 40:9 |
51:7 50:9 |
0.4971 0.666 |
|
AFP
China: (μg/L), <20:≥20 EU: |
42:29 n.d. |
16:42 n.d. |
0.0004 n.d. |
|
Etiology:
China: HBsAg, positive:negative EU: alc:viral:other |
55:16 15:25:9 |
53:5 18:28:13 |
0.0332 0.884 |
|
Diameter
China: <3cm:3-5cm:>5cm EU: median cm (range) |
31:14:26 2.5 (1-13) |
13:21:24 4.5 (1-24) |
0.0222 <0.001# |
|
Microvascular Invasion
China: yes:no EU: yes:no |
11:60 7:42 |
18:40 19:40 |
0.0354 0.030 |
|
TNM staging
China: I : II : III : (IV) EU: I : II : III : (IV) |
60:8:3 11:12:26:0 |
38:12:8 2:12:44:1 |
0.0339 0.008 |
|
Recurrence of HCC,
China: no:yes EU: no:yes |
45:26 49:0 |
22:36 29:30 |
0.0040 <0.001 |
AFP: alpha-Fetoprotein, TNM: classification of malignant tumors (Tumor, Node, Metastasis);
includes samples where tumor and tissue adjacent to carcinoma were negative for EGFR in Kupffer cells;
includes samples where either tumor or tissue adjacent to carcinoma (or both) were positive for EGFR in Kupffer cells;
Mann-Whitney test
In the Chinese cohort, about 73% of normal livers were negative (0) for EGFR expression in hepatocytes and there was no difference in EGFR expression levels (+, ++, +++) among tumor cells and hepatocytes of the adjacent non-cancerous tissue and normal livers (Table 1a, Supplementary Fig.3a). High expression of EGFR (++, +++) was more prevalent in HCC compared to normal tissue (Table 1a, Supplementary Fig.3a), which confirms previous reports20. Similar results were observed for EGFR expression in hepatocytes and tumor cells of the European cohort (Table 1a). For both cohorts, EGFR expression in tumor cells did not show significant prognostic value for patient’s overall (OS) or disease-free survival (DSF) after surgery (Supplementary Fig.3b-e, Table 1a) and no relationship between EGFR expression in hepatocytes and clinico-pathological characteristics was found.
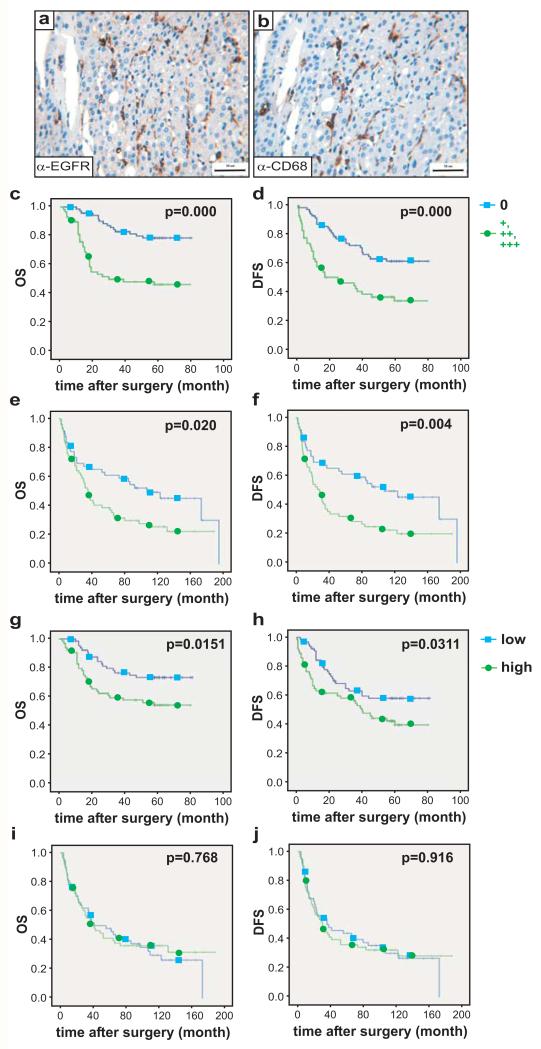
To analyze EGFR expression in liver macrophages we stained adjacent tissue sections for EGFR and the macrophage marker CD68. In “normal” liver tissue, all CD68-positive cells did not express EGFR (Table 1a). In contrast, 45% of Chinese and 34% of European samples harbored EGFR-expressing CD68-positive cells in the tumors ranging from + to +++ (Fig.4a,b, Supplementary Fig.3f, Table 1a). EGFR-expressing macrophages were also present within the non-cancerous tissue adjacent to the carcinoma: 12% for the Chinese and 27% for the European cohort. The higher number of EGFR-positive macrophages in adjacent tissue of the European samples might reflect the more advanced cirrhosis in these livers. The specificity of EGFR and CD68 co-expression was confirmed by co-staining in fresh frozen human HCC samples (Supplementary Fig.3g). EGFR-expressing macrophages in HCCs were associated with poor clinical outcome of Chinese patients after surgical tumor resection and of European patients after liver transplantation mirrored by significantly reduced OS and DFS (Fig.4c-f). α-fetoprotein (AFP), a HCC tumor marker, was significantly higher in HCC patients with EGFR-positive macrophages (Table 1b). In the European cohort, EGFR-positive macrophages were associated with more aggressive tumors and patients suffering from HCC recurrence after liver transplantation had EGFR-positive macrophages in their tumors (Table 1b). To analyze whether total numbers of CD68-positive cells – regardless of EGFR expression – had an impact on HCC prognosis, we grouped patient samples into low and high macrophage counts considering the median of the respective patient cohorts as cut-off. For the European cohort, the number of Kupffer cells/liver macrophages alone was not predictive for OS and DFS (Fig.4i-j). However, total numbers of macrophages negatively correlated with OS and DFS of Chinese patients (Fig.4g-h). Collectively, these data demonstrate that it is not the overall number of liver macrophages, but the number of EGFR-positive liver macrophages present in HCC, which is predictive for OS and DFS.
Figure 4. EGFR expression in Kupffer cells of HCC patients correlates with poor prognosis.
(a-b) Representative EGFR and CD68 staining (brown) on serial sections revealing Kupffer cells in human HCC samples. Scale bar: 50μm. (c-f) Overall survival (OS, c, e) and disease free survival (DFS, d, f) of HCC patients with (+, ++, +++) or without (0) EGFR expression in Kupffer cells/liver macrophages of the Chinese (c, d: 129 patients (n=71 negative for EGFR; n=58 positive for EGFR) and European cohort (e, f: 108 patients (n=49 negative for EGFR; n=59 positive for EGFR). (g-j) Overall survival (OS, g, i) and disease free survival (DFS, h, j) of HCC patients with low or high numbers of Kupffer cells/liver macrophages in tumors in the Chinese (g, h: 129 patients: n=52 with low and n=77 with high counts) and European cohort (i, j: 108 patients: n=50 with low and n=58 with high counts). The cut-off value for defining high and low was the median. For the respective patient cohorts, low was classified as values below or at the 50th percentile and high was classified as values above the 50th percentile. Scoring system: 0=negative staining (0%-10% positive), 1=weak signal (10%-20% positive), 2=intermediate signal (20%-50% positive) and 3=strong signal (>50% positive) as previously described30. Log-rank test was used to assess statistical significance.
EGFR-deficient Kupffer cells fail to produce IL-6
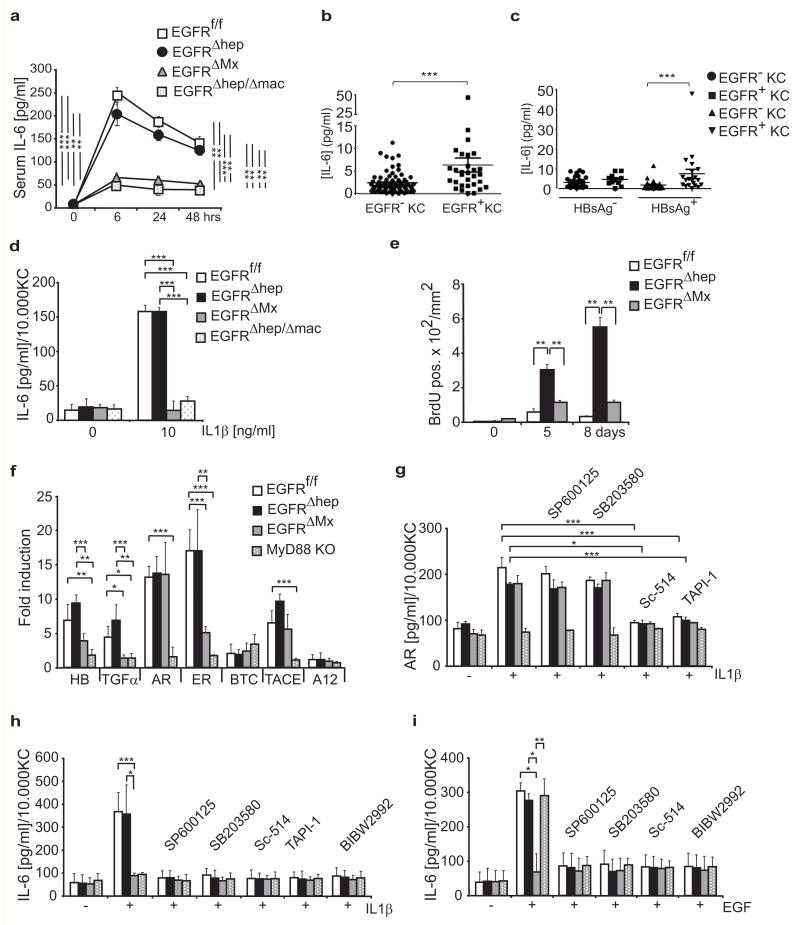
We next analyzed the mechanism whereby EGFR signaling in macrophages promotes tumorigenesis. IL-6 is produced at high levels by Kupffer cells in response to IL-1 derived from damaged hepatocytes, and stimulates compensatory hepatocyte proliferation through IL-6R activation10,11. We found IL-6 serum levels to be strongly induced after DEN injection in EGFRf/f and EGFRΔhep mice, but not in EGFRΔMx and EGFRΔhep/Δmac mice, which are deficient for EGFR in macrophages (Fig.5a). Importantly, we also found significantly higher IL-6 levels in the plasma of Chinese HCC patients displaying EGFR-positive Kupffer cells in their tumors (Fig.5b). This occurred in association with HBV infection (Fig.5c), suggesting that infections and inflammatory conditions lead to upregulation of EGFR in Kupffer cells with consequent increased IL-6 production. For the European cohort, patient plasma was not available. Thus, upon DEN-induced liver damage, EGFR in Kupffer cells/liver macrophages is required to induce expression of IL-6.
Figure 5. EGFR-dependent IL-6 production and release.
(a) ELISA showing IL-6 serum levels in EGFRf/f, EGFRΔhep, EGFRΔMx, and EGFRΔhep/Δmac mice (n=3 for each genotype and time point) 6, 24, and 48hrs after DEN injection in vivo. (bc) IL-6 plasma levels in HCC patients (n=104) of the Chinese cohort grouped according to (b) the presence of EGFR-positive (n=31) or EGFR-negative (n=73) Kupffer cells in tumors or additionally considering (c) positivity for hepatitis B surface antigen (HBsAg). HBsAg negative/EGFR negative (n=37); HBsAg negative/EGFR positive (n=11); HBsAg positive/EGFR negative (n=36) and HBsAg positive/EGFR positive (n=20). (d) IL-6 release into the supernatant by cultured Kupffer cells 24 hours after incubation with IL-1β in vitro. (n=3 Kupffer cell isolates per genotype). (e) Quantification of BrdU positive cells in liver sections of mice of the indicated genotypes 5 and 8 days after DEN injection (sum of counted HPF of n=3 mice for each genotype and time point). (f) qRT-PCR showing expression of heparin-binding EGF-like growth factor (HB-EGF) (HB), TGFα, AR, epiregulin (ER), BTC, ADAM17 (also known as TACE) and ADAM12 (A12) in Kupffer cells after IL-1β stimulation in vitro (n=4 Kupffer cell isolates per genotype and condition). Two pooled independent experiments. KO, knockout. (g) AR release into the supernatant of cultured Kupffer cells 24 hours after IL-1β stimulation in the presence of the indicated inhibitors. (n=3 Kupffer cell isolates per genotype, each n is the average of 3 technical replicates). Three pooled independent experiments. (h, i) IL-6 release by cultured Kupffer cells 24 hours after incubation with (h) IL-1β (n=3 Kupffer cell isolates per genotype, each n is the average of 3 technical replicates, three pooled independent experiments) or (i) EGF (n=3 Kupffer cell isolates per genotype, each n is the average of 3 technical replicates, three pooled independent experiments) in the presence of the indicated inhibitors. (g-i) Inhibitors: JNK=SP600125, p38=SB203580, IKK2=Sc-514, TACE=TAPI-1, EGFR=BIBW2992. Data (a-i) represent mean±s.d. Student‘s t-test for independent samples and unequal variances was used to assess statistical significance (*p<0,05, **p<0,01, ***p<0,001). Original data are provided in Supplementary Table 1.
To further investigate whether IL-1 can induce IL-6 production in liver macrophages in an EGFR-dependent manner, we quantified IL-6 levels after incubation of isolated Kupffer cells/liver macrophages with IL-1β in vitro. IL-1β was able to induce IL-6 secretion in EGFR-expressing, but not in EGFR-deficient liver macrophages (Fig.5d). IL-6 production in Kupffer cells could be prevented by treatment with EGFR inhibitors in a dose-dependent manner (Supplementary Fig.4a,b). Inflammatory cytokines such as IL-17A, IL-22 and IL-23 were not detectable in the supernatants of EGFRf/f, EGFRΔMx as well as MyD88−/− Kupffer cells following IL-1β or EGF stimulation (Supplementary Fig.4d). IL-6 production was comparable between EGFR-expressing and -deficient Kupffer cells after stimulation with Toll-Like Receptor (TLR) agonists such as PolyIC, Imiquimod and LPS (Supplementary Fig.4c) showing that EGFR-deficient Kupffer cells are not intrinsically impaired in IL-6 production. Thus, EGFR-deficient Kupffer cells cannot produce IL-6 in response to IL-1, indicating that EGFR-dependent IL-6 production is downstream of IL-1R signaling.
Since IL-6 stimulates compensatory proliferation, we next analyzed hepatocyte proliferation after DEN treatment in mice (Fig.5e). The strongest BrdU incorporation was observed in EGFRΔhep livers likely because DEN-induced damage is high (because hepatocytes lack EGFR) and EGFR-expressing Kupffer cells/liver macrophages produce IL-6 to stimulate proliferation. In contrast, proliferation was lower in EGFRΔMx and control livers, likely because of impaired IL-6 production by EGFR-negative Kupffer cells/liver macrophages, and less severe DEN-induced damage in control (EGFR-expressing) hepatocytes, respectively (Fig.5e). Together, these results show that increased compensatory proliferation correlates with increased IL-6 levels and increased HCC formation.
Mechanism of IL-1β-induced IL-6 production by EGFR
To investigate the molecular mechanism by which EGFR signaling leads to IL-1-induced IL-6 production in Kupffer cells, we measured EGFR ligand expression following IL-1β stimulation and found that except for betacellulin (BTC), all other EGFR ligands were significantly induced (Fig.5f). TACE/ADAM17, a metalloprotease proteolytically releasing EGFR ligands31, was also induced by IL-1β (Fig.5f). TACE and EGFR ligands were not expressed in MyD88−/− Kupffer cells, indicating that their induction is under direct control of IL-1R signaling (Fig.5f). EGFR ligands and TACE were also induced by IL-1β in Kupffer cells lacking EGFR (EGFRΔMx) suggesting EGFR-independent transcriptional regulation (Fig.5f). Consistent with this, the release of amphiregulin (AR) into the culture medium of IL-1β stimulated Kupffer cells was significantly increased in both EGFR-expressing and -deficient Kupffer cells, but did not occur in MyD88−/− Kupffer cells or when inhibiting TACE (TAPI-1) (Fig.5g). AR release could be blocked by inhibiting IKK (Sc-514), but not by inhibiting JNK (SP600125) or p38 (SB203580) signaling (Fig.5g), demonstrating that release of AR occurs via IKK-dependent activation of TACE.
These results demonstrate that IL-1R signaling in Kupffer cells controls expression of EGFR ligands and TACE, which likely leads to EGFR activation and downstream IL-6 production. To test this, we analyzed IL-6 production in isolated Kupffer cells/liver macrophages in the presence of various inhibitors. TACE-1 and EGFR inhibitors blocked IL-1β induced release of IL-6 in EGFR-expressing Kupffer cells to a similar extent as observed in MyD88−/− and EGFR-deficient Kupffer cells (Fig.5h). This demonstrates a linear pathway from IL-1R signaling via MyD88, TACE, and EGFR ligands to EGFR signaling and IL-6 production. Based on these results, we hypothesized that direct stimulation of EGFR would induce IL-6 in Kupffer cells. Indeed EGF was equally potent as IL-1β in inducing IL-6 production in EGFR-expressing Kupffer cells (Fig.5h,i). Importantly, EGF, but not IL-1β, was able to fully restore IL-6 production in MyD88−/− Kupffer cells (Fig.5i). These results indicate that EGFR activation and IL-6 production are downstream of IL-1R/MyD88 signaling. Consistent with this, IL-6 production by Kupffer cells/liver macrophages induced by either EGF or IL-1β was prevented in the absence of EGFR. Pre-incubation of EGFR-positive liver macrophages with JNK-, p38- or IKK-inhibitors, also inhibited IL-6 production induced by either EGF or IL-1β, indicating the importance of JNK, p38 and NFκB signaling downstream of EGFR in mediating IL-6 production (Fig.5h,i).
To demonstrate activation of EGFR, we analyzed EGFR phosphorylation in isolated Kupffer cells. IL-1β stimulation was able to induce EGFR phosphorylation in EGFR-expressing Kupffer cells to a similar extent as EGF. This did not occur in EGFR-deficient (EGFRΔMx) Kupffer cells (Supplementary Fig.4e,f). IL-1β-induced EGFR transactivation was blocked by p38 inhibition but not by JNK or IKK inhibition, suggesting that activation of p38 is necessary for EGFR activation (Supplementary Fig.4e). A similar requirement for p38 for EGFR transactivation by LPS has been recently described32. JNK, p38, IKKα/β and NFκB phosphorylation following IL-1β stimulation of EGFR-expressing and -deficient Kupffer cells could be efficiently blocked by respective inhibitors (Supplementary Fig.4e). Except for IKK and NFκB, JNK and p38 were also activated following EGF stimulation of EGFRf/f Kupffer cells (Supplementary Fig.4f). Together, our data show that IL-1β stimulation of Kupffer cells leads to induction of EGFR ligands and ADAM17 with subsequent p38-dependent EGFR transactivation required for IL-6 production via JNK, p38 and IKK (Supplementary Fig.5).
DISCUSSION
EGFR is frequently overexpressed in human HCC, but its relevance for malignant progression is poorly understood. Our finding on the tumor-promoting role of EGFR in Kupffer cells might provide a possible explanation for the poor response of unstratified advanced stage HCC patients to EGFR targeted therapies. Based on our results we would predict that only HCC patients with EGFR expression in liver macrophages will show a therapeutic effect with EGFR inhibitors (provided that liver macrophages are targeted by EGFR inhibitors). If EGFR is expressed only in tumor cells of HCC we anticipate that EGFR inhibitors may even promote tumorigenesis, since our genetic results revealed that loss of EGFR in hepatocytes promotes HCC. Clinical follow-up studies are needed to re-evaluate the use of EGFR inhibitors in HCC, and to consider the possibility of targeting specific cell populations. At this stage, it is also possible that EGFR-expressing Kupffer cells play a tumor-promoting role only in the early stages of HCC development. Should this be true, patients with advanced stage HCC would likely not benefit from EGFR targeted therapies. However, patients with HBV or HCV infections might benefit from EGFR inhibitor treatment in early disease stages to prevent HCC development. It will therefore be interesting to explore the predictive power of EGFR-expression in Kupffer cells also in patients in more advanced disease stages. Treatment of patients with EGFR-positive Kupffer cells with EGFR inhibitors selectively targeting Kupffer cells could allow for improved HCC treatment in preselected patient populations.
Our results also highlight the complexity and provide mechanistic insights of EGFR signaling in hepatocarcinogenesis (Supplementary Fig.5). We show that EGFR plays a hepatoprotective role during DEN-induced liver damage, as absence of EGFR renders hepatocytes more susceptible to DEN-induced damage leading to increased IL-1β secretion and subsequent enhanced stimulation of Kupffer cells. This occurs in both mouse models lacking EGFR in parenchymal cells (EGFRΔhep and EGFRΔMx mice). However, IL-6 production in Kupffer cells is strictly dependent on EGFR expression and occurs in a bimodal way involving first IL-1R/MyD88 signaling followed by TACE/EGFR-L production and p38-dependent EGFR transactivation. In mice lacking EGFR only in hepatocytes (EGFRΔhep), IL-6 secretion and compensatory proliferation are elevated leading ultimately to increased HCC formation. Additional deletion of EGFR in Kupffer cells/macrophages (EGFRΔMx and EGFRΔhep/Δmac mice) impairs HCC development, despite increased liver damage, as EGFR-deficient Kupffer cells cannot produce IL-6 to stimulate compensatory proliferation (Supplementary Fig.5). In conclusion, we have discovered a crucial role for EGFR signaling in Kupffer cells/macrophages during inflammation-driven HCC formation, demonstrating that EGFR signaling plays a tumor-promoting function in non-tumor cells. Thus, EGFR-positive Kupffer cells might constitute a future prognostic marker and could potentially represent a target for HCC therapy.
METHODS
Mice and genotyping
EGFRf/f, EGFRΔhep, and EGFRΔMx mice have been previously described25. EGFRΔmac and EGFRΔhep/Δmac mice were generated by crossing EGFRf/f or EGFRΔhep mice to LysM-Cre33 transgenic mice. Male mice used in this study were kept in the facilities of the Medical University of Vienna in accordance with institutional policies and federal guidelines. Inducible EGFR deletion in EGFRΔMx mice was achieved by 3 consecutive intraperitoneal (i.p.) injections with pIpC (400μg) every third day in adult mice or by 3 consecutive i.p. injections with pIpC (150μg) at day 9, 11, and 13 after birth. To genotype Cre transgenic mice a forward primer (Cre-F (5′CAT ACC TGG AAA ATG CTT CTG TCC 3′) and a reverse primer situated in the Cre transgene (Cre-R (5′-CCCAGAAATGCCAGATTACG-3′) were combined. To genotype Alfp-Cre; LysM-Cre double transgenic mice, promoter specific primers for the particular transgenes (Alb (5′-GCAAACATACGCAAGGGATT-3′) or LysM (5′-GAGGGATGAAATTCCTGCAA-3′)) were combined with the Cre-R primer. The primers Δ-EGFR-F (5′-GCCTGTGTCCGGGTCTCGTCG-3′) and Δ-EGFR-R (5′-CAACCAGTGCACCTAGCCTGGC-3′) were used to detect deletion of EGFR (Δ-EGFR). The mice were of mixed 129/Sv × C57BL/6 × CBA/J genetic background and in all experiments EGFR expressing littermates (EGFRf/f or Cre+ or EGFR+/+) served as controls to the respective EGFR deleted mice. For tumor experiments, age of mice is indicated in the relevant results section and the figure legends. For all short-term experiments, mice were between 8 and 12 weeks of age.
Liver tumor induction in mice by DEN/PB
Liver tumors were induced by chemical carcinogenesis in male mice according to the scheme shown in Supplementary Fig. 1a. Mice were sacrificed when liver tumors were visible in EGFRf/f, EGFRf/+ or EGFR+/+ Cre+ littermate control mice, which occurred around 36 weeks in the Alfp-Cre, around 46 weeks in the Mx-Cre and around 63 weeks in the Alfp-Cre; LysMCre double transgenic background. The genetic background of the mice was mixed (C57BL/6 × 129/Sv × CBA/J), but varied between the different Cre lines thus explaining the difference in the timing of tumor development. For each experiment, we show EGFRf/f littermates as controls. To exclude Cre-mediated effects we confirmed that EGFRf/+ or EGFR+/+ Alfp-Cre, Mx-Cre, LysM-Cre mice developed the same defects as EGFRf/f controls. Liver injury after DEN injection (100mg/kg body weight) was determined by measuring the circulating transaminases AST/ALT (Reflotron, Roche) and by quantifying necrotic areas using H&E stained sections at the time points indicated.
Histology
Liver tissue was fixed in 4% paraformaldehyde for 24 hours, dehydrated, embedded in paraffin and sectioned (5μm). Sections were stained with haematoxylin and eosin (Sigma) for quantification of necrosis after DEN-induced damage and for quantification of liver tumors. Images were obtained with a Nikon eclipse 80i microscope and quantification was done by Adobe Photoshop CS4 (Adobe). Quantification of liver tumors was performed on two H&E sections per liver, which were at least 200μm apart as previously described25.
Immunohistochemistry and Immunofluorescence
Protocols for Ki67 staining, BrdU in situ detection, TUNEL and immunoblotting have been previously described34-37. In brief, for antigen retrieval, paraffin-embedded tissue was treated with Target Retrieval Solution (Dako) unless otherwise stated in the manufacturer’s instructions and further processed for immunohistochemistry according to the manufacturer’s recommendation. To analyze cell proliferation, mice were injected intraperitoneally with 100μg/g body weight of BrdU. Stainings for BrdU (Caltag, Burlingame, CA) and Ki67 were performed using the ABC staining kit (Vector Laboratories, Burlingame, CA). An in situ cell-death detection kit (Roche, Indianapolis, IN) was used for TUNEL staining. The number of positive cells was determined by manual counting of the indicated number of high-power fields. For immuonofluorescent staining, livers were embedded in optimal cutting temperature compound (Sakura) for frozen section preparation (4μm) and fixed according to the manufacturers’ suggestions for each respective antibody. For immunohistochemistry and immunofluorescent stainings, antibodies against the following antigens were used: EGFR (CST #4267; clone D38B1; 1/50), F4/80 (Serotec MCA497R, clone CI:A3-1; 1/100 and eBioscience 14-4801; clone BM8; 1/100), Ki67 (Novo Castra NCL-Ki67p; 1/1000), CD68 (abcam ab955; clone KP1; 1/200), HMGB1 (CST #3935; 1/100), active Caspase 3 (R&D Systems AF835; 1/2000). Secondary antibodies were purchased from Molecular Probes and Vector Laboratories. Confocal images were taken with a Zeiss-LSM 700 microscope and evaluated using the ZEN2010 software. For mean fluorescence intensity measurements, confocal images were analyzed with ImageJ. Single F4/80-positive cells were selected using the ellipsoid selection tool and analyzed for mean fluorescence intensity of EGFR expression (Alexa 488) by using the Histogram tool (only value of green channel was used).
Western Blot Analysis
Protein lysates were prepared according to standard protocols. Lysed protein was resuspended in denaturing protein-loading buffer. Proteins were separated by SDS-PAGE and transferred to PVDF membranes (Millipore). The following antibodies were used: pEGFR (Tyr1068 CST #3777; clone D7A5; 1/1000; Tyr1173 CST#4407; clone 53A5; 1/1000), EGFR (CST #4267; clone D38B1; 1/500), pJNK (CST #9255; clone G9; 1/2000), JNK (CST #9252; 1/1000), pp38 (CST #4631; clone 12F8; 1/1000), p38 (CST #9212; 1/1000), pIKKα/β (CST #2697; clone 16A6; 1/1000), IKKβ (CST #2370; clone 2C8; 1/1000), pNF-κB (CST #3033; clone 93H1; 1/1000), NF-κB (CST #3034; 1/1000), pStat3 (CST#9145; clone D3A7; 1/2000), Stat3 (sc-7179; 1/1000), Actin (Sigma A2066; 1/1000), α-Tubulin (Sigma T9026; clone DM1A; 1/500).
ELISA and MTT assay
Supernatants from Kupffer cell cultures were collected at the indicated time points after stimulation and ELISA kits were employed according to the manufacturer’s instructions. ELISA for IL-6, IL17A, IL22, IL23 (eBioscience), amphiregulin (R&D Systems), CCL2, IL-1β and IL-1α (BD Biosciences-Pharmigen) were performed. MTT assay (EZ4U, Biomedica) was performed to quantify the number of viable Kupffer cells in the wells. The cytokine value was normalized to the number of viable Kupffer cells.
RT-PCR
Total RNA from cultured hepatocytes, cultured Kupffer cells or livers and liver tumors was isolated with TRIzol Reagent (Invitrogen). cDNA synthesis was performed with SuperScript First-Strand Synthesis System (Invitrogen) according to the manufacturer’s instructions. qRTPCR reactions were carried out using SYBR Green Mix (Applied Biosystems), according to the manufacturer’s instructions, with primers detecting: IL-1β-F (GGGCCTCAAAGGAAAGAATC), IL-1β-R (TACCAGTTGGGGAACTCTGC), IL-1α-F (CACCTTACACCTACCAGAGTGATTTG), IL-1α-R (TGTTGCAGGTCATTTAACCAAGTG), IL-6-F (TTCCATCCAGTTGCCTTCTTGG), IL-6-R (TTCTCATTTCCACGATTTCCCAG), HB-EGF-F (ACCAGTGGAGAATCCCCTATAC), HB-EGF-R (GCCAAGACTGTAGTGTGGTCA), TGFα-F (TCTGGGTACGTGGGTGTTC), TGFα-R (ACAGGTGATAATGAGGACAGCC), AR-F (GGTCTTAGGCTCAGGCCATTA), AR-R (CGCTTATGGTGGAAACCTCTC), ERF (CACCGAGAAAGAAGGATGGA), ER-R (TCACGGTTGTGCTGATAACTG), BTC-F (GACGAGCAAACTCCCTCCT), BCT-R (ATCAAGCAGACCACCAGGAT), TACE-F (ACCACTTTGGTGCCTTTCGT), TACE-R (GTCGCAGACTGTAGATCCCT), ADAM12-F (AGACGTGCTGACTGTGCAAC), ADAM12-R (CCGTGTGATTTCGAGTGAGAGA), EGF-F (CCCAGGCAACGTATCAAAGT), EGF-R (GGTCATACCCAGGAAAGCAA), Tubulin-F (AGAAGCATGGGGAGGACTACA), Tubulin-R (GTCGTTGTTCATCACTGGCG). PCRs were performed on a 7500 Fast Real-Time PCR System (Applied Biosystems, California USA) under the following conditions: an initial incubation at 50°C for 20 s and 95°C for 10 min followed by 40 cycles of 95°C for 15 s, 54°C for 1 min. Relative quantification of RNA was calculated by ΔΔ Ct method. Omission of cDNA was used as a negative control.
Hepatocyte and Kupffer cell culture
Hepatocytes and Kupffer cells were isolated after liver perfusion according to previously published protocols25. In brief, livers were perfused at 7ml/min via the portal vein with perfusion buffer containing collagenase (Gibco). The resulting cell suspension was passed through a 70μm cell strainer (BD Falcon Biosciences). Cell fractions were separated by Percoll gradients. Cells were either directly used for further analysis or cultured as follows: Hepatocytes were plated onto collagen pre-coated cell culture dishes and cultured in HepatoZyme-SFM (Gibco) supplemented with 10% FCS, 2mM glutamine and 1% penicillin-streptomycin (P/S). Hepatocytes were either treated with the indicated amounts of DEN for 24 hours or with 10ng/ml TNFα (eBioscience) in the presence of cycloheximide (100μg/ml, Sigma) for 12 hours. Kupffer cells were plated onto uncoated cell culture dishes (50.000 per well for 96-well plates) in RPMI 1640 supplemented with 10% FCS, 2mM glutamine and 1% P/S for 24 hours followed by overnight starvation (in 0.5% FCS containing medium for cytokine induction; serum-free medium for signaling experiments). Kupffer cells were stimulated with IL-1β (10ng/ml, eBioscience), EGF (10ng/ml, Lonza), polyIC (20μg/ml, GE Healthcare), Imiquimod (12μg/ml, Invivogen) and LPS (10ng/ml, Invivogen) for the time period indicated. Whenever indicated, Kupffer cells were pre-incubated for 5 hours (for cytokine secretion) or 1 hour (for signaling experiments) prior to stimulation with the following inhibitors: BIBW2992 (0.005-20μM, Selleck), Cetuximab (0.01-1μg/ml, Merck), TAPI-1 (10μM, Peptides International), SP600125 (25μM, Calbiochem), SB203580 (10μM, Cell Signaling), and SC-514 (100μM, Calbiochem). Since the number of Kupffer cells, which can be recovered from one mouse is very low, it was technically impossible to perform Western Blot analysis for all indicated signaling molecules on one single batch of Kupffer cells. Usually Kupffer cells from 2 livers of the same genotype were pooled together to obtain around 20-30 μg of protein lysate, which was sufficient for 1 Western blot.
Patient material and immunohistochemistry
Human samples were collected following informed consent according to the established protocol approved by the Ethic Committee of the Eastern Hepatobiliary Surgery Hospital and the Medical University of Vienna. The 129 HCC patients of the Chinese cohort were randomly taken from the pool of HCC patients undergoing curative resection in the Eastern Hepatobiliary Surgery Hospital (Shanghai, China) between January 2002 and June 2006. None of these HCC patients received preoperative anticancer treatment. The normal liver tissues were collected from the distal normal liver sections of patients undergoing surgery for liver hemangioma, who did not show any evidence of chronic liver disease.
The 108 HCC patients of the European cohort were randomly taken from a pool of patients who underwent orthotopic liver transplantation for HCC at the Medical University of Vienna as previously published38, 39. None of these HCC patients received preoperative anticancer treatment. Immunohistochemistry and quantitative analyses (scoring) on all human tissues (Chinese and European cohorts) were performed by the same laboratory (China) in a blinded manner following standard procedures.
The slides were incubated with the following primary antibodies: anti-EGFR (CST #4267; clone D38B1; 1/50); anti-CD68 (macrophage marker, abcam ab955; clone KP1; 1/200). Staining for EGFR in hepatocytes or Kupffer cells for both the European and Chinese patient cohort was semi-quantitatively examined and blindly scored by 2 independent observers using the following scale: 0 = negative staining (0%-10% positive), + = weak signal (10%-20% positive), ++ = intermediate signal (20%-50% positive) and +++ = strong signal (>50% positive) as previously described40. There were only slight variations between the scoring results of the 2 pathologists and in such cases; the lower scoring was taken to generate Table 1.
Statistical analysis
Mouse experiments
The mouse experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment. Sample size calculation: For tumor studies we considered 10 mice per group to detect a relevant difference in means of 1.5 within-group standard deviations at a two-sided significance level of 0.05 and a power of 90%, which ensures 80% power in case of a 20% drop-out rate. For the EGFRΔhep/Δmac experiment 6 mice per group were considered, which ensures a 90% power to detect a difference in means of 2 standard deviations at the same significance level of 0.05. Experiments in mice (various injections, ex vivo cell and tissue isolations etc.) were performed as indicated in the figure legends. Quantifications on histological samples were performed by counting/measuring microscopic fields (HPF where indicated) as indicated in the legends. Data are presented as mean±s.d or mean±s.e.m. Student’s t-test for independent samples and unequal variances was used to assess statistical significance (*p<0,05, **p<0,01, ***p<0,001). Each tumor measurement contributed by one animal is the mean value over several liver sections. Based on the central limit theorem, we can assume a normal distribution of these animal-specific means even if the underlying variable is not perfectly normally distributed. All statistical analyses were performed with the SPSS18.0 software. A two-sided P < 0.05 was considered statistically significant.
Human material
The statistical analyses were performed by the respective Chinese and Viennese laboratories. The experiments were not randomized. Staining for EGFR in hepatocytes or Kupffer cells of human patient material for both – the European and the Chinese patient cohort was semi-quantitatively examined and blindly scored by two independent observers. Overall survival (OS) in both patient cohorts was defined as the time between the dates of surgery and death or the last follow-up. Disease-free survival (DFS) was defined as the time between the dates of surgery and recurrence. If recurrence was not diagnosed, patients were classified on the date of death or the last follow-up. Survival curves were calculated using the Kaplan–Meier method. Median survival times (OS) and their 95% confidence intervals (CIs) were reported. The survival in the European cohort is low for a current transplant population because many patients were transplanted with tumors that were too large (outside the now accepted Milan criteria) as previously described30. The log-rank test was used to assess the effects of patient variables on DFS and OS.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Latifa Bakiri, Denise P. Barlow, Robert Eferl, Martin Oft, and Erwin F. Wagner for critical reading of the manuscript. We thank Temenuschka Baykuscheva-Gentscheva and Sarah Bardakji for genotyping and Florian Hucke and Georg Heinze for excellent statistical support. This work was supported by the EC program LSHC-CT-2006-037731 (Growthstop), and the Austrian Science Fund (FWF) grant SFB F3518-B20 (to M.S.) and F3517 (to M.T.), the FWF-DK W1212 and P25925, and the Austrian Federal Government’s GEN-AU program “Austromouse” (GZ 200.147/1-VI/1a/2006 and 820966). H.W. acknowledges funding by the National Natural Science Foundation of China 30921006.
Footnotes
AUTHOR INFORMATION The authors declare no competing financial interests.
REFERENCES
- 1.Jemal A, et al. Global cancer statistics. CA. 2011;61:69–90. doi: 10.3322/caac.20107. doi:10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989–5005. doi: 10.1038/onc.2010.236. doi:10.1038/onc.2010.236. [DOI] [PubMed] [Google Scholar]
- 3.Laurent-Puig P, Zucman-Rossi J. Genetics of hepatocellular tumors. Oncogene. 2006;25:3778–3786. doi: 10.1038/sj.onc.1209547. doi:10.1038/sj.onc.1209547. [DOI] [PubMed] [Google Scholar]
- 4.EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. Journal of hepatology. 2012;56:908–943. doi: 10.1016/j.jhep.2011.12.001. doi:10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Finkin S, Pikarsky E. NF-kappaB in liver cancer: the plot thickens. Current topics in microbiology and immunology. 2011;349:185–196. doi: 10.1007/82_2010_104. doi:10.1007/82_2010_104. [DOI] [PubMed] [Google Scholar]
- 6.Luedde T, et al. Deletion of NEMO/IKKgamma in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell. 2007;11:119–132. doi: 10.1016/j.ccr.2006.12.016. doi:10.1016/j.ccr.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 7.Sakurai T, Maeda S, Chang L, Karin M. Loss of hepatic NF-kappa B activity enhances chemical hepatocarcinogenesis through sustained c-Jun N-terminal kinase 1 activation. Proc Natl Acad Sci U S A. 2006;103:10544–10551. doi: 10.1073/pnas.0603499103. doi:10.1073/pnas.0603499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vainer GW, Pikarsky E, Ben-Neriah Y. Contradictory functions of NF-kappaB in liver physiology and cancer. Cancer Lett. 2008;267:182–188. doi: 10.1016/j.canlet.2008.03.016. doi:10.1016/j.canlet.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 9.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. doi:10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. doi:10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakurai T, et al. Hepatocyte necrosis induced by oxidative stress and IL-1 alpha release mediate carcinogen-induced compensatory proliferation and liver tumorigenesis. Cancer Cell. 2008;14:156–165. doi: 10.1016/j.ccr.2008.06.016. doi:10.1016/j.ccr.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naugler WE, et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. doi:10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 13.Hui L, Zatloukal K, Scheuch H, Stepniak E, Wagner EF. Proliferation of human HCC cells and chemically induced mouse liver cancers requires JNK1-dependent p21 downregulation. The Journal of clinical investigation. 2008;118:3943–3953. doi: 10.1172/JCI37156. doi:10.1172/JCI37156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li HP, et al. miR-451 inhibits cell proliferation in human hepatocellular carcinoma through direct suppression of IKK-beta. Carcinogenesis. 2013;34:2443–2451. doi: 10.1093/carcin/bgt206. doi:10.1093/carcin/bgt206. [DOI] [PubMed] [Google Scholar]
- 15.Wang SN, Lee KT, Tsai CJ, Chen YJ, Yeh YT. Phosphorylated p38 and JNK MAPK proteins in hepatocellular carcinoma. European journal of clinical investigation. 2012;42:1295–1301. doi: 10.1111/eci.12003. doi:10.1111/eci.12003. [DOI] [PubMed] [Google Scholar]
- 16.Das M, Garlick DS, Greiner DL, Davis RJ. The role of JNK in the development of hepatocellular carcinoma. Genes Dev. 2011;25:634–645. doi: 10.1101/gad.1989311. doi:10.1101/gad.1989311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinrichsdorff J, Luedde T, Perdiguero E, Nebreda AR, Pasparakis M. p38 alpha MAPK inhibits JNK activation and collaborates with IkappaB kinase 2 to prevent endotoxin-induced liver failure. EMBO reports. 2008;9:1048–1054. doi: 10.1038/embor.2008.149. doi:10.1038/embor.2008.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hui L, et al. p38alpha suppresses normal and cancer cell proliferation by antagonizing the JNK-c-Jun pathway. Nat Genet. 2007;39:741–749. doi: 10.1038/ng2033. doi:10.1038/ng2033. [DOI] [PubMed] [Google Scholar]
- 19.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. doi:10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Buckley AF, Burgart LJ, Sahai V, Kakar S. Epidermal growth factor receptor expression and gene copy number in conventional hepatocellular carcinoma. Am J Clin Pathol. 2008;129:245–251. doi: 10.1309/WF10QAAED3PP93BH. doi:10.1309/WF10QAAED3PP93BH. [DOI] [PubMed] [Google Scholar]
- 21.Feitelson MA, Pan J, Lian Z. Early molecular and genetic determinants of primary liver malignancy. Surg Clin North Am. 2004;84:339–354. doi: 10.1016/S0039-6109(03)00226-3. doi:10.1016/S0039-6109(03)00226-3. [DOI] [PubMed] [Google Scholar]
- 22.Hopfner M, et al. Targeting the epidermal growth factor receptor by gefitinib for treatment of hepatocellular carcinoma. J Hepatol. 2004;41:1008–1016. doi: 10.1016/j.jhep.2004.08.024. doi:10.1016/j.jhep.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 23.Schiffer E, et al. Gefitinib, an EGFR inhibitor, prevents hepatocellular carcinoma development in the rat liver with cirrhosis. Hepatology. 2005;41:307–314. doi: 10.1002/hep.20538. doi:10.1002/hep.20538. [DOI] [PubMed] [Google Scholar]
- 24.Mallarkey G, Coombes RC. Targeted therapies in medical oncology: successes, failures and next steps. Therapeutic advances in medical oncology. 2013;5:5–16. doi: 10.1177/1758834012467829. doi:10.1177/1758834012467829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Natarajan A, Wagner B, Sibilia M. The EGF receptor is required for efficient liver regeneration. Proc Natl Acad Sci U S A. 2007;104:17081–17086. doi: 10.1073/pnas.0704126104. doi:10.1073/pnas.0704126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eferl R, et al. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112:181–192. doi: 10.1016/s0092-8674(03)00042-4. [DOI] [PubMed] [Google Scholar]
- 27.He G, Karin M. NF-kappaB and STAT3 - key players in liver inflammation and cancer. Cell Res. 2011;21:159–168. doi: 10.1038/cr.2010.183. doi:10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sell S. Cellular origin of hepatocellular carcinomas. Semin Cell Dev Biol. 2002;13:419–424. doi: 10.1016/s1084952102001295. [DOI] [PubMed] [Google Scholar]
- 29.Degryse B, et al. The high mobility group (HMG) boxes of the nuclear protein HMG1 induce chemotaxis and cytoskeleton reorganization in rat smooth muscle cells. J Cell Biol. 2001;152:1197–1206. doi: 10.1083/jcb.152.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, et al. Overexpression of aspartyl-(asparaginyl)-beta-hydroxylase in hepatocellular carcinoma is associated with worse surgical outcome. Hepatology. 2010;52:164–173. doi: 10.1002/hep.23650. doi:10.1002/hep.23650. [DOI] [PubMed] [Google Scholar]
- 31.Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6:32–43. doi: 10.1038/nrm1548. doi:10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 32.McElroy SJ, et al. Transactivation of EGFR by LPS induces COX-2 expression in enterocytes. PloS one. 2012;7:e38373. doi: 10.1371/journal.pone.0038373. doi:10.1371/journal.pone.0038373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 34.Lichtenberger BM, et al. Autocrine VEGF signaling synergizes with EGFR in tumor cells to promote epithelial cancer development. Cell. 2010;140:268–279. doi: 10.1016/j.cell.2009.12.046. doi:10.1016/j.cell.2009.12.046. [DOI] [PubMed] [Google Scholar]
- 35.Sibilia M, et al. The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell. 2000;102:211–220. doi: 10.1016/s0092-8674(00)00026-x. [DOI] [PubMed] [Google Scholar]
- 36.Drobits B, et al. Imiquimod clears tumors in mice independent of adaptive immunity by converting pDCs into tumor-killing effector cells. The Journal of clinical investigation. 2012;122:575–585. doi: 10.1172/JCI61034. doi:10.1172/JCI61034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smedsrod B, Pertoft H. Preparation of pure hepatocytes and reticuloendothelial cells in high yield from a single rat liver by means of Percoll centrifugation and selective adherence. Journal of leukocyte biology. 1985;38:213–230. doi: 10.1002/jlb.38.2.213. [DOI] [PubMed] [Google Scholar]
- 38.Sieghart W, et al. Osteopontin expression predicts overall survival after liver transplantation for hepatocellular carcinoma in patients beyond the Milan criteria. J Hepatol. 2011;54:89–97. doi: 10.1016/j.jhep.2010.06.030. doi:10.1016/j.jhep.2010.06.030. [DOI] [PubMed] [Google Scholar]
- 39.Mazzaferro V, et al. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. The New England journal of medicine. 1996;334:693–699. doi: 10.1056/NEJM199603143341104. doi:10.1056/NEJM199603143341104. [DOI] [PubMed] [Google Scholar]
- 40.Wang K, et al. Overexpression of aspartyl-(asparaginyl)-beta-hydroxylase in hepatocellular carcinoma is associated with worse surgical outcome. Hepatology. 2010;52:164–173. doi: 10.1002/hep.23650. doi:10.1002/hep.23650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.